Abstract
We examined salt tolerance responsive genes in Pak-choi under salt stress and analyze their potential function. The mRNA differential display was used to screen the transcript derived fragments (TDFs) related to salinity tolerance in tolerant and moderately tolerant Pak-choi germplasm. Seventy-eight primer combinations generated 101 differential cDNA fragments, which were divided into 10 expression types. Seven cDNA sequences (GenBank accession Nos. DQ006915~DQ006921) obtained and sequenced were highly homologous to some known expression genes or the genes related to the signaling pathways in plants under different abiotic stress.
Keywords: Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee, Salinity tolerance, Gene differential expression, cDNA fragments, Basic local alignment search tool (BLAST) analysis
INTRODUCTION
In China, there is a saline soil area of 6.67 million hm2, accounting for about 7% of total farmland areas (Zhao, 1994). Salt harm to vegetable production is not only from the natural saline soil in open field but also from the secondary saline soil in greenhouse. Although various methods such as reclamation, irrigation, and drainage have been used to reduce soil salinity, they are not always economic or practical. One of optional strategies is to develop salinity-tolerant varieties.
Although most vegetables are very sensitive to salinity, some varieties were found to be salinity-tolerant, such as water cress (Nasturtium officinale R. Br.) (Du et al., 1999); celery (Apium graveolens L.), beet [Beta vulgaris (L.) Koch, Cicla group], New Zealand spinach (Tetragonia expansa Murray), and spinach (Spinacia oleracea L.) (Li, 2001). Pak-choi [Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee] originated in China, and in recent decades, has been introduced to other Asian countries and European and American countries. Its annual yield accounts for 30%~40% of total vegetable production in China (Xiong, 2004). Although highly tolerant germplasm had been found in Pak-choi through salt tolerance identification (Zhi, 2003), no investigation at molecular level on salt tolerance in Pak-choi has been reported before. In the present study, we examined salt tolerance responsive genes in elite Pak-choi germplasm under salt stress in order to develop salinity-tolerant vegetables and to further understand the mechanism of salinity tolerance.
MATERIALS AND METHODS
Plant materials and salt stress treatment
The parental Pak-choi, salt-tolerant germplasm II2B 1183 and moderately salt-tolerant germplasm II2B 548, were from our previous study (Zhi, 2003). They were planted in an experimental field at the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China, in January 2004.
To analyze differential expression of salinity-tolerant genes, the salt-tolerant germplasm (1183) and moderately salt-tolerant germplasm (548) were sown in nursery pots. When their seedlings had two true leaves, they were transplanted into a plastic container (680 mm×460 mm×165 mm) with 1/4 Hongland liquid and the suspending foam slab with holes for water cultivation. The 1/4 Hongland liquid was renewed once every two days for aerating. Seven days later, the seedlings were transplanted into the 1/4 Hongland liquid containing 1.6% (w/v) NaCl for 48 h for RNA isolation. The same set of materials was cultured in 1/4 Hongland liquid without salt stress as control.
RNA isolation and cDNA synthesis
Total RNA was extracted from tender leaves of tested seedlings using the TRIzol reagent kit provided by GIBCO (USA) and the manufacturer’s instruction was followed. Complementary DNA (cDNA) was generated using SuperScript™ First-Strand Synthesis System for reverse transcription-polymerase chain reaction (RT-PCR; Invitrogen, USA) according to the manufacturer’s protocol. OligodT12A, oligodT12G, and oligodT12C were used as anchored primers. The sequences of arbitrary primers were from the Shanghai Biology Inc., China. All primers were synthesized by Shanghai Biology Inc. (Table 1).
Table 1.
Sequences of anchored and arbitrary primers used in the study*
| Primer | Sequence |
| B0301 | 5′d(TAC AAC GAG G)3′ |
| B0302 | 5′d(TGG ATT GGT C)3′ |
| B0303 | 5′d(CTT TCT ACC C)3′ |
| B0304 | 5′d(TTT TGG CTC C)3′ |
| B0305 | 5′d(GGA ACC AAT C)3′ |
| B0306 | 5′d(AAA CTC CGT C)3′ |
| B0307 | 5′d(TCG ATA CAG G)3′ |
| B0308 | 5′d(TGG TAA AGG G)3′ |
| B0309 | 5′d(TCG GTC ATA G)3′ |
| B0310 | 5′d(GGT ACA TTG G)3′ |
| B0311 | 5′d(TAC CTA AGC G)3′ |
| B0312 | 5′d(CTG CTT GAT G)3′ |
| B0313 | 5′d(GTT TTC GCA G)3′ |
| B0314 | 5′d(GAT CAA GTC C)3′ |
| B0315 | 5′d(GAT CCA GTA C)3′ |
| B0316 | 5′d(GAT CAC GTA C)3′ |
| B0317 | 5′d(GAT CTG ACA C)3′ |
| B0318 | 5′d(GAT CTC AGA C)3′ |
| B0319 | 5′d(GAT CAT AGC C)3′ |
| B0320 | 5′d(GAT CAA TCG C)3′ |
| B0321 | 5′d(GAT CTA ACC G)3′ |
| B0322 | 5′d(GAT CGC ATT G)3′ |
| B0323 | 5′d(GAT CTG ACT G)3′ |
| B0324 | 5′d(GAT CAT GGT C)3′ |
| B0325 | 5′d(GAT CAT AGC G)3′ |
| B0326 | 5′d(GAT CTA AGG C)3′ |
| dT12A | 3′d(TTT TTT TTT TTT A)5′ |
| dT12G | 3′d(TTT TTT TTT TTT G)5′ |
| dT12C | 3′d(TTT TTT TTT TTT C)5′ |
From Shanghai Biology Inc., China
Differential display
Extract telomerase and detect its activity according to the manufacturer’s instruction in the telomerase PCR-enzyme-linked immunosorbent assay (ELISA) kit (Boehringer Mannheim Company, Germany). The synthesized cDNA was used as template for differential display RT-PCR. The reaction included 10× buffer, 1.5 mmol/L MgCl2, 0.05 mmol/L dNTP (Sino-American Biotechnology Company), 0.96 μmol/L anchored primer, 0.48 μmol/L random primer (B0301~B0326, Table 1). Shanghai Biology Inc., Shanghai, China), 1.25 U Taq DNA polymerase (Promega, Madison, WI, USA), and 2 μl cDNA template in a total volume of 25 μl. The PCR cycling parameters were 40 cycles of 94 °C for 2 min, 94 °C for 30 s, 40 °C for 1 min, and 72 °C for 1 min, followed by 72 °C for 4 min. The amplified cDNA was separated by electrophoresis in 6% (w/v) denaturing polyacrylamide gels.
Northern blot analysis
Differentially displayed cDNA fragments were re-amplified followed by Northern blotting using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Switzerland). Random prime DNA was labeled with digoxigenin-dUTP Alkali-labile and hybrids were detected by enzyme immunoassay according to the manufacturer’s instruction. The cDNA fragments from 6% (w/v) denaturing polyacrylamide gels were dissolved in 50 μl water. The re-amplification reaction system was the same as differential display PCR expect 0.96 μmol/L random primer (B0301~B0326; Shanghai Biology Inc., Shanghai, China) and 2 μl cDNA fragments in a total volume of 50 μl. The PCR cycling parameters were the same as differential display PCR.
Cloning and sequencing of the target differential cDNA
Differential cDNA fragments after re-amplification were separated on a 1% (w/v) agarose gel and purified with TaKaTa agarose gel DNA purification lot CA401 (TaKaTa, Japan) according to the manufacturer’s instruction. The differentially displayed cDNAs were ligated into pGEM-T easy vector (Promega, Madison, USA) and sequenced (Sangon, China).
Basic local alignment search tool (BLAST) analysis
BLAST analysis on homology of target fragments in this study was conducted through GenBank (www.ncbi.nlm.nih.gov) and Brassica database (www.arabidopsis.org). Homology comparison of putative fragments in the study was done by DNAsis software.
RESULTS
Differential expression of the salinity tolerance related genes
It was indicated that differential expression of salinity tolerance related genes in Pak-choi for 48 h under 1.6% (w/v) NaCl salt stress was stable and significant based on the results of the mRNA differential display under salt stress and non-salt stress for different time courses by the primer combination T12A+B0325 in our previous study (Qiu, 2004). The mRNA differential display was used to screen the related transcript derived fragments (TDFs) of salt-tolerant germplasm 1183 and moderate salt-tolerant germplasm 548 (Fig.1). Seventy-eight primer combinations generated 101 differential cDNA fragments that were divided into 10 expression types (Table 2).
Fig.1.
mRNA differential display in two Pak-choi germplasms under salt stress and non-salt stress by primer combination T12A+B0301
The five lanes are marker (M), 1183 under salt stress, 1183 control (CK), 548 under salt stress, and 548 CK, respectively
Table 2.
Differential expression types and their frequency distribution under salt stress
| Differential expression type* | Numbers of bands corresponding to each expression type |
| 1 0 0 0 | 25 |
| 1 0 1 0 | 1 |
| 0 1 0 0 | 16 |
| 0 1 1 1 | 8 |
| 0 0 0 1 | 15 |
| 1 1 1 0 | 2 |
| 0 0 1 0 | 15 |
| 0 1 1 0 | 15 |
| 1 0 1 1 | 1 |
| 0 1 0 1 | 3 |
| Total | 101 |
“1” stands for band present; “0” stands for band absent
The order of four lanes is 1183 under salt stress, 1183 control (CK), 548 under salt stress, and 548 CK. The enhanced expression types under salt stress were 1000, 1010, 1110, 0010, and 1011, the inhibited expression types under salt stress were 0001, 0111, 0100, 0111, and 0101
Identification of cDNA differential expression fragments
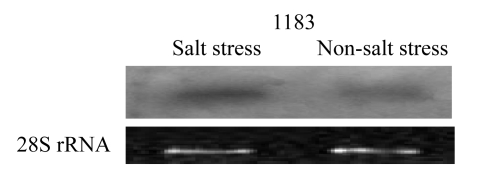
The cDNA fragments of stable and strong differential expression were proved to be positive by repeated PCR. cDNA fragment of No. 85 was proved to be positive by Northern blotting (Fig.2). Among them, seven cDNA fragments were re-amplified, cloned, and sequenced. cDNA fragments of No. 38 (GenBank accession No. DQ006920) and No. 68 (GenBank accession No. DQ006917) from 1183 belonged to the type of enhanced expression. It was found that they were respectively 90% and 95% homologous to the expression genes in Brassica napus under frost stress and in Raphanus sativus under drought stress by BLAST analysis from GenBank and Brassica database. The cDNA fragments of No. 03 (GenBank accession No. DQ006919) and No. 27 (GenBank accession No. DQ006915) both from 1183 were also high expressed. They were 85% and 86% homologous, respectively, to the mRNA of calcium-dependent protein kinase in Arabidopsis thaliana. cDNA fragment No. 73 (GenBank accession No. DQ006921) from 1183 was 90% homologous to putative kinase interactor mRNA in Arabidopsis thaliana. cDNA fragment No. 79 (GenBank accession No. DQ006916) restrained under salt stress in 548 was 90% homologous to the conserved mitogen activated protein kinase (MAPKK) kinase mRNA that is related to the negative regulation of defense responses and the mitogen activated protein kinase kinase kinase (MAPKKK) mRNA in Arabidopsis thaliana. The cDNA fragment No. 85 (GenBank accession No. DQ006918) boosted up in 1183 was 99% homologous to Homo sapiens genomic DNA fragment of anti-oncogene of hepatocellular colorectal and non-small cell lung cancers.
Fig.2.
Northern blotting analysis of cDNA fragment No. 85
cDNA fragment No. 85 was more strongly induced under salt stress than under non-salt stress. 2 μg of total RNA of 1183 under salt stress and non-salt stress was used for the detection of cDNA fragment No. 85. The gel was stained with ethidium bromide as a loading control (28S rRNA)
DISCUSSION
There were many differential fragments related to salt tolerance found in the current study. In different germplasms, the changes of the number of enhanced or restrained fragments were irregular. It was shown that the mechanism of salt tolerance in Pak-choi is very complicated and cannot be explained simply by the total fragment number of enhanced or restrained expression. From our previous genetic analysis results (Qiu, 2004), salinity tolerance may be related to the role of every related cDNA fragment. By BLAST analysis, it was found that the induced cDNA fragments Nos. 38 and 68 from 1183 were homologous to the expression genes in Brassica napus under frost stress and in Raphanus sativus under drought stress, respectively. These two fragments may be directly related to the salt-tolerant genes in Pak-choi as major genes. It also could be inferred that some similar mechanism in different plants may exist for stress tolerance under different abiotic stress.
Mitogen-activated protein kinases (MAPKs) have been shown to play an important role in conducting extracellular signals to cellular response (Terada et al., 1999). In plants, numerous Ca2+-stimulated protein kinase activities occur through calcium-dependent protein kinases (CDPKs). These novel calcium sensors are likely to be crucial mediators of responses to diverse endogenous and environmental cues (Cheng et al., 2002). CDPKs were encoded by polygenes that regulated gene expression during the growth and developmental processes. Many abiotic stress signals, such as wounding, cold, high salinity, and drought, are known to elicit fluctuations by CDPK mediation (Ludwig et al., 2004). It was reported that CDPKs are involved in the Ca2+-mediated inward K+ channel regulation in guard cells (Wang et al., 1998). For example, cold treatment enhanced the activity of a membrane-bound rice CDPK (Martin and Busconi, 2001). cDNA fragments Nos. 03, 27 and 73, all from 1183, were highly homologous to positive defense response and signal transmission genes in other organisms. The expression of cDNA fragment No. 79 from 548 was inhibited under salt stress and was shown to be related to the negative regulation of defense responses. These four fragments seemed to be involved in signalling pathways in plants under stress.
The cDNA fragment No. 85 from 1183 was 99% homologous to Homo sapiens genomic DNA of anti-oncogene of hepatocellular colorectal and non-small cell lung cancers. This novel gene related to salt stress found in plant implies that there may be similar stress-tolerant mechanism between totally different species. Compared with the sequences of known genes or cDNA fragments, our study provides valuable evidence to further understand the mechanism of salt resistance in plant although the cDNA fragment was not related to some known salinity tolerance related genes. It is necessary to obtain the full cDNAs and their open reading frames (ORFs) and to characterize their functions.
Acknowledgments
We acknowledge Dr. Zhi-jun CHENG, Institute of Corps Science, Chinese Academy of Agricultural Sciences, Beijing, China, for helpful advice during the preparing the paper.
Footnotes
Project supported by the National “the Tenth Five-Year-Plan” Key Program (No. 2004BA525B08) of China and the Key Laboratory of Vegetable Genetics and Physiology, Ministry of Agriculture, China
References
- 1.Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129(2):469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du LQ, LI YX, Li HJ, Guo BH, Zhu ZQ, Zhou BC. Screening of salt tolerant watercress variants on natural seawater contained medium. Acta Bot Sin. 1999;41(6):633–639. [Google Scholar]
- 3.Li YX. Improvement of Salt Tolerance through Biotechnology and 1/3 Seawater Hydroponic of the Salt Tolerant Vegetables. Beijing: Institute of Botany, Chinese Academy of Sciences Press; 2001. PhD Thesis. (in Chinese) [Google Scholar]
- 4.Ludwig AA, Romeis T, Jones JDG. CDPK-mediated signaling pathways: specificity and cross-talk. J Exp Bot. 2004;395(55):181–189. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- 5.Martin ML, Busconi L. A rice membrane-bound calcium-dependent protein kinase is activated in response to low temperature. Plant Physiol. 2001;125(3):1442–1449. doi: 10.1104/pp.125.3.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu Y. Genetic Analysis of Salinity Tolerance and Differential Expression Study of the Salinity Tolerance Related Genes in Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee. Beijing: Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences Press; 2004. PhD Thesis. (in Chinese) [Google Scholar]
- 7.Terada Y, Nakashima O, Inoshita S, Kuwahara M, Sasaki S, Marumo F. Mitogen-activated protein kinase cascade and transcription factors: the opposite role of MKK3/6-p38K and MKK1-MAPK. Nephrol Dial Transplant. 1999;14(90001):45–47. doi: 10.1093/ndt/14.suppl_1.45. [DOI] [PubMed] [Google Scholar]
- 8.Wang XQ, Wu WH, Zhang JS. Evidences for regulation of the inward K+ channels by CDPK in vicia faba guard cells. Acta Bot Sin. 1998;40(1):1001–1009. (in Chinese) [Google Scholar]
- 9.Xiong B. How to plant the Pak-choi better in summer and gain high yield to supply off-season. Mod Contemp Vegetable. 2004;7:20–21. (in Chinese) [Google Scholar]
- 10.Zhao MF. Current study on world saline soil. World Forestry Res. 1994;7(1):84–86. (in Chinese) [Google Scholar]
- 11.Zhi HY. Evaluation for Salt Tolerance and Study on Mechanism of Salt Tolerance in Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee. Taigu, China: Shanxi Agricultural University Press, Shanxi Agricultural University; 2003. MS Thesis. (in Chinese) [Google Scholar]