Summary
In many cell types, the formation of membrane protrusions and directional migration depend on the spatial and temporal regulation of the actin-binding protein cofilin. Cofilin, which is important for the regulation of actin-polymerization initiation, increases the number of actin free barbed ends through three mechanisms: its intrinsic actin-nucleation activity; binding and severing of existing actin filaments; and recycling actin monomers from old filaments to new ones through its actin-depolymerization activity. The increase in free barbed ends that is caused by cofilin initiates new actin polymerization, which can be amplified by the actin-nucleating ARP2/3 complex. Interestingly, different cell systems seem to have different mechanisms of activating cofilin. The initial activation of cofilin in mammary breast tumors is dependent on PLCγ, whereas cofilin activation in neutrophils is additionally dependent on dephosphorylation, which is promoted through Rac2 signaling. Although the literature seems to be confusing and inconsistent, we propose that all of the data can be explained by a single activity-cycle model. In this Opinion, we give an overview of cofilin activation in both tumor cells and inflammatory cells, and demonstrate how the differences in cofilin activation that are observed in various cell types can be explained by different starting points in this single common activity cycle.
Keywords: Cofilin activation cycle, Inflammation, Metastasis
Introduction
Coordinated cell migration is a crucial event for numerous normal physiological processes, ranging from embryonic development all the way through to immunological surveillance in the mature adult. However, the molecular machinery that regulates the complex cellular events that lead to cell translocation can also be `hijacked' such that cells that are not `supposed' to move do migrate, which can lead to cancer metastasis. In addition, a number of inflammatory diseases involve the over-recruitment, through aberrant cell migration, of white blood cells to sites of inflammation.
Cell migration is physically mediated by the actin cytoskeleton. The assembly of polarized actin filaments at the leading-edge membrane propels the cell edge forward, resulting in net forward displacement of the cell. Actin polymerization occurs mainly at one end of the filament, named the `barbed end', and it is through the regulation of the number of barbed ends that the cell is able to control the assembly of filamentous actin (F-actin). As has been reviewed elsewhere (e.g. Chhabra and Higgs, 2007; Condeelis et al., 2005; Kedrin et al., 2007; Pak et al., 2008; Pollard, 2007), a large array of actin-binding proteins is responsible for regulating the actin-polymerization and -depolymerization cycle by changing the number of free actin barbed ends (Fig. 1A). One of the key proteins in this process is the actin-binding protein cofilin, which can increase the number of barbed ends by severing existing F-actin (Ichetovkin et al., 2002) and by its intrinsic actin-nucleation activity (Andrianantoandro and Pollard, 2006). Although severing can occur at cofilin concentrations in the nM to low-μM range, nucleation occurs only at high concentrations of active and non-actin-bound cofilin (10 μM) in vitro. This concentration can hypothetically occur in vivo when active cofilin, which is only a small fraction of the total cofilin (10 μM) in the cytosol, becomes concentrated into clusters. However, direct nucleation by cofilin has not been formally observed in vivo, so this model remains hypothetical at this time. In addition to generating new free barbed ends, the actin-severing event also results in depolymerization of older actin filaments, thereby increasing the number of actin monomers that are now free to add onto the newly available free barbed ends (Bamburg, 1999; Pantaloni et al., 2001; Wang et al., 2007a) (Fig. 1A). As a result, in the presence of physiological actin-monomer concentrations, cofilin activity can both increase the rate of actin-filament turnover and increase the net polymerization of actin (Fig. 1A). The central role for cofilin in the actin-polymerization process places it as a key regulator of events that require actin assembly, ranging from the migration of tumor and inflammatory cells to neural development and growth-cone guidance (Bellenchi et al., 2007; Wang et al., 2007a; Wen et al., 2007).
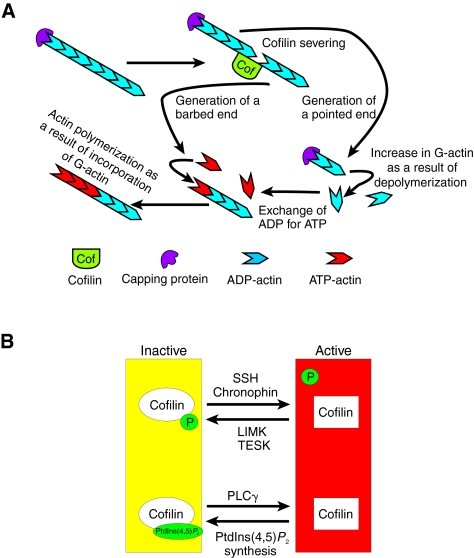
Fig. 1.
The actin-severing and -depolymerization activity of cofilin is regulated by several mechanisms. (A) The barbed ends of filamentous actin (F-actin) can be capped by capping proteins, preventing further polymerization. Cofilin binds to and severs F-actin, creating both barbed and pointed ends. From the pointed ends, the filaments depolymerize and thereby increase the concentration of monomeric globular actin (G-actin). The new `free' barbed ends can be extended by incorporation of G-actin. Therefore, cofilin can induce both the polymerization and depolymerization of F-actin. (B) The binding of cofilin to actin is inhibited by the phosphorylation of cofilin at Ser3, and by the binding of PtdIns(4,5)P2 to cofilin. Upon dephosphorylation of cofilin, or release of cofilin from PtdIns(4,5)P2 by PLCγ-dependent hydrolysis, cofilin becomes active.
In this Opinion we compare and contrast the available information about how inflammatory cells and cancer cells use cofilin to regulate cell migration. We show that cofilin is a regulator of cell migration in inflammatory cells, as well as a key regulator of cancer-cell metastasis. These two cell types, at first glance, seem to have distinct mechanisms for regulating cofilin activity. In this Opinion, however, we propose a single common activity cycle for cofilin that is consistent with the literature describing cofilin activity in both cell types.
Main players in cofilin regulation
The activity of cofilin is tightly regulated in the cell by several mechanisms, of which the most important and best-studied include phosphorylation (Pak et al., 2008) and the binding of phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] (van Rheenen et al., 2007) (Fig. 1B). Cofilin is phosphorylated at serine 3 (Ser3) by LIM kinase (LIMK) and testis-specific kinase 1 and 2 (TESK1 AND TESK2), which are regulated by Rho GTPases (Arber et al., 1998; Bernard, 2007; Pak et al., 2008). The phosphorylation site is located in the actin-binding domain and inhibits the binding of cofilin to F-actin, which therefore renders phospho-cofilin inactive towards F-actin (Arber et al., 1998; Bamburg, 1999; Bernard, 2007; Moriyama et al., 1996). Interestingly, it was recently found that phospho-cofilin, which was thought to be the inactive form of cofilin, has a stimulatory effect on phospholipase D1 (PLD1) and might control a wide variety of cellular functions through this enzyme (Han et al., 2007). Nevertheless, actin severing by cofilin, and therefore its actin-polymerization and -depolymerization activity, is inhibited upon phosphorylation of cofilin. The dephosphorylation of cofilin by the phosphatases Slingshot homolog (SSH) and chronophin (CIN) circumvent this inhibition, thereby stimulating the actin-severing and -depolymerization activity of cofilin (Huang et al., 2006; Niwa et al., 2002). The activity of SSH phosphatases is regulated by multiple signaling pathways, including those activated by phosphoinositide 3-kinase (PI3K) (for a review, see Huang et al., 2006). Once cofilin is dephosphorylated, its activity can still be inhibited by the phospholipid PtdIns(4,5)P2. The cofilin-PtdIns(4,5)P2 interaction sequesters the actin-binding residues of cofilin. Therefore, in contrast to what is sometimes assumed, the phosphorylation status of cofilin should not be taken as a direct measure of cofilin activity (Fig. 1B). For example, in resting mammary-tumor cells, phospho-cofilin accounts for only 18% of the total cofilin level in the cell, but cofilin activity in the cell overall is at its lowest. Within 1 minute of epidermal growth factor (EGF) stimulation, phospho-cofilin levels rise by at least twofold at a time at which the actin-severing activity of cofilin is at its highest level (Song et al., 2006). We have shown that this disconnection results from the fact that cofilin is activated in a different location than where it is phosphorylated (described below and in Fig. 2).
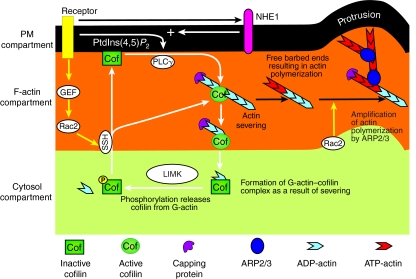
Fig. 2.
The common cofilin activity cycle in invasive tumor cells and inflammatory cells. Cofilin molecules cycle through three compartments in the cell: the cytosol, F-actin and the PM compartments. Cofilin is inactivated at the PM by binding to PtdIns(4,5)P2, and in the cytosol by either low pH levels or phosphorylation. Upon activation, cofilin translocates from the PM and cytosolic compartments to the F-actin compartment. The translocation of cofilin from the PM to the F-actin compartment is induced by a PLCγ-mediated decrease in PtdIns(4,5)P2 levels, which is amplified by an increase in pH (mediated by the Na+-H+ exchanger NHE1), which reduces the affinity of cofilin for PtdIns(4,5)P2, whereas the translocation from the cytosol to the PM compartment is induced by dephosphorylation of cofilin. At the F-actin compartment, cofilin binds to and severs actin filaments, which results in filaments with free barbed ends and the formation of cofilin–G-actin complexes. These complexes cannot bind to actin or the PM, and therefore diffuse to the cytosol compartment, where cofilin is phosphorylated and released from the cofilin–G-actin complex by LIMK. A new cycle is started when cofilin is dephosphorylated (by SSH, for example), which results in the re-entry of cofilin to the PM or F-actin compartment. The generation of actin free barbed ends by cofilin initiates the polymerization of new actin filaments to which the ARP2/3 complex prefers to bind, which amplifies cofilin-induced actin polymerization and results in the formation of cellular protrusions. `+' indicates pH increase.
In addition to phosphorylation and PtdIns(4,5)P2 binding, cofilin activity is also regulated by unrelated mechanisms that involve pH changes (Bernstein, 2000; Frantz et al., 2008; Yonezawa et al., 1985) and interaction with proteins such as actin-interacting protein (Aip1) and adenylyl cyclase-associated protein (CAP; also known as Srv2). The re-annealing of actin filaments that are being severed by cofilin is prevented by an Aip1-cofilin complex that caps the barbed ends of F-actin. Moreover, cofilin activity is increased by the CAP complex (Aip1, CAP and cofilin), and it is thought that this complex recycles cofilin by disassociating it from ADP-actin (Balcer et al., 2003). In this Opinion, we focus on the regulation of cofilin activity by the phosphorylation and PtdIns(4,5)P2 binding of cofilin, as these two regulatory mechanisms are conserved among and have been characterized in several different migration models.
Activation of cofilin in tumor cells
Remodeling of the F-actin cytoskeleton is crucial for tumor-cell invasion and metastasis (Wang et al., 2007a). It is therefore not surprising that the cofilin pathway has been found to be affected in many tumor types. For example, altered cofilin expression has been found in clinical tumor samples of renal-cell carcinoma (Unwin, 2003), ovarian cancer (Martoglio et al., 2000) and oral squamous-cell carcinomas (Turhani et al., 2006), and also in cancer cell lines such as C6 (rat glioblastoma) (Gunnersen et al., 2000), A549 (human lung cancer) (Keshamouni, 2006), HeLa (human cervical carcinoma), KM12 (colon), HepG2 (liver) and COS1 (kidney) (Nebl et al., 1996). The expression of cofilin and other proteins in the cofilin pathway, such as LIMK, is upregulated in the invasive subpopulation of tumor cells in mammary tumors (Wang et al., 2004), and the activation status of cofilin is directly linked to invasion, intravasation and metastasis (Wang et al., 2006). Although all of these examples illustrate that cofilin has an important role in many types of cancer, methodical studies of cofilin regulation and function have only been carried out in mammary tumors; therefore, we will focus on cofilin function in these tumors.
Cofilin activation in mammary-tumor cells is dependent on PLCγ, but not on cofilin dephosphorylation
Stimulation of many cell types – including mammary-tumor cells, Dictyostelium discoideum, and mammalian fibroblasts, neutrophils and macrophages – with various growth factors results in two transient increases of actin barbed ends (Wang et al., 2007a). In mammary-tumor cells, the first transient of barbed ends is dependent on cofilin and phospholipase Cγ (PLCγ) (Mouneimne et al., 2004) but not on the dephosphorylation of cofilin (Song et al., 2006), and initiates actin polymerization that results in the formation of a small membrane protrusion and promotes directional sensing during chemotaxis towards EGF (Mouneimne et al., 2006). During this first transient, cofilin activity increases and is responsible for causing more than half of the barbed ends of the transient (Mouneimne et al., 2004).
Cofilin moves between three different compartments during the first transient – the plasma membrane (PM), cytosol and F-actin compartments (van Rheenen et al., 2007). The PM and the F-actin compartments are in close proximity and cannot be resolved directly by conventional light microscopy, which has until recently prevented direct visualization of these compartments. Using fluorescence resonance energy transfer (FRET) and fluorescence recovery after photobleaching (FRAP) experiments, we were able to distinguish these cofilin compartments in living mammary-tumor cells (van Rheenen et al., 2007). Cofilin, but not a phospho-mimicking mutant form of cofilin (S3E), binds to PtdIns(4,5)P2 (Song et al., 2006), and the unphosphorylated, but not the phosphorylated, form of cofilin colocalizes with PtdIns(4,5)P2 in the PM compartment (van Rheenen et al., 2007). Therefore, unphosphorylated cofilin is present in both the PM and F-actin compartments, whereas phospho-cofilin is localized in the cytosol compartment. At the PM compartment, unphosphorylated cofilin binds to and is inactivated by PtdIns(4,5)P2 (Gorbatyuk et al., 2006; Ojala et al., 2001; van Rheenen et al., 2007; Yin and Janmey, 2003; Yonezawa et al., 1990). Stimulation of mammary-tumor cells by EGF results in an activation of PLCγ (Mouneimne et al., 2004) and a subsequent decrease in the level of PtdIns(4,5)P2 (van Rheenen et al., 2007). The inactive and unphosphorylated cofilin molecules that are bound to PtdIns(4,5)P2 in the PM compartment are released and diffuse to the nearby F-actin compartment (van Rheenen et al., 2007).
Initially, it was thought that PLCγ was not able to release cofilin from the PM compartment, because in vitro assays using lipid micelles that contained PtdIns(4,5)P2 suggested that, once cofilin had bound to PtdIns(4,5)P2, it inhibited hydrolysis of this PtdIns(4,5)P2 molecule by PLCγ (Yonezawa et al., 1991). However, we showed recently through FRAP experiments in real biological membranes that cofilin-PtdIns(4,5)P2 binding is transient (lasting a few seconds) with fast on- and off-rates (van Rheenen et al., 2007). PLCγ-mediated translocation of cofilin from the PM compartment is therefore not the result of a direct release of cofilin from PtdIns(4,5)P2, but rather of a lower on-rate [as a result of a lower PtdIns(4,5)P2 concentration] combined with the normal fast off-rate. Therefore, a reduction of PtdIns(4,5)P2 levels, not the hydrolysis of cofilin-bound PtdIns(4,5)P2 by PLCγ, releases cofilin from the PM compartment. In support of this conclusion, decreasing PtdIns(4,5)P2 levels by PLCγ- and EGF-independent mechanisms, such as rapamycin-induced membrane targeting of a PtdIns(4,5)P2 phosphatase, also causes the translocation of cofilin from the PM compartment (van Rheenen et al., 2007).
Once in the F-actin compartment, cofilin binds to and severs actin filaments, thereby increasing the number of free barbed ends and leading to the first barbed-end transient (van Rheenen et al., 2007). In addition, the number of barbed ends might be increased by the intrinsic actin-nucleation activity of cofilin (Andrianantoandro and Pollard, 2006), although the relative contributions of actin severing vs intrinsic nucleation activity of cofilin have not been determined. The actin filaments that bind to cofilin during the first transient are very close to the PM (within the diffraction limit of the light microscope), which results in part from the inhibition of cofilin binding to F-actin by tropomyosin in regions of the cytoplasm that are farther from the PM (Des Marais et al., 2004). Thus, in EGF-stimulated mammary-tumor cells, a PLCγ-mediated decrease in PtdIns(4,5)P2 levels induces a translocation of cofilin molecules from the PM to the actin compartment, where cofilin binds and severs actin filaments to give rise to a subsequent increase in actin polymerization.
Localized activation combined with global inactivation leads to highly localized cofilin activity
In mammary-tumor cells, LIMK is activated nearly simultaneously with PLCγ, through a PI3K-, Rho- and ROCK-dependent pathway (Song et al., 2006). Although LIMK phosphorylates and inactivates cofilin, phosphorylation of cofilin occurs uniformly throughout the cytosol and does not keep up with cofilin activation by PLCγ near the PM; consequently, cofilin activity increases on the side of the cell nearest to the EGF stimulus (Song et al., 2006; Wang et al., 2007a). In EGF-stimulated cells, activated cofilin molecules released from PtdIns(4,5)P2 by PLCγ sever F-actin, and thereby increase the number of barbed and pointed ends. However, some cofilin molecules that diffuse away from the initial activation site at the PM compartment into the cytosol compartment are inactivated through phosphorylation by LIMK (Song et al., 2006; van Rheenen et al., 2007). Hence, as the cofilin is being activated the level of phospho-cofilin in the cell also increases. The localized activation, combined with the global inactivation, of cofilin confines its severing activity to an asymmetrical region of the cell, thereby defining the site of protrusion formation. As described for models of chemotaxis (Devreotes and Janetopoulos, 2003), cofilin signaling seems to follow a local-excitation–global-inhibition (LEGI) model (Wang et al., 2007a), in which the local activation of cofilin by PtdIns(4,5)P2 hydrolysis and its global inhibition by LIMK-mediated phosphorylation leads to highly localized activation of cofilin. This idea is supported by experiments that show that the localized activation of cofilin by photoactivation of caged cofilin (Ghosh et al., 2004) generates the same response as localized EGF stimulation by a micropipette (Mouneimne et al., 2006; van Rheenen et al., 2007), which results in localized activation of cofilin, actin polymerization and directional cell protrusion.
Output of the cofilin pathway changes in different ways according to the altered expression of single or multiple genes
Interestingly, in the invasive subpopulation of tumor cells in mammary tumors, multiple genes in the cofilin pathway are up- and down-regulated (Goswami et al., 2004; Sidani et al., 2007; Wang et al., 2004; Wang et al., 2005; Wang et al., 2002). The output of the cofilin pathway in mammary-tumor cells, which is measured as the number of barbed ends generated upon cofilin severing within the first minute of EGF stimulation, is the sum of the activities of all gene products of the cofilin pathway. Hence, changing the expression of only one gene of the pathway can cause a phenotype that is totally different to the phenotype observed following changes in the expression of another gene or of several genes simultaneously (Wang et al., 2007a). For example, overexpression of LIMK in mammary-tumor cells leads to an increased number of phosphorylated cofilin molecules and decreased cofilin activity. Decreasing cofilin activity inhibits actin polymerization and motility in vitro (Zebda, 2000), and inhibits invasion, intravasation and metastasis of mammary tumors in vivo (Wang et al., 2006). By contrast, in the invasive subpopulation of tumor cells in different types of mammary tumors, LIMK is spontaneously overexpressed together with either cofilin [MTLn3 tumors (Wang et al., 2004)] or the cofilin activator SSH [PyMT tumors (Wang et al., 2007b)]. Similar findings were obtained in other cell systems; for example, in rat ascites hepatoma (MM1) cells, the knockdown of SSH inhibits migration and invasion, and knockdown of LIMK has the same effect (Horita et al., 2008). The simultaneous overexpression of both the inhibitory and stimulatory components of the cofilin pathway sharpens the local activation of cofilin in these cells (LEGI model) and therefore increases the transient barbed-end output of the cofilin pathway in response to EGF (thereby increasing chemotaxis and motility) (Wang et al., 2006; Wang et al., 2007b).
These results might explain why alternating the expression of a single gene in the cofilin pathway [e.g. by overexpression or RNA interference (RNAi)] can have different effects on different types of tumor cells according to which other genes in the pathway are elevated, and why metastatic phenotype does not correlate with the expression of only a single protein, such as LIMK (Sidani et al., 2007; Wang et al., 2007a). Moreover, these results emphasize that the phenotypes resulting from the alteration in cofilin expression are cell-type dependent. Although cofilin activation is regulated by various conserved mechanisms, variations in the expression of one or multiple genes of the cofilin pathway might cause one of the mechanisms of cofilin regulation to become dominant. For example, in metastatic amoeboid tumor cells in which migration is PLCγ- and cofilin-dependent (Wang et al., 2007a), cofilin activation occurs predominantly through PtdIns(4,5)P2 hydrolysis. By contrast, in tumor cells with a less-metastatic mesenchymal phenotype, in which migration is PLCγ-independent, cofilin regulation might rely more on other mechanisms, such as dephosphorylation and changes in pH (Sidani et al., 2007; Wang et al., 2007a).
Cofilin-mediated chemotaxis in inflammatory cells
Leukocytes are highly motile cells that undergo rapid directed migration to sites of infection (Fenteany and Glogauer, 2004). There is a significant body of research that clearly identifies cofilin as a crucial regulator of actin assembly and subsequent chemotaxis in leukocytes (Heyworth et al., 1997; Hirayama et al., 2007). However, as most of these studies have used different cell types, including primary cells and cell lines, little effort has been made to reconcile the data to definitively identify the activation cycle of cofilin during leukocyte migration.
Cofilin is required for leukocyte chemotaxis
The importance of cofilin in leukocyte chemotaxis has been demonstrated through the downregulation of cofilin expression using small interfering RNA (siRNA) in neutrophil-like HL-60 cells (Hirayama et al., 2007) and T-cell-like Jurkat cells (Nishita, 2005). When cofilin expression levels were significantly reduced in these leukocyte-like cell lines, a complete block of chemoattractant-mediated chemotaxis was noted. A similar chemotactic defect was observed in primary murine neutrophils that were deficient in the Rho-family small GTPase Rac2. Although these Rac2-null neutrophils had normal cofilin expression levels, there was a block in dephosphorylation of cofilin at Ser3 [which is required for cofilin-mediated filament severing; see above (Sun et al., 2007)]. The grouping of this data, which involve various sources of leukocytes, implicates cofilin as an essential regulator of chemotaxis in these rapidly migrating cells.
Cofilin activation in leukocytes is dependent on dephosphorylation of cofilin
Chemoattractant-initiated dephosphorylation of cofilin at Ser3 (which occurs within seconds of chemoattractant-receptor activation) is required to initiate actin-mediated chemotaxis in leukocytes (Boldt et al., 2006; Sun et al., 2007). Cofilin activity is also tightly regulated in leukocytes, as seen by the rephosphorylation of cofilin at Ser3 (which occurs 3-5 minutes after initiation of the activation cycle). Hirayama and co-workers used HL-60 cells to study the cofilin activation cycle, and were able to demonstrate a clear activation cycle using the chemoattractant interleukin-8 (Hirayama et al., 2007). Treatment with this agonist immediately results in cofilin dephosphorylation but, within 5 minutes of stimulation, the total amount of phosphorylated cofilin increases to well above baseline pre-stimulated levels (Hirayama et al., 2007). In order to understand the significance of the rephosphorylation step, the authors used a cell-membrane-permeable peptide (S3-R) derived from the N-terminus of cofilin (MASGVAVSDGVIKVFN) linked to a polymer of eight arginine residues (Hirayama et al., 2007). This peptide, which inhibits the rephosphorylation step, causes a dramatic increase in the velocity of cell chemotaxis. This suggests that tight regulation of the activation state of cofilin is required for normal cell migration and that the rephosphorylation step acts as an essential downregulator of chemotaxis. It is important to note that this downregulation of cofilin activity through phosphorylation has also been noted in primary neutrophils that were stimulated with the chemoattractant formyl-methionyl-leucyl-phenylalanine (fMLP) (M.G., unpublished data).
Translocation of dephosphorylated cofilin to the leading edge
Chemotaxis requires rapid actin-filament polymerization that results in the development of an actin-rich leading edge (Glogauer et al., 2000). A number of studies have demonstrated that dephosphorylated cofilin localizes to the leading edge of migrating leukocytes, where actin polymerization is ongoing (Adachi et al., 2000; Nishita, 2005). The important role of cofilin in regulating actin assembly during chemotaxis is also highlighted by the observation that leukocytes with defects in cofilin dephosphorylation (downstream of Rac2) or in which cofilin is knocked down are unable to generate free F-actin barbed ends downstream of chemoattractant receptors (Jovceva et al., 2007; Sun et al., 2007). Overexpressing the phosphatase CIN, which dephosphorylates cofilin at Ser3 (see above) (Gohla et al., 2005), in Rac2-null neutrophils (which are unable to dephosphorylate cofilin) rescues the dephosphorylation defect and restores actin assembly (Sun et al., 2007).
As has been seen in tumor cells, the regulatory mechanisms that are responsible for controlling cofilin activity and actin assembly require tight regulation of the phosphorylation state of cofilin at Ser3. Using Jurkat cells stimulated with a chemokine, it has been demonstrated that leading-edge actin dynamics are mediated by cofilin under the regulation of LIMK and SSH (Nishita, 2005). Suppression of LIMK-mediated phosphorylation of cofilin inhibited lamellipodium formation and cell migration in these leukocytes, whereas suppression of SSH activity and cofilin dephosphorylation led to multiple lamellipodia and impaired directional migration (chemotaxis). This further confirms the idea that cofilin is a key initiator of the leading-edge lamellipodium and that tight regulation of its initiator role in actin assembly is required to maintain a single leading edge (Nishita, 2005).
The contribution of cofilin to leading-edge actin assembly in leukocytes
As has been reviewed elsewhere, actin free-barbed-end generation and filament assembly is mediated locally in the leading lamellipodium by either de novo nucleation [through the ARP2-ARP3 (ARP2/3) complex, formins and/or the intrinsic nucleation activity of cofilin], by uncapping of the barbed ends of existing filaments, or by the generation of free actin barbed ends following cofilin-mediated actin-filament severing (DesMarais et al., 2005; Pollard, 2007; Sarmiento et al., 2008). In addition to supplying free actin monomers for ongoing polymerization (Fig. 1A) (Kiuchi et al., 2007), cofilin severing has a crucial role in initiating free-barbed-end generation at the leading edge in response to chemoattractants, as has been demonstrated by recent work (Sun et al., 2007; Wang et al., 2007a). Although cofilin only generates 10% of the total free barbed ends that are formed in response to fMLP in neutrophils, this relatively small number constitutes the first `seed' free barbed ends; these are required for the action of the ARP2/3 complex, which accelerates and propagates the leading-edge F-actin complex or network (Sun et al., 2007). In the absence of cofilin-mediated severing, no actin polymerization occurs, whereas restoring cofilin-mediated severing recovers 100% of all actin polymerization (Sun et al., 2007). Similar results have been obtained in mammary-tumor cells, in which cofilin is required to initiate and support ARP2/3-mediated actin polymerization during EGF-stimulated cell migration and chemotaxis (DesMarais et al., 2005; Ichetovkin et al., 2002; Wang et al., 2007a).
The cofilin activity cycle
Although the sequence of events of cofilin activation differs in invasive tumor cells and inflammatory cells (that is, the initial activation of cofilin in mammary-tumor cells is dependent on PLCγ, whereas in inflammatory cells cofilin activation is additionally dependent on dephosphorylation through Rac2), we propose that these different initiation events are simply different starting points in a common cofilin activity cycle (Fig. 2). In addition, we propose that all of the observations recorded in different cell types and model systems can be tied together by this common cofilin activity cycle. During the cofilin activity cycle, cofilin moves between the three compartments described above – the PM, cytosol and F-actin compartments (van Rheenen et al., 2007). Inactive cofilin is localized in the PM and cytosol compartments. Cofilin is inactivated at the PM by binding to PtdIns(4,5)P2, whereas inactivation of cofilin in the cytosol can result from either low pH (not shown) or phosphorylation (Wang et al., 2007a). Upon activation, cofilin translocates from the PM and cytosol compartments to the F-actin compartment, where it binds to and severs F-actin, resulting in filaments with free barbed ends (van Rheenen et al., 2007). Cofilin exits the severing reaction as a cofilin–G-actin complex. As this complex cannot bind to F-actin, it diffuses to the cytosol compartment (van Rheenen et al., 2007); there, cofilin is phosphorylated, which releases it from the cofilin–globular-actin (G-actin; monomeric) complex (Wang et al., 2005). Upon dephosphorylation in the cytosol, cofilin translocates to the PM compartment or actin compartment at the cell periphery. At the PM compartment, cofilin binds to and is inactivated by PtdIns(4,5)P2 (Gorbatyuk et al., 2006; Ojala et al., 2001; van Rheenen et al., 2007; Yin and Janmey, 2003; Yonezawa et al., 1990). In the F-actin compartment, cofilin binds only to PM-proximal actin filaments, because these filaments are tropomyosin-free (as noted above, cofilin binding to F-actin is inhibited by tropomyosin) (DesMarais et al., 2002; Gunning et al., 2008). The unphosphorylated and inactive pool of cofilin at the PM can be activated by a PLCγ-mediated reduction of PtdIns(4,5)P2 levels (Mouneimne et al., 2006; Mouneimne et al., 2004; van Rheenen et al., 2007). The increase in pH that results from the stimulation of receptors involved in the cofilin activity cycle decreases the affinity of cofilin for PtdIns(4,5)P2 and enhances PLCγ-mediated activation of PM cofilin (Frantz et al., 2008). The activation of cofilin results in the formation of new barbed ends (Fig. 2) and an increased G-actin concentration (Fig. 1A) as a result of actin severing (Andrianantoandro and Pollard, 2006), which supplies a population of newly growing actin filaments that are preferred for dendritic nucleation by the ARP2/3 complex (Fig. 2) (DesMarais et al., 2005; Ichetovkin et al., 2002).
New insights provided by the cofilin activity cycle
A cofilin activity cycle that is common to tumor and inflammatory cells explains many observations about the behavior and activation of cofilin in cells, including the following examples: (1) The rapid movement of cofilin between the cytosol, PM and F-actin compartments means that cofilin can appear to be uncoupled from events in any one of these individual compartments (Lai et al., 2008). Because of a lack of resolution, cofilin localization in the PM compartment cannot be distinguished from localization in the F-actin compartment using standard light microscopy (van Rheenen et al., 2007). Distinguishing the localization or movement of cofilin in these different compartments requires high-resolution imaging methods such as FRET (van Rheenen et al., 2007). (2) The localization of cofilin, and especially the unphosphorylated form of cofilin, in protrusions or at the leading edge of migrating cells is often interpreted as being the result of F-actin localization. However, it has been overlooked that a fraction of leading-edge cofilin is bound to and inactivated by PtdIns(4,5)P2 in the PM compartment, and that this cofilin cannot be optically resolved from that in the F-actin compartment. Hence, interpreting all dephosphorylated cofilin molecules as active is potentially incorrect and depends on the cell type (Sidani et al., 2007; Wang et al., 2007a), the pH of the cytoplasm (Bernstein, 2000) and the compartment that is being imaged (van Rheenen et al., 2007). (3) The binding of cofilin to PtdIns(4,5)P2 at the PM gives the cell the extra ability to control cofilin activity locally because PtdIns(4,5)P2 binding might reduce or slow the phosphorylation of cofilin, or cause the local accumulation of cofilin molecules that are then available to be released in a future PLCγ-dependent event and to induce actin polymerization near the PM. Thus, although the primary activation of cofilin is through dephosphorylation in leukocytes, the binding of PtdIns(4,5)P2 might be the key to regulating the activity locally at the cell periphery. By contrast, in tumor cells, the reduction in PtdIns(4,5)P2 levels by PLCγ, with the subsequent release and activation of cofilin, is the key determinant of the initial time and place of cofilin activity after receptor stimulation. In this case, the LIMK-mediated phosphorylation of cofilin keeps cofilin activation local (LEGI model). Although the regulation of cofilin seems to differ for inflammatory and tumor cells, all observations to date can be explained by different starting points for cofilin activation in a common activity cycle.
Conclusions and perspectives
We have proposed the existence of a cofilin activity cycle that is common to invasive tumor cells and leukocytes. Although different cell types might use different starting points in the cofilin activity cycle, the output signal is the same – local cofilin activity that initiates local actin polymerization, membrane protrusion, directional migration and even chemotaxis. Chemotaxis is the key common feature of invasive tumor cells and inflammatory cells, making the model of a common cofilin activity cycle attractive for explaining the behavior and interactions of these two cell types in vivo. It will therefore be important to study the localization and activation status of cofilin in all three compartments (PM, cytosol and F-actin) in several model systems. Additional studies must be performed to establish the contribution of all these mechanisms to the local and temporal control of cofilin activity in different cell types and migration models.
We thank all of the members of our departments for their helpful discussions. J.v.R. was supported by the Cell Migration Consortium; J.C. by an NIGMS RO1; and M.G. by a CIHR operating grant. Deposited in PMC for release after 12 months.
References
- Adachi, R., Matsui, S., Kinoshita, M., Nagaishi, K., Sasaki, H., Kasahara, T. and Suzuki, K. (2000). Nitric oxide induces chemotaxis of neutrophil-like HL-60 cells and translocation of cofilin to plasma membranes. Int. J. Immunopharmacol. 22, 855-864. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro, E. and Pollard, T. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Mol. Cell 24, 13-23. [DOI] [PubMed] [Google Scholar]
- Arber, S., Barbayannis, F. A., Hanser, H., Schneider, C., Stanyon, C. A., Bernard, O. and Caroni, P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805-809. [DOI] [PubMed] [Google Scholar]
- Balcer, H. I., Goodman, A. L., Rodal, A. A., Smith, E., Kugler, J., Heuser, J. E. and Goode, B. L. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159-2169. [DOI] [PubMed] [Google Scholar]
- Bamburg, J. R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185-230. [DOI] [PubMed] [Google Scholar]
- Bellenchi, G. C., Gurniak, C. B., Perlas, E., Middei, S., Ammassari-Teule, M. and Witke, W. (2007). N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev. 21, 2347-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, O. (2007). Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39, 1071-1076. [DOI] [PubMed] [Google Scholar]
- Bernstein, B. W. (2000). Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. Cytoskeleton 47, 319-336. [DOI] [PubMed] [Google Scholar]
- Boldt, K., Rist, W., Weiss, S. M., Weith, A. and Lenter, M. C. (2006). FPRL-1 induces modifications of migration-associated proteins in human neutrophils. Proteomics 6, 4790-4799. [DOI] [PubMed] [Google Scholar]
- Chhabra, E. S. and Higgs, H. N. (2007). The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110-1121. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., Singer, R. H. and Segall, J. E. (2005). The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 21, 695-718. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., Ichetovkin, I., Condeelis, J. and Hitchcock-DeGregori, S. E. (2002). Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649-4660. [DOI] [PubMed] [Google Scholar]
- Des Marais, V., Macaluso, F., Condeelis, J. and Bailly, M. (2004). Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117, 3499-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais, V., Ghosh, M., Eddy, R. and Condeelis, J. (2005). Cofilin takes the lead. J. Cell Sci. 118, 19-26. [DOI] [PubMed] [Google Scholar]
- Devreotes, P. and Janetopoulos, C. (2003). Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 278, 20445-20448. [DOI] [PubMed] [Google Scholar]
- Fenteany, G. A. and Glogauer, M. B. (2004). Cytoskeletal remodeling in leukocyte function. Curr. Opin. Hematol. 11, 15-24. [DOI] [PubMed] [Google Scholar]
- Frantz, C., Barreiro, G., Dominguez, L., Chen, X., Eddy, R., Condeelis, J., Kelly, M. J. S., Jacobson, M. P. and Barber, D. L. (2008). Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183, 865-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M., Song, X., Mouneimne, G., Sidani, M., Lawrence, D. S. and Condeelis, J. S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743-746. [DOI] [PubMed] [Google Scholar]
- Glogauer, M., Hartwig, J. and Stossel, T. (2000). Two pathways through Cdc42 couple the N-formyl receptor to actin nucleation in permeabilized human neutrophils. J. Cell Biol. 150, 785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla, A., Birkenfeld, J. and Bokoch, G. M. (2005). Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7, 21-29. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk, V. Y., Nosworthy, N. J., Robson, S. A., Bains, N. P. S., Maciejewski, M. W., dos Remedios, C. G. and King, G. F. (2006). Mapping the phosphoinositide-binding site on chick cofilin explains how PIP2 regulates the cofilin-actin interaction. Mol. Cell 24, 511-522. [DOI] [PubMed] [Google Scholar]
- Goswami, S., Wang, W., Wyckoff, J. B. and Condeelis, J. S. (2004). Breast cancer cells isolated by chemotaxis from primary tumors show increased survival and resistance to chemotherapy. Cancer Res. 64, 7664-7667. [DOI] [PubMed] [Google Scholar]
- Gunnersen, J. M., Spirkoska, V., Smith, P. E., Danks, R. A. and Tan, S. S. (2000). Growth and migration markers of rat C6 glioma cells identified by serial analysis of gene expression. Glia 32, 146-154. [PubMed] [Google Scholar]
- Gunning, P., O'Neill, G. and Hardeman, E. (2008). Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 88, 1-35. [DOI] [PubMed] [Google Scholar]
- Han, L., Stope, M. B., de Jesús, M. L., Oude Weernink, P. A., Urban, M., Wieland, T., Rosskopf, D., Mizuno, K., Jakobs, K. H. and Schmidt, M. (2007). Direct stimulation of receptor-controlled phospholipase D1 by phospho-cofilin. EMBO J. 26, 4189-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth, P. G., Robinson, J. M., Ding, J., Ellis, B. A. and Badwey, J. A. (1997). Cofilin undergoes rapid dephosphorylation in stimulated neutrophils and translocates to ruffled membranes enriched in products of the NADPH oxidase complex. Evidence for a novel cycle of phosphorylation and dephosphorylation. Histochem. Cell Biol. 108, 221-233. [DOI] [PubMed] [Google Scholar]
- Hirayama, A., Adachi, R., Otani, S., Kasahara, T. and Suzuki, K. (2007). Cofilin plays a critical role in IL-8-dependent chemotaxis of neutrophilic HL-60 cells through changes in phosphorylation. J. Leukoc. Biol. 81, 720-728. [DOI] [PubMed] [Google Scholar]
- Horita, Y., Ohashi, K., Mukai, M., Inoue, M. and Mizuno, K. (2008). Suppression of the invasive capacity of rat ascites hepatoma cells by knockdown of Slingshot or LIM kinase. J. Biol. Chem. 283, 6013-6021. [DOI] [PubMed] [Google Scholar]
- Huang, T. Y., DerMardirossian, C. and Bokoch, G. M. (2006). Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18, 26-31. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., Grant, W. and Condeelis, J. (2002). Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79-84. [DOI] [PubMed] [Google Scholar]
- Jovceva, E., Larsen, M. R., Waterfield, M. D., Baum, B. and Timms, J. F. (2007). Dynamic cofilin phosphorylation in the control of lamellipodial actin homeostasis. J. Cell Sci. 120, 1888-1897. [DOI] [PubMed] [Google Scholar]
- Kedrin, D., van Rheenen, J., Hernandez, L., Condeelis, J. and Segall, J. (2007). Cell Motility and Cytoskeletal Regulation in Invasion and Metastasis. J. Mammary Gland Biol. Neoplasia 12, 143-152. [DOI] [PubMed] [Google Scholar]
- Keshamouni, V. G. (2006). Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 5, 1143-1154. [DOI] [PubMed] [Google Scholar]
- Kiuchi, T., Ohashi, K., Kurita, S. and Mizuno, K. (2007). Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177, 465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, F. P. L., Szczodrak, M., Block, J., Faix, J., Breitsprecher, D., Mannherz, H. G., Stradal, T. E. B., Dunn, G. A., Small, J. V. and Rottner, K. (2008). Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio, A. M., Tom, B. D., Starkey, M., Corps, A. N., Charnock-Jones, D. S. and Smith, S. K. (2000). Changes in tumorigenesis- and angiogenesis-related gene transcript abundance profiles in ovarian cancer detected by tailored high density cDNA arrays. Mol. Med. 6, 750-765. [PMC free article] [PubMed] [Google Scholar]
- Moriyama, K., Iida, K. and Yahara, I. (1996). Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1, 73-86. [DOI] [PubMed] [Google Scholar]
- Mouneimne, G., Soon, L., DesMarais, V., Sidani, M., Song, X., Yip, S. C., Ghosh, M., Eddy, R., Backer, J. M. and Condeelis, J. (2004). Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166, 697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne, G., DesMarais, V., Sidani, M., Scemes, E., Wang, W., Song, X., Eddy, R. and Condeelis, J. (2006). Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16, 2193-2205. [DOI] [PubMed] [Google Scholar]
- Nebl, G., Meuer, S. C. and Samstag, Y. (1996). Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J. Biol. Chem. 271, 26276-26280. [DOI] [PubMed] [Google Scholar]
- Nishita, M. (2005). Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171, 349-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, R., Nagata-Ohashi, K., Takeichi, M., Mizuno, K. and Uemura, T. (2002). Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 108, 233-246. [DOI] [PubMed] [Google Scholar]
- Ojala, P. J., Paavilainen, V. and Lappalainen, P. (2001). Identification of yeast cofilin residues specific for actin monomer and PIP2 binding. Biochemistry 40, 15562-15569. [DOI] [PubMed] [Google Scholar]
- Pak, C. W., Flynn, K. C. and Bamburg, J. R. (2008). Actin-binding proteins take the reins in growth cones. Nat. Rev. Neurosci. 9, 136-147. [DOI] [PubMed] [Google Scholar]
- Pantaloni, D., Le Clainche, C. and Carlier, M. F. (2001). Mechanism of actin-based motility. Science 292, 1502-1506. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D. (2007). Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451-477. [DOI] [PubMed] [Google Scholar]
- Sarmiento, C., Wang, W., Dovas, A., Yamaguchi, H., Sidani, M., El-Sibai, M., DesMarais, V., Holman, H. A., Kitchen, S., Backer, J. M. et al. (2008). WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol. 180, 1245-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidani, M., Wessels, D., Mouneimne, G., Ghosh, M., Goswami, S., Sarmiento, C., Wang, W., Kuhl, S., El-Sibai, M., Backer, J. M. et al. (2007). Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 179, 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X., Chen, X., Yamaguchi, H., Mouneimne, G., Condeelis, J. S. and Eddy, R. J. (2006). Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 119, 2871-2881. [DOI] [PubMed] [Google Scholar]
- Sun, C. X., Magalhaes, M. A. O. and Glogauer, M. (2007). Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179, 239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turhani, D., Krapfenbauer, K., Thurnher, D., Langen, H. and Fountoulakis, M. (2006). Identification of differentially expressed, tumor-associated proteins in oral squamous cell carcinoma by proteomic analysis. Electrophoresis 27, 1417-1423. [DOI] [PubMed] [Google Scholar]
- Unwin, R. D. (2003). Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3, 1620-1632. [DOI] [PubMed] [Google Scholar]
- van Rheenen, J., Song, X., van Roosmalen, W., Cammer, M., Chen, X., DesMarais, V., Yip, S.-C., Backer, J. M., Eddy, R. J. and Condeelis, J. S. (2007). EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Xi, L., Hunt, J. L., Gooding, W., Whiteside, T. L., Chen, Z., Godfrey, T. E. and Ferris, R. L. (2004). Expression pattern of chemokine receptor 6 (CCR6) and CCR7 in squamous cell carcinoma of the head and neck identifies a novel metastatic phenotype. Cancer Res. 64, 1861-1866. [DOI] [PubMed] [Google Scholar]
- Wang, W., Wyckoff, J. B., Frohlich, V. C., Oleynikov, Y., Huttelmaier, S., Zavadil, J., Cermak, L., Bottinger, E. P., Singer, R. H., White, J. G. et al. (2002). Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 62, 6278-6288. [PubMed] [Google Scholar]
- Wang, W., Goswami, S., Sahai, E., Wyckoff, J., Segall, J. and Condeelis, J. (2005). Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 15, 139-145. [DOI] [PubMed] [Google Scholar]
- Wang, W., Mouneimne, G., Sidani, M., Wyckoff, J., Chen, X., Makris, A., Goswami, S., Bresnick, A. R. and Condeelis, J. S. (2006). The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 173, 395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Eddy, R. and Condeelis, J. (2007a). The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 7, 429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Wyckoff, J. B., Goswami, S., Wang, Y., Sidani, M., Segall, J. E. and Condeelis, J. S. (2007b). Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 67, 3505-3511. [DOI] [PubMed] [Google Scholar]
- Wen, Z., Han, L., Bamburg, J. R., Shim, S., Ming, G. L. and Zheng, J. Q. (2007). BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J. Cell Biol. 178, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H. L. and Janmey, P. A. (2003). Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65, 761-789. [DOI] [PubMed] [Google Scholar]
- Yonezawa, N., Nishida, E. and Sakai, H. (1985). pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410-14412. [PubMed] [Google Scholar]
- Yonezawa, N., Nishida, E., Iida, K., Yahara, I. and Sakai, H. (1990). Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265, 8382-8386. [PubMed] [Google Scholar]
- Yonezawa, N., Homma, Y., Yahara, I., Sakai, H. and Nishida, E. (1991). A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J. Biol. Chem. 266, 17218-17221. [PubMed] [Google Scholar]
- Zebda, N. (2000). Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151, 1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]