Abstract
Prevotella intermedia is a major periodontopathogen contributing to human gingivitis and periodontitis. Such pathogens release proteases as virulence factors that cause deterrence of host defences and tissue destruction. A new cysteine protease from the cysteine-histidine-dyad class, interpain A, was studied in its zymogenic and its self-processed mature form. The latter consists of a bivalved moiety made up by two subdomains. In the structure of a catalytic cysteine-to-alanine zymogen variant, the right subdomain interacts with an unusual prodomain, thus contributing to latency. Unlike the catalytic cysteine residue, already in its competent conformation in the zymogen, the catalytic histidine is swung out from its active conformation and trapped in a cage shaped by a backing helix, a zymogenic hairpin and a latency flap in the zymogen. Dramatic rearrangement of up to 20Å of these elements triggered by a tryptophan switch occurs during activation and accounts for a new activation mechanism for proteolytic enzymes. These findings can be extrapolated to related potentially pathogenic cysteine proteases such as Streprococcus pyogenes SpeB and Porphyromonas gingivalis periodontain.
Periodontal disease (PD) affects the tissues that surround and support the teeth and may lead to loosening and eventual loss of teeth if untreated. It is caused by bacteria and affects mildly 90% and severely 10% of the population worldwide (1,2). In addition, symptoms of PD appear in a series of systemic diseases due to its inflammatory and infective character (2,3). Present day treatment and curettage of severe PD includes the mechanical cleansing of the affected area and is efficient in general. However, it is costly, time consuming and painful and needs frequent repetition. In addition, it may entail the indiscriminate usage of antibiotics, which contributes to the spread of antibiotic-resistant strains (2,4). Consequently, there is a need for innovative and specific therapeutic approaches against PD.
Prevotella intermedia is a major bacterial periodontal pathogen in humans together with Porphyromonas gingivalis among others (5,6). Such bacteria colonise the gingival crevice and produce virulence factors that cause disease. Bacterial infection leads to the bacterial secretion or induction of host overproduction of proteolytic enzymes such as bacterial collagenases, matrix metalloproteases and serine and cysteine proteases (CPs) (2,7,8). These proteases destroy host tissue and compromise host defences. In addition, proteases may give rise to fibrinolytic activity and inactivate components of the blood-coagulation cascade such as the protease inhibitors, α1-proteinase inhibitor and α2-macroglobulin. Proteolysis further covers alimentary requirements since most of bacterial nutrition is obtained from degraded periodontal tissue and tissue fluid (9).
Most studies on the bacterial proteolytic armamentarium in PD have been performed with P. gingivalis (9). In contrast, the factors governing P. intermedia infection, a black-pigmented Gram-negative obligate anaerobic non-motile rod bacterium, are poorly understood (7). In humans, Prevotella sp. have frequently been recovered from subgingival plaque in patients suffering from acute necrotising gingivitis, pregnancy gingivitis and adult periodontitis (10). In addition, Prevotella species easily acquire resistance towards antibiotics, which hamper their elimination (11). A deep molecular knowledge of how infection and resistance occur is crucial for the development of alternative treatments. In P. intermedia, several proteases have been described, among them trypsin-like serine proteases, a dipeptidyl peptidase IV and CPs (12–14), but no structural studies are available that could help to understand their particular mode of action or facilitate the design of specific drugs. The structures of some clan-A papain-like CPs (according to MEROPS database; (15)) from other infective bacteria are known, namely those of staphopain A and B from Staphylococcus aureus (16,17), the avirulence putative peptidase AvrPphB from Pseudomonas syringae (18) and streptopain (alias streptococcal pyrogenic exotoxin B and SpeB) and IdeS endopeptidase, both from Streptococcus pyogenes (19,20). Together with other bacterial enzymes such as bleomycin hydrolase from Lactococcus lactis and a calpain-like enzyme from P. gingivalis, they may be among the ancestral enzymes that gave rise to the 20 families currently identified within this clan of proteases (15,21). They display a relatively broad substrate specificity but are restricted to a small group of related bacterial species or are even limited to a single species, thus constituting attractive targets for the selective design of antibiotics (22). All these proteases have been identified as or proposed to be secreted virulence factors that elicit nutrient generation, evasion of the adaptive immune system response through inactivation of immunoglobulins or release of bacterial proteins from the cell surface (23).
For more than 60 years, SpeB, a protein secreted by Streptococcus pyogenes (24), was considered a unique CP, unrelated to plant papains or vertebrate cathepsins, and the founding member of family C10 within clan CA (15). A recent analysis of bacterial genomes identified genes encoding potential SpeB orthologues in several species, predominantly Bacteroidetes (31). Interestingly, two forms of genes are common, either short orthologues encoding an SpeB-like protein with an N-terminal pro-domain and a catalytic CP domain or large orthologues with an additional large C-terminal extension, which shares no similarity with any other proteins sequenced. The latter orthologues are present in bacteria that are involved in pathogenicity of periodontal disease in humans. With this in mind, a genome search within P. intermedia 17 was undertaken and three open-reading-frames potentially encoding CPs were identified (22). We studied the first of these potential proteases, interpain A (InpA), encoded by locus PIN0048. This gene encodes a long SpeB-orthologue of 868 residues including a 44-residue signal peptide, a pro-domain (Ala1-Asn111, see Fig. 1), a catalytic domain (Val112-Pro359) and a further 465 C-terminal residues arranged in distinct domains, with putative regulatory and secretory functions (25). We cloned, overexpressed, purified and functionally analysed protein variants comprising the first two domains, the wild-type (wt) form and a variant, in which the active-site Cys154 had been mutated to alanine (C154A), hereafter termed pro-cd-InpA and pro-cd-InpA C154A, respectively. We further analysed the 3D structures of a major fragment of pro-cd-InpA C154A and of the wt catalytic domain, cd-InpA. Unexpectedly, these studies have uncovered a hitherto undescribed activation mechanism for cysteine proteases and helped us to understand a family of virulence factors produced by human pathogens.
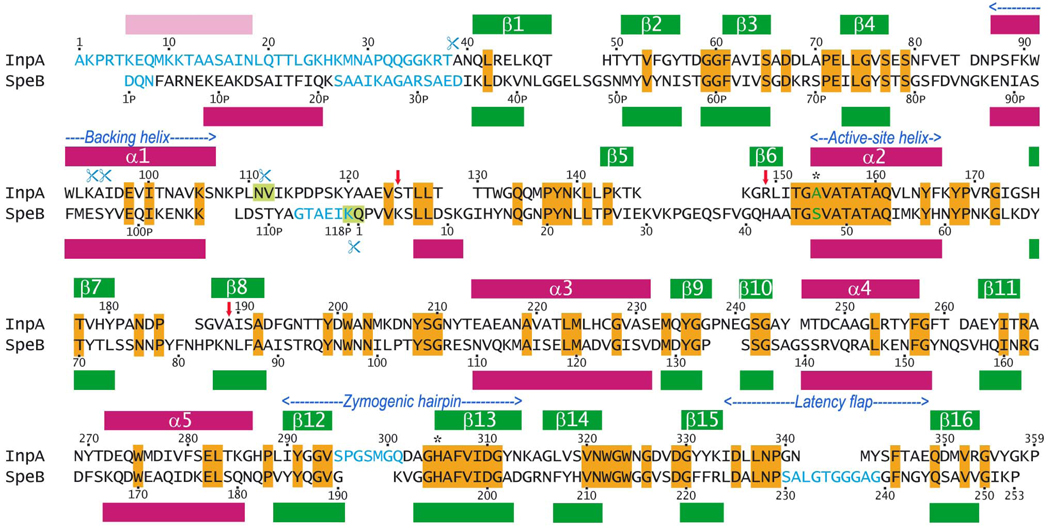
Figure 1. Sequence of pro-cd-InpA.
Sequence of the pro-domain (Ala1-Asn111) and the catalytic domain (Val112-Pro359) of pro-interpain A with interstrain differences (our unpublished results; vertical red arrows) and alignment with S. pyogenes pro-SpeB (pro-domain: Asp1PSPE-Lys118PSPE; mature protease domain: Gln1SPE-Pro253SPE; numbering according to PDB entry 1dki; (19)). Identical residues are displayed over orange background (28% sequence identity). Amino-acid residues not present in the respective 3D structures are depicted in blue. The four autolytic cleavage points of pro-cd-InpA are indicated by blue scissors. The main cleavage points of either protein leading to the stable mature forms are characterised by blue scissors and light green background. The presently studied protein, ΔN1pro-cd-InpA C154A, includes all the residues from Ala39 onwards and a predicted N-terminal helix in the N-terminal missing region is shown in pink.
Experimental procedures
Expression, mutant construction and purification of pro-interpain A
Genomic DNA of P. intermedia was extracted from strain ATCC 25611. The structural gene region of InpA comprising the pro-domain and the catalytic domain, pro-cd-InpA, was amplified by PCR using forward primer 5'-ATGCCATGGCAAAGCCACGCACAAAGGAACAG-3' with an NcoI recognition site and reverse primer 5'-ATGCTCGAGTGGTTTTCCGTAAACACCC-3' with an XhoI recognition site. Because the NcoI site encompasses the ATG start codon, two bases (CA) were introduced into the forward primers immediately after the NcoI site for in-frame translation of the target protein. This genetic manipulation inserted a methionine before the N-terminal alanine residue of InpA. In addition, the reverse primer introduced two additional codons (CTC GAG) for a leucine and a glutamate following the C-terminal proline residue of pro-cd-InpA. The PCR product was purified and cloned into the NcoI/XhoI site of pET24d(+) expression vector (Novagen), which provides the coding sequence for a C-terminal hexahistidine-tag (6xHis). The recombinant plasmid was transformed into Escherichia coli strain BL21(DE3) pLysS under the control of the T7 promoter. The wt construct was used to produce mutation C154A using overlap extension PCR (26). The correctness of the constructs was verified by double-stranded DNA sequencing.
Protein production and purification were essentially the same for the wt and the mutant protein. Cells freshly transfected with the expression plasmid were grown at 37°C to an optical density (A600) of 0.7–0.8 in 1L Luria-Bertani medium supplemented with 2% glucose and kanamycine sulfate (50µg/mL). The culture was induced with isopropyl-1-thio-β-D-galactopyranoside to a final concentration of 0.1 mM and further incubated at 26°C for 2–3h for protein production. Cells were harvested, washed with PBS buffer and resuspended in binding buffer A (20mM sodium phosphate, 500mM NaCl, 20mM imidazole, pH 7.4) supplemented with 1.5mM 4’,4’-dithiodipyridine (a reversible CP inhibitor), 6mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 10µM phenylmethylsulphonyl fluoride, 1mM HgCl2 and 1mM 1,4-dithio-DL-threitol (DTT). The latter compounds were added to prevent protein aggregation and autolysis. Cells were lysed by ultrasonication on ice for ~5’ and cell lysates were cleared by centrifugation, filtered through 0.45µM-pore-size filters and mixed with Fast Flow Ni-NTA Sepharose resin slurry (2mL) previously equilibrated with buffer B (buffer A implemented with 1mM HgCl2). After 1h at room temperature (or overnight at 4°C), the slurry was poured into a column and first washed in buffer B until the baseline (OD280) was stable and then in 2mL of buffer B supplemented with 60mM imidazole. The protein was eluted stepwise with buffer B further containing 100, 200, 300, 400, and 500mM imidazole, respectively. The fractions collected were analyzed by SDS-PAGE and those containing a single band attributable to the target protein were pooled, dialyzed at 4°C overnight against 10mM Tris·HCl, 1mM HgCl2, pH 7.5, passed through 0.45µM-filters and concentrated using Centricon-10 (Millipore). The last purification step comprised ion-exchange chromatography (Amersham Biosciences) with a MonoQ column equilibrated with 20mM Tris·HCl, 1mM HgCl2 , pH 7.5.
Activity assay
Activity was determined with the fluorigenic substrate Boc-Val-Leu-Lys-AMC. Briefly, recombinant pro-cd-InpA protein was activated at 37°C in 0.1M Tris·HCl, 5mM EDTA, pH 7.5, freshly supplemented with 2mM DTT. The reaction was started by adding substrate (10mM; final concentration in the reaction mixture, 250µM) and the release of AMC was recorded by measuring the increase in fluorescence using a micro-titer fluorescent plate reader.
Autocatalytic assay
A total of 100µg of pro-cd-InpA protein, alone or with 0.7µg of active cd-InpA protein, was preincubated at 37°C in buffer C (0.1M Tris·HCl, 1mM HgCl2, 2mM DTT, pH 7.6). The autocatalytic reaction was initiated by diluting the sample with buffer D (buffer C but with 5mM EDTA instead of 1mM HgCl2) at 37°C (final pro-cd-InpA and cd-InpA concentrations were 10µM and 0.1µM, respectively). Aliquots were taken at distinct time intervals and mixed with E-64 inhibitor (N-[N-{L-trans-carboxyoxiran-2-carbonyl}-L-leucyl]-agmatine) to stop the reaction. At the same time intervals, samples of the incubation mixture were assessed for activity against the above fluorigenic substrate and the initial rate of substrate turnover was determined. As a negative control, the same experiments were carried out using buffer C. To ascertain whether pro-cd-InpA autoactivation was an intra- or an inter- molecular process, the zymogen was incubated as described above at 10µM, 2µM and 0.4µM, respectively, with samples withdrawn from the above activation reaction mixture at the mentioned time intervals.
Processing of pro-cd-InpA C154A by wt cd-InpA
Pro-cd-InpA C154A was tested as a substrate for wt cd-InpA in a reaction mixture containing 0.1 µM of the latter and 10µM of the former protein in buffer D at 37°C. Aliquots of 10µL were withdrawn from the reaction mixture at distinct time intervals and the reaction was quenched by addition of E-64. Results were analyzed by 12% SDS-PAGE.
Generation of N-terminally truncated pro-cd-InpA C154A
Pro-cd-InpA C154A (25mg/ml) in 20mM Tris-HCl, pH 7.6, was incubated with 1.7 µg of DTT-activated wt cd-InpA overnight at 21°C. The reaction was terminated by addition of E-64 to 100 µM final concentration and the protein purified by ionic-exchange chromatography employing a NaCl gradient. Fractions containing the N-terminally truncated 36-kDa form of pro-cd-InpA C154A were pooled, concentrated, and dialyzed against a buffer suitable for protein crystallization. N-terminal sequencing, mass spectrometry and western blot analyses revealed that this protein variant (ΔN1pro-cd-InpA C154A) encompassed residues Ala39-Pro359 plus the C-terminal expression vector-derived leucine-glutamate dipeptide but was lacking the 6xHis-tag.
N-terminal sequence analysis
Wild-type pro-cd-InpA, the C154A mutant protein, their truncated variants, as well as their cleavage products were analyzed by 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were stained with 0.2% amido black and subjected to Edman-degradation using a Procise 494-HT protein sequencer.
Crystallisation and data collection and processing
ΔN1pro-cd-InpA C154A was crystallised from sitting drops containing protein (22 mg/mL) and 10% polyethylene glycol (PEG) 3,000, 0.2M magnesium chloride, 0.1M sodium cacodylate, pH6.5. Wt cd-InpA protein comprising residues Val112-Pro359 plus the C-terminal dipeptide was crystallised from drops comprising protein solution (16 mg/mL) and 28% PEG 4,000, 0.2M magnesium chloride, 30% xylitol, 0.1M Tris·HCl, pH8.5. Diffraction data were collected at 110K at ESRF (Grenoble, France) beam line ID29 using an ADSC Q315 CCD area detector. ΔN1pro-cd-InpA C154A crystals diffracted beyond 1.5Å resolution and belonged to space group C2 with one molecule per asymmetric unit. Wt cd-InpA crystals diffracted to 3.2Å, belonged to the tetragonal space group P41212, and contained two molecules per asymmetric unit. Diffraction data were indexed and integrated with program XDS (27) and scaled and reduced with program SCALA within the CCP4 suite (28). Statistics on data collection and processing are presented in Table 1. Wt cd-InpA crystals diffracted very weakly. This led to a high value for the merge indicator Rr.i.m. but to an acceptable Rp.i.m. value due to the almost eight-fold average multiplicity of the data. In any case, these data were accurate enough to yield valid structural information. In the case of the ΔN1pro-cd-InpA C154A crystals, diffraction data were strong and of excellent quality, leading to low values for both Rp.i.m. and Rr.i.m. (see Table 1).
Table 1.
Crystallographic data collection and refinement
| Dataset | ΔN1pro-cd-InpA (C154A) | cd-InpA |
| Space group | C2 | P41212 |
| Space group / cell constants (a, b, c, in Å; β in °) | 128.87, 38.81, 78.14, 127.3 | 129.24, 129.24, 81.97, 90.0 |
| Wavelength (Å) | 0.9763 | 0.9760 |
| No. of measurements / unique reflections | 344,943 / 49,377 | 90,923 / 13,017 |
| Resolution range (Å) (outermost shell)a | 39.07–1.50 (1.58–1.50) | 47.25-3.22 / 3.40-3.22 |
| Completeness (%) | 99.5 (97.4) | 99.0 (98.2) |
| Rr.i.m.= Rrmeas b / Rp.i.m b | 0.086 (0.448) / 0.033 (0.180) | 0.352 (0.959) / 0.120 (0.330) |
| Rmerge b | 0.080 (0.409) | 0.329 (0.897) |
| Average intensity (<[<I> / σ(<I>)]>) | 16.4 (5.3) | 6.2 (1.9) |
| B-Factor (Wilson) (Å2) / Average multiplicity | 13.9 / 7.0 (6.1) | 46.8 / 7.9 (7.9) |
| Crystallographic Rfactor (free Rfactor) c | 0.156 (0.193) | 0.207 (0.261) |
| Estimated overall co-ordinate error based on free Rfactor (Å) | 0.079 | 0.492 |
| No. of protein atoms d / solvent molecules | 2,452 / 370 | 3,798 / 0 |
| Rmsd from target values | ||
| bonds (Å) / angles (°) | 0.013 / 1.40 | 0.012 / 1.31 |
| bonded B-factors (main chain / side chain)(Å2) | 1.36 / 2.80 | 0.277 / 0.654 |
| Average B-factors for protein solvent molecule atoms (Å2) e | 14.7 / 23.6 | 66.7 / - |
| Main-chain conformational angle analysis f | ||
| Residues in favoured regions / outliers / all residues | 303 / 1 / 310 | 464 / 0 / 496 |
Values in parentheses refer to the outermost resolution shell if not otherwise indicated.
Rr.i.m.= Σhkl(nhkl /[nhkl−1] 1/2)Σi |Ii(hkl) − <I(hkl)>| / ΣhklΣi Ii(hkl) and Rp.i.m.= Σhkl(1/[nhkl−1]1/2)Σi |Ii(hkl) − <I(hkl)>| / ΣhklΣi Ii(hkl), where Ii(hkl) is the i-th intensity measurement and nhkl the number of observations of reflection hkl, including symmetry-related reflections, and <I(hkl)> its average intensity. Rmerge= ΣhklΣi |Ii(hkl) − <I(hkl)>| / ΣhklΣi Ii(hkl).
Crystallographic Rfactor = Σhkl ‖Fobs| − k |Fcalc‖ / Σhkl |Fobs|, with Fobs and Fcalc as the observed and calculated structure factor amplitudes; free Rfactor, same for a test set of reflections not used during refinement.
Including atoms in alternate conformation.
The last refinement step included anisotropic B-factor refinement of the protein atoms in the case of ΔN pro-cd-InpA (C154A).
According to program MOLPROBITY (47).
Structure solution and analysis
The structure of ΔN1pro-cd-InpA C154A was solved with program PHASER (29) using all diffraction data and the co-ordinates of S. pyogenes pro-SpeB protein (Protein databank (PDB) access codes 1pvj and 1dki; (19)) as a searching model. A refined final solution was found at 170.8, 60.0, 294.0 for α, β, γ (in Eulerian angles) and 0.145, −1.007, 0.734 for x, y, z (in fractional cell co-ordinates) with a Log-likelihood gain value of 44.2. The appropriately rotated and translated search-model co-ordinates were subjected to crystallographic refinement albeit with no positive result. Accordingly, the model was given 200 cycles of refinement with program SHELXL (30) starting from a resolution of 3Å and increasing it by 0.01Å with every cycle until full data resolution (1.5Å). No data were set aside as a free Rfactor set. The resulting model phases were subjected to a density modification step with program SHELXE (31). Amplitudes and phases for non-measured reflections were thereafter extrapolated from the current map to fill in the missing reflections within the experimental resolution limits and beyond, to a resolution of 1.0Å. 60 cycles of density modification and 5 iterations of 20 cycles each of main-chain tracing with SHELXE followed. Combination of the resulting partial main-chain model with the original phases was succeeded by a new density modification run that eventually led to a partial backbone model for 192 of the residues and a set of phases with a figure-of-merit (fom) of 0.76. An electron density map was computed and subjected to a final density modification step with program DM within CCP4. This step improved the fom to 0.87 and enabled straightforward model completion and refinement. Manual model completion with TURBO- Frodo alternated with crystallographic refinement using REFMAC5 within the CCP4 suite. The final ΔN1pro-cd-InpA C154A model comprised all residues from Ala39 to Pro359 (Fig. 1) except Ser295-Gln301.
The structure of wt cd-InpA was solved with program AMoRe (32) using the co-ordinates corresponding to residues Ala121-Pro359 from ΔN1pro-cd-InpA C154A and structure-factor amplitudes in the range 15–3.5 Å. These calculations unambiguously confirmed P41212 as the correct space group and a unique solution was found at 48.5, 88.4, 223.7, 0.1172, 0.5977, 0.1152 (α, β, γ, x, y, z; refined values after rigid-body refinement; see (32)) and 41.7, 88.6, 115.6, 0.1940, 0.0880, 0.7122 for each of the two molecules A and B in the asymmetric unit, respectively, with a combined score CCF/crystallographic Rfactor, according to (32), of 54.4%/41.4%. The appropriately rotated and translated co-ordinates were subjected to rigid-body and positional refinement applying strong non-crystallographic-symmetry (ncs) restraints with program CNS v. 1.2 (33). Despite the weakness and low resolution of the wt cd-InpA diffraction data, the resulting electron density maps clearly disclosed the entire polypeptide chain of the mature protease moiety. It contained an unambiguous trace for the first eight residues (Val112-Tyr120) of the polypeptide chain that had not been included in the search model, a proof of concept for data quality which ruled out model bias. Careful model building alternated with crystallographic refinement under application of strong ncs-restraints with programs CNS and REFMAC5 (at the final stages). The final wt cd-InpA model comprised residues Val112-Gly357 for molecule A and Val112-Pro359 plus two residues from the C-terminal tag (termed Leu360 and Glu361) for molecule B. These two molecules were almost equivalent in practice. Accordingly, results and discussion considered model A unless otherwise stated. Table 1 provides statistics on the final refinement steps and parameters of the quality of the resulting models.
Miscellaneous
Figures were prepared with programs TURBO-Frodo, SETOR (34) and MOLMOL (35). Structures were superimposed with TURBO-Frodo. Bioinformatic amino-acid sequence similarity searches were undertaken within MEROPS database (merops.sanger.ac.uk) and with the PSI-BLAST server (www.ncbi.nlm.nih.gov/blast). Structural similarity searches were performed with program DALI (www.ebi.ac.uk/msd) and secondary structure predictions with program JPRED (www.compbio.dundee.ac.uk/~www-jpred). Close contacts and interaction surfaces (with a probe radius of 1.4Å) were calculated with CNS taking the half of the total surface buried at the interface. The final co-ordinates of ΔN1pro-cd-InpA C154A and wt cd-InpA have been deposited with the Protein Data Bank at the Research Collaboratory for Structural Bioinformatics (www.rcsb.org/pdb) with access codes XXXX and YYYY.
Results and discussion
Protein purification and characterisation
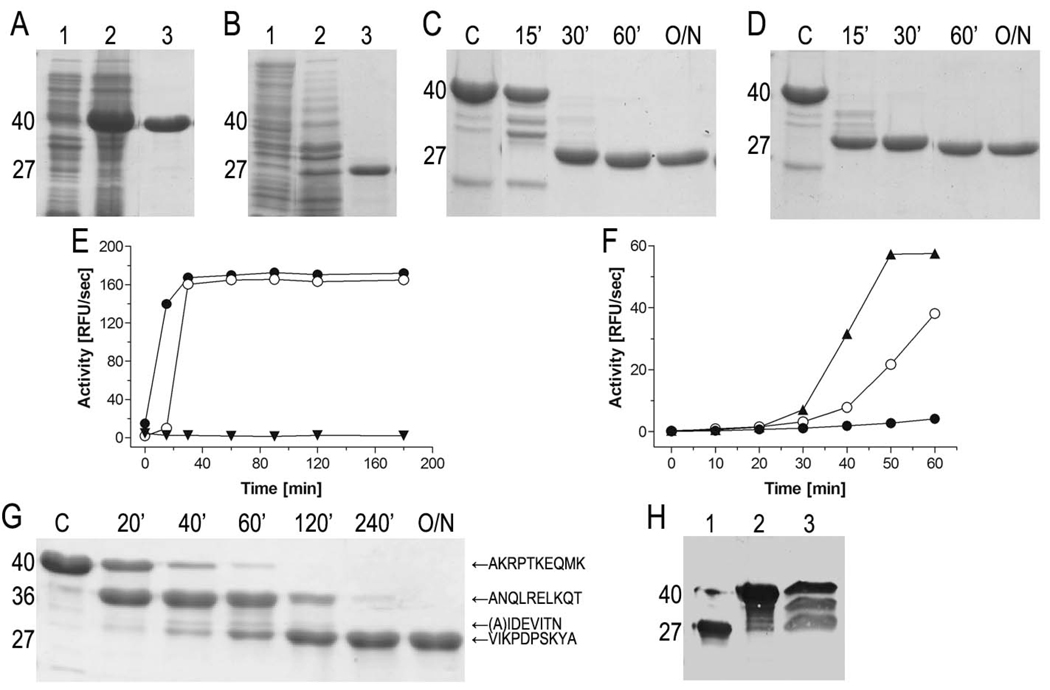
Pro-cd-InpA and pro-cd-InpA C154A were overexpressed as 40-kDa proteins and purified to homogeneity. The wt zymogen was readily converted into the fully-processed mature 27-kDa catalytic domain during purification so that the zymogenic form could only be obtained if reversible CP inhibitors were included during homogenization of bacterial cells and purification (Fig. 2abc). Subsequent inhibition release resulted in time-dependent autocatalytic processing of the zymogen with the concurrent release of activity (Fig. 2ce). Processing and activity release were accelerated by catalytic amounts of active cd-InpA (Fig. 2de). This, together with the finding that the initial rate of activity generation was dependent on the zymogen concentration (Fig. 2f), suggested that the autocatalytic maturation of pro-cd-InpA occurred in trans (inter-molecularly). Pro-cd-InpA C154A was produced to elucidate the sequence of cleavage events during activation and for structural purposes. As in the case of other CPs, pro-cd-InpA C154A was enzymatically inert and did not undergo autoprocessing. Analysis of concentration- and time-dependent proteolysis of pro-cd-InpA C154A by the active protease revealed that the process occurred stepwise through a main 36-kDa intermediate (ΔN1pro-cd-InpA C154A) generated by hydrolysis of peptide bond Thr38-Ala39. Accordingly, ΔN1pro-cd-InpA C154A lacks the first 38 residues of the full-length zymogen (see Fig. 1). In addition, minor cleavages were mapped to Lys94-Ala95 and Ala95-Ile96 (Fig. 1 and Fig 2g). Finally, limited proteolysis of the accumulating 36-kDa intermediate at the Asn111-Val112 peptide bond released the 27-kDa mature protein which was resistant to further degradation. The same activation pathway may operate in vivo since a similar band pattern representing variably processed InpA species was detected in the P. intermedia culture medium (Fig. 2h). The maturating self processing of InpA resembles pro-SpeB with respect to formation of one major intermediate and several cleavages within the remaining part of the N-terminal pro-domain (36). Such a mechanism provides regulation of proteolytic activity independent of other secreted and host proteases, thus ensuring that the activity is developed when required.
Figure 2. Expression and activity of InpA.
(A) Expression and purification of pro-cd-InpA C154A. Lanes 1 and 2, E. coli homogenate before and 3h after protein expression induction, respectively. Lane 3, recombinant protein after affinity chromatography purification. Molecular masses of the distinct protein species (40 and 27 kDa) are shown on the left. (B) Same for wt pro-cd-InpA. (C) Time-course analysis of autocatalytic processing and activation of wt pro-cd-InpA (final concentration 10µM) incubated with 1mM HgCl2. The reaction was initiated by adding EDTA (5mM final concentration) as a Hg2+-chelator, i.e. by releasing metal-mediated inhibition. Samples were withdrawn at the time intervals specified (O/N, overnight incubation; lane C, pro-cd-InpA alone). (D) Same as (C) but after addition of active InpA (10nM final concentration) to the reaction mixture. In this case, the reaction proceeded much faster. (E) A subset volume of the withdrawn aliquots from (C) and (D) was used to quantify the activity released from wt pro-cd-InpA in the absence (○) and presence (●) of catalytic amounts of wt cd-InpA. As a control, pro-cd-InpA spiked with InpA but without EDTA was incubated in parallel (▼). (F) Concentration dependent autoactivation of pro-cd-InpA. The reaction was initiated by releasing Hg2+-mediated inhibition in mixtures containing 0.04µM (●), 2µM (○) and 10µM (▲) zymogen. At indicated time points, 50µl (●), 10µl (○), and 2µl (▲) were withdrawn from each reaction mixture and directly assayed for activity. (G) SDS-PAGE of the digestion of pro-cd-InpA C154A by wt cd-InpA. The zymogen (final concentration of 10µM) was incubated with cd-InpA (0.1µM) for time intervals as specified (lane C, control pro-cd-InpA C154 incubated alone). N-terminal amino acid sequences of pro-cd-InpA derived fragments are indicated on the right. (H) Western blot analysis of culture supernatant of P. intermedia using InpA-specific rabbit antiserum (lane 3). Wt cd-InpA and pro-cd-InpA (C154A) were loaded on lane 1 and 2, respectively, for comparison.
Structure solution employing a novel approach
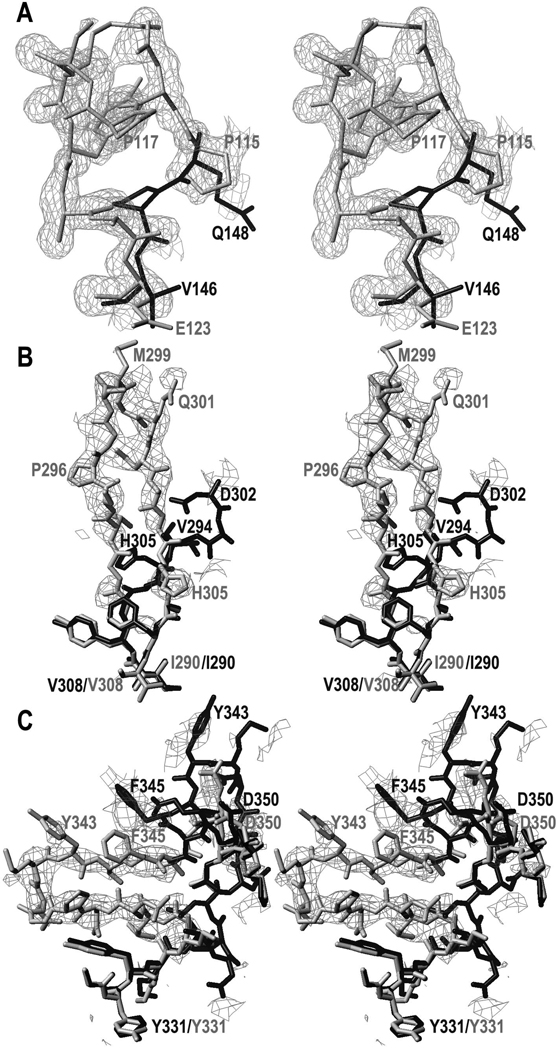
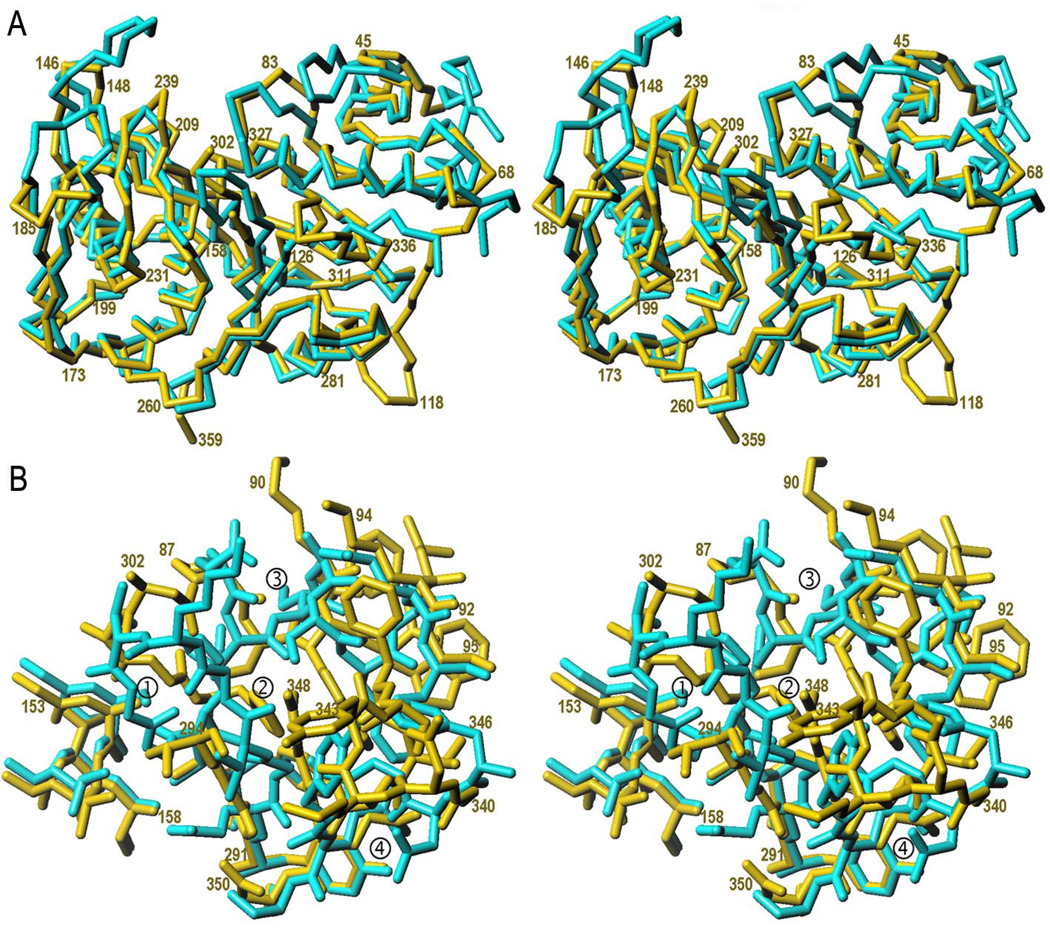
Contrary to the intact mutant protein, the ΔN1pro-cd-InpA C154A variant crystallized. Its structure was solved by Patterson-search methods using maximum-likelihood criteria. This approach improves the definition of the target for the search by removing the contribution of unknown variables. This means that the errors attributable to lack of completeness of a search model are better estimated. In practice, this entails a larger radius of convergence (i.e. it yields a solution for structurally more distant searching models) than conventional search methods, which failed in the present case. Unfortunately, the current crystallographic refinement programs have a shorter radius of convergence. This restricted model refinement and led us to develop a novel approach based on a further development of the SHELX suite of programs (37). It consists of the application of the “free-lunch algorithm”, whose theoretical bases had been developed by Giacovazzo and co-workers (38), combined with auto-tracing, model refinement and density modification. This process essentially envisaged that the initially (poorly) refined model, displaying a weighted mean-phase error (wMPE) of 64° with respect to the final refined model (as determined a posteriori), was used to calculate an electron density map that was subjected to density modification. With this map, missing structure-factor amplitudes and phases were estimated within the resolution range of the experimental data. Further values were extrapolated to a nominal resolution of 1.0Å. Subsequently, density modification (wMPE=33°), main-chain auto-tracing and phase-combination (wMPE=27°) eventually produced an accurate partial model for ~60% of the residues. In addition, the resulting electron density map was excellent, even in those parts where the original search model showed a different chain trace (Fig. 3). This permitted straightforward manual tracing of the entire molecule and successful refinement, enabling to ascertain three differences in comparison to the sequence of the PIN0048 ORF in the TIGR data base that were subsequently confirmed by sequencing at the DNA level (see Fig. 1).
Figure 3. Experimental electron density maps.
(A) Representative example of the Fobs-type map obtained after multistep-density modification, contoured at 1σ above threshold and superimposed with the model placed in accordance to the Patterson search calculations (black stick model; residues 145–148, see PDB 1pvj) and the final refined co-ordinates of ΔN1pro-cd-InpA C154A (light-grey stick model; residues 115–123). (B) (mFobs-DFcalc)-type σA-weighted omit map contoured above +1.75σ above threshold illustrating the distinct chain trace around the zymogenic hairpin of molecule A (final model in light-grey) as compared with the ΔN1pro-cd-InpA model employed for phasing (black stick model). Some residues of either structure are labelled in the respective grey-tone for reference. (C) Same as (B) but showing the latency flap.
Structure of InpA zymogen
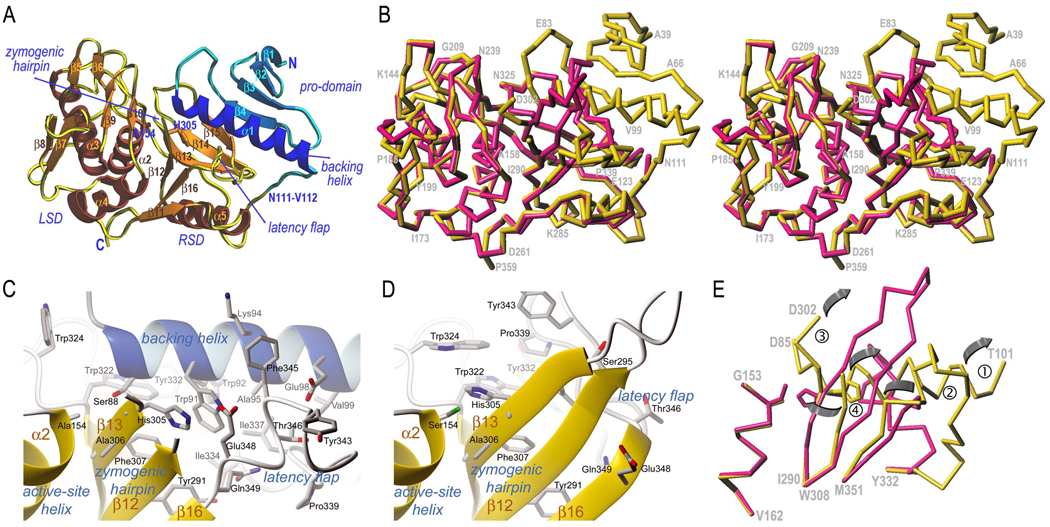
The protein has an elongated shape with an N-terminal pro-domain (Ala39-Asn111) and a C-terminal papain-like CP domain (Val112-Pro359), which bifurcates into a right subdomain (RSD) and a left subdomain (LSD) (see Fig. 4a). RSD and LSD interact through a surface of 1,332Å2 establishing 69 contacts (<4Å), among them 11 hydrogen bonds (<3.4Å) and 22 hydrophobic interactions (Table 2). The pro-domain contacts laterally the top of the CP moiety through a surface of 1,177Å2, with 54 contacts (<4Å), among them 12 hydrogen bonds and 19 hydrophobic interactions (Fig. 4a and Table 2). The pro-domain is stabilised by a central hydrophobic core and evinces an open-faced sandwich with a twisted antiparallel four-stranded β-sheet (sheet I; strands β1–β4) of simple up-and-down connectivity mediated by short loops. After β4, a segment in extended conformation (loop joining strands β4 and α1, Lβ4α1) leading to helix α1. The N-terminal part of the helix approaches the active-site cleft, thus contributing to latency, and is hereafter termed “backing helix”. The polypeptide reaches the molecular surface after α1 and undergoes a sharp turn, folding back along the surface and entering a connecting segment that links the pro-domain with the CP domain. This segment adopts an extended conformation from Asn107 to Pro117, i.e.optimal for binding to and cleavage by an active-site cleft of a protease (39). This stretch includes the activation cleavage point, Asn111-Val112 (Fig. 4a), which is superficial and accessible for processing.
Figure 4. Structures of ΔN1pro-cd-InpA C154A and wt cd-InpA.
(A) Richardson diagram of ΔN1pro-cd-InpA C154A in standard orientation. The pro-domain is displayed in blue/cyan and the mature protein moiety (subdivided into a right and a subdomain) in yellow/brown. The subdomains, the regular secondary structure elements (see Fig. 1), the N- and the C-terminus, the primary activation point (at Asn111-Val112) and the structure regions responsible for latency maintenance are marked and labelled. (B) Superimposition of the Cα-carbon traces of ΔN1pro-cd-InpA C154A (yellow) and wt mature cd-InpA (red) in standard orientation. Some residues of ΔN1pro-cd-InpA C154A are labelled for reference. (C) Close-up view of the active site of ΔN1pro-cd-InpA C154A. Orientation as in (B) after a horizontal rotation of ~45°. (D) Same as in (C) but for wt active cd-InpA. (E) Cα-trace of the structure of ΔN1pro-cd-InpA C154A (yellow) and wt mature cd-InpA (red) around the active site including ①, the catalytic cysteine residue (Cys154; mutated to alanine in ΔN1pro-cd-InpA C154A), imbedded in active-site helix α2; ②, the zymogenic hairpin encompassing the catalytic histidine (His305) (undefined from Ser295 to Gln301 in ΔN1pro-cd-InpA C154A); ③, the backing helix α1 (absent in cd-InpA); ④, the latency-flap, displayed from Tyr332 to Met351 for either structure. The grey arrows indicate the displacements of the keynote structural elements upon zymogen activation as explained in the text.
Table 2.
Inter-domain (pro-domain/protease domain) and inter-subdomain (RSD/LSD) interactions in ΔN1pro-cd-InpA.
| Pro-domain | Protease moiety |
Dist.(Å) | RSD | LSD | Dist.(Å) |
|---|---|---|---|---|---|
| van-der-Waals interactions | van-der-Waals interactions | ||||
| Phe61 Cζ | Val328 Cγ1 | 3.59 | Leu128 Cδ2 | Gln161 Cβ | 3.97 |
| Pro71 Cγ | Lys333 Cε | 3.72 | Leu128 Cδ1 | Tyr165 Cβ | 3.82 |
| Pro71 Cγ | Leu336 Cδ2 | 3.79 | Thr130 Cδ2 | Trp132 Cε2 | 3.50 |
| Leu73 Cδ2 | Leu317 Cδ2 | 3.68 | Ala262 Cβ | Leu253 Cβ | 3.90 |
| Gly75 Cα | Tyr332 Cδ1 | 3.81 | Tyr264 Cδ2 | Met246 Cβ | 3.66 |
| Phe81 Cδ1 | Val328 Cγ1 | 3.90 | Tyr264 Cδ1 | Thr247 Cβ | 3.70 |
| Pro87 Cδ | Trp324 Cε3 | 3.50 | Pro288 Cγ | Tyr165 Cδ2 | 3.71 |
| Phe89 Cγ | Val328 Cγ2 | 3.63 | Pro288 Cδ | Phe166 Cζ | 3.73 |
| Trp91 Cζ3 | Phe307 Cε2 | 3.90 | Ile290 Cγ1 | Val162 Cγ2 | 3.79 |
| Trp91 Cη2 | Leu337 Cδ1 | 3.91 | Ile290 Cγ2 | Met246 Cε | 3.93 |
| Trp91 Cζ2 | Gln349 Cγ | 3.78 | Ile290 Cδ1 | Leu253 Cδ1 | 3.94 |
| Trp92 Cβ | Trp322 Cη2 | 3.60 | Val294 Cγ2 | Tyr245 Cε2 | 3.58 |
| Trp92 Cη2 | Tyr332 Cδ1 | 3.58 | Ala306 Cβ | Met246 Cε | 3.74 |
| Trp92 Cδ1 | Leu337 Cδ1 | 3.97 | Val308 Cγ1 | Val162 Cγ2 | 3.67 |
| Lys94 Cδ | Phe345 Cε1 | 3.57 | Asp350 Cβ | Met246 Cγ | 3.73 |
| Ala95 Cα | Phe345 Cβ | 3.97 | Val352 Cγ2 | Met246 Sδ | 3.91 |
| Ile96 Cδ1 | Leu336 Cδ1 | 3.73 | Val352 Cγ1 | Leu253 Cδ1 | 3.93 |
| Val99 Cγ2 | Leu337 Cδ2 | 3.95 | Val355 Cγ1 | Val162 Cγ1 | 3.86 |
| Val99 Cγ1 | Thr346 Cγ2 | 3.35 | Val355 Cγ1 | Leu163 Cδ1 | 3.88 |
| Hydrogen bonds | Val355 Cγ2 | Leu253 Cδ1 | 3.80 | ||
| Leu74 O | Lys333 N | 2.87 | Val355 Cγ1 | Phe259 Cε1 | 3.75 |
| Val76 N | Tyr331 O | 2.79 | Tyr356 Cα | Phe166 Cζ | 3.62 |
| Ser77 Oγ | Asp329 O | 2.72 | Hydrogen bonds | ||
| Thr84 Oγ1 | Asp327 Oδ1 | 2.75 | Thr129 Oγ1 | Asp206 Oδ1 | 2.85 |
| Asn86 Oδ2 | Asp327 O | 3.19 | Thr130 Oγ1 | Gln161 Oε1 | 2.75 |
| Ser88 Oγ | Ala306 N | 3.01 | Thr131 N | Asp206 O | 2.97 |
| Trp91 Nε1 | Phe345 O | 2.92 | Thr260 N | Gly258 O | 2.99 |
| Trp92 Nε1 | Lys333 O | 2.87 | Asp261 N | Phe259 O | 3.05 |
| Glu98 Oε1 | Phe345 N | 2.83 | Ala262 O | Arg254 Nη1 | 2.78 |
| Glu98 Oε2 | Thr346 N | 3.04 | Ala262 N | Phe259 O | 3.03 |
| Asn111 Nδ2 | Asp272 Oδ2 | 2.48 | Asn321 Oδ1 | Gln161 Nε2 | 2.97 |
| Asn111 Oε1 | Asp335 O | 3.17 | Trp324 N | Trp132 O | 3.33 |
| Asn325 N | Trp132 O | 2.97 | |||
| Lys358 Nζ | Tyr168 Oη | 2.80 | |||
At Val112, the polypeptide chain enters the RSD of the mature enzyme moiety, which is a split subdomain (Val112-Leu127 + Thr260-Pro359) with an open-faced sandwich topology created by a six-stranded twisted antiparallel β-sheet (sheet II; strands β11 to β16). The sheet extends from the bottom of the molecule (outermost strand β11) to the interface with the pro-domain at β15 (Fig. 4a). The twist gives rise to a concave and a convex face and the latter mediates the main interaction with the LSD. The main contact between the pro-domain and the CP part is formed by the outermost strand of sheet I, β4, and the lateral strand of sheet II, β15. This gives rise to a continuous ten-stranded β-sheet that completely traverses the zymogen from its upper right to the bottom centre (Fig. 4a). After the inset of the LSD (see below), the polypeptide chain rejoins the RSD at strand β11 of sheet II, which runs outward approximately perpendicular to the view in Fig. 4a. After this strand, a short loop leads to helix α5, which nestles in the concave side of sheet II, followed by the next four strands of sheet II (β12–β15), inserted with simple up-and-down connectivity. These strands are connected by loops, which contribute to the substrate-binding cleft and the active site. The polypeptide chain is very well defined for the whole protein moiety except for the tip of a β-hairpin structure created by strands β12 and β13 and the enclosed loop, the “zymogenic hairpin” in the following. The hairpin is rigid at its trunk, as it is stabilised by six β-sheet interactions between β12 and β13 but flexible at its tip (between Ser295 and Gln301; Fig. 4). After β15, the polypeptide runs below the backing helix α1 and gives rise to what will now be referred to as the “latency-flap”, Lβ15β16, which spans the 16 residues from Ile334 to Gln349. This structure displays a unique conformation and is stabilised by a series of internal contacts. It consists of two sequential dextrohelical elements, Ile334-Asn338 and Ser344-Gln349, connected by two residues in extended conformation (Pro339-Gly340) and a tight 1,4-turn of type I (Asn341 O-Ser344 N, 3.13Å), which protrudes from the molecular surface. The bottom of the first dextrohelical segment is anchored to Lβ11α5 through a bidentate interaction of its main chain with the completely buried side chain of Arg267 and includes another tight 1,4-turn of type I (Ile334 O-Leu337 N; 2.94Å). In addition to this arginine anchor, the structure of the latency flap is galvanized by a total of nine internal hydrogen bonds that confer an extraordinary rigidity to this structural element. After this flap, the protein chain enters the second strand of sheet II, β16, and leads to the surface C-terminus of the molecule at Pro359, whose position permits additional downstream domains in the full-length InpA protein (Fig. 4a).
The LSD (Leu128-Phe259) is inserted into the RSD and is characterised by a central three-helical bundle made up by helices α2, called the “active-site helix”, as well as α3 and α4, which traverse the subdomain from the back to the front (Fig. 4a). In addition, three β-hairpins are found on the front side of the LSD, β5β6, β7β8, and β9β10. After α4, the polypeptide chain rejoins the RSD at Thr260 and leads to β11. As observed for the pro-domain, the LSD is held together by a large central hydrophobic cluster that reaches the subdomain surface at the bottom and at the left of the molecule and accommodates active-site helix α2.
Substrate-binding crevice and active site
The active-site cleft of InpA is in a crevice formed by loops connecting strands of sheet II at its carboxy end. The walls of the crevice are provided by RSD and LSD (see Fig. 4). Classic CPs like papain, cathepsin B and staphopain have a short, four-residue segment connecting the two residues that are topologically equivalent to Gln134 and Gly153 of InpA, respectively, as contributors to the left-side rim of the cleft on its primed side. In contrast, InpA displays between the latter two residues an 18-residue insertion that forms a unique upper-left region of the molecule. This entails that the zone ascribable to substrate binding would be reduced in InpA to Gly133-Gln135 and Thr152-Gly153, immediately preceding the catalytic cysteine, Cys154. The former stretch includes Gln134, whose position is absolutely conserved among CPs and which, by analogy, would be involved in the formation of an oxyanion hole together with the amide nitrogen of Cys154, which would bind the scissile carbonyl (21,40). On the non-primed side of the cleft, Ser242-Met246 and Tyr264 would also contribute to the left rim. Again in contrast to classic CPs, InpA possesses a much longer connection between helices, which shapes part of the front surface and gives rise to a unique β-hairpin, novel for CPs (β9β10). This entails that the residues from Pro238 to Gly241 should further assist Ser242-Met246 in shaping the cleft rim. In even greater contrast to classic CPs, the segments shaping the right-hand rim of the cleft on its primed side may be restricted to the side chains of the strongly conserved Trp324 from Lβ14β15, which becomes rearranged upon activation, and the previously mentioned Gln134 (21). Regarding the right rim on the non-primed side of the cleft, binding may be provided by the main chain of the rearranged zymogenic hairpin, in particular His305-Ala306 and Tyr291-Gly293, as well as Asp350.
Structures related to InpA
As might have been expected, a search for structural relatives of ΔN1pro-cd-InpA identified pro-SpeB as the closest homologue, with an rms deviation of 2.0Å over 275 topological equivalent residues (PDB 1dki and 1pvj; (19)). This protein is secreted as a zymogen and no structural information on the mature protein is currently available. InpA and SpeB are the only members of the catalytic-dyad enzymes, i.e. those lacking a catalytic asparagine, structurally studied to date (19). As P. intermedia has been shown to degrade connective-tissue constituents and to interfere with the tightly regulated defence mechanism of the host (9), like SpeB in S. pyogenes (22), it is tempting to speculate that InpA is a virulence factor equivalent to SpeB in P. intermedia. In addition, P. gingivalis was shown to harbour a further CP, periodontain (41), that is closely related to SpeB and InpA. Accordingly, we conclude that P. gingivalis, P. intermedia and S. pyogenes may have inherited these homologous genes from a common ancestor and that they may undergo a similar activation mechanism (42).
Overall, the core of the protease and the pro-domain of InpA conform to the pro-SpeB fold (Fig. 5a). However, the difficulties encountered during ΔN1pro-cd-InpA structure solution employing pro-SpeB as a search model for phasing and a sequence identity of just 28%, i.e. in the twilight zone of protein sequence alignments (43), already pointed to significant differences in structure. The pro-SpeB crystal structure displays unconnected electron density for a helix nestling on the concave side of sheet I of the pro-domain. Several secondary structure prediction algorithms consistently predicted an α-helix to similarly run from Lys6 to Asn18 in pro-cd-InpA (Fig. 1). However, differences in length in the loop connecting this (putative) first helix with strand β1 (nomenclature of ΔN1pro-cd-InpA, see Fig. 1 for equivalences), as well as in Lβ1β2, lead sheet I to have a bulge on its left-hand side in the streptococcal enzyme, which is compensated in ΔN1pro-cd-InpA by a different chain trace of Lβ4α1 (around residue Val83, see Fig. 5b). The pro-SpeB pro-domain is undefined for segment Ala112PSPE-Gln1SPE (residues of pro-SpeB are subscripted SPE; see Fig. 1 for the complete pro-SpeB sequence), which includes the primary activation cleavage site at Lys118PSPE-Gln1SPE. Accordingly, this region, which is instrumental for understanding a latent structure, is defined in the prevotellaceal zymogen but not in the streptococcal protein.
Figure 5. Comparison of ΔN1pro-cd-InpA C154A with pro-SpeB.
(A) Superimposition of the Cα-carbon traces of ΔN1pro-cd-InpA C154A (blue) and pro-SpeB (yellow) in standard orientation revealing the large-scale structural deviations of the activation segments. Some residues of ΔN1pro-cd-InpA C154A are labelled for reference. (B) Detail of the region around the active site including segments shown under Fig. 4e, i.e. ①, the catalytic cysteine residue (Cys154; alanine in ΔN1pro-cd-InpA C154A), within active-site helix α2, depicted for its residues Gly153-Ala158 (pro-cd-InpA numbering, see Fig. 1); ②, the zymogenic hairpin including the catalytic histidine (His305), shown for Tyr291-Phe307; ③, the backing helix α1 from the pro-domain, shown for Pro87-Ala95; ④, the segment containing the latency-flap, displayed from Gly340 to Asp350.
There are also important differences in the surface structures of pro-cd-InpA and pro-SpeB, which affect activation and substrate binding in the mature enzymes. At the end of the first segment of the RSD, at Leu128-Thr129, a three-residue insertion creates a bulge in pro-SpeB leading to structural differences in the loop structure preceding α3, with a maximal difference at Gly210 (Ser105SPE) of 2.8Å. Further downstream of the polypeptide chain, pro-SpeB has ten extra residues preceding the active site helix and does not display a hairpin equivalent to β5β6 of ΔN1pro-cd-InpA. This, together with the flipped Lβ7β8 loop (three residues more in pro-SpeB), has implications in shaping the left rim of the substrate-binding crevice and for the interaction with other proteins (Fig. 5). In addition, the tip of hairpin Lβ9β10, possibly involved in the left rim of the crevice in InpA (see above), also diverges due to the three extra residues in the latter proteinase. The greatest differences, however, affect regions surrounding the active site, in particular the zymogenic hairpin and the latency flap. The former, comprising the catalytic histidine in both proteins, is flexible and five residues longer in ΔN1pro-cd-InpA, where it adopts a different orientation. In turn, the segment equivalent to the latency flap is completely disordered between Ser230SPE and Gly239SPE and has six additional residues in pro-SpeB (Fig. 1 and Fig 5), following a completely different path. Hence, it does not contribute significantly to interactions with the zymogenic hairpin or the backing helix to maintain the zymogenic structure. Furthermore, in pro-SpeB the polypeptide preceding Ser230SPE invades the space occupied by the segment connecting the pro-domain with the mature moiety in pro-cd-InpA, thus pointing to differences in the segment flanking the primary activation cleavage point.
In contrast to these differences, there are also similarities in detail. As in ΔN1pro-cd-InpA, latency is achieved in pro-SpeB through a catalytically incompetent conformation of the catalytic histidine, His195SPE, while the catalytic cysteine is probably in a functional position. It is conceivable, extrapolating from our structures, that the zymogenic hindrance is exerted in the streptococcal enzyme by a simple ~90°-rotation around the χ1 angle of His195SPE. This movement swings the imidazole side chain away from its cysteine-binding position and establishes a van-der-Waals interaction with Val192SPE Cγ2 within the hairpin segment equivalent to the zymogenic hairpin in ΔN1pro-cd-InpA. The competent imidazole position is occupied in pro-SpeB by a unique asparagine, Asn89PSPE from the pro-domain, which establishes a highly-specific key hydrogen bonding network with Trp214SPE Nε1, Ala196SPE O and Trp212SPE O (19). The position equivalent to Asn89PSPE is occupied in ΔN1pro-cd-InpA by Ser88, which establishes one of these three interactions (with Ala306 O) in the InpA zymogen as one of the elements likewise leading to an incompetent histidine conformation. Accordingly, the major differences in the structure of the zymogens do not preclude that the novel activation mechanism described below may also be valid with variations for pro-SpeB and, possibly, periodontain activation.
A novel mechanism for latency maintenance and activation
The mature enzyme structure confirms that the pro-domain, including the backing helix, is removed upon activation and that it does not sterically block access to the substrate-binding cleft in ΔN1pro-cd-InpA. There are two parallels between this zymogen and other CP zymogens such as human and rat pro-cathepsin B (44), K (45), and pro-staphopain B (16). In the latter, the pro-segment packs against a surface loop of the C-terminal domain termed the pro-segment binding loop. This loop is absent in ΔN1pro-cd-InpA but its pro-domain binds in the same place. A further common feature is that the association between the pro-domain and the protease domain is based on hydrophobic residues. However, in the mentioned CP zymogens the pro-domain segments run the full length of the cleft in the opposite direction to a peptidyl substrate, and block the crevice. In the InpA zymogen, in contrast, backing helix α1 and the preceding loop Lβ4α1 are inserted laterally like a wedge (Fig. 4a). Trp324, from Lβ14β15 within the CP domain, stops the wedge with its side chain (Fig. 4c). The relative antipodal disposition on the molecular surface of the active site and Val122 (Fig. 4), which are ~26Å apart, supports kinetic data (see above) suggesting that autolytic activation of InpA is likely to occur in trans. The CP domain is similar in both the mature enzyme and the zymogen (239 out of 248 common Cα atoms show an rmsd of 0.82Å; see Fig. 4b). Interestingly, activation is not correlated with significant displacement of the newly formed N-terminus at Val122 (Fig. 4b). Despite this similarity, detailed comparison of the two structures reveals that selected structure elements display complete different chain traces (Fig. 3).
In CPs, function requires a correct spatial arrangement of the catalytic cysteine provided by the active-site helix within LSD and the catalytic histidine of the RSD to render a functional thiolate-imidazolium ion pair (46) (Fig. 4cde). Unlike InpA, most other CPs also have an asparagine with a supportive role (46). The position and conformation of the active-site helix and the cysteine, Cys154, are maintained in both InpA structures. In contrast, the catalytic histidine, His305, undergoes major rearrangement. In the mature enzyme it is oriented to favour the interaction with Cys154 Sγ through a hydrogen bond, His305 Nε2-Trp322 O, and is further stabilised in this position by a hydrophobic environment created by Trp324, Phe307, Phe345, and Trp322 (Fig. 4d). In the zymogen, however, the histidine is swung out from its active position. It requires a rotation of ~45° around bond Ala306 Cα-N and of ~180° around its χ1 angle to adopt an active conformation (Fig. 4cde). The position of His305 in the zymogen is stabilised by three of elements that contribute to a compact “histidine cage” structure (Fig. 4c): the backing helix, the zymogenic hairpin, and the latency flap. The first one is completely removed and the further two undergo major rearrangement upon activation (Fig. 4d).
As mentioned, the zymogenic hairpin is only defined until residue Gly294 and from Asp302 onwards in the ΔN1pro-cd-InpA structure so that the enclosed region Lβ12β13 is disordered. The position of the hairpin base is kept by interactions with surrounding elements. Strand β12 establishes three inter-main-chain contacts with β16. In turn, β13 interacts with the backing helix through a hydrogen bond (Ala306 N-Ser88 Oγ, 3.01Å) and face-to-face ring stacking between His305 and Trp91. Removal of the backing helix leads to a rearrangement of the zymogenic hairpin due to a rotation of ~45° producing a maximal displacement of ~ 7Å (measured at Ala303 Cα). In addition, the hairpin becomes rigid and fully defined by electron density (Fig. 4cde). The hairpin-constituting β-strands are extended to Gly297 (β12) and from Gln301 onwards (β13) and give rise to a new intra-main-chain interaction (Ser295 N–Ala303 O). These changes carry along the activatory reorientation of His305 (Fig. 4e).
Another important element shaping the histidine cage in the zymogen is the latency flap, which anchors the catalytic histidine in the non-competent position through a hydrogen bond (Glu348 Oε1–His305 Nε2). In addition, the latency flap interacts with the backing helix through three hydrogen bonds, a hydrophobic interaction, and a small hydrophobic cluster made up by the side chains of Ala95, Val99, Leu337 and Thr346. This cluster extends below the side chains of His305 and Trp91 and further incorporates Tyr291, Phe307, Trp322, Tyr332, Ile334 and Trp92. Upon removal of the backing helix, the latency flap undergoes a large rearrangement and displacement caused by a ~110°-rotation that causes maximal displacement of ~22Å. Simultaneously, the latency flap adopts a β-hairpin structure with two extended segments, Leu336-Pro339 and Tyr343-Phe345, paralleling each other and establishing hydrogen bonds (see Fig. 4cde). These two extended segments are joined at the top and form a small hairpin. In its new position, the latency flap resides on a hydrophobic pillow created by the region preceding the flap, β14-Lβ14β15-β15. This region accommodates the backing helix in the zymogen and remains unchanged after activation, only the interacting partners change. In addition to these interactions, the latency flap provides a physical support to the zymogenic hairpin in the active enzyme through four main-chain and a side-chain/main-chain interactions. A last finding concomitant with the removal of the backing helix is the reorientation of the side chain of stopper Trp324 through two rotations of ~100° and ~60° around its angles χ1 and χ2, respectively (see Fig. 4cd). In this way, the side chain of this residue joins the previously mentioned hydrophobic pillow acting as a “tryptophan switch” and participating in the activating rearrangement.
In summary, we have described a new cysteine protease from a highly-active pathogenic bacterium, InpA, which undergoes autolytic activation in vitro and, possibly, in vivo. The structural features reported reveal a new mechanism of activation/latency maintenance within CPs, distinct from cathepsins and plant CPs, which may also be valid for related proteins such as S. pyogenes SpeB and P. gingivalis periodontain. This mechanism starts when the backing helix is removed after proteolytic cleavage at the Asn111-Val112 scissile peptide bond (step 1 in Fig. 4e). This liberates a space that enables stopper Trp324, actually a tryptophan switch, to reorient (see Fig. 4cd) and contribute to a hydrophobic pillow created by the apolar side chains of segment β14-Lβ14β15-β15 of the CP moiety. This segment participates in a large hydrophobic core with the backing helix in the zymogen and remains unchanged in the active enzyme. The movement of Trp324 correlates with the large displacement and internal rearrangement observed for the latency flap which, by pivoting around Met351 and Tyr332, causes this segment to adopt a β-hairpin-like structure and to occupy the space released by the backing helix (step 2 in Fig. 4e). Consequently, the zymogenic hairpin becomes rigid at its top and folds back through a ~60°-rotation pivoting around the anchor points Gly292 and Ala306, thus liberating the substrate-binding cleft of the enzyme (step 3 in Fig. 4e). This rearrangement correlates with a ~180°-rotation of the His305 side chain to a competent position to interact with the catalytic cysteine (step 4 in Fig. 4e).
Acknowledgments
This study was supported by the following grants: BIO2004-20369-E and BIO2003-06653 from the former Spanish Ministry for Science and Technology; BIO2006-02668, BIO2006-14139, BFU2006-09593 and CONSOLIDER-INGENIO 2010 Project “La Factoría de Cristalización” (CSD2006-00015) from the Spanish Ministry for Education and Science; EU FP6 Integrated Project LSHC-CT-2003-503297 “CANCERDEGRADOME”; EU FP6 Strep Project 18830 “CAMP”; and by “AVON-Project” 2005X0648 from the Spanish Association Against Cancer. Additional funding was obtained by J.J.E. from the Danish National Science Research Council and by J.P. from MNiSW (Warsaw, Poland) and an NIH grant DE 09761. Funding for synchrotron diffraction data collection was provided by the European Synchrotron Radiation Facility and the European Union.
M.S. is a beneficiary of the “Ramón y Cajal” Program of the Spanish Ministry for Science and Education. We thank Robin Rycroft and Mary Kopecki for helpful contributions to the manuscript and George M. Sheldrick for providing an α-version of program SHELXE.
The abbreviations used are
- 6xHis
hexahistine tag
- AMC
aminomethylcoumarin
- Boc
di-tertbutyl dicarbonate
- cd-InpA
catalytic domain of interpain A
- CP
cysteine protease
- DTT
1,4-dithio-DL-threitol
- E-64
N-[N-{L-trans-carboxyoxiran-2-carbonyl}-L-leucyl]-agmatine
- fom
mean figure-of-merit
- LSD
left subdomain
- MMP
matrix metalloprotease
- ncs
non-crystallographic symmetry
- PBS
phosphate buffer saline
- PD
periodontal disease
- PDB
protein data bank
- PEG
polyethylene glycol
- pro-cd-InpA
prodomain+catalytic domain of interpain A
- RSD
right subdomain
- wMPE
weighted mean-phase error
- wt
wild-type
References
- 1.AAPHD-SPP. J. Public Health Dent. 1983;43:106–117. doi: 10.1111/j.1752-7325.1983.tb01885.x. [DOI] [PubMed] [Google Scholar]
- 2.Pihlstrom BL, Michalowicz BS, Johnson NW. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 3.Jordan RC. Periodontol. 2000. 2004;34:217–229. doi: 10.1046/j.0906-6713.2002.003433.x. [DOI] [PubMed] [Google Scholar]
- 4.Haffajee AD, Socransky SS, Gunsolley JC. Ann. Periodontol. 2003;8:115–181. doi: 10.1902/annals.2003.8.1.115. [DOI] [PubMed] [Google Scholar]
- 5.Tanner AC, Izard J. Periodontol. 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 6.Fine DH, Kaplan JB, Kachlany SC, Schreiner HC. Periodontol. 2000. 2006;42:114–157. doi: 10.1111/j.1600-0757.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura S, Ueda O, Shibata Y, Hirai K. FEMS Microbiol. Lett. 2003;219:305–309. doi: 10.1016/S0378-1097(03)00048-X. [DOI] [PubMed] [Google Scholar]
- 8.Pike R, McGraw W, Potempa J, Travis J. J. Biol. Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 9.Eley BM, Cox SW. Periodontol. 2000. 2003;31:105–124. doi: 10.1034/j.1600-0757.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- 10.Loesche WJ, Syed SA, Laughon BE, Stoll J. J. Periodontol. 1982;53:223–230. doi: 10.1902/jop.1982.53.4.223. [DOI] [PubMed] [Google Scholar]
- 11.Walker CB. Periodontol. 2000. 1996;10:79–88. doi: 10.1111/j.1600-0757.1996.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 12.Shibata Y, Miwa Y, Hirai K, Fujimura S. Oral Microbiol. Immunol. 2003;18:196–198. doi: 10.1034/j.1399-302x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 13.Guan SM, Nagata H, Shizukuishi S, Wu JZ. Anaerobe. 2006;12:279–282. doi: 10.1016/j.anaerobe.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Deschner J, Singhal A, Long P, Liu CC, Piesco N, Agarwal S. Arch. Microbiol. 2003;179:430–436. doi: 10.1007/s00203-003-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawlings ND, Tolle DP, Barrett AJ. Nucl. Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filipek R, Szczepanowski R, Sabat A, Potempa J, Bochtler M. Biochemistry. 2004;43:14306–14315. doi: 10.1021/bi048661m. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann B, Schomburg D, Hecht H-J. Acta Cryst. sect. A. 1993;49 suppl.:C102–C102. [Google Scholar]
- 18.Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. Proc. Natl. Acad. Sci. USA. 2004;101:302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagawa TF, Cooney JC, Baker HM, McSweeney S, Liu M, Gubba S, Musser JM, Baker EN. Proc. Natl. Acad. Sci. USA. 2000;97:2235–2240. doi: 10.1073/pnas.040549997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wenig K, Chatwell L, von Pawel-Rammingen U, Bjorck L, Huber R, Sondermann P. Proc. Natl. Acad. Sci. USA. 2004;101:17371–17376. doi: 10.1073/pnas.0407965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berti PJ, Storer AC. J. Mol. Biol. 1995;246:273–283. doi: 10.1006/jmbi.1994.0083. [DOI] [PubMed] [Google Scholar]
- 22.Potempa J, Golonka E, Filipek R, Shaw LN. Mol. Microbiol. 2005;57:605–610. doi: 10.1111/j.1365-2958.2005.04714.x. [DOI] [PubMed] [Google Scholar]
- 23.Collin M, Olsen A. Infect. Immun. 2001;69:7187–7189. doi: 10.1128/IAI.69.11.7187-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott SD. J. Exp. Med. 1945;81:573–592. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen KA, Travis J, Potempa J. J. Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiyar A, Xiang Y, Leis J. Meth. Mol. Biol. 1997;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 28.CCP4. Acta Crystallogr. sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr. sect. D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick GM, Schneider TR. Meth. Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 31.Sheldrick GM. Z. Kristallogr. 2002;217:644–650. [Google Scholar]
- 32.Navaza J. Acta Crystallogr. sect. A. 1994;50:157–163. [Google Scholar]
- 33.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Evans SV. J. Mol. Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 35.Koradi R, Billeter M, Wüthrich K. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen CY, Luo SC, Kuo CF, Lin YS, Wu JJ, Lin MT, Liu CC, Jeng WY, Chuang WJ. J. Biol. Chem. 2003;278:17336–17343. doi: 10.1074/jbc.M209038200. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick GM. In: International tables for crystallography - Volume F: Crystallography of biological macromolecules. Rosmann MG, Arnold E, editors. Dordrecht/Boston/London: Kluwer Academic Publishers for the International Union of Crystallography; 2001. pp. 734–743. [Google Scholar]
- 38.Caliandro R, Carrozzini B, Cascarano GL, De Caro L, Giacovazzo C, Siliqi D. Acta Crystallogr. sect. D. 2005;61:556–565. doi: 10.1107/S090744490500404X. [DOI] [PubMed] [Google Scholar]
- 39.Tyndall JDA, Nall T, Fairlie DP. Chem. Rev. 2005;105:973–999. doi: 10.1021/cr040669e. [DOI] [PubMed] [Google Scholar]
- 40.Drenth J, Kalk KH, Swen HM. Biochemistry. 1976;15:3731–3738. doi: 10.1021/bi00662a014. [DOI] [PubMed] [Google Scholar]
- 41.Nelson D, Potempa J, Kordula T, Travis J. J. Biol. Chem. 1999;274:12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 42.Madden TE, Clark VL, Kuramitsu HK. Infect. Immun. 1995;63:238–247. doi: 10.1128/iai.63.1.238-247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost B. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 44.Cygler M, Sivaraman J, Grochulski P, Coulombe R, Storer AC, Mort JS. Structure. 1996;4:405–416. doi: 10.1016/s0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 45.Sivaraman J, Lalumière M, Ménard R, Cygler M. Protein. Sci. 1999;8:283–290. doi: 10.1110/ps.8.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polgár L. In: Handbook of proteolytic enzymes. 2nd Ed. Barrett AJ, Rawlings ND, Woessner JF Jr, editors. Vol. 2. London: Elsevier Academic Press; 2004. pp. 1072–1079. 2 vols. [Google Scholar]
- 47.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Bryan Arendall W, 3rd, Snoeyink J, Richardson JS, Richardson DC. Nucl. Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]