SUMMARY
Signaling pathways that respond to DNA damage are essential for the maintenance of genome stability and are linked to many diseases, including cancer. Here, a genome-wide siRNA screen was employed to identify novel genes involved in genome stabilization by monitoring phosphorylation of the histone variant H2AX, an early mark of DNA damage. We identified hundreds of genes whose down-regulation led to elevated levels of H2AX phosphorylation (γH2AX) and revealed new links to cellular complexes and to genes with unclassified functions. We demonstrate a widespread role for mRNA processing factors in preventing DNA damage, which in some cases is caused by aberrant RNA-DNA structures. Furthermore, we connect increased γH2AX levels to the neurological disorder, Charcot-Marie-Tooth (CMT) syndrome, and we find a role for several CMT proteins in the DNA damage response. These data indicate that preservation of genome stability is mediated by a larger network of biological processes than previously appreciated.
INTRODUCTION
The ability of cells to maintain and regulate genome stability is critical for homeostasis, and defects in the maintenance of genome stability underlie a number of developmental disorders and human diseases, including cancer and premature aging (Aguilera and Gomez-Gonzalez, 2008; Kolodner et al., 2002; McKinnon and Caldecott, 2007; Rass et al., 2007). Genome instability can take the form of a variety of genetic alterations that range in complexity from point mutations to the loss or gain of whole chromosomes or gross chromosomal rearrangements (GCRs). Some of these alterations are a direct result of DNA damage and a failure to repair that damage in an error-free manner. Indeed, translocations, deletions, inversions, and duplications are all forms of GCRs that arise from the formation of DNA double-strand breaks (DSB).
Cells have evolved elaborate mechanisms to respond to DNA damage, at the heart of which is a signaling pathway known as the DNA damage checkpoint (Branzei and Foiani, 2007; Cimprich and Cortez, 2008; Harper and Elledge, 2007; Kolodner et al., 2002). This pathway coordinates many aspects of the DNA damage response, including effectors that regulate the cell cycle, DNA repair, transcription, cellular senescence, and apoptosis. Central to the DNA damage response and checkpoint are the phosphatidylinositol-kinase related protein kinases (PIKKs), ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related), which effectively sense DNA lesions caused by DNA damage and replication stress and respond in turn by activating downstream effectors and other kinases. Because the genome is under constant assault from endogenous and exogenous sources of stress, loss of the checkpoint or other components of the DNA damage response leads to increased basal DNA damage and a loss of genome stability.
The DNA damage response also serves as a barrier to cancer progression (Bartkova et al., 2005; Gorgoulis et al., 2005). For example, cell proliferation driven by oncogene activation creates replication stress and activates the DNA damage checkpoint as a means of terminally arresting cellular growth through senescence (Bartkova et al., 2006; Di Micco et al., 2006). Similarly, the disruption of proteins involved in preventing rereplication also causes activation of the DNA damage response (Saxena and Dutta, 2005). Thus, when the DNA damage response itself is deregulated, oncogenes or tumor suppressors could become particularly problematic, driving tumor progression by promoting the loss of genomic integrity. These findings argue that a better understanding of the DNA damage response and the events that lead to its activation is critical to understanding cancer initiation and progression.
Phosphorylation of the histone variant H2AX serves as an early mark of DNA damage (Fernandez-Capetillo et al., 2004; Stucki and Jackson, 2006). H2AX is phosphorylated at Ser-139 by the PIKKs, ATM, ATR, and DNA-PKcs (DNA-dependent protein kinase, catalytic subunit). The phospho-H2AX signal (also known as γH2AX) spreads throughout a 2MB region on either side of the DSB, and it has multiple functions, acting to amplify and maintain the checkpoint as well as recruit downstream repair proteins. Modification of H2AX occurs primarily following DSB formation, although it may also occur during replication stress. As such, it is indicative of the tumorigenic events that occur early in the progression of many cancer types and an early marker of genome instability. Indeed, H2AX phosphorylation is frequently observed in premalignant lesions (Bartkova et al., 2005; Gorgoulis et al., 2005).
Here, we performed a genome-wide siRNA screen in human cells using H2AX phosphorylation as a readout to obtain a global understanding of the different molecular pathways that prevent genome instability and whose loss activates the checkpoint. Using an approach that integrates bioinformatics and functional analysis of our hits, we have uncovered a variety of new genes and molecular networks that maintain genome stability. Our findings suggest that diverse mechanisms with little previous connection to the DNA damage response are needed to maintain genome stability, and they link genome instability with a number of human diseases.
RESULTS AND DISCUSSION
The siRNA Screen
We carried out our siRNA screen in HeLa cells using the Thermofisher siGenome library. The library targets ~21,000 genes and was arrayed in pools of four individual siRNA duplexes per gene. Cells were stained with antibodies to γH2AX to measure DNA damage and with propidium iodide (PI) to determine cell cycle distribution. Images were collected on a laser scanning fluorimeter, allowing analysis of cell number and quantification of γH2AX intensity and DNA content on a single-cell basis. The screen was performed in duplicate with a non-targeting siRNA pool as the negative control and a siRNA pool targeting the replication checkpoint kinase Chk1 as the positive control (Fig. 1A, B). Pools were used at 25 nM total siRNA concentration to minimize off-target effects.
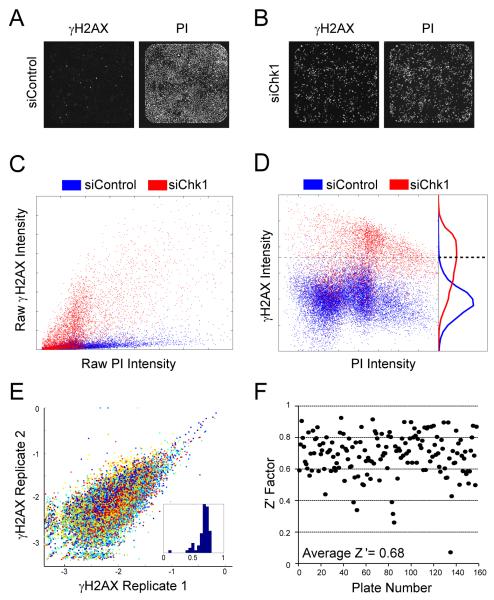
Figure 1. siRNA screen for genes suppressing H2AX phosphorylation.
Images of siControl (A) or siChk1 (B) transfected cells stained with anti-γH2AX antibodies and PI acquired on the Isocyte™. (C) Scatter plot of the raw γH2AX and PI intensity for negative (siControl, blue) and positive (siChk, red) controls. Each dot represents the γH2AX intensity of a single cell as a function of PI intensity. (D) The same cells from panel C shown after normalization. Right portion shows the histograms of each population. The dotted horizontal line reflects the adjusted γH2AX intensity cutoff used to designate a cell as γH2AX positive. (E) Deviation between duplicates of the screen is shown by plotting the first replicate against the second for each siRNA tested. Individual colors indicate the day in which the siRNA pool was tested. Inset shows the histogram of correlation coefficients for all plates analyzed. (F) The Z’ factor for each plate.
The data were normalized to account for plate-to-plate and day-to-day variation in the screen. Because there is a slight increase in γH2AX intensity with increasing DNA content (Mirzoeva and Petrini, 2003), we normalized the raw γH2AX intensity (Fig. 1C) to correct for DNA content on a single-cell basis. Normalization was done by first transforming the γH2AX into log-scale to account for the tail in γH2AX distributions. Second, we fitted a regression line for the negative control cells in each plate to estimate the expected γH2AX signal for each PI intensity. Finally, for each cell we adjusted the observed γH2AX by subtracting the estimated γH2AX value obtained by linear regression for a given PI intensity. As a result of this data normalization, we obtained a single value per cell (Fig. 1D, y-axis) referred to as the adjusted γH2AX intensity. The percentage of γH2AX positive cells (γH2AX+) was then calculated for each siRNA tested using an intensity threshold determined using the eight replicates of the negative and positive control cells from each plate (Fig. 1D & Table S1). Further details are provided in the supplemental methods. Duplicate measurements resulted in little variation, with an average correlation coefficient between replicas of 0.66 ± 0.11 (mean ± SD) (Fig. 1E). The overall Z’ factor calculated for the screen was 0.68 (Fig. 1F), suggesting that our assay had a robust signal-to-noise ratio.
To assign significance to individual genes, we took into account the proportion of γH2AX+ cells and the reproducibility between duplicates. The negative controls from the multiple plates analyzed on a given day allowed us to control for the background γH2AX staining, as well as the variation observed within a single day. From these measurements, we calculated a p-value for each well using a statistical method we developed (Table S2). Because the large number of statistical tests performed in genome-wide siRNA screens creates the potential for a large number of false positives (Wollman and Stuurman, 2007), we also implemented a four-tier method to account for the degree of reproducibility between the replicates by defining two p-value cutoffs using a false discovery rate (FDR) correction.
Genes in the most significant level (group 4, 581 genes) have a p-value for both replicates lower than the FDR corrected level (p-value < 0.0042). Genes in the next level (group 3, 206 genes) have p-values that are significant at the FDR level for one replicate and that fall below the traditional level of p = 0.05 for the second replicate. Genes in the third level (group 2, 1451 genes) have a p-value < 0.05 for only one replicate. Genes within these three levels also have a γH2AX signal that scored within the top 25% of the genome. We also created a final group containing those genes which have a strong, albeit statistically insignificant signal, since the stringent statistical procedure we used is likely to omit true biological hits. This group includes all genes not included in other groups with an average γH2AX signal in the top 5% of the genome (group 1, 164 genes). The γH2AX signals observed, cell viability, and cell cycle distributions for all genes tested can be found in Table S1, and those falling into the top four significance groups can be found in Table S2.
Bioinformatic Analysis
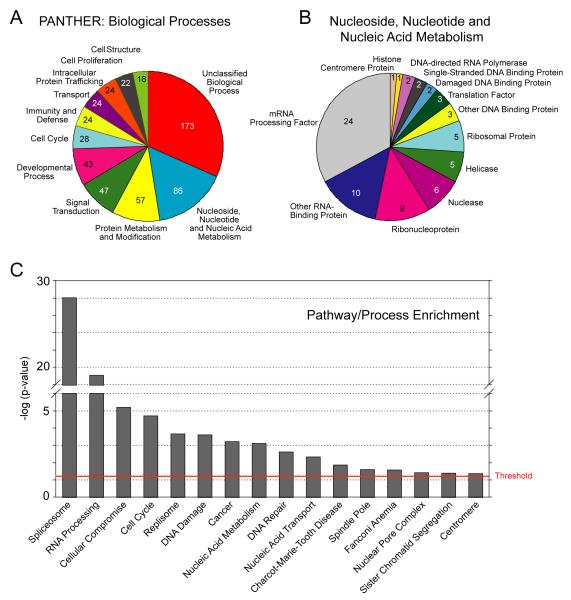
To survey the spectrum of biological functions within our candidate genes, we utilized PANTHER (Protein Analysis through Evolutionary Relationships) (Thomas et al., 2003) on the genes found within our top significance group (group four). siRNA pools that caused extensive cell death (<400 cells at 72 h, a value <50% of the cells originally plated) (Table S3) were eliminated from this and subsequent analyses. The predominant categories of genes we found included those with roles in nucleoside, nucleotide, and nucleic acid metabolism, as might be expected for effectors of genome stability, protein metabolism/modification, and signal transduction, as well as many genes with unclassified functions (Fig. 2A, B).
Figure 2. Functional classification of statistically significant gene set.
(A) Genes identified by statistical methods described in the text were grouped by biological process using PANTHER (http://www.pantherdb.org). (B) Detailed protein categorization of the nucleoside, nucleotide, and nucleic acid metabolism category from (A). (C) Classification enrichment was determined using the David bioinformatic database and Ingenuity Pathway Analysis, and the right-tailed Fisher’s exact test. The threshold of significance was applied for —log (p=0.05).
To determine if our strongest γH2AX effectors (group 4) were enriched for any groups of genes involved in known biological processes in a statistically significant manner, we functionally categorized our hits using the DAVID bioinformatics database (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003) and Ingenuity pathway analysis (Ingenuity Systems, www.ingenuity.com). The genes were categorized according to GO (gene ontology) terms (biological process, cellular complex, molecular function), protein information resource keywords, or the OMIM/Genetic Association disease datasets. As might be expected, we found that genes involved in the cell cycle, cancer, DNA replication and repair were enriched in our data set, providing confidence in our results. Surprisingly, we also found that genes involved in RNA post-transcriptional modification and splicing represented the most significantly enriched categories of genes (Fig. 2C, Table S4).
Validation
Next, we chose ~350 genes to validate using multiple individual siRNAs (deconvolution) (Table S5). The genes were chosen based on our significance analysis, relevant literature information, and/or functional categorization. Additionally, we selected a few siRNA targets with borderline γH2AX signals that were of biological interest or that functioned in pathways, processes or complexes found among the genes deemed significant. Several genes known to cause an increase in γH2AX upon knockdown, such as TopBP1 and ATR, had low signals, suggesting that use of a low concentration of pooled siRNA (25 nM) may have led to incomplete knockdown resulting in false negatives (Cimprich and Cortez, 2008). Therefore, we rationalized that further exploration of genes displaying lower levels of γH2AX could also reveal true hits.
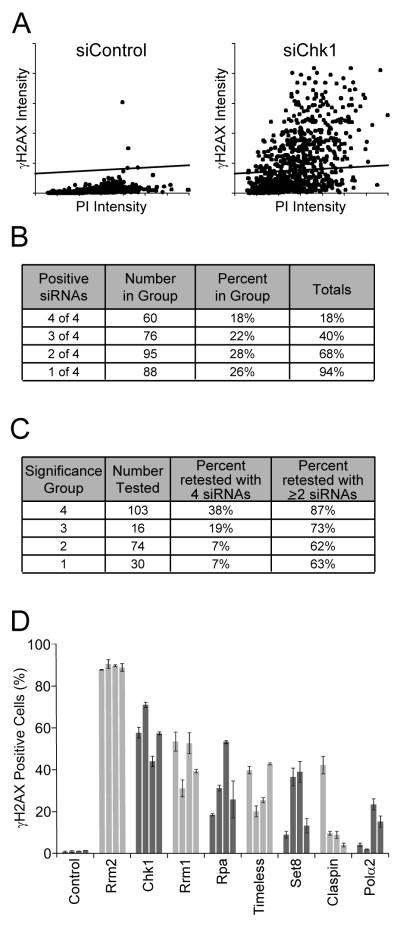
For the chosen genes, the four individual siRNAs comprising the original screening pool were individually tested at 25nM using the same platform as the primary screen. A cell was considered positive if its γH2AX intensity was greater than a linear intensity threshold set approximately three times the mean γH2AX intensity observed in siControl-treated cells (Fig. 3A). After applying the threshold, the median percentage of γH2AX+ cells was calculated from all siControl wells tested (1.6%), and siRNAs that displayed a value at least two standard deviations greater than the control value were considered positives (≥3%). This analysis revealed that 94% of the genes retested scored positive with at least one siRNA while 68% (231 genes) scored positive with two or more siRNAs (Fig. 3B). The majority of genes in the top two significance groups retested with multiple siRNAs (Fig. 3C).
Figure 3. Screening validation.
(A) A representative well showing the γH2AX signal as a function of PI intensity for siControl- and siChk1-transfected cells. The percentage of γH2AX positive cells per well was calculated by applying a γH2AX intensity cutoff (horizontal line). (B) Table representing the effects of the four individual siRNAs tested during deconvolution. siRNAs were considered positive if the percentage of γH2AX+ cells was siControl-transfected wells. (C) Table demonstrating the retest rate for genes chosen from the primary screening significance groups. (D) Bar graph showing the effect of targeting genes involved in DNA replication and checkpoint activation. Each bar represents an individual siRNA tested, and error bars indicate variation between duplicates.
Of the genes that scored positive when targeted by multiple siRNAs, those causing the largest increase in γH2AX+ cells were primarily proteins involved in DNA replication or checkpoint signaling (Fig. 3D). Both of these processes are known to have roles in preserving genomic stability, adding confidence to our validated data set (Aguilera and Gomez-Gonzalez, 2008; Kolodner et al., 2002). Other genes that scored with multiple siRNAs included proteins involved in a broad spectrum of functions including cell cycle control, DNA binding, ion flux, gene regulation, and RNA processing (Table 1 & Table S5).
Table 1.
Genes Scoring With Four siRNAs
| Category | Symbol | Comments |
|---|---|---|
| Apoptosis | BIRC5 | Baculoviral IAP repeat-containing 5 |
| CASP8AP2 | CASP8 associated protein 2 | |
| Cell Cycle | CDCA5 | Cell division cycle associated 5 |
| CENPE | Centromere protein E | |
| STK6 | Aurora kinase A | |
| Channel | SLC28A2 | Solute carrier family 28 |
| LRP1B | Low density lipoprotein-related protein 1B | |
| Cytoskeletal-Related | CKAP5 | Cytoskeleton associated protein 5 |
| KRTAP12-3 | Keratin associated protein 12-3 | |
| KIF11 | Kinesin family member 11 | |
| DNA Binding | SON | SON DNA binding protein |
| PABPC4 | Poly(A) binding protein, cytoplasmic 4 | |
| DNA replication/repair | ERCC6 | Excision repair cross-complementation group 6 |
| XAB2 | XPA binding protein 2 | |
| DBF4/ASK | DBF4 homolog; activator of S phase kinase | |
| CHEK1/CHK1 | Checkpoint kinase 1 homolog | |
| CLSPN | Claspin homolog | |
| MCM10 | Minichromosome maintenance complex protein 10 | |
| RPA1 | Replication protein A1, 70kDa | |
| RPA2 | Replication protein A2, 32kDa | |
| RRM1 | Ribonucleotide reductase M1 polypeptide | |
| RRM2 | Ribonucleotide reductase M2 polypeptide | |
| SETD8/SET8 | SET domain containing 8 | |
| TIMELESS | Timeless homolog | |
| Other | COPZ1 | Coatomer protein complex, subunit zeta 1 |
| CSA2 | Cytokine, down-regulator of HLA II | |
| ITGB1 | Integrin, beta 1 | |
| BRD8 | Bromodomain containing 8 | |
| NCAN/CSPG3 | Neurocan | |
| FAM24A | Familywith sequence similarity 24, member A | |
| FKBP6 | FK506 binding protein 6 | |
| PSMD3 | Proteasome 26S non-ATPase subunit 3 | |
| Polymerase | POLR2G | Polymerase (RNA) II (DNA-directed) polypeptide G |
| POLR2I | Polymerase (RNA) II (DNA-directed) polypeptide I | |
| RNA Splicing | PRPF19/PRP19 | PRP19/PSO4 pre-mRNA processing factor 19 homolog |
| CRNKL1 | Crooked neck pre-mRNA splicing factor-like 1 | |
| HNRPC | Heterogeneous nuclear ribonucleoprotein C | |
| CWC22/KIAA1604 | CWC22 spliceosome-associated protein homolog | |
| LSM2 | LSM2 homolog, U6 small nuclear RNA-associated | |
| PRPF8 | PRP8 pre-mRNA processing factor 8 homolog | |
| SART1 | Squamous cell carcinoma antigen recognized by T cells | |
| SF3A1 | Splicing factor 3a, subunit 1 | |
| SF3A2 | Splicing factor 3a, subunit 2 | |
| SF3B2 | Splicing factor 3b, subunit 2 | |
| SF3B4 | Splicing factor 3b, subunit 4 | |
| SKIIP | SNW domain containing 1; SKI-interacting protein | |
| SNRPA1 | Small nuclear ribonucleoprotein polypeptide A1 | |
| SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 | |
| SNRPD1 | Small nuclear ribonucleoprotein D1 polypeptide | |
| SNRPD3 | Small nuclear ribonucleoprotein D3 polypeptide | |
| WBP11 | WW domain binding protein 11 | |
| AQR | Aquarius homolog | |
| CDC40 | Cell division cycle 40 homolog | |
| CDC5L | CDC5 cell division cycle 5-like | |
| Transcription Factor | ZNF157 | Zinc finger protein 157 |
| ZNF16 | Zinc finger protein 16 |
Network Analysis
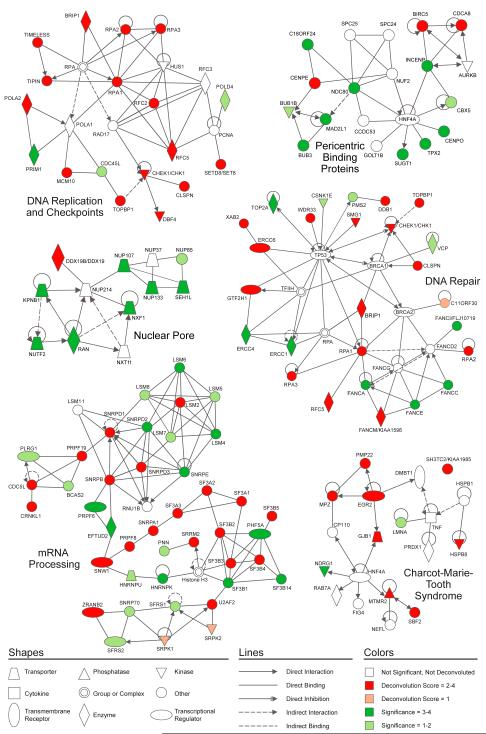
To determine if we had identified and validated groups of genes that were common to previously characterized pathways or complexes, we used the statistically significant functional categories (Fig. 2C, Table S4) identified from the genes in group four of the original screen to define networks of interacting proteins. Once a network of interacting proteins had been defined, we mapped our list of deconvoluted genes as well as genes found within significance groups 3, 2, or 1 onto these networks. Several interaction modules were identified encompassing expected pathways and those that had not been previously linked to genome maintenance (Fig. 4, Fig. S1, Table S6).
Figure 4. Network modeling of screen hits identifies new functional groups linked to genome maintenance.
Networks of interacting proteins identified using DAVID Bioinformatics Database and Ingenuity Pathway Analysis. Color indicates strength of statistical significance (green) or strength of deconvolution results (red). If a statistically significant gene was deconvoluted, the deconvolution result is preferentially shown.
DNA Replication, Checkpoint and Repair Modules
As we expected, our screen identified genes involved in DNA replication and checkpoint activation (Fig. 4). Among the genes we found to be involved in these processes are several components of the replication machinery, including the replication factor C (RFC) complex, the single-strand DNA binding protein, replication protein A (RPA), the DNA primase-DNA polymerase alpha complex, and MCM10, a minichromosome maintenance protein. We also identified Timeless and Tipin, a complex of two proteins needed to activate the replication checkpoint (Kondratov and Antoch, 2007), as well as Set8, a histone methyltransferase needed for DNA replication (Jorgensen et al., 2007; Tardat et al., 2007). Other checkpoint proteins identified that play a role in S phase progression include Chk1, Claspin, TopBP1, and Dbf4, a regulator of the Cdc7 kinase.
Many DNA repair proteins were also well represented amongst our hits, including components of homologous recombination (HR) and nucleotide excision repair (NER) processes (Fig. 4). The HR portion of the module contains many proteins from the BRCA/Fanconi anemia (FA) pathway, which is required for double-strand break and cross-link repair. These proteins include the BRCA1-interacting protein, BRIP1(FANCJ), the BRCA2(FANCD1)-interacting protein C11ORF30/EMSY, and the additional FA components FANCM, FANCI, FANCC, FANCE and FANCA. Components of the ATR checkpoint, TopBP1, Claspin and Chk1, which regulate the FA/BRCA pathway, are also linked to this module. The NER portion of this module includes excision repair cross complementation group 6 (ERCC6/CSB), an ERCC6 and Xeroderma Pigmentosa A binding protein (XAB2), the interacting nucleases ERCC4 and ERCC1, and GTF2H1, a component of TFIIH. Altogether, the significant representation of replication, checkpoint and repair genes among our validated hits provides confidence in our screening data.
mRNA Processing Module
Interestingly, the most significantly enriched interaction network contains proteins involved in mRNA processing. These hits are involved in different stages of mRNA processing, including RNA splicing, spliceosome assembly, mRNA surveillance, and mRNA export, with the majority having roles in RNA splicing (Fig 4., Table S7). Recent studies have linked some mRNA processing genes to genome maintenance both in yeast and mammals (Aguilera and Gomez-Gonzalez, 2008; Li and Manley, 2006). Based on these and a limited number of additional observations, we expected to identify a few mRNA processing proteins within our screen (Azzalin and Lingner, 2006; Brumbaugh et al., 2004; Hossain et al., 2007; Li and Manley, 2005; Moumen et al., 2005; Xiao et al., 2007). Strikingly, however, our studies revealed that mRNA processing is involved in preserving genomic integrity on a much broader scale.
Charcot-Marie-Tooth Disease Module
Genes involved in Charcot-Marie-Tooth disease (CMT) were also statistically enriched within our most stringent gene set, and although the γH2AX signal was relatively low for this category of genes, we confirmed many of these hits through deconvolution (Table S8). CMT is a clinically and genetically heterogeneous set of disorders of a relatively high prevalence that cause demyelinating and axonal neuropathies (Berger et al., 2002; Szigeti and Lupski, 2009). Among the genes we found are peripheral myelin protein (Pmp22), whose mutation or altered gene dosage accounts for nearly 70% of all cases of hereditary neuropathies; gap junction protein beta 1 (GJB1), another commonly mutated gene in CMT patients; early growth response protein 2 (EGR2), a transcription factor that regulates the expression of myelin proteins including PMP22; SH3TC2, a protein of unknown function with a putative SH3 domain and tetracopeptide repeats, myotubularin-related protein 2 (MTMR2), a phosphatidylinositol phosphatase, and its interacting protein CMT4B2/MTMR13 (Berger et al., 2002; Niemann et al., 2006; Szigeti and Lupski, 2009). Although there is no reported connection between CMT and genome instability, defects in the DNA damage response have been linked to other neurodegenerative disorders (Rass et al., 2007).
Additional Modules
Our screen and significance analysis also identified several protein interaction networks with less defined links to H2AX phosphorylation that have yet to be extensively deconvoluted (Fig. 4). One of these networks contains pericentric binding proteins including components of the kinetochore, centromere and spindle assembly checkpoint. Defects in formation of the kinetochore or centromere could lead to defects in spindle assembly and the mitotic checkpoint. Thus, the increase in γH2AX could be caused by premature mitosis and chromosome breaks due to incomplete decatenation (Cleveland et al., 2003; Damelin and Bestor, 2007). While additional work will be needed to validate this gene set, it is noteworthy that a screen for genes causing Rad52 foci in yeast also led to identification of several mitotic checkpoint genes (Alvaro et al., 2007).
Components of the nuclear pore comprised another interesting group of genes significantly enriched among our hits. This link is of particular interest in light of studies linking the nuclear pore complex (NPC) and components of the nuclear periphery to DNA repair and DNA damage responses in several organisms (Davuluri et al., 2008; De Souza et al., 2006; Loeillet et al., 2005; Nagai et al., 2008; Palancade et al., 2007). Indeed, Nup107, a conserved component the NPC among our hits, appears to regulate repair of DSBs in yeast (Nagai et al., 2008) where recruitment of telomeres and persistent breaks to the nuclear pore and nuclear periphery may suppress potentially dangerous chromosomal rearrangements and promote certain types of repair (Gartenberg, 2009). While it appears as if some types of DSBs have limited mobility within the nucleus of mammalian cells, specific types of DNA damage, such as deprotected telomeres, do exhibit increased mobility and may be able to undergo relocalization (Misteli and Soutoglou, 2009). Thus, while further studies are clearly required, our data are consistent with the idea that the NPC may have a role in DNA damage processing in higher eukaryotes.
Other modules of genes we identified by deconvolution include a group of proteins involved in circadian rhythms, several genes involved in Wnt signaling, and several components of the GABA receptor (Fig. S1). Previous links between circadian rhythms and circadian rhythm proteins to cancer and tumor progression, and direct connections between proteins involved in circadian oscillations with the DNA damage response make this an interesting cluster (Chen-Goodspeed and Lee, 2007; Kondratov and Antoch, 2007).
Interestingly, the Nup107-Nup160 complex and other components of the NPC appear to link several of the modules we identified (Fig. S2). Nup107-Nup160 along with the interacting nucleoporins Seh1L and Elys/Mel-28, which were identified within the screen, interact with the kinetochore during mitosis (Loiodice et al., 2004; Rasala et al., 2006). In addition, Elys interacts with the MCM2-7 helicase complex, and its loss sensitizes cells to replication stress (Davuluri et al., 2008; Gillespie et al., 2007). It is therefore tempting to speculate that some of the effects of NPC perturbations are due to problems with replication and the resolution or repair of stalled or collapsed forks, a hypothesis consistent with the model that the Nup84Nup107-Slx5/8 complex resolves DNA damage at collapsed forks in yeast (Nagai et al., 2008). Finally, it is worth noting that NPC components also interact with several mRNA processing proteins. Clearly further work will be required to validate the many hypotheses that arise from these hits, but the extensive nature of this network and many interconnections between known mediators of genome stability and the unexpected modules we identified suggests the preservation of genome stability might be coordinated by a larger network of biological processes than previously appreciated.
Network Characterization
To further investigate how the genes identified in our screen led to increased H2AX phosphorylation and to test the idea that they mediate genome stability, we chose the RNA processing and CMT related genes for further characterization.
mRNA Processing
The mRNA processing cluster was the most significantly enriched group of genes to arise from our screen with 86 genes inducing significant H2AX phosphorylation (Table S7). While mRNA processing could affect genome stability indirectly by altering protein levels, recent studies have eluded to a more direct mechanism. In S. cerevisiae, when genes involved in mRNA processing are mutated, defects arise in the packaging of nascent mRNAs (Aguilera and Gomez-Gonzalez, 2008; Li and Manley, 2006). As a result the nascent mRNA hybridizes with the transcribed strand (RNA-DNA hybrid) creating an R loop and causing elevated recombination. Furthermore, the genome instability arising from the disruption of cotranscriptional processes is suppressed upon removal of the RNA structures suggesting that R loops formed by lack of proper mRNA processing are a direct source of genome instability. Similar events have also been observed in mammalian cells with loss of the splicing factor ASF/SF2 (Li and Manley, 2005).
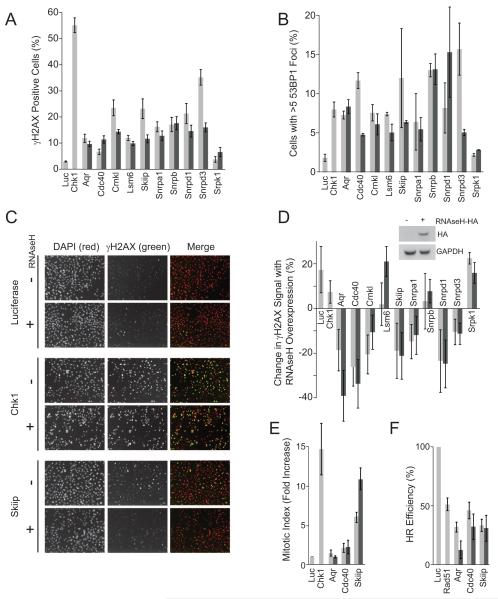
To test whether the mRNA processing genes we identified were indeed causing DNA damage, we assessed the formation of a distinct marker for DNA damage by analyzing 53BP1 foci for several of the factors that caused an increase in γH2AX (Fig. 5A). Consistent with the idea that increased γH2AX is due to increased DNA damage, knockdown of most splicing factors caused an increase in cells with multiple 53BP1 foci (Fig. 5B). To determine if the observed DNA damage may involve the cotranscriptional formation of R loops, we then created a cell line stably expressing RNaseH and analyzed γH2AX before and after expression (Fig. 5C, D). This approach has been shown to prevent R loop formation in yeast and mammalian cells, and it reverses the increases in DSB formation and G2 arrest caused by knockdown of the splicing factor ASF/SF2 (Li and Manley, 2005; Li and Manley, 2006). We found that expression of RNase H caused a slight increase in γH2AX for our control, suggesting that it may be generally toxic to cells. Despite this effect, H2AX phosphorylation was reduced in several of the samples (Fig. 5C, D). These observations suggest that the cotranscriptional formation of R loops may be a broad source of genome instability which is prevented by efficient mRNA processing.
Figure 5. Functional assays for mRNA processing genes affecting γH2AX.
(A) Effect on γH2AX after targeting genes involved in mRNA processing. (B) Percent of cells exhibiting greater than five 53BP1 foci after siRNA treatment. (C) Representative images of change in γH2AX signal following RNaseH expression. (D) Quantitation of effect represented in C for the indicated genes. Inset shows RNaseH-HA protein expression. (E) G2/M checkpoint assay after knockdown of the indicated genes and IR treatment. Fold increase of the mitotic index (% mitotic cells post-IR / % mitotic cells nontreated) in cells transfected with targeting siRNA relative to control is shown. (F) HR repair frequency at an induced double-strand break after knockdown of indicated gene. Samples were normalized to the HR frequency in the siControl-transfected cells. All graphs shown are mean ± SE for n=3. Duplicate bars indicate the effects of two different siRNAs.
How R loops might lead to genome instability is unclear. One possibility is that the displaced ssDNA is more susceptible to DNA damage, ultimately leading to DSB formation and recombination. Alternatively, disrupted mRNA splicing may create replication fork barriers via R loop formation or by preventing timely removal of the transcription machinery during replication. These structures could cause fork arrest and collapse or could be subject to aberrant processing (Li and Manley, 2006). Indeed, interference between transcription and replication is a major source of replication stress and studies show that H2AX is preferentially phosphorylated at gene rich regions when genes suppressing R loop formation are lost (P. Pasero, personal communication). Whether the R loop formation caused by loss of our genes is causing genome instability specifically in S phase remains to be determined.
Not all of the effects of mRNA processing genes on H2AX phosphorylation were decreased by RNAse H expression (Table S9), and why these genes but not others affect R loop formation is not yet apparent. Because R loop formation is thought to be suppressed by cotranscriptional packaging of the mRNA (Aguilera and Gomez-Gonzalez, 2008; Li and Manley, 2006), our observations may suggest the proteins we identified play crucial or early roles in this process. However, it is also quite likely there are additional and multiple mechanism by which these genes affect genome stability.
To test the idea that some of these mRNA processing proteins might have other roles in the DNA damage response, we analyzed activation of the G2/M checkpoint and homologous recombination upon knockdown of three genes: Cdc40/Prp17, a splicing factor linked to S phase progression in yeast; Skiip/Snw1, a transcriptional coactivator that also affects splicing, and Aqr, a putative RNA helicase related to Dna2 (Ben Yehuda et al., 1998; Folk et al., 2004; Sam et al., 1998). Interestingly, Cdc40 and Skiip showed a defect in activation of the checkpoint, while all three genes led to a decrease in homologous recombination, similar to that caused by loss of Rad51 (Fig. 5E, F). This suggests the overall process is complex and that there may also be more direct roles for these RNA processing genes in checkpoint and repair responses.
Charcot-Marie-Tooth Disease
To further investigate the link between Charcot-Marie-Tooth (CMT) genes and γH2AX phosphorylation, we selected several CMT genes for further study. Since the main roles of most CMT genes have been characterized in neurons, we reconfirmed the effects seen on H2AX phosphorylation in our original screen both in HeLa and U2OS cells with at least two siRNAs targeting each gene (Fig. 6A, Table S8, S10). Knockdown was confirmed by Q-PCR. Analysis of γH2AX after knockdown of several CMT genes also revealed that its localization was nuclear and focal (Fig. 6B), consistent with the idea that DNA damage is elevated when these genes are lost.
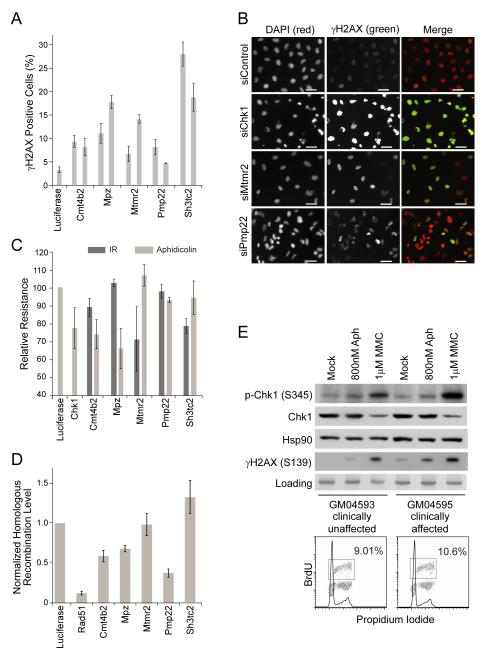
Figure 6. Loss of Charcot-Marie-Tooth disease genes leads to increased DNA damage and repair defects.
(A) Percentage of γH2AX+ cells after knockdown of the indicated genes. Duplicate bars indicate effects of individual siRNAs tested. (B) Images of γH2AX signal 72h after knockdown of the indicated gene. Scale bar indicates 50μm. (C) Sensitivity to aphidicolin (100nM) or IR treatment (2Gy). Samples were corrected for the effect of the indicated siRNA on growth rate and then normalized to the siLuciferase-transfected sample. Details can be found in the supplemental methods. (D) HR repair frequency at an induced double-strand break after knockdown of the indicated gene. Samples were normalized to the HR frequency in the siLuciferase-transfected cells. (E) p-Chk1 and γH2AX response in GJB1 patient cell lines. Samples were collected 24 hrs after drug addition. BrdU plots indicate there is no significant difference in the cell cycle distribution of the mock treated cells. All graphs shown are mean ± SE for n=3.
To better understand how these CMT genes might be affecting the DNA damage response, we assessed the effect of their knockdown on cell survival after exposure to ionizing radiation or aphidicolin, which induces replicative stress (Fig. 6C). Knockdown of several genes caused a dramatic cellular sensitivity to these treatments, suggesting CMT genes may be needed for DNA damage processing. To further test this idea, we looked at homologous recombination efficiency after CMT gene knockdown by measuring the effect on repair of a chromosomal double-strand break induced by the I-SceI nuclease (Fig. 6D). Interestingly, we also observed defects in homologous recombination for several of the genes we tested. These findings strongly suggest the increase in H2AX phosphorylation observed in the original screen results from an increase in DNA damage. Further, they indicate that the CMT genes may play a role in the DNA damage response.
Since the pathology of this disease has not previously been linked to DNA damage, we also asked if increased H2AX phosphorylation was observed in patients with CMT. To do so, we analyzed γH2AX in two fibroblast cell lines, one derived from a CMT patient with a mutation in GJB1 and another derived from an unaffected family member (Fig. 6E). Importantly, we observed increased basal levels of γH2AX in the patient line as well as higher levels post-damage. Furthermore, we observed elevated levels of Chk1 phosphorylation following DNA damage in the patient lines, suggesting that checkpoint signaling is increased. Taken together, these results suggest the increased genomic instability resulting from the down-regulation of these genes may be a common phenotype of the CMT disorder.
Other proteins involved in the DNA damage response have profound effects on neuronal development and function (Brooks et al., 2008; McKinnon and Caldecott, 2007; Rass et al., 2007). For example, ataxia, axonal neuropathy, progressive neurodegeneration, and myelination defects are some of the characteristic features observed in individuals bearing mutations in crucial DNA damage response genes. Interestingly, most of the CMT genes we found to induce γH2AX cause demyelinating forms of CMT, suggesting there may be connections between DNA damage and myelination defects. Indeed, many of the CMT genes affecting γH2AX that we found include components of myelin, regulators of its production, and proteins involved in vesicle mediated transport, a process that affects myelination (Niemann et al., 2006). While further work will be required to understand the molecular links between the function of these genes, DNA damage accumulation, and pathogenesis of CMT, potential mechanisms may include improper membrane division in mitosis, altered nuclear structure affecting DNA replication/repair, or activation of the unfolded protein response.
PERSPECTIVE
A proper response to DNA damage is critical for the maintenance of genome stability, and it serves as a key barrier to the prevention of cancer. The screen described in this study has led to the identification of several hundred genes that when lost, induce the phosphorylation of H2AX, a robust and reliable marker for DNA damage. We expect there is a strong likelihood of identifying genes with functions in the DNA damage response pathway amongst these hits, as both CMT and splicing genes exhibited either DNA repair or checkpoint defects. This is the first DNA damage response screen of its kind that has been reported in higher eukaryotes, and the results provide new insight into processes that prevent the formation and accumulation of DNA damage. Indeed, many of the genes and processes identified have not been previously linked to the formation of DNA damage, suggesting the events that contribute to genome instability may be more widespread than previously realized.
A number of the genes we identified exhibited a relatively high level of H2AX phosphorylation when knocked down, particularly those known to be involved in DNA replication and DNA damage responses. The genes involved in RNA splicing also caused a high level of H2AX phosphorylation. However, several hundred genes consistently led to low, but reproducible and significant levels of phosphorylation when targeted. Thus, while the high effectors are of obvious importance, those causing low levels of H2AX phosphorylation may also be of interest. Indeed, loss of function of these genes may be tolerated by the cell/organism and could drive genome instability and transformation, while those causing high levels of γH2AX seem more likely to cause cell death or senescence. In this respect, it is interesting that the level of genome instability linked to the CMT genes is relatively low.
For a majority of the genes identified in our screen, it seems likely the increase in γH2AX observed is due to increased spontaneous DNA damage. However, spontaneous or unrepaired DNA damage may not be the only reason for increased γH2AX. For example, H2AX phosphorylation could result from loss of the phosphatases that dephosphorylate γH2AX. In fact, we did identify subunits of the PP2A and PP4 phosphatase complexes that are involved in dephosphorylating γH2AX (Chowdhury et al., 2005; Chowdhury et al., 2008; Nakada et al., 2008). Some of the genes identified may also cause an increase in γH2AX via apoptosis; however, this category was largely eliminated by removing genes that caused overt and widespread cell death, as well as by setting nuclear area parameters to eliminate the identification of nuclear fragments.
Other screens assessing different aspects of the DNA damage response have been carried out in various organisms. For example, the formation of Rad52 foci was examined in Saccharomyces cerevisiae (Alvaro et al., 2007), and several screens were also carried out in Caenorhabditis elegans to identify genes affecting radiation sensitivity (van Haaften et al., 2006; van Haaften et al., 2004). Of the genes identified in these screens, many were also found in our data set suggesting that some of the properties measured by previous screens may be linked to increased γH2AX (Table S11). A proteomic analysis designed to identify the targets of the DNA damage protein kinases has also been carried out using mammalian cells (Matsuoka et al., 2007). Although this approach is orthogonal to ours, we found significant overlap in the genes and pathways identified by this method and our dataset (110 genes, p = 1.5 × 10-2) (Table S11). Interestingly, beyond specific gene overlap between screens, greater commonality was observed between the biological processes and pathways found, suggesting that while individual hits may vary from screen to screen, the enriched pathways observed may provide greater biological insight. For example, mRNA processing genes were also enriched in this proteomic analysis. Nevertheless, the majority of genes found in all studies were not found in the other, and we identified many additional genes and pathways of diverse function not previously linked to the DNA damage response. This indicates that our knowledge of this process is still incomplete and that the screens are not yet saturating. Further, it suggests that a systems biology approach utilizing many genomic datasets could ultimately prove useful in understanding the mechanisms underlying genomic stability.
Altogether, the results of our study indicate the pathways and processes affecting genome stability are much broader than anticipated, and our data provide new unexpected links between the maintenance of genome stability and the kinetochore, the nuclear pore, mRNA processing and Charcot-Marie-Tooth disease. We expect there will be important roles for these genes and pathways in the DNA damage response, cancer, neurodegeneration, aging, and other human diseases, and the nature of these links will be of great interest for future study.
EXPERIMENTAL PROCEDURES
siRNA Screen
The siRNA screen was performed using the siARRAY human genome siRNA library from ThermoFisher Scientific. HeLa cells were reverse-transfected using Dharmafect 1 transfection reagent. After 72h the cells were fixed and stained with phospho-H2AX antibody (Cell Signaling Cat# 2577) and propidium iodide. Cells were imaged on the IsoCyte™ (MDS Analytical Technology) laser scanning platform at 10×10 μm2 sampling. The total fluorescence intensity of each cell was calculated for both channels by integrating the pixel values associated with the cell and subtracting the average background intensity of the well. Additional details can be found in the Supplemental Data.
Data Analysis and Statistical Analysis
Genome data were analyzed and normalized to account for two sources of variability in the data: per-plate and per-day variations. A statistical analysis was carried out to estimate the significance of a given result. A detailed procedure is provided in the Supplemental Data.
Enrichment Analysis and Bioinformatics
Functional classification was determined using PANTHER, Ingenuity Pathway Analysis (IPA), and the DAVID bioinformatic database on those genes whose γH2AX signal scored with the highest significance (group 4). Statistically enriched categories were then input into IPA software for protein-protein interaction network identification. Gene functions in tables and text were assigned using resources from the above programs and the Biobase Knowledge Library (www.biobase.de). Additional details can be found in the Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Cimprich lab for a careful reading of this manuscript and helpful discussions. This work was supported by an NSF predoctoral fellowship awarded to RDP, and by grants to KAC from the National Institutes of Health (ES016486, ES016867) and the California Breast Cancer Research Program (131B-0029). KAC is a Leukemia and Lymphoma Scholar.
RDP and DVS performed the primary screen and validated the hits. RDP managed the data, designed the figures, performed all bioinformatic analyses, and contributed to Fig 5A-D, and Fig 6B, C, E. DVS and AG contributed to Fig 5E, F. RW performed statistical analyses for the screen. AH designed figures and contributed to Fig 6 A, C-D. MCY performed the qPCR and contributed to Fig 6 A, C-D. JAH, SCM, EFC, DES-C, and TM helped optimize the screen. KAC and RDP wrote the manuscript. KAC conceived of and guided the project.
Evan F. Cromwell and Jayne Hesley are employees of MDS Analytical Technologies.
Footnotes
SUPPLEMENTAL DATA Supplemental data includes detailed experimental procedures, 2 figures and 11 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Yehuda S. Ben, Dix I, Russell CS, Levy S, Beggs JD, Kupiec M. Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA. 1998;4:1304–1312. doi: 10.1017/s1355838298980712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger P, Young P, Suter U. Molecular cell biology of Charcot-Marie-Tooth disease. Neurogenetics. 2002;4:1–15. doi: 10.1007/s10048-002-0130-z. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Chen-Goodspeed M, Lee CC. Tumor suppression and circadian function. J Biol Rhythms. 2007;22:291–298. doi: 10.1177/0748730407303387. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Damelin M, Bestor TH. The decatenation checkpoint. Br J Cancer. 2007;96:201–205. doi: 10.1038/sj.bjc.6603537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4:e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Horn KP, Osmani SA. A point mutation in the Aspergillus nidulans sonBNup98 nuclear pore complex gene causes conditional DNA damage sensitivity. Genetics. 2006;174:1881–1893. doi: 10.1534/genetics.106.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lemicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Folk P, Puta F, Skruzny M. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell Mol Life Sci. 2004;61:629–640. doi: 10.1007/s00018-003-3215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR. Life on the edge: telomeres and persistent DNA breaks converge at the nuclear periphery. Genes Dev. 2009;23:1027–1031. doi: 10.1101/gad.1805309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA Damage Response: Ten Years After. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hossain MN, Fuji M, Miki K, Endoh M, Ayusawa D. Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa cells to various stresses. Mol Cell Biochem. 2007;296:151–157. doi: 10.1007/s11010-006-9308-2. [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol. 2007;17:311–317. doi: 10.1016/j.tcb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- Loeillet S, Palancade B, Cartron M, Thierry A, Richard GF, Dujon B, Doye V, Nicolas A. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair (Amst) 2005;4:459–468. doi: 10.1016/j.dnarep.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res. 2003;1:207–218. [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–1026. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann A, Berger P, Suter U. Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:217–242. doi: 10.1385/nmm:8:1-2:217. [DOI] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell. 2007;18:2912–2923. doi: 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Sam M, Wurst W, Kluppel M, Jin O, Heng H, Bernstein A. Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev Dyn. 1998;212:304–317. doi: 10.1002/(SICI)1097-0177(199806)212:2<304::AID-AJA15>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Saxena S, Dutta A. Geminin-Cdt1 balance is critical for genetic stability. Mutat Res. 2005;569:111–121. doi: 10.1016/j.mrfmmm.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Szigeti K, Lupski JR. Charcot-Marie-Tooth disease. Eur J Hum Genet Epub ahead of print. 2009 doi: 10.1038/ejhg.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Romeijn R, Pothof J, Koole W, Mullenders LH, Pastink A, Plasterk RH, Tijsterman M. Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Curr Biol. 2006;16:1344–1350. doi: 10.1016/j.cub.2006.05.047. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Vastenhouw NL, Nollen EA, Plasterk RH, Tijsterman M. Gene interactions in the DNA damage-response pathway identified by genome-wide RNA-interference analysis of synthetic lethality. Proc Natl Acad Sci U S A. 2004;101:12992–12996. doi: 10.1073/pnas.0403131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Stuurman N. High throughput microscopy: from raw images to discoveries. J Cell Sci. 2007;120:3715–3722. doi: 10.1242/jcs.013623. [DOI] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.