Abstract
One intradermal injection of incomplete Freund’s adjuvant-oil induces a T cell-mediated inflammatory joint disease in DA rats. Susceptibility genes for oil-induced arthritis (OIA) are located both within and outside the major histocompatibility complex (MHC, Oia1). We have searched for disease-linked non-MHC loci in an F2 intercross between DA rats and MHC-identical but arthritis-resistant LEW.1AV1 rats. A genome-wide scan with microsatellite markers revealed two major chromosome regions that control disease incidence and severity: Oia2 on chromosome 4 (P = 4 × 10−13) and Oia3 on chromosome 10 (P = 1 × 10−6). All animals homozygous for DA alleles at both loci developed severe arthritis, whereas all those homozygous for LEW.1AV1 alleles were resistant. These results have general implications for situations where nonspecific activation of the immune system (e.g., incomplete Freund’s adjuvant-oil) causes inflammation and disease, either alone or in conjunction with specific antigens. They may also provide clues to the etiology of inflammatory diseases in humans, including rheumatoid arthritis.
Experimental inflammatory joint diseases can be induced in genetically susceptible rats and mice by immunization with mineral oils containing cartilage autoantigens (1–4) or immunological adjuvants (5–7). The mineral oils are common denominators in these diseases, exerting a decisive but yet unclear arthritogenic role. Their pathogenic actions may be elucidated in oil-induced arthritis (OIA) (8, 9), an experimental situation where mineral oil alone causes disease in arthritis-prone DA rats (8–16). Intradermal injection of incomplete Freund’s adjuvant-oil (IFA) induces transcription of mRNA for proinflammatory cytokines such as interferon-γ and tumor necrosis factor-α (17). The arthritis development depends on T lymphocytes (18, 19); a cell type specialized at antigen recognition. This is intriguing because the arthritis-inducing oil is generally considered to be nonantigenic; it is often used to enhance nonspecifically the immune responses to antigens used for immunization.
Understanding the arthritogenic mechanisms of inflammatory oils is of general importance because several environmental agents, both chemical and microbial, are nonspecific stimulators of the immune system. We have therefore performed a genetic analysis of OIA that may provide clues to the etiology of inflammatory joint diseases in humans, including rheumatoid arthritis. The aim was to identify chromosome regions harboring susceptibility genes localized outside the major histocompatibility complex (MHC), a previously described susceptibility locus (9) that we denote Oia1. MHC effects were excluded by analysis of an F2 intercross between inbred strains that share the arthritis-permissive MHC haplotype (RT1av1), i.e., DA rats and MHC-congenic but arthritis-resistant LEW.1AV1 rats (10, 20).
MATERIALS AND METHODS
Experimental Animals.
Inbred DA and LEW.1AV1(DA) rats were originally derived from Hans J. Hedrich at the Zentralinstitut für Versuchstierzucht, Hannover, Germany. The parental strains and crosses were bred and kept at the BioMedical Center in Uppsala, Sweden. The crosses were as follows (female first): (DA × LEW.1AV1)F1, DA × F1, and (DA × LEW.1AV1)F2. The animals were free from rat pathogens as tested for in a health monitoring program at the National Veterinary Institute in Uppsala. They were kept in a 12-h light/12-h dark cycle and housed in polystyrene cages containing aspen wood shavings, with free access to water and autoclaved rodent chow (Lactamin R3, Vadstena, Sweden). All animal procedures were in accordance with national regulations on animal experiments.
Induction and Evaluation of Arthritis.
The animals, 14–21 weeks old, were anesthesized with methoxyflurane and immunized intradermally at the base of the tail with IFA [1 μl/g (body weight); Difco]. Arthritis was assessed by using a scale from 0 to 108, where 108 represents full-blown arthritis as in DA rats with collagen-induced arthritis (20). Each of the four paws were evaluated from 0 to 27 as follows: swelling of the ankle, 0–9 (an estimation of inflammation based on the area and degree of swelling where 0 = no swelling and 9 = “complete” swelling); swelling of intratarsal and/or metatarsal joints, 0–9 (as for the ankle); swelling of one or more interphalangeal joints, 0–9 (one point for each inflamed finger joint). The rats were examined every second to fourth day, from day 10 to day 30 after immunization. Arthritis severity data are based on the highest arthritis score for each animal.
Genotyping and Linkage Analysis.
Genomic DNA was purified from tail tips by a standard protocol (21). Genotyping was performed by PCR amplification of tandemly repeated DNA sequences (microsatellites) that were polymorphic between the two parental strains, essentially as described (22), except [γ-33P]ATP was used to label one of the primers in each pair. A linkage map was constructed by using the mapmaker computer package (23). The percentage of the rat genome lying within 10 centimorgans of the 160 markers used was 85%. For each chromosome, the coverage was as follows: 1, 71%; 2, 75%; 3, 67%; 4, 96%; 5, 100%; 6, 58%; 7, 82%; 8, 86%; 9, 95%; 10, 98%; 11, 99%; 12, 85%; 13, 100%; 14, 100%; 15, 71%; 16, 100%; 17, 87%; 18, 83%; 19, 100%; 20, 94%; and 97% for the X chromosome. The alleles at each marker locus were determined and denoted D for DA-specific alleles and L for LEW.1AV1 alleles (the corresponding genotypes were DD, DL, and LL).
Arthritis-associated traits were regressed on the rat linkage map. This was performed by analyzing the 45 F2 progeny with the highest disease scores (scores 10–92) for departing from the expected 1:2:1 (DD/DL/LL) genotype distribution for markers not linked to arthritis, by using a χ2 test (1 degree of freedom). To avoid accepting a false null hypothesis (type II error), all microsatellite markers indicating linkage (P = 0.05), as well as nearby markers, were used to genotype all F2 progeny. Genotypes for the most representative marker at each locus were analyzed for arthritis susceptibility (all animals with scores 0–92), incidence (all animals), and severity (arthritic animals with scores 1–92). Incidence was evaluated by χ2 tests. Susceptibility and severity were estimated by using test statistics calculated by a nonparametric analysis of variance method (Kruskall–Wallis), because the semiquantitative scores were not normally distributed.
RESULTS
Disease Development in DA, LEW.1AV1, and Intercross Progeny.
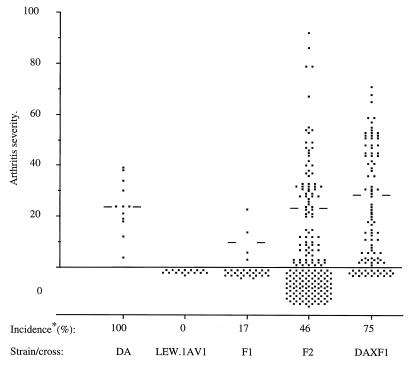
IFA was injected into DA rats, LEW.1AV1 rats, and offspring from crosses between the two parental strains. Development of joint inflammation was monitored for 30 days by using a scale from 0–108. Arthritis was evident from day 11 after injection in 100% of DA rats, 0% of LEW.1AV1 rats, 17% of (DA × LEW.1AV1)F1 rats, 46% of (DA × LEW.1AV1)F2 rats, and 75% of DA × F1 rats (Fig. 1). Notably, there were marked differences in arthritis onset between affected (DA × LEW.1AV1)F2 and DA × F1 progeny. Furthermore, several F2 and DA × F1 progeny developed a more severe disease than rats of the parental DA strain. This indicates a genetic contribution to susceptibility from the LEW.1AV1 rat and, thus, a complex inheritance of susceptibility that makes it difficult to predict the number of genes involved. The arthritis susceptibility of F2 rats (scores, 0–92) was influenced by sex (P = 0.03) but not by environmental factors such as cage size or cage location. The sex difference, males being more susceptible than females, may relate to the weight-adjusted dosage of arthritogenic oil because there was a tendency for body weight and age to influence susceptibility.
Figure 1.
Arthritis scores for the parental rat strains and F1 and F2 progeny: DA (no. 12), LEW.1AV1 (no. 16), (DA × LEW.1AV1)F1 (no. 29), (DA × F1)Bx (no. 96), and (DA × LEW.1AV1)F2 (no. 189). Bars indicate the median arthritis score of the affected animals in each group. The incidence is calculated as the percent of animals with arthritis scores between 1 and 92 (the animals affected with arthritis).
Strategy to Identify Arthritis-Linked Loci.
To identify informative genomic markers, DNA from parental DA and LEW.1AV1 rats was analyzed with 452 microsatellites. This procedure identified 246 polymorphic markers (54%) from which a set of 160 were selected for even spacing over the chromosomes, covering 85% of the genome. These markers were used for a two-step procedure to identify arthritis-linked loci. (i) A genome-wide scan was performed on 45 F2 rats selected for severe disease. (ii) Loci indicating potential arthritis linkage (nominal P ≤ 0.05) were investigated further by genotyping the remaining 144 F2 progeny.
Identification of Arthritis-Linked Loci.
For the genome scan, 45 F2 rats were selected to include the 23 males and 22 females that developed the most severe disease (scores, 10–92). Arthritis-resistant animals were not included in the scan, because a lack of arthritis is not necessarily genetically determined because it can be due to incomplete disease penetrance (sometimes less than 100% in DA rats). The genotypes of the 45 arthritic F2 rats were determined for each of the 160 markers and categorized as DA homozygous (DD), heterozygous (DL), or LEW.1AV1 homozygous (LL). The genotype proportions for each marker were compared with an expected 1:2:1 ratio (DD/DL/LL) for arthritis-unlinked markers, by using χ2 analysis.
Two major disease-linked loci were evident: D4Mgh10 (28:17:0, P < 9 × 10−9) and D10Mgh1 (24:14:7, P < 1 × 10−5), but deviations below a nominal P value of 0.05 were observed at 15 additional loci (data not shown). Additional markers covering these 2 + 15 loci were used to genotype the remaining 144 F2 progeny. The subsequent characterization of each of these arthritis-associated loci was performed by three analyses: (i) differences in arthritis susceptibility between the three genotypes (all 189 F2 progeny with scores 0–92), (ii) differences in arthritis incidence between the three genotypes (all 189 F2 progeny, categorized as affected, if scores were above 0, and as unaffected, if they scored 0), and (iii) differences in arthritis severity (for the 87 F2 progeny with scores above 0). These procedures confirmed the two major susceptibility loci D4Mgh10 and D10Mgh1, and five additional loci remained linked to arthritis with a nominal P < 0.05 for at least one of the traits. Table 1 includes these potentially disease-linked chromosome regions to allow verification in future studies, although nominal P < 0.05 is expected to occur frequently in genome-wide scans due to multiple tests of the null hypothesis (24, 25). Interestingly, the genetic analysis yielded three loci (D4Mit12 on chromosome 4, D8Mgh1 on chromosome 8, and D10Mgh1 on chromosome 10) in the vicinity of reported susceptibility loci for collagen-induced arthritis in DA rats (26). These shared loci may relate to the use of IFA for arthritis induction in both models. Furthermore, for two loci (D2Mit15 at chromosome 2 and D6Mit1 at chromosome 6) the disease-linked allele were from the LEW.1AV1 rat. This may partly explain the extraordinary severe disease in some F2 progeny (Fig. 1) and relate to the poor antiinflammatory glucocorticoid response of LEW rats (27–29).
Table 1.
Arthritis-linked loci in (DA × LEW.1AV1)F2 progeny
| Chromosome/marker | Distance, cM |
P values
|
Disease genotype | ||
|---|---|---|---|---|---|
| Susceptibility | Incidence | Severity | |||
| 2/D2Mit15 | 4 × 10−2 | 2 × 10−2 | 4 × 10−2 | LL | |
| 4/D4Mit12 | 28.0 | 2 × 10−2 | NS | 1 × 10−2 | DD |
| 4/D4Mgh10 | 4 × 10−13 | 9 × 10−11 | 1 × 10−4 | DD | |
| 6/D6Mgh7 | 35.4 | 2 × 10−2 | NS | NS | DL |
| 6/D6Mit1 | 3 × 10−2 | NS | NS | DD | |
| 8/D8Mgh1 | 8 × 10−3 | 2 × 10−2 | NS | DD | |
| 10/D10Mgh1 | 2 × 10−6 | 2 × 10−4 | 8 × 10−3 | DD | |
| 14/D1BR | NS | NS | 3 × 10−2 | DD | |
| 19/D19Mgh1 | NS | NS | 1 × 10−2 | DD | |
Chromosome regions linked to disease with P < 0.05 and the marker with the lowest P value are presented for each region. NS, nonsignificant; cM, centimorgan(s). Susceptibility* is the maximum arthritis score of all animals (0–92) in relation to genotypes (DD = DA homozygous, LL = LEW.1AV1 homozygous, and DL = heterozygous) analyzed by the nonparametric Kruskall–Wallis test (2 df). Incidence is all animals divided into the categories affected (scores > 0) or unaffected (score = 0) in relation to genotypes and analyzed by χ2 test.
Severity denotes arthritis severity in affected animals only (scores > 0) in relation to genotypes analyzed by the Kruskall–Wallis test (2 df).
Characterization and High-Resolution Mapping of Two Major Susceptibility Loci: Oia2 on Chromosome 4 and Oia3 on Chromosome 10.
The genome-wide scan identified two major susceptibility loci (Table 1) that are denoted Oia2 represented by the marker D4Mgh10 on chromosome 4 (P = 4 × 10−13) and Oia3 represented by the marker D10Mgh1 on chromosome 10 (P = 1 × 10−6). Both loci fulfill the stringent criteria suggested for claims of significant linkage of F2 intercross progeny (P = 5.2 × 10−5) (24). The locations and arthritis linkages of markers flanking D4Mgh10 and D10Mgh1 are given in Table 2.
Table 2.
Two arthritis-linked loci in (DA × LEW.1AV1)F2 progeny with genome-wide significance for linkage
| Chromosome | Marker | Gene | Distance, cM | P values |
|---|---|---|---|---|
| 4 | D4Mit24 | NPY | 3 × 10−2 | |
| 9.4 | ||||
| 4 | D4Mit12 | Fabp1 | 2 × 10−2 | |
| 6.2 | ||||
| 4 | D4Mgh17 | Spr | 1 × 10−4 | |
| 14.4 | ||||
| 4 | D4Mgh7 | 4 × 10−6 | ||
| 12.9 | ||||
| 4 | D4Mgh10 | 4 × 10−13 | ||
| 0.8 | ||||
| 4 | D4Mit21 | 3 × 10−13 | ||
| 0.3 | ||||
| 4 | EN3C | Eno2 | 5 × 10−13 | |
| 8.0 | ||||
| 4 | D4Mit27 | 1 × 10−7 | ||
| 7.8 | ||||
| 4 | D4Mgh21 | 2 × 10−6 | ||
| 10 | D10Mit13 | 2 × 10−3 | ||
| 3.9 | ||||
| 10 | IGFBP4 | lgfbp4 | 2 × 10−3 | |
| 6.3 | ||||
| 10 | BAND3A | Band3A | 2 × 10−5 | |
| 4.8 | ||||
| 10 | D11Mit58* | 1 × 10−4 | ||
| 21.3 | ||||
| 10 | D10Mgh1 | 2 × 10−6 |
Arthritis susceptibility (all F2 rats with scores of 0–92, Fig. 1) in relation to genotypes (DD, DL, or LL) at different locations (markers) were calculated with the nonparametric Kruskall–Wallis test. Distances between consecutive markers were calculated by mapmaker.
A mouse marker.
Effects of Oia2 and Oia3 on Arthritis Incidence and Severity.
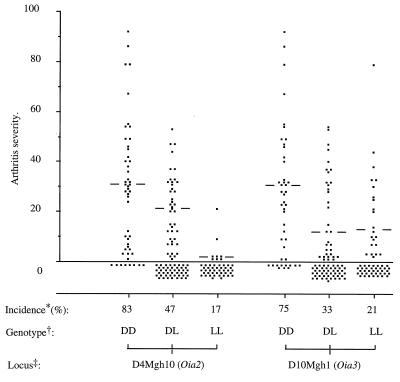
An analysis of the influences of Oia2 and Oia3 on arthritis incidence and severity (Fig. 2) revealed that both loci influenced arthritis incidence (Oia2, P = 4 × 10−10; Oia3, P = 2 × 10−4) and severity (Oia2, P = 1 × 10−4; Oia3, P = 8 × 10−3). For Oia2 (D4Mgh10), progeny of the three genotypes differed in both incidence and severity (DD > DL > LL), demonstrating that each allele contribute to the phenotype; i.e., they are additive (Fig. 2). In contrast, the DA allele at Oia3 (D10Mgh1) is recessive; it only promotes incidence and severity when present in the DA homozygous state (DD > DL = LL; Fig. 2).
Figure 2.
Arthritis scores of the (DA × LEW.1AV1)F2 rats in relation to genoypes at the two Oia loci, as defined by the markers D4Mgh10 (Oia2) and D10Mgh1 (Oia3). Bars indicate the median arthritis severity of affected animals in each group. The Oia2 locus on chromosome 4 exerts additive effects on incidence (DD > DL, P = 1 × 10−4; DL > LL, P = 7 × 10−5) and on severity (DD > DL, P = 5 × 10−3; DL > LL, P = 1 × 10−4), whereas the Oia3 locus on chromosome 10 exerts recessive effects on incidence (DD > DL, P = 3 × 10−4; DL = LL) and on severity (DD > DL, P = 2 × 10−2; DL = LL).
* Percent animals affected with arthritis (scores, 1–92).
† Genotypes: DD = DA homozygous; LL = LEW.1AV1 homozygous; DL = heterozygous. ‡ The markers D4Mgh10 and D10Mgh1 define loci Oia2 and Oia3.
Combined Effects of Oia2 and Oia3 on Arthritis Susceptibility.
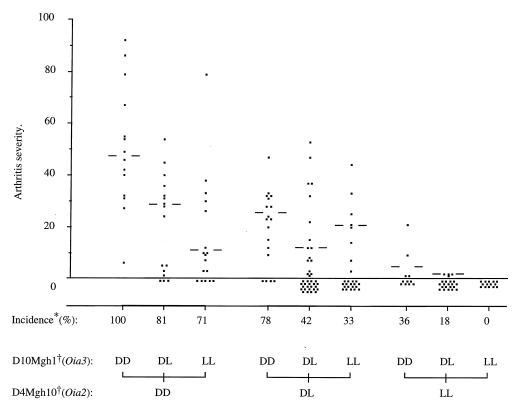
In combination, Oia2 and Oia3 decisively influenced the disease incidence and severity in F2 offspring (Fig. 3). All animals homozygous for LEW.1AV1 alleles at both loci were arthritis-resistant, whereas all those homozygous for DA alleles developed arthritis [score (mean ± SD), 50 ± 23] that was significantly more severe than that of parental DA rats (score 24 ± 12, P = 1 × 10−4, Student’s t test). This suggests disease-promoting effects of susceptibility alleles from the OIA-resistant LEW.1AV1 strain. Influences from additional susceptibility loci are also indicated by the higher incidence in F2 progeny heterozygous at Oia2 and Oia3 (42%) compared with F1 rats (17%) that are heterozygous for the whole genome.
Figure 3.
Arthritis scores for (DA × LEW.1AV1)F2 rats in relation to combinations of genotypes at the two major arthritis loci defined by the markers D4Mgh10 (Oia2) and D10Mgh1 (Oia3). Bars indicate the median arthritis severity of affected animals in each group.
* Percent animals with arthritis (scores, 1–92).
† Markers: D4Mgh10 (Oia2) and D10Mgh1 (Oia3). Genotypes: DD = DA homozygous; LL = LEW.1AV1 homozygous; and DL = heterozygous animals.
DISCUSSION
Rheumatoid arthritis, ankylosing spondylitis, and psoriatic arthritis are inflammatory joint diseases for which there are at present no treatments to definitely halt disease progression. Development of effective therapies is complicated by an incomplete understanding of the causative disease mechanisms. Increased knowledge of their pathogeneses may by achieved by identifying arthritis-associated genes (“disease pathway mapping”). This is problematic, however, in heterogenous diseases where susceptibility is believed to depend on complex interactions between environmental factors and multiple genes with low penetrance. To date, genetic studies have revealed MHC associations (30) but disease-associated non-MHC genes remain to be determined (31, 32).
This may be facilitated by identification of susceptibility loci in animal models (33, 34). We therefore performed a genome-wide scan of OIA. This model was selected because it clarifies the arthrogenicity of IFA, a common pathogenic denominator in many experimental arthritides.
Several potentially arthritis-linked loci were identified in this genome scan of progeny from an F2 intercross between arthritis-susceptible DA rats and arthritis-resistant but MHC-identical LEW.1AV1 rats. Interestingly, three of these loci were near or identical to previously reported susceptibility loci for collagen-induced arthritis in DA rats (26). These shared susceptibility elements support our hypothesis that OIA-linked loci exert decisive effects also in more complex arthritis models where additional arthritogenic substances are mixed with the IFA, e.g., cartilage antigens, such as collagens (1–3) and proteoglycan (4), or immunological adjuvants, such as mycobacteria (7, 11), muramyl-dipeptide (6), and avridine (5, 14).
Furthermore, some loci may have general implications also for other organ-specific diseases where IFA is used for disease induction. This possibility can be investigated in the disease-prone DA rat (8–16, 35–37) in which such adjuvant-response loci (38) could influence the susceptibility to experimental autoimmune encephalomyelits (10, 35), neuritis (10), uveitis (36), and thyroiditis (37).
Our genetic dissection of OIA defined two major susceptibility loci controlling both arthritis incidence and disease severity, Oia2 on chromosome 4 (P = 4 × 10−13) and Oia3 on chromosome 10 (P = 1 × 10−6). Severe arthritis developed in all F2 progeny homozygous for DA alleles at both loci, whereas all those homozygous for LEW.1AV1 genes were resistant. These remarkably strong effects of Oia2 and Oia3 will facilitate future efforts to define the susceptibility genes they harbor. This involves establishing congenic strains for the chromosome regions, localization of susceptibililty genes by fine mapping in intraregion recombinant strains followed by positional cloning. At present we can only speculate on the identity of susceptibility genes, but Oia2 and Oia3 or the conserved syntenic loci in humans do offer some candidates. These include genes for a tissue inhibitor of metalloproteinase-2 (39) and the α-chain of protein kinase C (40) within Oia3, whereas genes for CD4 (41), tumor necrosis factor receptors (42), CD69 (43), and natural killer (NK) cell receptors are candidates for Oia2. That Oia2 harbor the NK cell receptor genes Nkrp1 and -2, Ly49, and CD94 (44) is intriguing because DA rats have an aberrant NK cell function (45), which is linked to these genes (46). Interestingly, aberrant NK cell function is also reported for the disease-prone SJL/J mouse strain (47). Other traits linked to chromosomal intervals near Oia2 and Oia3 include collagen-induced arthritis (26) in the DA rat (Cia3 and Cia5, respectively). It is likely that Oia3 (D10Mgh1) and Cia5 (D10Arb22) are identical loci because D10Mgh1 and D10Arb22 are colocalized on our map. We have thus confirmed Cia5 as an arthritis-linked locus, but we demonstrate that it harbors susceptibility genes for the pathogenic effects of adjuvant-oil. In contrast, tightly arthritis-linked markers within Oia2 (D4Mgh10) and Cia3 (D4Arb24) are separated by at least 30 centimorgans. We suggest therefore that Cia3 do not correspond to Oia2, but that it may correspond to a minor OIA-linked chromosome region centromeric to Oia2 (OiaW).
In conclusion, our genetic dissection of adjuvant-oil-induced arthritis revealed two disease-linked non-MHC loci, Oia2 and Oia3, that may have general implications for experimental autoimmune diseases. Their identification may give clues to the mechanisms whereby nonspecific activation of the immune system induces inflammation and disease. The two loci provide interesting candidates for studies in human inflammatory diseases.
Acknowledgments
We thank Dr. Howard J. Jacob for fruitful discussions and generous supply of markers and Robert Harris for linguistic advice. This investigation was supported in parts by grants from NovoNordisk, the Swedish Medical Research Council, the Swedish Association Against Rheumatism, Arbetsmarknadens Försäkrings Aktiebolag, the Swedish Diabetes Association, the Swedish Medical Association, and the following foundations: Petrus och Augusta Hedlund, Jonsson, Berth von Kantzow, King Gustaf V and Queen Victoria, Emil and Wera Cornell, Magnus Bergvall, Ulf Widengrens Minne, and Gamla Tjänarinnor.
ABBREVIATIONS
- IFA
incomplete Freund’s adjuvant
- OIA
oil-induced arthritis
- MHC
major histocompatibility complex
- NK
natural killer
References
- 1.Trentham D E, Townes A S, Kang A H. J Exp Med. 1977;146:857–868. doi: 10.1084/jem.146.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan K, Evans H B, Firth S A, Smith M N, Ayad S, Weiss J B, Holt P J L. Ann Rheum Dis. 1983;42:680–683. doi: 10.1136/ard.42.6.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossier M-C, Chiocchia G, Roziere M-C, Herbage D, Fournier C. Arthritis Rheum. 1990;33:1–8. doi: 10.1002/art.1780330101. [DOI] [PubMed] [Google Scholar]
- 4.Glant T T, Mikecs K, Arzoumanian A, Poole A R. Arthritis Rheum. 1987;30:201–212. doi: 10.1002/art.1780300211. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y H, Pearson C M, Abe C. Arthritis Rheum. 1980;23:62–71. doi: 10.1002/art.1780230111. [DOI] [PubMed] [Google Scholar]
- 6.Kohashi O, Aihara K, Ozawa A, Kotani S, Azuma I. Lab Invest. 1982;47:27–36. [PubMed] [Google Scholar]
- 7.Pearson, C. M. & Chang, Y. H. (1979) Ann. Rheum. Dis. 38 (supplement), 102–110. [DOI] [PMC free article] [PubMed]
- 8.Kleinau S, Erlandsson H, Holmdahl R, Klareskog L. J Autoimmunity. 1991;4:871–880. doi: 10.1016/0896-8411(91)90050-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon G W, Woods M L, Clayton F, Griffiths M M. J Rheumatol. 1993;20:7–11. [PubMed] [Google Scholar]
- 10.Lorentzen J C, Olsson T, Klareskog L. Transplant Proc. 1995;27:1532–1534. [PubMed] [Google Scholar]
- 11.Battisto J R, Smith R N, Beckman K, Sternlicht M, Welles W L. Arthritis Rheum. 1982;25:1194–1200. doi: 10.1002/art.1780251008. [DOI] [PubMed] [Google Scholar]
- 12.Samuelson C O, Jr, Griffiths M M, Mathews J L, Clegg D O, Ward J R. Arthritis Rheum. 1984;27:689–693. doi: 10.1002/art.1780270613. [DOI] [PubMed] [Google Scholar]
- 13.Binder A, Gartner K, Hedrich H J, Hermanns W, Kirchoff H, Wonigeit K. Infect Immun. 1990;58:1584–1590. doi: 10.1128/iai.58.6.1584-1590.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vingsbo C, Jonsson R, Holmdahl R. Clin Exp Immunol. 1995;99:359–363. doi: 10.1111/j.1365-2249.1995.tb05558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths M M. Int Rev Immunol. 1988;4:1–15. doi: 10.3109/08830188809044766. [DOI] [PubMed] [Google Scholar]
- 16.Larsson P, Kleinau S, Holmdahl R, Klareskog L. Arthritis Rheum. 1990;33:693–701. doi: 10.1002/art.1780330512. [DOI] [PubMed] [Google Scholar]
- 17.Müssener Å, Klareskog L, Lorentzen J C, Kleinau S. Scand J Immunol. 1995;42:128–134. doi: 10.1111/j.1365-3083.1995.tb03635.x. [DOI] [PubMed] [Google Scholar]
- 18.Kleinau S, Klareskog L. J Autoimmunity. 1993;6:449–458. doi: 10.1006/jaut.1993.1037. [DOI] [PubMed] [Google Scholar]
- 19.Holmdahl R, Goldschmidt T J, Kleinau S, Kvick C, Jonsson R. Immunology. 1992;76:197–202. [PMC free article] [PubMed] [Google Scholar]
- 20.Lorentzen J C, Klareskog L. Scand J Immunol. 1996;44:592–598. doi: 10.1046/j.1365-3083.1996.d01-354.x. [DOI] [PubMed] [Google Scholar]
- 21.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacob H J, Lindpaintner K, Lincoln S E, Kusumi K, Bunker R K, Mao Y P, Ganten D, Dzau V J, Lander E S. Cell. 1992;67:213–224. doi: 10.1016/0092-8674(91)90584-l. [DOI] [PubMed] [Google Scholar]
- 23.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 24.Lander E, Kruglyak L. Nat Genet. 1994;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 25.Thomson G. Nat Genet. 1994;8:108–110. doi: 10.1038/ng1094-108. [DOI] [PubMed] [Google Scholar]
- 26.Remmers E F, Longman R E, Du Y, O′hare A, Cannon G W, Griffiths M M, Wilder R L. Nat Genet. 1996;14:82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- 27.Sternberg E M, Hill J M, Chrousos G P, Kamilaris T, Listwak S J, Gold P W, Wilder R L. Proc Natl Acad Sci USA. 1989;86:2374–2378. doi: 10.1073/pnas.86.7.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sternberg E M, Young W S, III, Bernardini R, Calogero A E, Chrousos G P, Gold P W, Wilder R L. Proc Natl Acad Sci USA. 1989;86:4771–4775. doi: 10.1073/pnas.86.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calogero A E, Sternberg E M, Bagdy G, Smith G, Bernardini R, Aksentijevich S, Wilder R L, Gold P W, Chrousos G P. Neuroendocrinology. 1992;55:600–608. doi: 10.1159/000126173. [DOI] [PubMed] [Google Scholar]
- 30.Nepom B S, Nepom G T. In: Textbook of Rheumatology. 4th Ed. Kelley W N, Harris E D Jr, Ruddy S, Sledge C B, editors. I. Philadelphia: Saunders; 1993. pp. 89–104. [Google Scholar]
- 31.Rigby A S, Silman A J, Voelm L, Gregory J C, Ollier W E, Khan M A, Nepom G T, Thomson G. Genet Epidemiol. 1991;8:153–175. doi: 10.1002/gepi.1370080303. [DOI] [PubMed] [Google Scholar]
- 32.Silman A J, Hennesy E, Ollier B. Br J Rheumatol. 1992;31:365–368. doi: 10.1093/rheumatology/31.6.365. [DOI] [PubMed] [Google Scholar]
- 33.Sundvall M, Jirholt J, Yang H-T, Jansson L, Engström Å, Pettersson U, Holmdahl R. Nat Genet. 1995;10:313–317. doi: 10.1038/ng0795-313. [DOI] [PubMed] [Google Scholar]
- 34.Kuokkanen S, Sundvall M, Terwilliger J D, Tienari P T, Wikström J, Holmdahl R, Pettersson U, Peltonen L. Nat Genet. 1996;13:477–480. doi: 10.1038/ng0896-477. and comment 377–378. [DOI] [PubMed] [Google Scholar]
- 35.Lorentzen J C, Issazadeh S, Storch M, Mustafa M I, Lassman H, Linington C, Klareskog L, Olsson T. J Neuroimmunol. 1995;63:193–205. doi: 10.1016/0165-5728(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 36.Lalic N M, Latkovic Z V, Mostarica-Stojkovic M, Lukic M L. Period Biol. 1983;85:77–78. [Google Scholar]
- 37.Rose N R. Cell Immunol. 1975;18:360–364. doi: 10.1016/0008-8749(75)90064-7. [DOI] [PubMed] [Google Scholar]
- 38.Sudweeks J D, Todd J A, Blankenhorn E P, Wardell B B, Woodward S R, Meeker N D, Estes S S, Teuscher C. Proc Natl Acad Sci USA. 1993;90:3700–3704. doi: 10.1073/pnas.90.8.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Clerck, Y., Szpirer, C., Aly, M. S., Cassiman, J.-J., Eeckhout, Y. & Rousseau, G. Genomics 14, 782–784. [DOI] [PubMed]
- 40.Summar M L, Phillips J A, Krishnamani M R, Keefer J, Trofatter J, Schwartz R C, Tsipouras P, Willard H, Ullrich A. Genomics. 1989;5:163–165. doi: 10.1016/0888-7543(89)90104-3. [DOI] [PubMed] [Google Scholar]
- 41.Dissen E, Fossum S. Immunogenetics. 1996;44:312–314. doi: 10.1007/BF02602563. [DOI] [PubMed] [Google Scholar]
- 42.Fuchs P, Strehl S, Dworzak M, Himmler A, Ambros P F. Genomics. 1992;13:219–224. doi: 10.1016/0888-7543(92)90226-i. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Cabrera M, Santis A G, Fernandez-Ruiz E, Blacher R, Esch F, Sanchez-Mateos P, Sanchez-Madrid F. J Exp Med. 1993;178:537–547. doi: 10.1084/jem.178.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dissen E, Berg S F, Westgaard I H, Fossum S. Eur J Immunol. 1997;27:2080–2086. doi: 10.1002/eji.1830270836. [DOI] [PubMed] [Google Scholar]
- 45.Rolstad B, Fossum S. Immunology. 1987;60:151–157. [PMC free article] [PubMed] [Google Scholar]
- 46.Dissen E, Ryan J C, Seamann W E, Fossum S. J Exp Med. 1996;183:2197–2207. doi: 10.1084/jem.183.5.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaminsky S G, Nakamura I, Cudkowics G. J Immunol. 1985;135:665–671. [PubMed] [Google Scholar]