Abstract
Aggregatibacter actinomycetemcomitans is a Gram-negative opportunistic pathogen and the proposed causative agent of localized aggressive periodontitis. A. actinomycetemcomitans is found exclusively in the mammalian oral cavity in the space between the gums and the teeth known as the gingival crevice. Many bacterial species reside in this environment where competition for carbon is high. A. actinomycetemcomitans utilizes a unique carbon resource partitioning system whereby the presence of L-lactate inhibits uptake of glucose, thus allowing preferential catabolism of L-lactate. Although the mechanism for this process is not fully elucidated, we previously demonstrated that high levels of intracellular pyruvate are critical for L-lactate preference. As the first step in L-lactate catabolism is conversion of L-lactate to pyruvate by lactate dehydrogenase, we proposed a model in which the A. actinomycetemcomitans L-lactate dehydrogenase, unlike homologous enzymes, is not feedback inhibited by pyruvate. This lack of feedback inhibition allows intracellular pyruvate to rise to levels sufficient to inhibit glucose uptake in other bacteria. In the present study, the A. actinomycetemcomitans L-lactate dehydrogenase was purified and shown to convert L-lactate, but not D-lactate, to pyruvate with a Km of approximately 150 µM. Inhibition studies reveal that pyruvate is a poor inhibitor of L-lactate dehydrogenase activity, providing mechanistic insight into L-lactate preference in A. actinomycetemcomitans.
Introduction
Aggregatibacter actinomycetemcomitans is a Gram-negative, nonmotile, opportunistic pathogen that resides exclusively in the mammalian oral cavity [1] and has been proposed to be the primary cause of the tooth and gum disease known as localized aggressive periodontitis [2], [3]. Within the oral cavity, A. actinomycetemcomitans resides in the gingival crevice, defined as the microaerophilic region bounded by the tooth surface and the epithelium lining the gingiva. The gingival crevice is bathed in gingival crevicular fluid (GCF) which provides nutrients for a robust and complex community of microorganisms. As a serum exudate, GCF likely contains several potential carbon sources to support this microbial community, including glucose, lactate, and fructose [4], [5]. Competition for resources is high in this environment and the rate of consumption of carbohydrates is extremely rapid [6], [7], [8], likely due to the large number of oral streptococci.
We recently showed that although A. actinomycetemcomitans divides faster and achieves higher cell yields when catabolizing glucose, L-lactate is preferentially utilized [9]. Interestingly, L-lactate addition to a chemically defined medium inhibited A. actinomycetemcomitans uptake of glucose, a process referred to as PTS substrate exclusion [9]. Glucose transport in A. actinomycetemcomitans utilizes the phosphotransferase system (PTS). The PTS involves transport of glucose across the cytoplasmic membrane through a sugar-specific channel and concomitant phosphorylation upon entry into the cell to produce glucose-6-phosphate. The phosphoryl group originates from the phosphodonor, phosphoenolpyruvate (PEP), and is subsequently passed through a series of PTS proteins and ultimately to glucose (Fig. 1). The first step in PTS transport involves protein EI, which undergoes autophosphorylation in the presence of PEP to yield pyruvate and EI∼P. The phosphoryl group is then transferred to HPr, followed by a sugar-specific EII protein, which then phosphorylates the incoming sugar [10]. The intracellular ratio of PEP:pyruvate plays a crucial role in PTS transport. Indeed, as the PEP:pyruvate ratio declines, the model Gram-negative organism Escherichia coli displays reduced uptake of several PTS carbohydrates [11]. Interestingly, L-lactate-grown A. actinomycetemcomitans produces extremely elevated intracellular levels of pyruvate [9], supporting a model in which elevated intracellular levels of pyruvate during catabolism of L-lactate inhibit glucose transport via reduction of the PEP:pyruvate ratio (Fig. 1).
Figure 1. PTS substrate exclusion model.
Lactate enters the cell through the lactate permease (LctP) and is converted to pyruvate by L-lactate dehydrogenase (LctD). Intracellular levels of pyruvate increase and prevent autophosphorylation of protein EI, thus inhibiting PTS-mediated carbohydrate transport. PEP is phosphoenolpyruvate.
One of the intriguing questions regarding this model is how the extremely high levels of intracellular pyruvate (approximately 50 mM) are produced during growth with L-lactate. In this study, we hypothesized that potential clues might be gained by examining the A. actinomycetemcomitans enzyme required for the first step in L-lactate catabolism, namely L-lactate oxidation to pyruvate. We show that the gene AA02749 (lctD) encodes for an NAD-independent L-lactate dehydrogenase that is critical for growth of A. actinomycetemcomitans with L-lactate. Interestingly, inhibitor studies reveal that unlike homologous enzymes, A. actinomycetemcomitans LctD maintains significant enzymatic activity, even at extremely high pyruvate levels (50 mM).
Results
AA02769 Is Required for Growth with L-Lactate
A. actinomycetemcomitans resides within the gingival crevice where it likely encounters levels of L-lactate ranging from 1–5 mM [5], and previous work in our lab demonstrated that this bacterium preferentially catabolizes L-lactate [9]. The first step in L-lactate catabolism is the conversion of L-lactate to pyruvate via the enzyme L-lactate dehydrogenase. Examination of the A. actinomycetemcomitans HK1651 genome sequence revealed two genes with high homology to lactate dehydrogenases: AA02769 (gene designations from www.oralgen.lanl.gov) putatively encodes a protein with 75% identity (E value<10−169 using BLASTp) to the catabolic L-lactate dehydrogenase (LctD) from E. coli K12 and AA02749 putatively encodes a protein with 73% identity (E value<10−141 using BLASTp) to the fermentative D-lactate dehydrogenase (LdhA) from E. coli K12. Based on the fact that LdhA homologs are primarily utilized for lactate biosynthesis in fermentative reactions [12], [13], and that A. actinomycetemcomitans does not grow with D-lactate as the sole carbon source (data not shown), we hypothesized that AA02769 likely encodes the enzyme required for growth on L-lactate. To test this hypothesis, the ability of an A. actinomycetemcomitans strain containing an insertion in AA02769 to grow with L-lactate as the sole catabolizable carbon source was assessed. Indeed, inactivation of AA02769 eliminated the ability of A. actinomycetemcomitans to grow with L-lactate, but not glucose (Fig. 2). Based on these results, along with those described below, we will refer to AA02769 as lctD.
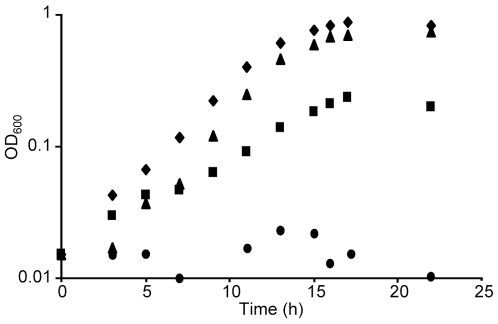
Figure 2. LctD is required for growth using L-lactate.
Growth curves of wild-type A. actinomycetemcomitans and the A. actinomycetemcomitans lctD insertion mutant in chemically defined medium (CDM) containing 20 mM glucose or 20 mM L-lactate as the sole source of energy. Glucose-grown A. actinomycetemcomitans (♦), L-lactate-grown A. actinomycetemcomitans (▴), glucose-grown A. actinomycetemcomitans lctD− (▪), L-lactate-grown A. actinomycetemcomitans lctD− (•).
Over-Expression and Purification of LctD
A. actinomycetemcomitans lctD putatively encodes a 42 kDa cytoplasmic protein that is a proposed member of a family of NAD-independent L-lactate dehydrogenases that convert L-lactate to pyruvate in a unidirectional manner [14]. Due to its high homology to the catabolic L-lactate dehydrogenase from E. coli and the observation that inactivation of lctD eliminated L-lactate-dependent growth of A. actinomycetemcomitans (Fig. 2), we hypothesized that A. actinomycetemcomitans LctD catalyzes the oxidation of L-lactate to pyruvate. To test this hypothesis, A. actinomycetemcomitans lctD was cloned into the pET21a(+) expression vector (pSB103) to create C-terminally his6-tagged LctD. Affinity purification using a nickel column resulted in nearly pure LctD-his6 as demonstrated by a prominent band at approximately 42-kDa on a Coomassie stained gel (Fig. 3A) and a single band in a Western blot using an anti-his6 antibody (Fig. 3B). As a control, a purification procedure was performed using cells containing pET21a(+), which resulted in no proteins readily apparent by SDS-PAGE analysis after purification and no L-lactate dehydrogenase enzyme activity (data not shown).
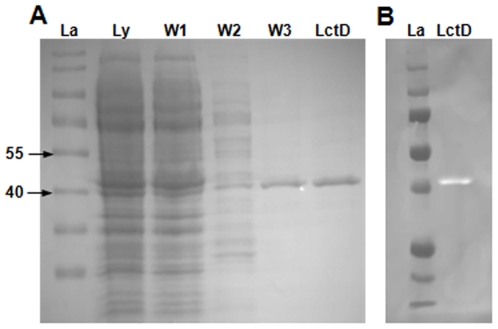
Figure 3. Purification of LctD-his6.
(A) SDS-PAGE analysis of LctD-his6. LctD-his6 was purified using a HisTrap nickel column and examined by SDS-PAGE and Coomassie staining. Lane designations above the gel are: (La), molecular weight ladder; (Ly), cell lysate; (W1), buffer A flow-through; (W2), buffer B with 0.15 M imidazole flow-through; (W3), buffer B with 0.5 M imidazole flow-through; (LctD), phosphate buffer-exchanged LctD-his6. Numbers to the left of the SDS-PAGE gel represent size standards in kilodaltons. Phosphate buffer-exchanged LctD-his6 was used for enzymatic activity studies. (B) Western blot analysis of purified LctD-his6. Purified LctD-his6 was separated on a 10% SDS-PAGE gel, transferred to nitrocellulose membrane, and detected using an anti-his6 antibody and chemiluminescence. Image represents an overlay of a white light image and a chemiluminescent image.
In addition to C-terminally his6-tagged LctD, N-terminally his6-tagged LctD was also constructed and purified. While both purified enzymes displayed enzymatic activity, they were extremely unstable and exhibited significant loss of enzymatic activity in a variety of buffers after overnight storage at 4, −20, or −80°C; therefore all assays were carried out using freshly purified protein. Of note, protein activity from independent purifications was consistent and reproducible. LctD-his6 was used for enzymatic characterization as its yields were somewhat higher than those of his6-LctD.
Kinetic Characterization of A. actinomycetemcomitans LctD
Once purified protein was obtained, the enzymatic activity of LctD-his6 was assessed. The final electron acceptor of many NAD-independent enzymes is unknown, as these enzymes are likely coupled to electron transport [14]. Therefore, LctD-his6 activity was determined in the presence of the electron carriers 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) and phenazine methosulfate (PMS) as previously described [15], [16], [17]. LctD-his6 catalyzed the oxidation of L-lactate to pyruvate (Fig. 4B); however, no activity was observed with the D-lactate isomer (Fig. 4C). Additionally, LctD-his6 was unable to catalyze the reverse reaction (pyruvate to lactate) even in the presence of the reduced substrate NADH (data not shown). These data indicate that, as expected, LctD-his6 is an NAD-independent L-lactate dehydrogenase that catalyzes the oxidation of L-lactate to pyruvate in a unidirectional manner.
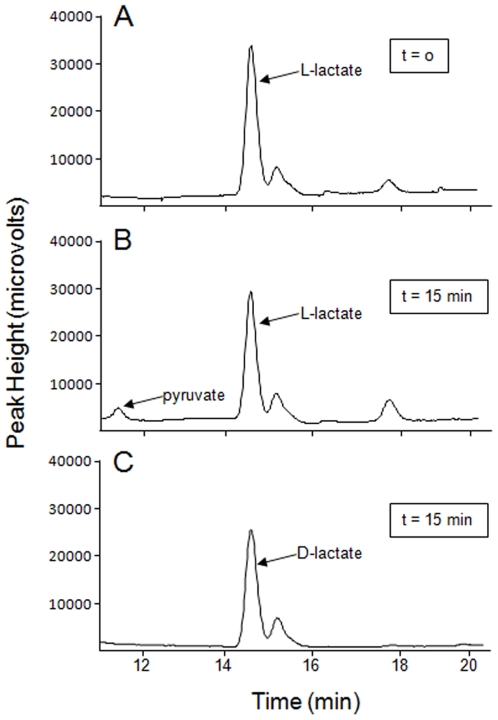
Figure 4. LctD-his6 catalyzes oxidation of L-lactate, but not D-lactate, to pyruvate.
HPLC chromatogram of LctD-his6 in vitro enzyme reactions with (A) L-lactate as the substrate at time 0, (B) L-lactate as the substrate after 15 min, and (C) D-lactate as the substrate after 15 min. Pyruvate and D- and L-lactate were detected using a refractive index detector, and data are displayed as peak height (microvolts). Using commercially available standards, it was determined that pyruvate is detected at approximately 11.6 to 12.1 min and lactate (D and L) is detected at approximately 14.6 to 15.2 min. Representative data are shown for experiments that were performed in duplicate or triplicate.
After determining that the product of LctD-his6 L-lactate oxidation was pyruvate, kinetic analysis was performed. For kinetic analysis, reduction of MTT (measured as the change in absorbance at 570 nm) was utilized to monitor LctD-his6 activity as previously described [15], [16], [17]. Activity assays were carried out in the presence of saturating substrate concentrations, and it was determined that with 8 nM enzyme, 4 mM lactate, and 60 µg/ml MTT, 240 µg/ml PMS achieved linear results for a 10 minute duration (Fig. 5A). To acquire a Km value, activity assays were performed in the presence of increasing lactate concentrations, and a Km value of approximately 150 µM was calculated using SigmaPlot 10.0.1 software (Fig. 5B).
Figure 5. Kinetic analysis of LctD-his6.
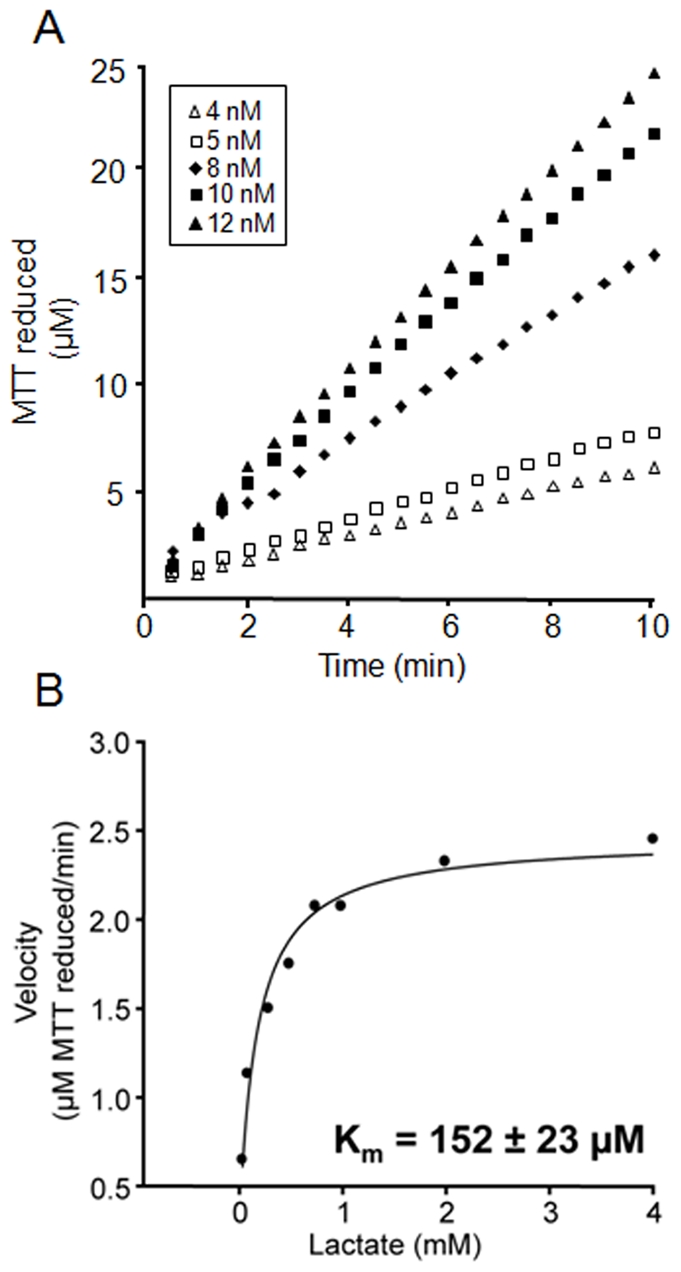
LctD-his6 was incubated with L-lactate and enzymatic activity assessed by monitoring reduction of MTT as described [15], [16], [17]. (A) Enzymatic activity was assessed over time for multiple LctD-his6 concentrations (4, 5, 8, 10, and 12 nM) in the presence of saturating substrate concentrations. For Km calculations, 8 nM LctD-his6 was used. Representative data are shown. (B) The LctD-his6 Km for lactate was calculated as 152±23 µM (average±standard deviation) by averaging values from quadruplicate experiments. Representative data are shown.
Pyruvate Is a Poor Inhibitor of LctD
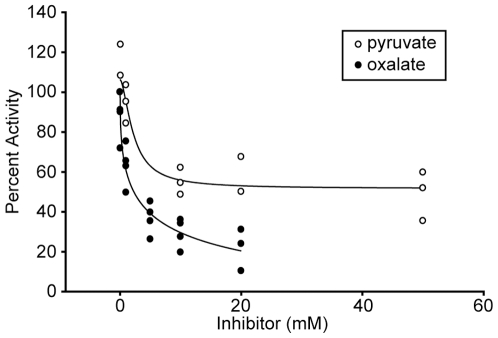
Our model for PTS substrate exclusion involves the production of high levels of intracellular pyruvate during growth with L-lactate (Fig. 1). Based on this model, we hypothesized that A. actinomycetemcomitans LctD-his6 would not be sensitive to feedback inhibition by product (pyruvate) accumulation; thus allowing intracellular accumulation of pyruvate to the high levels (50 mM) previously observed [9]. Interestingly, LctD homologs from other bacteria are often inhibited by relatively low levels (5 mM) of pyruvate [18]. To assess the impact of pyruvate on A. actinomycetemcomitans LctD-his6 activity, inhibition assays were performed in the presence of increasing concentrations of pyruvate as well as the common lactate dehydrogenase inhibitor oxalate [19], [20]. Pyruvate displayed poor inhibition of LctD-his6 activity, with approximately 90% activity remaining in the presence of 5 mM pyruvate and 50% activity remaining in the presence of 50 mM pyruvate (Fig. 6). As expected, oxalate was a potent inhibitor of LctD activity, with 50% inhibition observed at approximately 2 mM oxalate (Fig. 6).
Figure 6. Pyruvate is a poor inhibitor of LctD-his6.
The ability of pyruvate and oxalate to inhibit LctD-his6 enzymatic activity was assessed as described in Materials and Methods. 50% inhibition of LctD-his6 activity was observed with 10–50 mM pyruvate and 2.1 mM oxalate.
Discussion
A. actinomycetemcomitans is found exclusively in the mammalian oral cavity, a diverse environment where microbes likely compete for limited carbon sources including glucose, L-lactate, and fructose [4], [5]. While a large number of oral bacteria preferentially catabolize carbohydrates, previous results from our laboratory revealed that A. actinomycetemcomitans has evolved a preference for L-lactate despite its apparent inferiority as a carbon and energy source [9]. This unique mechanism for preferential L-lactate consumption, referred to as PTS substrate exclusion [9], may have benefits for A. actinomycetemcomitans as it may mitigate L-lactate-mediated acidification of the gingival crevice as well as reduce production of the antimicrobial H2O2 by oral streptococci [21]. Thus preferential consumption of L-lactate by A. actinomycetemcomitans may be a unique survival strategy allowing A. actinomycetemcomitans to compete with other members of the oral microbiota in vivo.
We recently proposed a model for PTS substrate exclusion in which intracellular levels of pyruvate inhibit glucose uptake in L-lactate-grown A. actinomycetemcomitans (Fig. 1). As this model is predicated on the accumulation of pyruvate to intracellular levels approximately 10 times higher than normally found in glucose growing bacteria [9], we hypothesized that the A. actinomycetemcomitans L-lactate dehydrogenase (LctD) is not feedback inhibited by high levels of pyruvate. To test this hypothesis, A. actinomycetemcomitans LctD was purified as a C-terminal his6 fusion protein and determined to be an NAD-independent L-lactate dehydrogenase that catalyzes irreversible oxidation of L-lactate to pyruvate (Fig. 4B). LctD-his6 displayed a Km for L-lactate of approximately 150 µM, a value within the range of other characterized NAD-independent lactate dehydrogenases [14], [18], [22], including the highly homologous E. coli LctD enzyme (75% identity) that has a Km value of 21–70 µM [22].
A. actinomycetemcomitans LctD-his6 was highly resistant to feedback inhibition by pyruvate, displaying 50% activity at 50 mM pyruvate (Fig. 6). This property is unique in that homologous enzymes in other bacteria display feedback inhibition by substantially lower levels of pyruvate. Indeed, the NAD-independent lactate dehydrogenase of Propionibacterium pentosaceum is inhibited by levels of pyruvate (1–5 mM) commonly observed intracellularly [18]. Our results reveal that LctD allows A. actinomycetemcomitans to specifically convert L-lactate to pyruvate in a unidirectional manner. Since this enzyme displays reduced inhibition by pyruvate, intracellular levels of pyruvate rise to levels known to inhibit PTS transport in other bacteria [11], [23], resulting in PTS substrate exclusion. While the PTS enzyme(s) affected by high levels of pyruvate is not known, we hypothesize that inhibition occurs at the first step of PTS transport, phosphorylation of protein EI by PEP (Fig. 1), and studies in our laboratory are currently addressing this hypothesis.
Materials and Methods
Bacterial Strains and Culture Conditions
A. actinomycetemcomitans strain VT1169 [24] and the A. actinomycetemcomitans ldhA mutant (referred to as the lctD mutant in this manuscript) [9] was grown in chemically defined Socransky's medium [25] lacking DL-mevalonic acid and hemin (referred to as Chemically Defined Medium, CDM) or tryptic soy broth with 0.5% yeast extract (TSBYE). Cultures were grown with shaking at 165 RPM at 37°C with a 10% CO2 atmosphere. For growth analysis, overnight TSBYE-grown cultures were washed three times with warm (37°C) CDM containing no catabolizable carbon source and diluted to an optical density at 600 nm (OD600 nm) of 0.015 in CDM supplemented with 20 mM L-lactate or glucose. E. coli DH5α and BL21(DE3) were grown in Luria broth (LB), shaking at 250 RPM at 37°C, with 75 µg/ml ampicillin for plasmid maintenance or 100 µg/ml for selection.
Construction of N- and C-Terminally Tagged LctD
The lctD gene from A. actinomycetemcomitans strain 1169 was amplified by PCR using primers lctDN-for (5′-GGAATTCCATATGATTATTTCGTCCGCTAACG-3′) and lctDN-rev (5′- CCGCTCGAGGCGTATATAAAATACGCCGTTTG-3′) or lctDN-for and lctDC-rev (5′- CTCGAGCTTACTTAAATCTACTAATGC-3′) to create the N-terminally or C-terminally his6-tagged constructs respectively. Forward primers contained an NdeI site and reverse primers contained an XhoI site. Products were digested with NdeI and XhoI and ligated into the NdeI/XhoI-digested pET15b expression vector (Novagen) for the his6 N-terminal tag or the NdeI/XhoI-digested pET21a(+) expression vector (Novagen) for the his6 C-terminal tag. The resultant plasmids were transformed into E. coli DH5α and sequenced using T7 promoter and SP6 terminator primers. Resulting plasmids were named pSB201 (his6 N-terminal tag) and pSB203 (his6 C-terminal tag). Plasmids were transformed into the expression strain E. coli BL21(DE3) for expression and purification.
Protein Purification
E. coli BL21(DE3) carrying pSB201 or pSB203 were grown in TB (EMD) containing 75 µg/ml ampicillin to an OD600 nm = 0.6, and induced overnight at 16°C with 100 µM IPTG in the presence of 100 µM riboflavin (Sigma). Induced cells were harvested by centrifugation for 15 minutes at 6100×g in a Beckman Coulter Avanti J–E centrifuge at 4°C. The pellet was resuspended in 3 ml buffer A (25 mM phosphate buffer, 0.5 M NaCl, 20 mM imidazole, 10 µM flavin adenine dinucleotide (FAD), pH 7.07) containing one-half tablet complete Mini protease inhibitor cocktail (Roche), 25 U Benzonase Nuclease (Novagen) and 10 µM FAD (Alfa Aesar). The cells were passed three times through a French press (American Instruments Company) at 20,000 pounds/square inch. The resulting lysate was centrifuged at 60,000×g in a Beckman-Coulter OptimaL 100 K Ultracentrifuge for one hour at 4°C to remove cellular debris and insoluble protein. The lysate was then applied to a HisTrap HP column (GE Healthcare). The column was washed with 3 ml cold buffer B (25 mM phosphate buffer, 0.5 M NaCl, 10 µM FAD, pH 7.07) containing 0.15 M imidazole, followed by elution with cold buffer B containing 0.5 M imidazole. The eluted protein was concentrated, and a buffer exchange was performed with 25 mM phosphate buffer containing 10 µM FAD. Samples were separated on a 10% SDS-PAGE gel and stained with Coomassie Brilliant Blue (Pierce). Bradford analysis to quantify protein was performed with the Bio-Rad Protein Assay as outlined by the manufacturer. Western blot analysis was performed as outlined [26] using a commercially available anti-his6 antibody (Sigma) and a stabilized goat anti-mouse HRP-conjugated secondary antibody (Pierce). Chemiluminescent detection was performed using the SuperSignal West Dura Extended Duration Substrate as outlined by the manufacturer (Thermo). Blots were imaged with a G-box gel documentation system (Syngene). As a control, a purification procedure was carried out as described above with BL21(DE3) cells containing the parent plasmid pET21a(+).
Characterization of LctD-his6 Enzymatic Activity
Activity assays were performed with phenazine methosulfate (PMS) (Acros Organics) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (Calbiochem) as previously described [15], [16], [17]. To initially examine enzymatic activity of LctD-his6, reactions (1.5 ml) were carried out in 0.08 M Tris buffer containing 120 µg/ml PMS, 60 µg/ml MTT, 2 mM substrate (L-lactate, D-lactate, or pyruvate), and 100 nM LctD-his6. Reactions containing pyruvate as the substrate also contained 1 mM NADH. Half of the reaction volume was removed at time zero, heated at 65°C for 5 minutes, chilled on ice, and stored at −80°C overnight. The remaining half of the reaction was processed in the same manner at 15 minutes, and all samples were filtered through a 0.2 µm Nanosep centrifugal device (Pall). Product analysis was carried out on a Varian HPLC using a Varian Metacarb 87H 300×6.5 mm column at 35°C. Samples were eluted with 0.025 N H2SO4 isocratic elution buffer with a flow rate of 0.5 ml/minute. A Varian refractive index (RI) detector at 35°C was used for product detection with commercially available L-lactate, D-lactate, and pyruvate as standards.
For kinetic analysis, assays were performed monitoring a PMS-coupled reduction of MTT as previously described [15], [16], [17]. The assay was performed in 0.08 M Tris-HCl (pH 8.6) containing 60 µg/ml MTT, 240 µg/ml PMS, and a range of purified enzyme. Absorbance changes in MTT were measured at 570 nm over 10 minutes. Absorbance values obtained from enzyme containing reactions were adjusted by subtracting background absorbance values from a reaction containing no enzyme. Calculations were performed using an extinction coefficient of 17 mM−1 cm−1 [27]. For inhibition studies, the activity assays described above were performed in the presence of increasing concentrations (0.05 to 50 mM) of pyruvate (Alfa Aesar) or oxalate (Fisher).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by the National Institutes of Health grant 5P20RR081741. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Norskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56:2135–2146. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- 2.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 3.Slots J, Reynolds HS, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuttall FQ, Khan MA, Gannon MC. Peripheral glucose appearance rate following fructose ingestion in normal subjects. Metabolism. 2000;49:1565–1571. doi: 10.1053/meta.2000.18553. [DOI] [PubMed] [Google Scholar]
- 5.Soyama K. Enzymatic determination of D-mannose in serum. Clin Chem. 1984;30:293–294. [PubMed] [Google Scholar]
- 6.Jensen ME, Polansky PJ, Schachtele CF. Plaque sampling and telemetry for monitoring acid production on human buccal tooth surfaces. Arch Oral Biol. 1982;27:21–31. doi: 10.1016/0003-9969(82)90172-8. [DOI] [PubMed] [Google Scholar]
- 7.Jensen ME, Schachtele CF. Plaque pH measurements by different methods on the buccal and approximal surfaces of human teeth after a sucrose rinse. J Dent Res. 1983;62:1058–1061. doi: 10.1177/00220345830620101001. [DOI] [PubMed] [Google Scholar]
- 8.Schachtele CF, Jensen ME. Comparison of methods for monitoring changes in the pH of human dental plaque. J Dent Res. 1982;61:1117–1125. doi: 10.1177/00220345820610100201. [DOI] [PubMed] [Google Scholar]
- 9.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, et al. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- 12.Bernard N, Ferain T, Garmyn D, Hols P, Delcour J. Cloning of the D-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 1991;290:61–64. doi: 10.1016/0014-5793(91)81226-x. [DOI] [PubMed] [Google Scholar]
- 13.Bunch PK, Mat-Jan F, Lee N, Clark DP. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143 (Pt 1):187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 14.Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futai M. Membrane D-lactate dehydrogenase from Escherichia coli. Purification and properties. Biochemistry. 1973;12:2468–2474. doi: 10.1021/bi00737a016. [DOI] [PubMed] [Google Scholar]
- 16.Futai M, Kimura H. Inducible membrane-bound L-lactate dehydrogenase from Escherichia coli. Purification and properties. J Biol Chem. 1977;252:5820–5827. [PubMed] [Google Scholar]
- 17.Kohn LD, Kaback HR. Mechanisms of active transport in isolated bacterial membrane vesicles. XV. Purification and properties of the membrane-bound D-lactate dehydrogenase from Escherichia coli. J Biol Chem. 1973;248:7012–7017. [PubMed] [Google Scholar]
- 18.Molinari R, Lara FJ. The lactic dehydrogenase of Propionibacterium pentosaceum. Biochem J. 1960;75:57–65. doi: 10.1042/bj0750057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horikiri S, Aizawa Y, Kai T, Amachi S, Shinoyama H, et al. Electron acquisition system constructed from an NAD-independent D-lactate dehydrogenase and cytochrome c2 in Rhodopseudomonas palustris No. 7. Biosci Biotechnol Biochem. 2004;68:516–522. doi: 10.1271/bbb.68.516. [DOI] [PubMed] [Google Scholar]
- 20.Markwell JP, Lascelles J. Membrane-bound, pyridine nucleotide-independent L-lactate dehydrogenase of Rhodopseudomonas sphaeroides. J Bacteriol. 1978;133:593–600. doi: 10.1128/jb.133.2.593-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnard JP, Stinson MW. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect Immun. 1999;67:6558–6564. doi: 10.1128/iai.67.12.6558-6564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kline ES, Mahler HR. The lactic dehydrogenases of E. coli. Ann N Y Acad Sci. 1965;119:905–919. doi: 10.1111/j.1749-6632.1965.tb47451.x. [DOI] [PubMed] [Google Scholar]
- 23.Weigel N, Kukuruzinska MA, Nakazawa A, Waygood EB, Roseman S. Sugar transport by the bacterial phosphotransferase system. Phosphoryl transfer reactions catalyzed by enzyme I of Salmonella typhimurium. J Biol Chem. 1982;257:14477–14491. [PubMed] [Google Scholar]
- 24.Mintz KP, Fives-Taylor PM. impA, a gene coding for an inner membrane protein, influences colonial morphology of Actinobacillus actinomycetemcomitans. Infect Immun. 2000;68:6580–6586. doi: 10.1128/iai.68.12.6580-6586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky SS, Dzink JL, Smith CM. Chemically defined medium for oral microorganisms. J Clin Microbiol. 1985;22:303–305. doi: 10.1128/jcm.22.2.303-305.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, et al. New York, N.Y.: John Wiley & Sons, Inc; 1997. Short Protocols in Molecular Biology. [Google Scholar]
- 27.Kistler WS, Lin EC. Anaerobic L- -glycerophosphate dehydrogenase of Escherichia coli: its genetic locus and its physiological role. J Bacteriol. 1971;108:1224–1234. doi: 10.1128/jb.108.3.1224-1234.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]