Abstract
An odorant-binding protein from the Southern house mosquito, Culex pipiens quinquefasciatus (Cqui-OBP1) binds to the mosquito oviposition pheromone (MOP), 6-acetoxy-5-hexadecanolide to facilitate the transport of MOP to membrane-bound odorant receptors. We report complete NMR chemical shift assignments of Cqui-OBP1 bound to the MOP pheromone obtained at pH 7.0 and 25°C (BMRB no. 16175).
Electronic supplementary material
The online version of this article (doi:10.1007/s12104-009-9173-5) contains supplementary material, which is available to authorized users.
Keywords: Odorant-binding protein, Pheromone signaling, Olfaction, NMR
Biological context
To find mates and successfully reproduce, insects rely heavily on pheromone detection. Odorant-binding proteins (OBPs) (Vogt and Riddiford 1981) and odorant receptors (ORs) (Clyne et al. 1999; Vosshall et al. 1999) are essential for the uptake, delivery and detection of sex pheromones (Leal et al. 2005). Molecular interactions amongst pheromone molecules, OBPs and ORs lead to a remarkable selectivity and sensitivity of the olfactory system in insects. The odorant-binding protein from the Southern house mosquito, Culex pipiens quinquefasciatus (CquiOBP1) binds tightly to the mosquito oviposition pheromone (MOP), 6-acetoxy-5-hexadecanolide to facilitate the transport of MOP to membrane-bound ORs. Cqui-OBP1 binds tightly to MOP at neutral pH and undergoes a pH-dependent conformational transition at low pH (Leal et al. 2008) to control release of MOP to the OR on the membrane surface, where the local pH is estimated to be low. Three-dimensional structures of a few OBPs are known at low pH (Damberger et al. 2007; Lautenschlager et al. 2007), but the atomic-level interaction of OBPs and ORs at neutral pH are not well understood. The three-dimensional structure of OBPs as a function of pH and bound ligand may unveil how ORs are activated by pheromones. We report here NMR assignments of CquiOBP1 bound to MOP at pH 7.0, as an important first step toward elucidating the atomic-level structural recognition of MOP binding to CquiOBP1.
Methods and experiments
Expression and purification of CquiOBP1
Uniformly 15N-labeled and 13C,15N-labeled CquiOBP1 was expressed in E. coli and purified by ion-exchange and gel-filtration chromatography as described previously (Damberger et al. 2007). Typically, 5 mg of purified protein was obtained from a 1 l culture. The identity and integrity of the final protein sample was confirmed by SDS–PAGE and LC-ESI/MS.
NMR spectroscopy
Samples for NMR analysis were prepared by dissolving 15N, or 15N/13C-labeled CquiOBP1 protein (0.5 mM) in 0.3 ml of a 95% H2O/5% D2O solution containing 10 mM phosphate at pH 7.4. Two equivalents of (5R, 6S)-MOP-6-acetoxy-5-hexadecanolide was added to saturate the protein with MOP. All NMR experiments were performed at 25°C on a Bruker Avance 600 MHz spectrometer equipped with a four channel interface and triple resonance cryogenic probe. The 15N–1H HSQC spectrum (Fig. 1) was recorded with the following parameters: the number of complex points and acquisition times were 256,180 ms for 15N (F1), and 512,64 ms for 1H(F2). Assignment of backbone and side-chain resonances were obtained by analyzing the following spectra: HNCACB, HN(CO)CACB,HNCO,CBCA(CO)NH, HBHA(CO)NH, C(CO)NH-TOCSY, H(CCO)NH-TOCSY,HCCH-TOCSY, and H(CCH)-COSY. The NMR data were processed using NMRPipe and analyzed using Sparky.
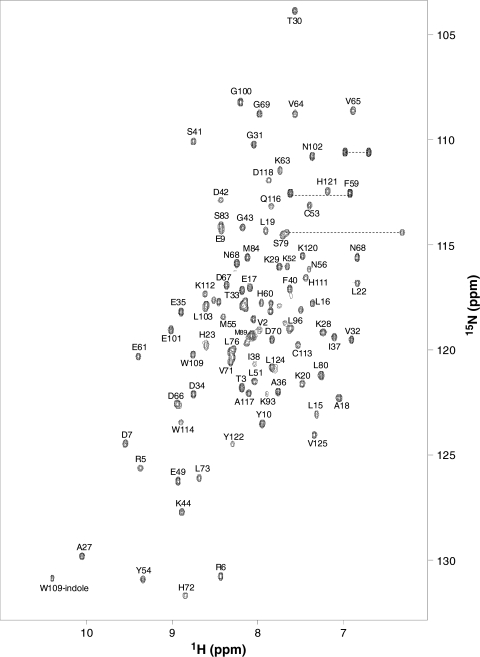
Fig. 1.
Two-dimensional 15N–1H HSQC spectrum of Cqui-OBP1 bound to MOP at pH 7.0 recorded at 600-MHz 1H frequency. Side-chain amide peaks are connected by the dotted lines. The protein sample was uniformly labeled with nitrogen-15. Resonance assignments are indicated and are reported in BMRB accession no. 16175
Assignments and data deposition
Figure 1 presents 1H/15N HSQC spectrum of CquiOBP1 bound to MOP at pH 7.0 to illustrate representative backbone resonance assignments. NMR assignments were based on 3D heteronuclear NMR experiments performed on 13C/15N-labeled Cqui-OBP1 (residues 1–125). The protein sample in this study consists of 125 native residues and does not contain any affinity tags or extra residues. The vast majority of these residues (114) exhibited strong NMR signals with uniform intensities, indicative of a well-defined three-dimensional protein structure. About 10% of these peaks are somewhat broadened, perhaps due to exchange interactions with the bound MOP. Interestingly, the apo Cqui-OBP1 at pH 7.0 (in the absence of MOP) appears to aggregate and form a mixture of conformers under NMR conditions. As a result, the NMR spectrum of apo Cqui-OBP1 at pH 7.0 exhibits severe spectral heterogeneity in stark contrast to the spectral homogeneity of monomeric Cqui-OBP1 bound to MOP (see supplemental data).
More than 95% of the backbone resonances (1HN, 15N, 13Cα, 13Cβ, and 13CO) and ~82% of aliphatic side chain resonances were assigned, including stereospecific assignment of valine and leucine methyl groups. A stretch of residues (M89–R94) was difficult to assign due to very weak NMR intensities. We suggest that the spectral broadening associated with these residues might reflect an exchange interaction with MOP in the target binding site. The chemical shift assignments (1H, 15N, 13C) of Cqui-OBP1 bound to MOP have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 16175.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Jeff de Ropp for technical support and help with NMR experiments and Jose (Pep) Rayo and Yunhong Li for assistance in protein purification. Work supported by NIH grants (EY012347) to J.B.A. and 5U01AI058267-05 to WSL, NSF (0234769), USDA-NRI (2003-35302-13648), the Almond Board of California, and UC Davis NMR facility.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/S0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Damberger FF, Ishida Y, Leal WS, Wuthrich K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J Mol Biol. 2007;373:811–819. doi: 10.1016/j.jmb.2007.07.078. [DOI] [PubMed] [Google Scholar]
- Lautenschlager C, Leal WS, Clardy J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: implications for pheromone recognition. Structure. 2007;15:1148–1154. doi: 10.1016/j.str.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Chen AM, Ishida Y, Chiang VP, Erickson ML, Morgan TI, Tsuruda JM. Kinetics and molecular properties of pheromone binding and release. Proc Natl Acad Sci USA. 2005;102:5386–5391. doi: 10.1073/pnas.0501447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE. 2008;3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/S0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.