Abstract
Mutations within either the SHH gene or its related pathway components are the most common, and best understood, pathogenetic changes observed in holoprosencephaly patients; this fact is consistent with the essential functions of this gene during forebrain development and patterning. Here we summarize the nature and types of deleterious sequence alterations among over one hundred distinct mutations in the SHH gene (64 novel mutations) and compare these to over a dozen mutations in disease-related Hedgehog family members IHH and DHH. This combined structural analysis suggests that dysfunction of Hedgehog signaling in human forebrain development can occur through truncations or major structural changes to the signaling domain, SHH-N, as well as due to defects in the processing of the mature ligand from its pre-pro-precursor or defective post-translation bi-lipid modifications with palmitate and cholesterol
Keywords: holoprosencephaly, mutation spectrum, SHH, protein processing
INTRODUCTION
Alterations in the SHH gene (Roessler et al., 1996, 1997a; Nanni et al., 1999; MIM# 600725; HPE3 #142945) attributable to chromosomal rearrangements (Roessler et al., 1997b), microdeletions (Lacbawan and Muenke, in preparation; Bendavid et al., 2005a, b), or more commonly point mutations account for at least 6-8% of heterozygous sequence-detectable holoprosencephaly (HPE) related genetic variation (Orioli et al., 2001; Dubourg et al., 2004, 2007). Additionally, in many cases the molecular alteration presents as a familial form, often including many generations, where its clinically relevant phenotypic spectrum is highly variable between mutation carriers. Clinical findings can extend from cyclopia, at the severe extreme, to less severe conditions that include several types of recognizable microforms such as the solitary median maxillary central incisor syndrome (SMMCI; see Nanni et al., 2001; Marini et al., 2003; Garavelli et al., 2004; reviewed in El-Jaick et al., 2007a), or even incomplete penetrance of obligate mutation carriers. Mutations have also been detected in related pathway factors such as PTCH1 (Ming et al., 2001; MIM# 601309; HPE7, MIM# 610828), the transcription factor gene GLI2 (Roessler et al., 2005; MIM# 165230; HPE9, MIM# 610829) and the putative ligand transporter DISP1 (Roessler et al., 2009; MIM# 607502). SHH is presently one of four genes, which also include SIX3 (Wallis et al., 1999; Domené et al., 2008; MIM# 603714; HPE2, MIM# 157170), ZIC2 (Brown et al., 1998; Roessler et al., 2009; MIM# 603073; HPE5, MIM# 609637) and TGIF (Gripp et al., 2000; El-Jaick et al., 2007b; MIM# 602630; HPE4, MIM# 142946), routinely screened as part of the molecular evaluation of new sporadic or familial HPE or SMMCI cases (reviewed in Muenke and Beachy, 2001; Cohen, 2006; Dubourg et al., 2007). Consequently, a large collection of mutations is presently available for a structural analysis.
The vertebrate hedgehog (hh) family of genes actively function during major developmental steps including pattern formation and growth of the brain, neural tube, myotome, skeletal structures and limb bud, as well as during tissue specification and growth of a variety of organs by acting as context-dependent morphogens and/or mitogens during early development (Cordero et al., 2004; see recent reviews by Ingham 2008; Bürglin 2009). An additional role in somatic cancer is also well documented. All known hedgehog proteins are synthesized as pre-pro-proteins that during their traffic through the endoplasmic reticulum undergo the removal of a signal peptide, followed by autocatalytic cleavage into a 19Kd secreted molecule (e.g. SHH-N, with all known biological activity) and a 25 Kd intein-like residual component [e.g. SHH-C, with the intrinsic ability to cleave the precursor into two fragments (due to the activity of the intein-like Hint motif) and also to add cholesterol to the C-terminus of its partner fragment (facilitated by the sterol-recognition region, SRR); for a review, see Muenke and Beachy 2001; Porter et al., 1995, 1996]. The biological activity of hh-like ligands, such as the human SHH-N, is further enhanced by the addition of palmitate to its N-terminus by the substrate-specific enzyme Hedgehog acyltransferase (Pepinsky et al., 1998; HHAT; MIM# 605743). This bi-lipidated ligand is actively secreted from the cell with the participation of additional co-factors, such as DISP1, that themselves can be targets of mutation in HPE cases (Ma et al., 2002; Roessler et al., 2009). The gradient of these signaling factors that will develop in a target field is influenced by many additional factors including heparin-sulfate proteoglycans, the assembly of multimeric complexes of ligands (Goetz et al., 2006; Singh et al., 2008), as well as both positive (GAS1 and CDO/BOC) and negative modulators (enhanced expression of PTCH1 and HIP) of the cell surface binding and response of target cells (see reviews by Krauss 2007 and Ingham 2008).
Over the past decade, molecular defects have been identified in all three paralogous genes of the prototypical Drosophila melanogaster hedgehog gene (hh) typically present in higher animals, such as humans: INDIAN HEDGEHOG (IHH with autosomal dominant Brachydactyly A-1, BDA1; MIM# 112500 and the autosomal recessive disorder acrocapitalfemoral dysplasia, ACFD; MIM# 607778; Gao et al., 2001; McCready et al., 2002; Liu et al., 2006; Lodder et al., 2008; reviewed in Byrnes et al., 2009), DESERT HEDGEHOG (DHH with gonadal dysgenesis; MIM# 605423; Umehara et al., 2000; Canto et al., 2004, 2005) and SONIC HEDGEHOG (with HPE and related disorders, see above). Fortunately, the past decade has also coincided with an extensive and detailed analysis of the biological effects and functional domains of the proto-typical hedgehog gene that can provide a foundation for predicting the effects of disease-related mutations (Hall et al., 1995, 1997; McLellan et al., 2008; Goetz et al., 2006; Singh et al., 2009; reviewed in Bürglin 2008). Based on these considerations it’s now possible to show that essentially all key steps in the generation of a secreted hedgehog molecule are targets for mutation in a collective retrospective analysis of cases.
MATERIALS AND METHODS
Study population
At the NIH, we analyzed approximately 600 HPE patients (collectively comprising the entire spectrum of HPE brain malformations and prospectively collected over 17 years) for potential sequence variations in the SHH gene under our NHGRI approved brain research protocol and newly established CLIA laboratory. In addition, we also studied 125 unrelated individual normal controls obtained as anonymous samples from the Coriell Institute for Medical Research that matched the predominant Northern European ethnicity of our HPE cases. In the cases extracted from literature reports the nature of the mutation was known to us only through these published sources (cited in Table 1). Similarly, in Rennes, 500 HPE patients were analyzed, prospectively collected over 12 years at the Laboratoire de Génétique Moléculaire (Rennes, France); we also include anonymous instances of mutations in the SHH gene shared with us from prospective studies performed under CLIA standards by GeneDx (Gaithersburg, MD), or from investigators in Maastricht, the Netherlands, as well as from Regensburg, Germany.
Table 1.
SHH mutations identified in this study. Novel variants are in bold type.
| Subject | cDNA | protein | function | Reference |
|---|---|---|---|---|
| 1 | c.9_12dup | p.Arg6GlyfsX59 | frameshift | Nanni 1999 |
| 2 | c.17G>C | p.Arg6Thr | Signal peptide | Dubourg 2004 |
| 3 | c.38_45del | p.Val13AlafsX48 | frameshift |
Nanni 1999; GeneDx confirmed |
| 4 | c.50T>C | p.Leu17Pro | Signal peptide | Kato 2000 |
| 5 | c.72C>A | p.Cys24X | Palmitoylation site |
Dubourg 2004; NIH new |
| 6 | c.77C>T | p.Pro26Leu |
Palmitoylation site and multimerization |
GeneDx new; see Goetz 2006 |
| 7 | c.80G>C | p.Gly27Ala | Palmitoylation site and multimerization |
Hehr 2004 |
| 8 | c.87delG | p.Gly29fsX11 | frameshift | Rennes new |
| 9 | c.91G>A | p.Gly31Arg | SHH-N | Roessler 1996 |
| 10 | c.116T>C | p.Leu39Pro | SHH-N | GeneDX new |
| 11 | c.108_109delinsG | p.Lys38SerfsX2 | frameshift | NIH new |
| 12 | c.119_120delCC | p.Pro41PhefsX22 | frameshift | Rennes new |
| 13 | c.136C>T | p.Gln46X | truncation | NIH new |
| 14 | c.148_166del | p.Asn50X | truncation | Maastrict new |
| 15 | c.157G>A | p.Glu53Lys | SHH-N | Rennes new |
| 16 | c.210dupC | p.Glu71ArgfsX4 | frameshift | GeneDx new |
| p.Gly290Asp | SHH-C | See 89 | ||
| 17 | c.211delG | p.Glu71SerfsX27 | frameshift | Dubourg 2004 |
| 18 | c.248A>T | p.Asp83Val | SHH-N | Rennes new |
| 19 | c.250A>T | p.Ile84Phe | SHH-N | NIH new |
| 20 | c.263A>T | p.Asp88Val | SHH-N |
Nanni 1999: Heussler 2002; Regensburg new |
| 21 | c.298delC | p.Gln100ArgfsX7 | frameshift | GeneDx new |
| 22 | c.298C>T | p.Gln100X | truncation |
Roessler 1996; Rennes new |
| 23 | c.300G>C | p.Gln100His | SHH-N |
Dubourg 2004; Bertolacini 2009 |
| 24 | c.304T>C | p.Cys102Arg | SHH-N |
Maastricht new |
| 25 | c.305G>A | p.Cys102Tyr | SHH-N | Rennes new |
| 26 | c.316_321del | p.Leu106_Asn107del | deletion | Dubourg 2004 |
| 27 | c.313A>T | p.Lys105X | truncation | Roessler 1996 |
| 28 | c.327G>T | p.Leu109Phe | SHH-N | GeneDx new |
| 29 | c.328G>A | p.Ala110Thr | SHH-N | GeneDx new |
| 30 | c.329C>A | p.Ala110Asp | SHH-N | Dubourg 2004 |
| 31 | c.331A>T | p.Ile111Phe | SHH-N | Nanni 2001 |
| 32 | c.332T>A | p.Ileu111Asn | SHH-N | El-Jaick 2005 |
| 33 | c.345C> A | p.Asn115Lys | SHH-N | Nanni 1999 |
| 34 | c.349T>G | p.Trp117Gly | SHH-N | Roessler 1996 |
| 35 | c.349T>C | p.Trp117Arg | SHH-N | Roessler 1996 |
| 36 | c.355G>T | p.Gly119X | truncation | NIH new |
| 37 | c.370G>A | p.Val124Met | SHH-N | GeneDx new |
| 38 | c.383G>A | p.Trp128X | truncation | Marini 2003 |
| 39 | c.388G>T | p.Glu130X | truncation |
Dubourg 2004; NIH new |
| 40 | c.406G>A | p.Glu136Lys | SHH-N | Bertolacini 2009 |
| 41 | c.415_416del | p.Leu139AlafsX13 | frameshift | NIH new |
| 42 | c.419A>C | p.His140Pro | SHH-N | Orioli 2001 |
| 43 | c.420C>G | p.His140Gln | SHH-N | Ribiero 2005 |
| 44 | c.428G>A | p.Gly143Asp | SHH-N | NIH new |
| 45 | c.431G>C | p.Arg144Pro | SHH-N | GeneDx new |
| 46 | c.439G>A | p.Asp147Asn | SHH-N | GeneDx new |
| 47 | c.449C>G | p.Thr150Arg | SHH-N |
Dubourg 2004; NIH new |
| 48 | c.449C>A | p.Thr150Lys | SHH-N | Rennes new |
| 49 | c.468C>A | p.Ser156Arg | SHH-N | NIH new |
| 50 | c.474C>G | p.Tyr158X | truncation | Dubourg 2004 |
| 51 | c.509T>G | p.Phe170Cys | SHH-N | NIH new |
| 52 | c.511G>C | p.Asp171His | SHH-N | Rennes new |
| 53 | c.525C>G | p.Tyr175X | SHH-N |
Regensburg new |
| 54 | c.526_534del | p.Glu176_Lys178del | deletion | Dubourg 2004 |
| 55 | c.547T>C | p.Cys183Arg | SHH-N | NIH new |
| 56 | c.548G>T | p.Cys183Phe | SHH-N | Orioli 2001 |
| 57 | c.548G>A | p.Cys183Tyr | SHH-N |
Regensburg new |
| 58 | c.551C>T | p.Ser184Leu | SHH-N | NIH new |
| 59 | c.562G>C | p.Glu188Gln | SHH-N | Dubourg 2004 |
| 60 | c.562+1G>A | Splice donor | frameshift | NIH new |
| 61 | c.584_598del | p.196_200del |
Cholesterol addition site |
NIH new; GeneDx confirmed |
| 62 | c.587delG | p.Gly196GlufsX20 |
Cholesterol addition site |
Rennes new |
| 63 | c.587G>A | p.Gly196Glu |
Cholesterol addition site |
Rennes new |
| 64 | c.590G>T | p.Gly197Val |
Cholesterol addition site |
NIH new |
| 65 | c.592T>A | p.Cys198Ser |
Cholesterol addition site |
Rennes new |
| 66 | c.593G>A | p.Cys198Phe | Cholesterol addition site |
Bertolacini 2009 |
| 67 | c.624_625delinsTT | p.Glu208AspfsX1 | frameshift | GeneDx new |
| 68 | c.625C>T | p.Gln209X | truncation | Nanni 1999 |
| 69 | c.653T>C | p.Leu218Pro | SHH-C | Richieri_Costa 2006 |
| 70 | c.664G>A | p.Asp222Asn | SHH-C | Dubourg 2004 |
| 71 | c.671T>A | p.Val224Glu | SHH-C | Roessler 1997 |
| 72 | c.676G>A | p.Ala226Thr | SHH-C | Roessler 1997 |
| 73 | c.692G>T | p.Gly231Val | SHH-C | NIH new |
| 74 | c.694C>G | p.Arg232Gly | SHH-C | GeneDx new |
| 75 | c.701T>C | p.Leu234Pro | SHH-C | GeneDx new |
| 76 | c.707G>A | p.Ser236Asn | SHH-C | Rennes new |
| 77 | c.708C>A | p.Ser236Arg | SHH-C | Nanni 1999 |
| 78 | c.721T>C | p.Phe241Val | SHH-C | Rennes new |
| 79 | c.723C>G | p.Phe241Leu | SHH-C | NIH new |
| 80 | c.764T>A | p.Ile255Asn | SHH-C | NIH new |
| 81 | c.766G>T | p.Glu256X | truncation |
Nanni 1999; GeneDx confirmed |
| 82 | c.787_807del | p.Arg263_Ala269del | deletion | Roessler 1997a |
| 83 | c.800C>T | p.Thr267Ile | SHH-C | Hehr 2004 |
| 84 | c.812T>C | p.Leu271Pro | SHH-C | Dubourg 2004 |
| 85 | c.824C>A | p.Ala275Glu | SHH-C | GeneDx new |
| 86 | c.839C>G | p.Ser280Trp | SHH-C |
Regensburg new |
| 87 | c.839C>A | p.Ser280X | truncation | Richieri-Costa 2006 |
| 88 | c.850G>T | p.Glu284X | truncation | Roessler 1997a; Hehr 2004 |
| 89 | c.869G>A | p.Gly290Asp | SHH-C |
Nanni 1999; Schell-Apacik 2009; GeneDx new; |
| 90 | c.887G>C | p.Gly296Ala | SHH-C | GeneDx new |
| 91 | c.928C>T | p.Arg310Cys | SHH-C | Rennes new |
| 92 | c.961C>A | p.Arg321Ser | SHH-C | NIH new |
| 93 | c.995T>C | p.Val332Ala | SHH-C |
Dubourg 2004; Garavelli 2004 |
| 94 | c.1015G>T | p.Glu339X | truncation | Rennes new |
| 95 | c.1037C>T | p.Ala346Val | SHH-C | NIH new |
| 96 | c.1040C>G | p.Pro347Arg | SHH-C |
NIH new, Rennes new |
| 97 | c.1040C>A | p.Pro347Gln | SHH-C | Dubourg 2004 |
| 98 | c.1040C>T | p.Pro347Leu | SHH-C | Rennes new |
| 99 | c.1051C>T | p.Gln351X | truncation | GeneDx new |
| 100 | c.1061T>C | p.Ile354Thr | SHH-C | Dubourg 2004 |
| 101 | c.1085C>T | p.Ser362Leu | SHH-C | NIH new |
| 102 | c.1088G>A | p.Cys363Tyr | SHH-C | Richieri-Costa 2006 |
| 103 | c.1091A>G | p.Tyr364Cys | SHH-C | Rennes new |
| 104 | c.1117G>A | p.Ala373Thr | SHH-C | NIH new; Hehr 2004 |
| 105 | c.1121A>G | p.His374Arg | SHH-C | NIH new |
| 106 | c.1127C>A | p.Ala376Asp | SHH-C | NIH new |
| 107 | c.1130T>C | p.Phe377Ser | SHH-C |
Regensburg new |
| 108 | c.1132_1140del | p.378_380del | deletion | Nanni 1999 |
| 109 | c.1142G>C | p.Arg381Pro | SHH-C | Dubourg 2004 |
| 110 | c.1145T>C | p.Leu382Pro | SHH-C |
Maastricht new |
| 111 | c.1147G>A | p.Ala383Thr | SHH-C | Roessler 1997a; Hehr 2004 |
| 112 | c.1171G>A | p.Ala391Thr | SHH-C |
Regensburg new |
| 113 | c.1204_1227del | p.402_409del | deletion | Schimmenti 2003 |
| 114 | c.1213_1227del | p.405_409del | deletion | Nanni 1999 |
| 115 | c.1231_1233dup | p.Gly411dup | duplication | NIH new |
| 116 | c.1246A>G | p.Thr416Ala | SHH-C | NIH new |
| 117 | c.1270C>G | p.Pro424Ala | SHH-C | Nanni 1999 |
| 118 | c.1303T>A | p.Tyr435Asn | SHH-C | NIH new |
| 119 | c.1307C>T | p.Ser436Leu | SHH-C | Nanni 1999 |
| 120 | c.1366G>C | p.Gly456Arg | SHH-C | NIH new |
| 121 | c.1370delT | p.Met457ArgfsX18 | frameshift | NIH new |
Descriptions of mutations are based on the NM_000193.2 reference sequence, follow HGVS recommendations (www.hgvs.org/mutnomen) and were checked using the Mutalyzer software for the predicted effects on the reference gene (www.lovd.nl/mutalyzer/1.0.1/). Position +1 refers to the A position of the ATG initiation codon for that gene.
Mutation screening, PCR amplification and DNA sequencing
A strategy for screening the SHH gene (available on request) has been modified from previously described ones (Roessler et al., 1996, 1997a; Dubourg et al., 2004) using only four pairs of primers resulting in four amplicons instead of the previous six (1F1/1R1, 2F2/2R1, 3F1/3R1 and 3F3/3R3). Descriptions of mutations (Table 1) are based on the NM_000193.2 reference sequence. Guidelines for the naming of the sequence variants conform to the recommendations of the human nomenclature committee (www.hgvs.org/mutnomen) and were checked using the Mutalyzer software for the predicted effects on the reference gene (www.lovd.nl/mutalyzer/1.0.1/). Position +1 refers to the A position of the ATG initiation codon for that gene.
Our current approach for CLIA testing of SHH also differs slightly from previous reports and is available upon request. Amplification of human genomic DNA is performed in a 35 μl reaction volume, using 60-100 ng of DNA template, 50 μM each of deoxynucleotide triphosphate, 20 pmol of each primer, 3.5 μl of 10X PCR Amplification buffer (Invitrogen, CA), 1.75 μl 10X PCR Enhancer solution (Invitrogen, CA), 1.5mM MgSO4 (Invitrogen, CA) and 2.5U of AmpliTaq DNA polymerase (Roche, IN). All reactions were performed using a PTC-255 thermocycler (MJ Research, MA). Typical PCR cycling parameters were 95°C for 4 minutes followed by 30 cycles at 95°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 1 minute, and a final extension step of 72°C for 7 minutes. One half of the PCR product was used for denaturing high-pressure liquid chromatography (dHPLC) analysis (Trangenomics WAVE™) and the remainder was retained for direct DNA sequencing. Amplicons displaying heterozygous profiles were purified using a high pure PCR purification kit (Roche, IN) and bi-directionally sequenced using the BigDye™ version 3.1 terminator cycle sequencing kit according to the manufacturer’s protocol (Applied Biosystems, CA) on an ABI 3100 automated sequencer.
Statistical analysis
To determinate whether the distribution of observed and expected number of mutations within SHH-N or SHH-C were the same, a classic two-tailed χ2 test was used. In order to detect domains (SHH-N vs. SHH-C) for which the observed number of mutations differed from the expected values, we conducted the following hypothesis test:
where pO,j and pE,j represent, respectively, the proportion of observed and expected mutations in the j domain ( j = Identical, Similar, Related, Unrelated or Biologically Equivalent). Considering that the proportion of observed or expected mutations in the Biologically Equivalent (BE) was calculated as pBE=1 - pU (U: Unrelated), testing H0,j : pO,BE = pE,BE is mathematically identical (but not biologically equivalent) to the test of H0,j : pO,U = pE,U.
We used R 2.9.0 Patched (R Development Core Team. 2009. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org) for the statistical analysis, the RODBC (Lapsley, M. and Ripley, B.D. 2009. RODBC: ODBC Database Access. http://cran.r-project.org/web/packages/RODBC/index.html) package to connect R and MS Excel®, and Tinn-R (Tinn-R Development Team 2004; http://www.sciviews.org/Tinn-R/ and http://sourceforge.net/projects/tinn-r) as editor for the R programs we wrote.
RESULTS
In order to derive insight into the important functions and domains of the SHH protein inferred from our mutational analysis, we have compiled into Table 1 all 121 of the known SHH variants detected exclusively among HPE cases and not in controls. When these alterations are aligned with other mutant Hedgehog proteins and annotated with the detailed structural and functional studies of the murine Shh protein, several important findings emerge from this compilation (broken down for convenience by mutation type in Figure 1A-C). Most of these types of variation have either been directly shown to reduce biological activity (Schell-Apacik et al., 2003; Traiffort et al., 2004; Maity et al., 2005; Goetz et al., 2006; Singh et al., 2009) or would be predicted to lose activity by analysis of the effects on key structural motifs of virtually translated protein products. However, the precise effects of individual mutations can rarely be predicted with certainty without detailed and often laborious functional analysis.
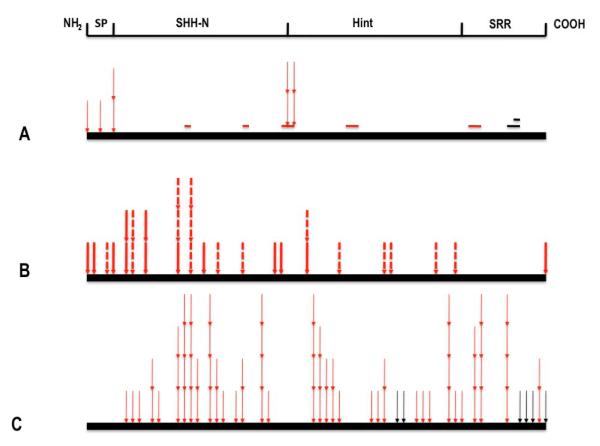
Figure 1.
Compilation of all of the mutations affecting the two principal SHH domains sorted by their position and category. A: deletions or insertions that retain frame occur most commonly in critical regions (red colored bars) but also non-conserved regions of unknown importance (black colored bars; see also Fig. 2); selected missense alterations (thin arrows) that occur in the signal peptide, palmitate addition region, cholesterol addition site are presented here to imply their likely effect on processing. B: nonsense and frameshift mutations truncate SHH-N or affect domains required for protein intein-like splicing or cholesterol addition. C: missense alterations (thin arrows) occur equally in both domains; however, the distribution of mutations is non-random and preferentially occurs within conserved domains (see Table 2).
Evidence for mutations likely causing processing defects of the SHH protein
Since any observed mutation in SHH-C would not contribute directly to the structure of the SHH-N ligand itself, and yet full biological activity is dependent on the success of many successive processing steps, it was originally postulated that defective maturation of the protein could the basis for the pathogenicity of some of these mutations (Roessler et al., 1997a; Hall et al., 1997). We can now affirm that mutations in HPE cases are non-randomly clustered, and commonly occur in precisely those key elements that contribute to successful processing, the absence of which is known to dramatically reduce protein function.
Interestingly, in addition to the single example of elimination of the initiator methionine of DHH (Umehara et al., 2000) there are two additional examples of missense changes detected in the signal peptide (p.Arg6Thr and p.Leu17Pro) of SHH that might interfere with entry into the endoplasmic reticulum; although neither variant has been analyzed functionally in transfected cells, the p.Leu17Pro variant segregates among affected individuals in a large pedigree. Similarly, a cluster of mutations are detected in the highly conserved CGPGR motif at the amino-terminus implicated in both the addition of palmitate (e.g. p.Pro26Leu and p.Gly27Ala) and the formation a multimeric high potency forms (Goetz et al., 2006; Singh et al., 2008). Experimental mutation of these or similar residues in the rat Shh protein alters its ability to be lipid modified, secreted and/or form full active multimer forms (Goetz et al., 2006, see also Fig.2). Another cluster of missense alterations and precise deletions eliminate the cholesterol addition site (p.Gly197; detailed in Fig. 1, 2) at the end of the ligand domain: p.196_200del, p.Gly196GlufsX20, p.Gly196Glu, p.Gly197Val, p.Cys198Phe and p.Cys198Ser. Thus, the mutational spectrum observed in patients directs our attention to essential motifs independently shown to be necessary for the production of a bioactive molecule.
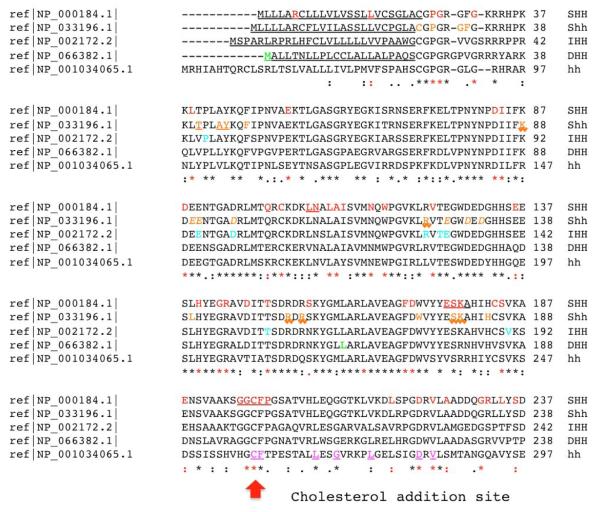
Figure 2. A detailed annotation of known mutations within closely related Hedgehog proteins.
A clustalW2 based (http://www.ebi.ac.uk/Tools/clustalw2/) single aminoacid code alignment of SHH, murine Shh, IHH, DHH and hh is presented. Protein changes are annotated by color and font. For example, HPE or SCMMI alterations are red mutations: normal font are typical missense, red underlined positions are in-frame deleted residues in functionally important motifs, while in-frame deletions or insertions of unknown effect are depicted as black lines above the alignment (p.402_409del and p.405_409del) or as a triangle (i.e. p.Gly411dup). For the mouse Shh alignment we use orange residues with normal font as synthetically engineered point mutations (see Hall et al., 1997; Goetz et al., 2006; Singh et al. 2008), and surface residues implicated in Shh-N-CDOFn3 (ligand-receptor) binding (McLellan et al. 2008) are underlined with a saw-tooth font. In contrast, residues implicated in binding to the fly paralog, Ihog, are orange and underlined. These authors also identified residues that co-ordinate with calcium and promote receptor binding that we present as orange and italic. In addition, IHH (Brachydactyly A-1 or acrofemoralcapital dysplasia; turquoise mutations), DHH (gonadal dysgenesis; green mutations) are presented and compared directly to the Drosophila hh reference sequence. Each paralog encodes a signal peptide shown as underlined normal font (the signal peptide of hh is not shown) and a Sterol Recognition Region (SSR) that is both underlined and italicized. Note that key motifs necessary for hh auto-processing [e.g. the Shh Hint motif (p.258_408) consists of functionally conserved core aminoacids that are show in purple and underlined; a subset of these have been shown to be functionally essential and are double underlined: D303, T326 and H329 (Hall et al. 1997).
The mutations observed in both the Hint module and SRR motifs are biologically plausible as loss-of-function alterations based on site-directed mutagenesis studies (Fig. 2, see also Hall et al., 1997) and are consistent with our general observation that they occur preferentially within conserved residues important for this structure (see chi squared analysis, Table 2). For example, the positions p.Asp222, p.Val224, p.Phe241, p.263_269del, p.Thr267, p.Leu271 and p.Ile354 correspond to positions that are highly conserved residues among intein-like elements across many phyla and it is statistically significant that they are also sites of mutation in HPE subjects (see also Traiffort et al., 2004 for examples of confirmed deleterious mutations in SHH-C; see Canto et al., 2004 for a similar mutation in DHH). This pattern of mutation within largely conserved residues applies to the majority of the mutations observed in the Hint motif (p.Leu218Pro, p.Ser236Asn, p.Ile255Asn, p.Val332Ala, p.Ala346Val, p.Pro347Gln, p.Pro347Arg, p.Pro347Leu, p.Ser362Leu, p.Cys363Tyr, p.Tyr364Cys). In fact, a similarly strong mutational signature is also observed in the SRR region that functions to bind cholesterol and uses it as a nucleophile in the autoprocessing reaction: p.Ala373Thr, p.His374Arg, p.Ala376Asp, p.Phe377Ser, p.378_380del, p.Arg381Pro, p.Leu382Pro and p.Ala383Thr. Note that the last position in this cluster is also the site of a nonsense mutation in DHH.
Table 2.
Chi squared analysis of mutations in different SHH domains.
| Domain | Observed | Expected | Statistics, p-value |
|---|---|---|---|
| Frequency (%) | Frequency (%) | ||
| N terminal | , p<0.00001 | ||
| Identical (*) | 28 (70) | 100 (50.8) | , p<0.05 |
| Similar (:) | 4 (10) | 34 (17.3) | , p>0.05 |
| Related (.) | 1 (2.5) | 13 (6.6) | , p >0.05 |
| Unrelated | 7 (17.5) | 50 (25.4) | , p >0.05 |
| Biologically Equivalent1 | 34 (82.5) | 147 (74.6) | , p >0.05 |
| Total | 40 (100) | 197 (100) | |
| C terminal | , p<0.00001 | ||
| Identical (*) | 20 (45.5) | 49 (18.5) | , p <0.001 |
| Similar (:) | 9 (20.5) | 29 (10.9) | , p >0.05 |
| Related (.) | 4 (9.1) | 18 (6.8) | , p >0.05 |
| Unrelated | 11 (25) | 169 (63.8) | , p<0.00001 |
| Biologically Equivalent1 | 33 (75) | 96 (36.2) | , p<0.00001 |
| Total | 44 (100) | 265 (100) | |
Includes Identical, Similar, and Related domains as defined by clustalW2 criteria.
Typical frameshift and truncation mutations
Nearly a quarter of all of the mutations detected (29/121) are frameshifts or truncations incompatible with a complete protein. All but one (pMet457ArgfsX18) would be expected to eliminate the intein-like Hint motif necessary for autocatalytic processing and would be considered as likely biological null alleles.
Missense alterations of both SHH-N and SHH-C occur in functionally defined domains
The distribution of HPE-associated mutations in the mature SHH-N domain continues to follow the pattern of conservation established for SHH-C (Table 2) although, counter-intuitively, the level of significance is less because of the overall high degree of conserved motifs; in addition, those mutations tend to be observed within centrally-located conserved residues thought to be important in ligand-receptor interactions or similar structurally relevant modules. For example, p.Glu176_Lys178del removes residues shown to be essential for the binding between Shh and the Fibronectin3 motif of the CDO receptor (McClellan et al., 2008). In general, mutations in IHH and DHH also occur in centrally located conserved residues (see Fig. 2; Byrnes et al., 2009); furthermore, it is unlikely to be a co-incidence that mutation of SHH-N residues p.Thr150 is also a site for mutation in IHH.
Unclassified mutations
Some variants do not fall into any clear category, other than that they are not generally conserved from flies to higher vertebrates: such as p.Gly290Asp, p.Gly296Ala, p.Ala391Thr, p.402_409del, p.405_409del, p.Gly411dup, p.Thr416Ala, p.Pro424Ala and p.Gly456Arg (Nanni et al., 1999; Schimmenti et al., 2001). It is probable that there will be instances of highly conserved residues being non-significant when functional tests are eventually performed (see Maity et al., 2005), as well as instances where seemingly un-obvious mutations have profound effects. For example, our alignment analysis fails to take into account human-specific features evolved in the primate lineage (Dorus et al., 2006). These authors note that SHH-C has undergone accelerated evolution within primates and the non-random acquisition of multiple serines and threonines that may serve as sites of post-translational modification. For example, the p.Gly290Asp mutation might affect the phosporylation state of p.Ser291 or the deletions p.402_409del or p.405_409del the postulated phosphorylation of p.Ser401. Additional studies would be needed to test this hypothesis. Hence, the best way for a clinician to establish the likelihood of a mutation in these genes being clinically relevant would be to carefully examine the family history and mutation status of additional family members.
DISCUSSION
Holoprosencephaly is the most common structural malformation of the developing forebrain in humans and its causation includes genetic and environmental components typical for a complex trait that must integrate both types of factors (Muenke and Beachy, 2001; Roessler et al., 2001, 2003, 2008; Cohen, 2006; Dubourg et al., 2007; Krauss 2007; Monuki 2007). Its genetic heterogeneity is supported by the fact that nearly 75% of cases are wild-type in their DNA sequence of the coding regions of the nine currently known HPE genes in patients with normal chromosomes. Its characteristic variable expressivity adds additional support for a model of combinatorial interactions among multiple factors (Ming and Muenke, 2002). Although much more needs to be elucidated regarding the potential roles of additional genes, we are beginning to see the emergence of a network of factors in the mouse that converge on the expression of Sonic hedgehog as a common final pathway in HPE etiology (Jeong et al., 2008; Geng et al., 2008; Warr et al., 2008).
ACKNOWLEDGMENTS
The authors wish to thank the patients who participated in this research and the many clinicians who referred them. This research was supported by the Division of Intramural Research of the National Human Genome Research Institute, National Institutes of Health, and by the GIS Institut des Maladies Rares, COREC (CHU, Faculté de Médecine, Rennes, France).
REFERENCES
- Bendavid C, Haddad BR, Griffin A, Huizing M, Dubourg C, Gicquel I, Cavalli LR, Pasquier L, Long R, Ouspenskaia M, Odent S, Lacbawan F, David V, Muenke M. Multicolor FISH and quantitative PCR can detect submicroscopic deletions in holoprosencephaly patients with a normal karyotype. J Med Genet. 2005a;43:496–500. doi: 10.1136/jmg.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Dubourg C, Gicquel I, Pasquier L, Saugler-Veber P, Durou M-R, Jaillard S, Frebourg T, Haddad BR, Henry C, Odent S, David V. Molecular evaluation of foetuses with holoprosencephaly shows high incidence of microdeletions in the HPE genes. Hum Genet. 2005b;119:1–8. doi: 10.1007/s00439-005-0097-6. [DOI] [PubMed] [Google Scholar]
- Bertolacini CDP, Richieri-Costa A, Ribeiro-Bicudo L. Sonic hedgehog mutation in patients within the spectrum of holoprosencephaly. Brain Dev. 2009 doi: 10.1016/j.braindev.2009.02.014. (in press) [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu C-y, Roeder ER, Stengel-Rutkowski S, Hennekam RCM, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. The Hedgehog protein family. Genome Biol. 2008;9:241. doi: 10.1186/gb-2008-9-11-241. (E-pub doi:10.1186/gb-2008-9-11-241] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes AM, Racacho L, Grimsey A, Hudgins L, Kwan AC, Sangalli M, Kidd A, Yaron Y, Lau Y-L, Nikkel SM, Bulman DE. Brachydactyly A-1 mutations restricted to the central region of the N-terminal active fragment of Indian Hedgehog. Euro J Hum Genet. 2009 doi: 10.1038/ejhg.2009.18. [e-pub: doi:10.1038/ejhg.2009.18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto P, Vilchis F, Söderlund D, Reyes E, Méndez JP. A heterozygous mutation in the desert hedgehog gene in patients with mixed gonadal dysgenesis. Mol Hum Reprod. 2005;11:833–836. doi: 10.1093/molehr/gah216. [DOI] [PubMed] [Google Scholar]
- Canto P, Söderlund D, Reyes E, Méndez JP. Mutations in the Desert hedgehog (DHH) gene in patients with 46,XY complete pure gonadal dysgenesis. The J Clin Endo Met. 2004;89:4480–4483. doi: 10.1210/jc.2004-0863. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr. Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res Part A Clin Mol Teratol. 2006;76:658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M, Helms JA. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–494. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domené S, Roessler E, El-Jaick KB, Snir M, Brown JL, Vélez JI, Bale S, Lacbawan F, Muenke M, Feldman B. Mutations in the human SIX3 gene in holoprosencephaly are loss of function. Hum Mol Genet. 2008;17:3919–3928. doi: 10.1093/hmg/ddn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Anderson JR, Vallender EJ, Gilbert SL, Zhang L, Chemnick LG, Ryder OA, Li W, Lahn BT. Sonic Hedgehog, a key development gene, experienced intensified molecular evolution in primates. Hum Mol Genet. 2006;15:2031–2037. doi: 10.1093/hmg/ddl123. [DOI] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphant J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le Duff F, Durou M-R, Odent S, David V. Molecular screening of SHH, ZIC2, SIX3 and TGIF genes in patients with features of holoprosencephaly spectrum: mutation review and genotype-phenotype correlations. Hum Mut. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Brunoni D, Castilla EE, Moreira MA, Orioli IM. SHH Ile111Asp in alobar holoprosencephaly in a proposita, whose mother had only a solitary median maxillary incisor. Am J Med Genet A. 2005;136:345. doi: 10.1002/ajmg.a.30624. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Fonseca RF, Moreira MA, Ribeiro MG, Bolognese AM, Dias SO, Pereira ET, Castilla EE, Orioli IM. Single median maxillary central incisor: new data and mutation review. Birth Defects Res (PartA) 2007a;79:573–580. doi: 10.1002/bdra.20380. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Powers SE, Bartholin L, Myers KR, Hahn J, Orioli IM, Ouspenskaia M, Lacbawan F, Roessler E, Wotton D, Muenke M. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007b;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Guo J, She C, Shu A, Yang M, Tan Z, Yang X, Guo S, Feng G, He L. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat Genet. 2001;28:386–388. doi: 10.1038/ng577. [DOI] [PubMed] [Google Scholar]
- Garavelli L, Zanacca C, Caselli G, Banchini G, Dubourg C, David V, Odent S, Gurrieri F, Neri G. Solitary median maxillary central incisor syndrome: Clinical case with a novel mutation of Sonic Hedgehog. Am J Med Genet. 2004;127A:93–95. doi: 10.1002/ajmg.a.20685. [DOI] [PubMed] [Google Scholar]
- Geng X, Speirs C, Laguitin O, Solnica-Krezel L, Jeong Y, Epstein D, Oliver G. Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell. 2008;15:236–247. doi: 10.1016/j.devcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ. A highly conserved amino-terminal region of Sonic hedgehog is required for the formation of its freely diffusible multimeric form. J Biol Chem. 2006;281:4087–4093. doi: 10.1074/jbc.M511427200. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richeri-Costa A, Zackai EH, Massague J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signaling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hall TMT, Porter JA, Beachy PA, Leahy DJ. A potential catalytic site revealed by the 1.7Å crystal structure of the amino-terminal signaling domain of Sonic hedgehog. Nature. 1995;378:212–216. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- Hall TMT, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Crystal structure of a hedgehog autoprocessing domain: homology between hedgehog and self-splicing proteins. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- Hehr U, Gross C, Diebold U, Wahl D, Beudt U, Heidemann P, Hehr A, Mueller D. Wide phenotypic variability in families with holoprosencephly and a sonic hedghog mutation. Eur J Pediatr. 2004;163:347–352. doi: 10.1007/s00431-004-1459-0. [DOI] [PubMed] [Google Scholar]
- Heussler HS, Suri M, Muenke M. Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephly. Arch Dis Child. 2002;86:293–296. doi: 10.1136/adc.86.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. Hedgehog signaling. Cur Biol. 2008;18:R238–R241. doi: 10.1016/j.cub.2008.01.050. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Leskow FC, El-Jaick K, Roessler E, Muenke M, Yocum A, Dubourg C, Li X, Geng X, Oliver G, Epstein DJ. Regulation of a remote Shh forebrain enhancer by the Six3 protein. Nat Genet. 2008;40:1348–1353. doi: 10.1038/ng.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Nanba E, Akaboshi S, Shiihara T, Ito A, Honma T, Tsuburaya K, Havasaka K. Sonic hedgehog signal peptide mutation in a patient with holoprosencephaly. Ann Neurol. 2000;47:514–516. [PubMed] [Google Scholar]
- Krauss RS. Holoprosencephaly: new models, new insights. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000440. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang X, Cai Z, Tang Z, Cao K, Liang B, Ren X, Liu JY, Wang QK. A novel heterozygous mutation in the Indian hedgehog gene (IHH) is associated with brachydactyly type A1 in a Chinese family. J Hum Genet. 2006;51:727–731. doi: 10.1007/s10038-006-0012-6. [DOI] [PubMed] [Google Scholar]
- Lodder EM, Hoogeboom AJM, Coert JH, de Graaff E. Deletion of 1 amino acid in Indian Hedgehog leads to Brachydactyly A1. Am J Med Genet. 2008;146A:2152–2154. doi: 10.1002/ajmg.a.32441. [DOI] [PubMed] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, Beachy PA. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of Dispatched. Cell. 2002;111:63–75. doi: 10.1016/s0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- Maity T, Fuse N, Beachy PA. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci, USA. 2005;102:17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini M, Cusano R, De Biasio P, Caroli F, Lerone M, Silengo M, Ravazzolo R, Seri M, Camera G. Previously undescribed nonsense mutation in SHH caused autosomal dominant holoprosencephaly with wide intrafamilial variability. Am J Med Genet. 2003;117A:112–115. doi: 10.1002/ajmg.a.10163. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologs is not conserved across different phyla. Nature. 2008;455:979–984. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready ME, Sweeney E, Fryer AE, Donnai D, Baig A, Racacho L, Warman ML, Hunter AGW, Bulman DE. A novel mutation in the IHH gene causes brachydactyly type A1: a 95-year-old mystery resolved. Hum Genet. 2002;111:368–375. doi: 10.1007/s00439-002-0815-2. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: digenic disease and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Monuki ES. The morphogen signaling network in forebrain development and holoprosencephaly. J Neuropath Exp Neurosci. 2007;66:566–575. doi: 10.1097/nen.0b013e3180986e1b. [DOI] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Holoprosencephaly. In: Scriver CR, et al., editors. The Metabolic & Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 6203–6230. [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, de Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Del Campo M, Martin RA, Meinecke P, Pierpoint MEM, Robin NH, Young ID, Roessler E, Muenke M. The mutational spectrum of the Sonic Hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Du Y, Hall RK, Aldred M, Bankier A, Muenke M. Shh mutation is associated with solitary median maxillary central incisor: A study of 13 patients and review of the literature. Am J Med Genet. 2001;102:1–10. doi: 10.1002/1096-8628(20010722)102:1<1::aid-ajmg1336>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Orioli IM, Castilla EE, Ming JE, Nazer J, de Aguiar MJ Burle, Lerena JC, Muenke M. Identification of novel mutations in SHH and ZIC2 in a South American (ECLAMC) population with holoprosencephaly. Hum Genet. 2001;109:1–6. doi: 10.1007/s004390100537. [DOI] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker S, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage is active in local and long-range signaling. Nature. 1995;374:363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Pepinsky RB, Zen C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowsky K, Taylor FR, Wang EA, Galdes A. Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem. 1998;273:14037–14045. doi: 10.1074/jbc.273.22.14037. [DOI] [PubMed] [Google Scholar]
- Ribiero LA, Richieri-Costa A. Single median maxillary incisor, hypophyseal tumor, and SHH mutation. Am J Med Genet. 2005;136A:346–347. doi: 10.1002/ajmg.a.30625. [DOI] [PubMed] [Google Scholar]
- Richieri-Costa A, Ribeiro LA. Holoprosencephaly-like phenotype: clinical and genetic perspectives. Am J Med Genet. 2006;140A:2587–2593. doi: 10.1002/ajmg.a.31378. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui L-C, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Vargas F, Scherer SW, Tsui L-C, Muenke M. Mutations in the C-terminal domain of Sonic Hedgehog cause holoprosencephaly. Hum Mol Genet. 1997a;6:1847–1853. doi: 10.1093/hmg/6.11.1847. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ward DE, Gaudenz K, Belloni E, Scherer SW, Donnai D, Siegel-Bartelt J, Tsui L-C, Muenke M. Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum Genet. 1997b;100:172–181. doi: 10.1007/s004390050486. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. Midline and laterality defects: left and right meet in the middle. BioEssays. 2001;23:888–900. doi: 10.1002/bies.1130. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. How a hedgehog might see holoprosencephaly. Hum Mol Genet. 2003;12(Spec No 1):R15–25. doi: 10.1093/hmg/ddg058. [DOI] [PubMed] [Google Scholar]
- Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dluglosz AA, Muenke M. A previously unidentified amino terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- Roessler E, Lacbawan F, Dubourg C, Paulussen A, Herbergs J, Hehr U, Bendavid C, Zhou N, Ouspenskaia M, Bale S, Odent S, David V, Muenke M. The full spectrum of holoprosencephaly-associated mutations within the ZIC2 gene in humans predict loss-of-function as the predominant disease mechanism. Hum Mutat. 2009;30:E541–544. doi: 10.1002/humu.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ouspenskaia MV, Karkera JD, Vélez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, Feldman B, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, Lacbawan F, Bendavid C, Dubourg C, Beachy PA, Muenke M. Truncating loss-of function mutations of DISP1 contribute to holoprosencephly-like microform features in humans. Hum Genet. 2009;125:393–400. doi: 10.1007/s00439-009-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell-Apacik C, Rivero M, Knepper JL, Roessler E, Muenke M, Ming JE. SONIC HEDGEHOG mutations causing human holoporsencephaly impair neural patterning activity. Hum Genet. 2003;113:170–177. doi: 10.1007/s00439-003-0950-4. [DOI] [PubMed] [Google Scholar]
- Schell-Apacik CC, Ertl-Wagner B, Panzel A, Klausner K, Rausch G, Muenke M, von Voss H, Hehr U. Maternally inherited heterozygous sequence change in the Sonic Hedgehog gene in a male patient with bilateral close-lip schizencephaly and partial absence of the corpus callosum. Am J Med Genet. 2009 doi: 10.1002/ajmg.a.32940. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmenti LA, de la Cruz J, Lewis RA, Karkera JD, Manligas GS, Roessler E, Muenke M. Novel mutaion in sonic hedgehog in non-syndromic colobomatous microphthalmia. Am J Med Genet. 2003;116A:215–221. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- Singh S, Tokhunts R, Baubet V, Goetz JA, Huang ZJ, Schilling NS, Black KE, MacKenzie TA, Dahmane N, Robbins DJ. Sonic hedgehog mutations identified in holoprosencephaly patients can act in a dominant negative manner. Hum Genet. 2009;125:95–103. doi: 10.1007/s00439-008-0599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traiffort E, Dubourg C, Faure H, Rognan D, Odent S, Durou M-R, David V, Ruat M. Functional characterization of Sonic Hedgehog mutations associated with holoprosencephaly. J Biol Chem. 2004;279:42889–42897. doi: 10.1074/jbc.M405161200. [DOI] [PubMed] [Google Scholar]
- Umehara F, Tate G, Itoh K, Yamaguchi N, Douchi T, Mitsuya T, Osame M. A novel mutation of desert hedgehog in a patient with 46, XY partial gonadal dysgenesis accompanied by minifascicular neuropathy. Am J Hum Genet. 2000;67:1302–1305. doi: 10.1016/s0002-9297(07)62958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M. Missense mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- Warr N, Powles-Glover N, Chappell A, Robson J, Norris D, Arkell RM. Zic2-associated holoprosencephaly is caused by a transient defect in the organizer region during gastrulation. Hum Mol Genet. 2008;17:2986–2996. doi: 10.1093/hmg/ddn197. [DOI] [PubMed] [Google Scholar]