Abstract
Glutathione peroxidase 4 (Gpx4) is a unique antioxidant enzyme that repairs oxidative damage to biomembranes. In the present study, we examined the effect of Gpx4 on the release of various apoptogenic proteins from mitochondria using transgenic mice overexpressing Gpx4 [Tg(GPX4+/0)] and mice deficient in Gpx4 (Gpx4+/− mice). Diquat exposure triggered apoptosis that occurred through intrinsic pathway and resulted in the mitochondrial release of cytochrome c (cyt. c), Smac/DIABLO, and Omi/HtrA2 in the liver of wild-type (Wt) mice. Liver apoptosis and cyt. c release were suppressed in Tg(GPX4+/0) mice but exacerbated in Gpx4+/− mice; however, neither the Tg(GPX4+/0) nor the Gpx4+/− mice showed any alterations in the levels of Smac/DIABLO or Omi/HtrA2 released from mitochondria. Submitochondrial fractionation data showed that Smac/DIABLO and Omi/HtrA2 existed primarily in the intermembrane space and matrix, while cyt. c and Gpx4 were both associated with inner membrane. In addition, diquat exposure induced cardiolipin peroxidation in the liver of Wt mice; the levels of cardiolipin peroxidation were reduced in Tg(GPX4+/0) mice but elevated in Gpx4+/− mice. These data suggest that Gpx4 differentially regulates apoptogenic protein release due to its inner membrane location in mitochondria and its ability to repair cardiolipin peroxidation.
Keywords: Gpx4, Phospholipid hydroperoxide glutathione peroxidase 4, Apoptosis, Cardiolipin, Lipid peroxidation, Oxidative stress
Introduction
Apoptosis (programmed cell death) is a process by which organisms eliminate injured or unwanted cells. In most cases (but not all), physiological cell death occurs by apoptosis as opposed to necrosis [1]. Therefore, apoptosis plays a critical role in cellular homeostasis. There are two well-documented apoptotic pathways: the extrinsic and intrinsic pathways [2]. The extrinsic pathway signals through death receptor such as Fas and the TNF receptor. Upon their ligand binding, death receptors recruit and oligomerize adaptor protein FADD, resulting in the recruitment and autoactivation of initiator caspase-8, which further activates downstream executioner casapse-3, 7. The intrinsic pathway is mediated by mitochondria. Upon activation by cellular stress, mitochondria release apoptogenic proteins into the cytosol, e.g., cyt. c, Smac/DIABLO, Omi/HtrA2, and apoptosis-inducing factor (AIF). These apoptogenic proteins function together to trigger apoptosis. Cyt. c interacts with monomeric apoptotic protease activating factor-1 (Apaf-1) to facilitate a conformational change in the latter. The conformation change leads to Apaf-1 oligomerization and the recruitment of initiator casapse-9 to form the apoptosome, resulting in caspase-9 activation and the subsequent activation of caspase-3, 7. Smac/DIABLO and Omi/HtrA2 bind to inhibitor of apoptosis protein (IAPs), blocking their ability to inhibit caspases and thus promoting apoptosis. AIF induces DNA fragmentation and chromatin condensation. In vertebrates, most (but not all) apoptosis is believed to occur through the intrinsic pathway [3].
A large body of literature suggests a close relationship between reactive oxygen species (ROS) and apoptosis. For example, apoptosis is induced by various pro-oxidants, such as hydroperoxide, diamide, etoposide and semiquinones [4,5]. In addition, other apoptotic stimuli, such as TNF-α [6], ceramide [7], glutamate [8], amyloid-β-peptide [9], alkaline conditions [10], and nerve growth factor withdrawal [11], elevate intracellular ROS levels. Moreover, the pseudo-apoptosis phenotype in yeast invariably involves ROS [12]. These data strongly suggest that ROS serve as a mediator for apoptosis. Thus, the inhibition of ROS by antioxidant enzymes could potentially suppress apoptosis.
Gpx4 is a unique antioxidant enzyme in that its major function is to repair oxidative damage to the biomembranes rather than to detoxify superoxide or hydrogen peroxide. It belongs to the glutathione peroxidase (Gpx) superfamily. All Gpxs can reduce hydrogen peroxide, alkyl peroxides, and fatty acid hydroperoxides; however, Gpx4 also reduces hydroperoxides in lipoproteins and complex lipids such as those derived from cholesterol, cholesteryl esters, and phospholipids. Gpx1 (as well as Gpx 2 and 3) is a homotetramer while Gpx4 is a 20–22 kDa monomer, which is rich in hydrophobic amino acid residues [13]. Its small size, hydrophobic surface, and unique substrate specificity for complex lipids have led to the view that Gpx4 plays a key role in the detoxification of membrane lipid hydroperoxides [14]. Using kinetic modeling, Antunes et al. [15] proposed that Gpx4 is more efficient in removing hydroperoxides from membranes than the phospholipase A2 (PLA2)/Gpx1 pathway (where PLA2 removes the fatty acid hydroperoxide from the membrane and Gpx1 reduces the hydroperoxide) because the affinity of Gpx4 for the membrane lipid hydroperoxides is more than 104-fold greater than PLA2.
Gpx4 is synthesized as a long form (23 kDa) and a short form (20 kDa), both of which arise from the same Gpx4 gene but with different translation initiation sites [16]. The long-form of Gpx4 is targeted to the mitochondria because it has a mitochondrial signal peptide. The short-form of Gpx4 is the non-mitochondrial Gpx4 and has been found in cytosol, nucleus, and endoplasmic reticulum of cells [17]. Experiments using cell lines that overexpress Gpx4 have shown that Gpx4 can reduce the toxicity of oxidative stress [18,19]. Interestingly, multiple reports also suggest that Gpx4 plays a role in the protection against apoptosis. The overexpression of Gpx4 inhibits the induction of apoptosis by oxidized, low-density lipoprotein in rabbit aortic smooth muscle cells [20], by photosensitizers in human breast cancer cells [21], and by cholesterol hydroperoxide in breast tumor cell lines [22]. In addition, overexpressing Gpx4 preferentially in the mitochondria protects RBL2H3 cells against apoptosis induced by various reagents, e.g., potassium cyanide, 2-deoxyglucose, etoposide, staurosporine, UV irradiation, cycloheximide; however, overexpressing non-mitochondrial Gpx4 had no effect on apoptosis [23]. Therefore, it appears that mitochondrial Gpx4 is important in protecting cells against the induction of apoptosis.
In this study, we used transgenic mice overexpressing Gpx4 [Tg(GPX4+/0)] and mice deficient in Gpx4 (Gpx4+/− mice) to examine the effect of Gpx4 on mitochondrial release of various apoptogenic proteins during oxidative stress induced apoptosis in vivo. Among all the apoptogenic proteins we studied, Gpx4 only protected against cyt. c release from mitochondria. This protection was correlated with reduced cardiolipin (CL) peroxidation.
Experimental Procedures
Animals Preparation
The Tg(GPX4+/0) and Gpx4+/− mice used in this study are previously described [24,25]. The wildtype (Wt) controls were littermates of the Tg(GPX4+/0) mice or Gpx4+/− mice. Mice were fed ad libitum a commercial diet and maintained under pathogen-free barrier conditions on a 12:12-h dark/light cycle. At 4–10 months of age, female mice were injected intraperitoneally with diquat dissolved in saline at a dose of 50 mg/kg. Six hours after injection, mice were euthanized by cervical dislocation and the liver collected. All procedures involving the mice were in accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at the University of Texas Health Science Center at San Antonio and the Audie L. Murphy Veterans Affairs Hospital.
Subcellular Fractionation
Liver tissue was homogenized in ice-old buffer I (250 mM mannitol, 75 mM sucrose, 500 µM EGTA, 100 µM EDTA, and 10 mM HEPES, pH 7.4) supplemented with protease inhibitor cocktail. The homogenates were centrifuged at 600 g for 10 min at 4°C to pellet nuclei and unbroken cells. The resulting supernatant was then centrifuged at 10,000 g for 15 min at 4°C to obtain the mitochondrial pellet. The supernatant was further centrifuged at 100,000 g for 60 min at 4°C to yield the cytosol. The mitochondrial pellets were washed once in buffer I containing 0.2% (w/v) bovine serum albumin (BSA) and twice in buffer I without BSA. The mitochondrial pellets obtained were used for measuring CL peroxidation. The cytosol obtained was used to measure mitochondrial release of cyt. c, second mitochondria-derived activator of caspases/direct IAP binding protein with low pI (Smac/DIABLO) and Omi/HtrA2 by Western blots.
Measurement of Apoptosis
Apoptotic cell counts in the liver were determined in situ by the presence of double-strand DNA breaks observed in paraffin-embedded tissue sections using in situ oligo ligation (ISOL) kit (Chemicon International, CA) with oligo B according to the manufacturer’s instructions. Compared to conventional TdT-mediated dUTP nick-end labeling (TUNEL) assay, the ISOL assay uses hairpin oligonucleotide probe to detect more specific DNA fragmentation caused by apoptosis, avoiding randomly damaged DNA [26]. Slides were visualized under light microscopy, and the number of positive cells was determined in 10 random fields at 400 × magnification for each liver. Data were expressed as the mean of the ratio of the number of positive cells and the total number of cells in all the 10 random fields.
Western Blots
Caspase-9 activation, Gpx4 protein expression, and mitochondrial release of apoptogenic proteins were measured by Western blots as described previously [25]. Caspase-9 activation and Gpx4 protein expression were detected in liver homogenates whereas mitochondrial release of apoptogenic proteins was detected in liver cytosols. Briefly, the protein concentrations were determined by the Bradford method [27]. Equal amounts of protein were then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, probed with appropriate antibodies, and detected by ECL Western blotting system. Data were normalized to loading control actin for liver homogenate or β-tubulin for cytosols. Antibodies used were: anti-caspase-9 antibody (#9508; Cell Signaling Technology, Beverly, MA); anti-Gpx4 antibody as described previously [24]; anti-cyt. c antibody (sc-13560; Santa Cruz Biotechnology, Santa Cruz, CA); anti-Smac/DIABLO antibody (ab9709; Abcam, Inc., Cambridge, MA); anti-Omi/HtrA2 antibody (AF1458; R&D systems, Inc., Minneapolis, MN); anti-actin antibody (#69100; MP Biomedicals Inc., Aurora, OH); anti-β-tubulin (32-2600; Zymed Laboratories, San Francisco, CA).
Detection of Cyt. c Release from Mitochondria by Immunohistochemistry
Cyt. c release was also measured using an immunohistochemistry method that only detects cyt. c present in the cytosol. Fixing tissue with formalin prevents the cyt. c antibody from reaching the mitochondrial intermembrane space. Therefore, only cells with cyt. c released into the cytosol will have cyt. c immunoreactivity [28]. Immunohistochemistry was performed using vector immunodetection M.O.M. kit according to the instructions. Briefly, histology slides were first deparaffinized and rehydrated. Slides were then treated to quench endogenous peroxidase activity and to unmask the antigen. After blocking for mouse IgG, slides were incubated with antibody to cyt. c and the corresponding secondary antibody. Slides were stained with substrate DAB and counterstained with methyl green. Cyt. c positive cells were detected under light microscopy, and the number of positive cells was determined in 10 random fields at 400 × magnification for each liver. Data were expressed as the mean of the ratio of the number of positive cells and the total number of cells in all the 10 random fields.
Subfractionation of Mitochondria
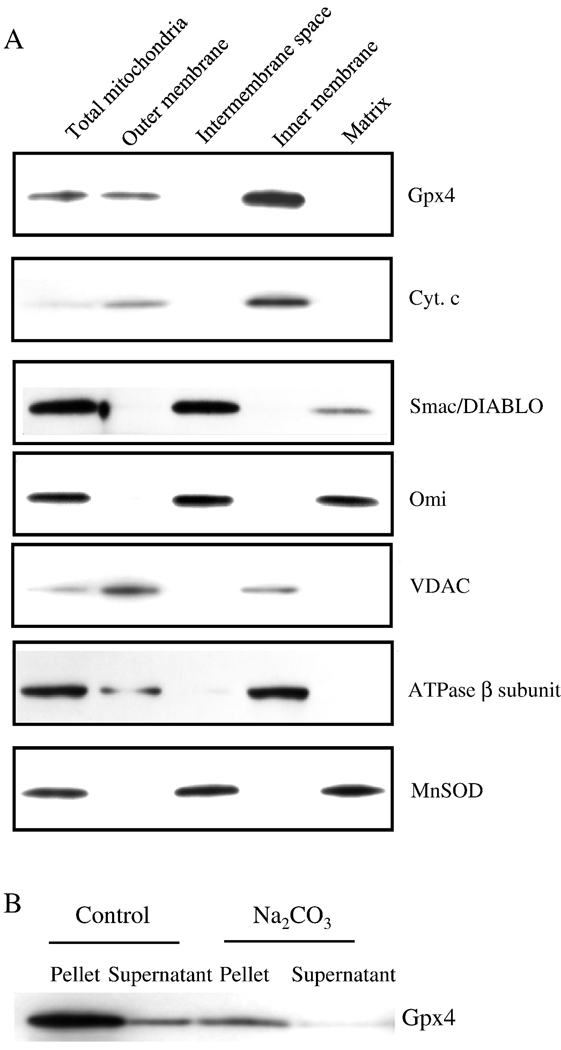
Liver mitochondria from Wt mice were subfractioned as previously described with some modifications [29]. Liver tissue was homogenized in ice-cold buffer A (0.25 M sucrose, 1 mM EDTA, 10 mM HEPES-NaOH, pH 7.4). The homogenates were centrifuged at 800 g for 10 min at 4°C. The resulting supernatant was then centrifuged at 3,000 g for 10 min at 4°C to obtain a crude heavy mitochondrial pellet. The crude heavy mitochondrial pellet was washed once and further purified by a discontinuous Nycodenz gradient [30]. Briefly, the heavy mitochondrial pellet re-suspended in 3.5 ml of 25% Nycodenz was placed over 1.5 ml of 40%, 1.5 ml of 34% and 2.5 ml of 30% Nycodenz, and this was topped off with 2.5 ml of 23% and 0.5 ml of 20% Nycodenz. The gradient was centrifuged at 95,000 g for 2 hrs at 4°C. Mitochondria were collected from the 25%/30% Nycodenz interface, washed, and re-suspended in a small amount of buffer A. A sample was taken and designated as total mitochondria. Digitonin was slowly added to the remaining mitochondria (1.5 mg of digitonin for every 10 mg of mitochondrial proteins). After incubation at 4°C for 15 min with gentle rocking, the suspension was diluted with 3 volumes of buffer A and then centrifuged at 15,000 g for 10 min at 5°C. The supernatant was further centrifuged at 144,000 g for 20 min at 5°C, and the resulting pellet and supernatant were designated as mitochondrial outer membrane and intermembrane space, respectively. The pellet from 15,000 g centrifugation spin, containing mitoplasts, was re-suspended in a small amount of buffer A and subjected to 3 freeze-thaw cycles. The mitoplast suspension was centrifuged at 144,000 g for 50 min at 5°C, and the resulting pellet and supernatant were designated as mitochondrial inner membrane and matrix, respectively. For the Na2CO3 treatment, the inner membrane fraction (40 ug) was incubated with 0.1 M Na2CO3 on ice for 30 min. Separation of the pellet from the released proteins were achieved by centrifugation at 160,000g for 30 min at 4°C. Pellets and supernatant were subjected to Western blotting for the detection of Gpx4.
Cardiolipin Peroxidation
10-N-nonyl-Acridin Orange (NAO, Molecular Probes, Eugene, OR) binding to mitochondria was used to estimate cardiolipin (CL) peroxidation as previously described [31] with some modifications. NAO binds to CL with high affinity, and the fluorochrome loses its affinity for peroxidized cardiolipin [32]. Because NAO binding to the mitochondria has been found to depend on mitochondrial membrane potential [33], experiments were performed using isolated liver mitochondria not energized with substrates, thereby eliminating the effect of the membrane potential on NAO binding. Freshly isolated liver mitochondria were resuspended in buffer A (125 mM KCl, 10 mM HEPES, 5 mM MgCl2, and 2 mM K2HPO4, pH 7.4). Ten 10 µg of mitochondrial protein was added to 200 µl of 20 µM NAO. The samples were incubated for 5 min and then centrifuged at 30,000 g for 5 min. Free dye in the supernatant was determined by measuring absorbance at 495 nm, and the NAO bound to the mitochondria calculated as the total minus free NAO.
CL Content
Phospholipids were extracted from mitochondria (~1 mg of mitochondrial protein) using chloroform/methanol/water as described previously [34]. The extracted phospholipids were dried and dissolved in chloroform:methanol mixture (2:1, v/v). CL was separated from other phospholipids by high performance liquid chromatography (HPLC) using a silicic acid column (5 µm Radial Pac Resolve Silica cartridge, 0.8 cm x 10 cm) as described by Nissen and Kreysel [35]. Briefly, the chromatographic system was programmed for gradient elution using two mobile phases: A. acetonitrile-water (80:20); B. acetonitrile. A linear solvent gradient from 87.5 to 25% B between 3 and 15 min was used, delivering a gradient of water running from 2.5 to 15% water. HPLC elution was performed at flow rate of 1 ml/min. Detection of CL elution was monitored at 203 nm with a Waters Absorbance Detector. The identity of eluted CL at particular time interval was confirmed by chromatography of CL standard. The CL content (nmol/mg mitochondrial protein) was calculated based on the integrated peak area under 203 nm by using standard curve.
Statistical Analysis
One-way analysis of variance was used for analysis, followed by Student-Newman-Keuls for multiple comparisons. Statistical significance was assumed at p<0.05.
Results
Effect of Gpx4 on the Induction of Apoptosis by Diquat
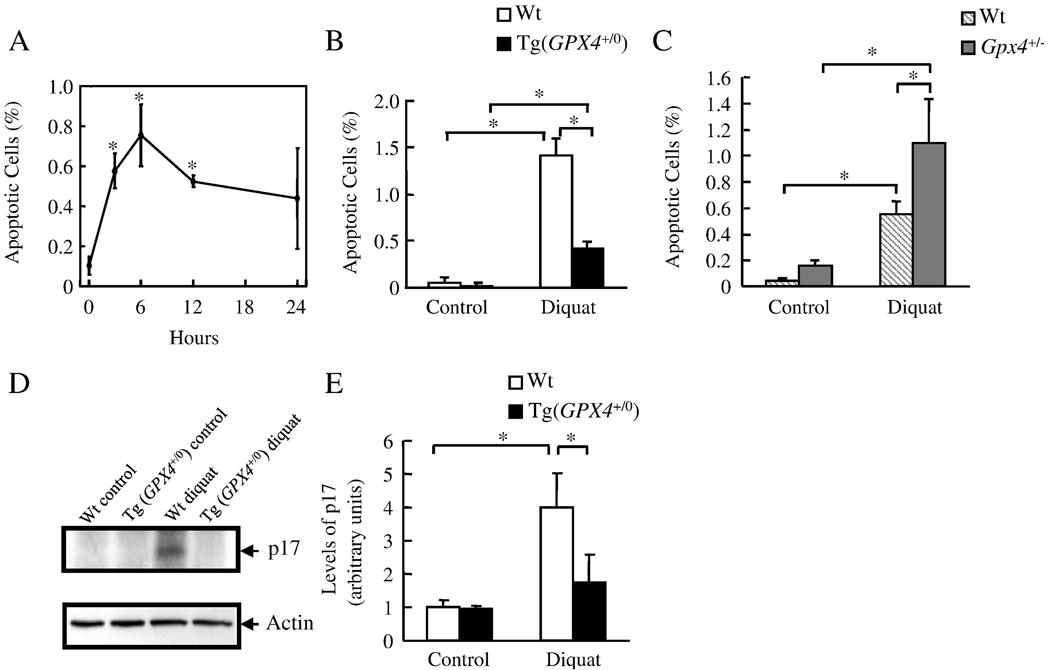
Diquat was used as a model of in vivo oxidative stress because bipyridyl herbicide diquat is a potent pro-oxidant that generates superoxide anions through redox cycling [36]. Diquat is targeted primarily to the liver and induces oxidative damage to the liver [37]. Figure 1A shows the time course of apoptosis induced by diquat in Wt mice. Endogenous apoptosis in the liver was low in untreated mice. Apoptosis was significantly increased 3 hours after diquat injection and was maximal at 6 hours. Therefore, in the following experiments, we studied the effect of Gpx4 on apoptosis 6 hours after diquat treatment. Figure 1B shows the levels of apoptosis in the Wt and Tg(GPX4+/0) mice before and after diquat treatment. There were very few apoptotic cells in untreated Wt (0.05% apoptotic cells) and Tg(GPX4+/0) mice (0.03% apoptotic cells). Diquat treatment dramatically increased the level of apoptosis in Wt mice; however, the induction of apoptosis in the Tg(GPX4+/0) mice was dramatically reduced (70%) compared to Wt mice. In contract, the induction of apoptosis was significantly increased (98%) in Gpx4+/− mice compared to Wt mice (Figure 1C). We also found that diquat induced apoptosis occurred through the intrinsic pathway of apoptosis because diquat activated caspase-9 as shown by a significant increase in the cleavage product in the Wt mice (Figure 1D and E). The activation of caspase-9 by diquat was greatly reduced (57%) in Tg(GPX4+/0) mice compared to Wt mice. Conversely, diquat did not induce the extrinsic pathway because of the lack of caspase-8 activation and Bid cleavage (data not shown). These data show that Gpx4 inhibits diquat induced apoptosis through intrinsic pathway of apoptosis in vivo.
Figure 1. The effect of Gpx4 on diquat induced apoptosis.
Mice were treated with diquat (50 mg/kg). The levels of liver apoptosis and caspase 9 activation were determined as described in the Experimental Procedures. Panel A: Time course for the induction of apoptosis in the livers of Wt mice. The asterisks show those values that are significantly different from untreated controls. Panel B: Level of apoptosis in the livers of Wt (open bars) and Tg(GPX4+/0) (solid bars) mice untreated or treated with diquat for 6 hours. Panel C: Level of apoptosis in the livers of Wt (hatched bars) and Gpx4+/− mice (shaded bars) mice untreated or treated with diquat for 6 hours. Panel D: Photograph of a representative western blot showing cleaved caspase-9 (p17) in the liver homogenates isolated from Wt and Tg(GPX4+/0) mice untreated or treated with diquat for 6 hours. Panel E: Quantification of cleaved caspase-9 (p17) from Wt (open bars) and Tg(GPX4+/0) (solid bars) mice as determined from the Western blots. All values are the mean ± SEM of data obtained from 3–4 mice. The asterisks in Panels B, C, and E show those values that are significantly different.
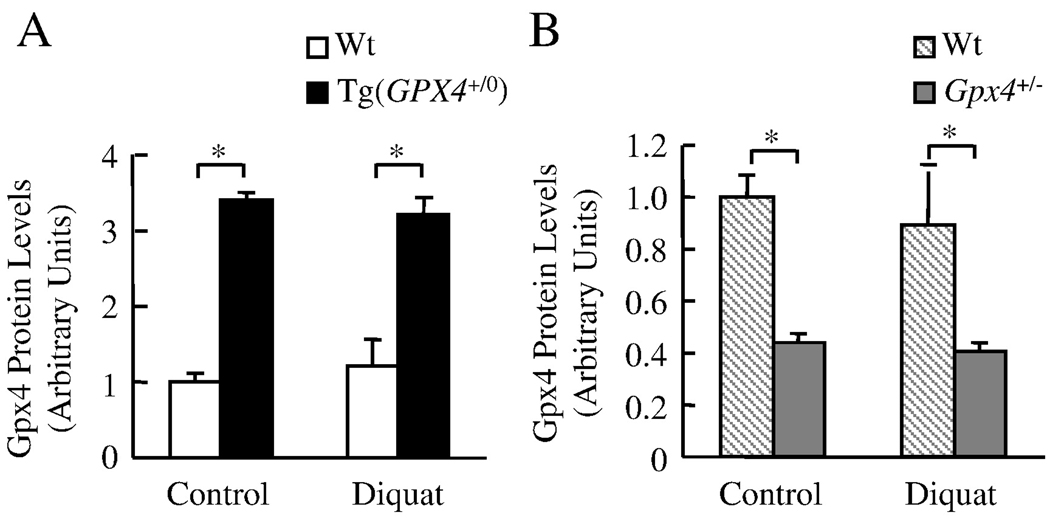
We measured Gpx4 protein expressions in the liver of Tg(GPX4+/0) mice and Gpx4+/− mice before and after diquat treatment (Figure 2). As expected, Tg(GPX4+/0) mice showed higher levels of Gpx4 proteins whereas Gpx4+/− mice showed lower levels of Gpx4 proteins when compared to their Wt controls. Moreover, diquat did not alter the level of Gpx4 in either Tg(GPX4+/0) mice or Gpx4+/− mice.
Figure 2. The levels of Gpx4 in Tg(GPX4+/0) and Gpx4+/− mice.
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat. Gpx4 protein levels in the liver were determined by Western blotting as described in the Experimental Procedures. Panel A: Quantification of Gpx4 protein levels in the liver of Tg(GPX4+/0) mice (solid bars) and their Wt littermates (open bars) as determined from the Western blotting. Panel B: Quantification of Gpx4 protein levels the liver of Gpx4+/− mice (shaded bars) and their Wt littermates (hatched bars) as determined from the Western blotting. Values are mean ± SEM of data obtained from 3–4 mice. The asterisks indicate those differences that are significantly different.
Effect of Gpx4 on mitochondrial release of apoptogenic proteins
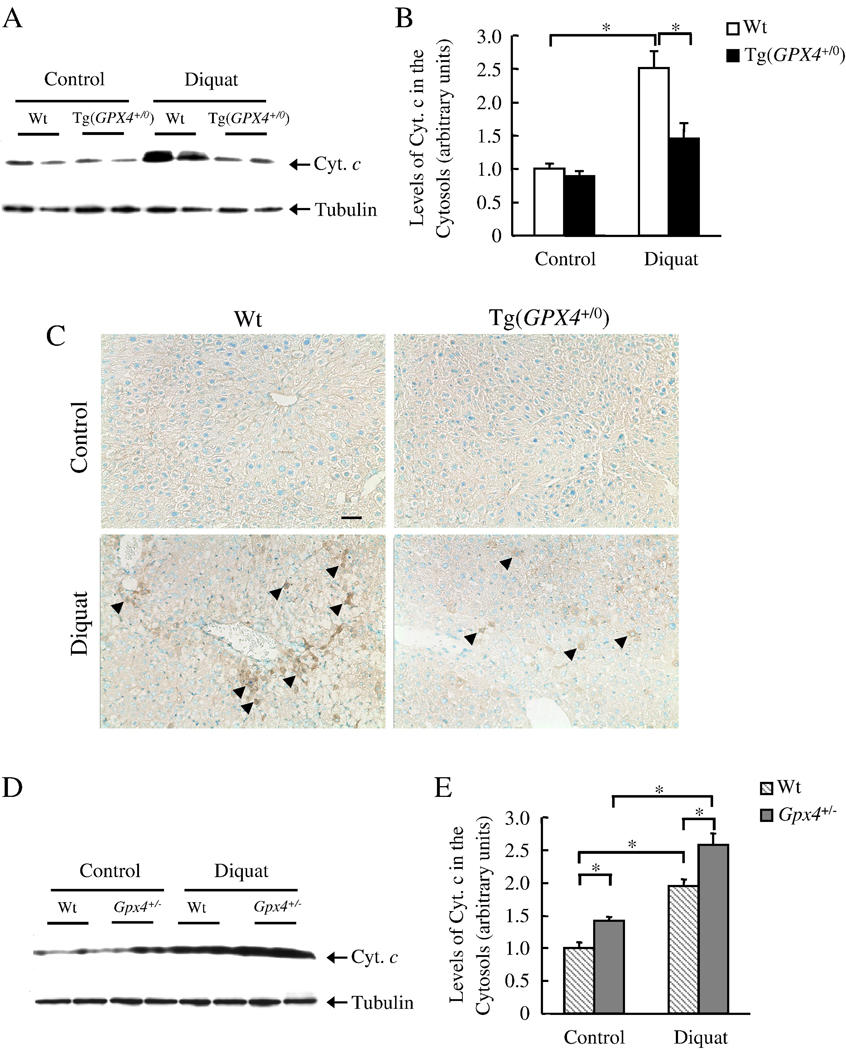
The intrinsic apoptotic pathway occurs through the release of mitochondrial apoptogenic proteins. We therefore studied the effect of Gpx4 on the release of apoptogenic proteins from mitochondria after diquat treatment. As shown by Western blots (Figure 3A and 3B), diquat treatment induced cyt.c release in Wt mice (2.5-fold increase), and the level of cyt. c release was significantly reduced (43%) in the Tg(GPX4+/0) mice. To elucidate the possibility that mitochondria rupture during isolation procedure, resulting in cyt. c release, we further measured cyt. c release by immunohistochemistry (Figure 3C). There was a significant number of cyt. c positive cells in diquat treated Wt mice, and therefore, the detection of cyt. c release by Western Blotting is not an artifact. In contrast, Tg(GPX4+/0) mice had less cyt. c positive cells suggesting that Tg(GPX4+/0) mice were truly protected against diquat induced cyt. c release. We also measured mitochondrial cyt. c release in Gpx4+/− mice (Figure 3D and 3E). The induction of cyt. c release was significantly greater (32%) in Gpx4+/− mice compared to Wt mice. These data show that Gpx4 inhibits cyt. c release induced by diquat.
Figure 3. Effect of Gpx4 on diquat-induced cyt. c release.
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat. Cyt. c release was determined by Western blotting and immunohistochemistry as described in the Experimental Procedures. Panel A: Photograph of a representative Western blotting showing cyt. c release into the cytosol in Tg(GPX4+/0) mice and their Wt littermates. Panel B: Quantification of cyt.c release in Tg(GPX4+/0) mice (solid bars) and their Wt littermates (open bars) as determined from the Western blotting. Panel C: Photograph of representative liver sections from Wt and Tg(GPX4+/0) mice. Sections were immunostained with the anti-cyt. c antibody. Arrows show examples of cyt. c positive cells. Original magnification: ×200. Scale bar=50 µm. Panel D: Photograph of a representative Western blotting showing cyt. c release into the cytosol in Gpx4+/− mice and their Wt littermates. Panel E: Quantification of cyt.c release in Gpx4+/− mice (shaded bars) and their Wt littermates (hatched bars) as determined from the Western blotting. Values are mean ± SEM of data obtained from 3–4 mice. The asterisks indicate those differences that are significantly different.
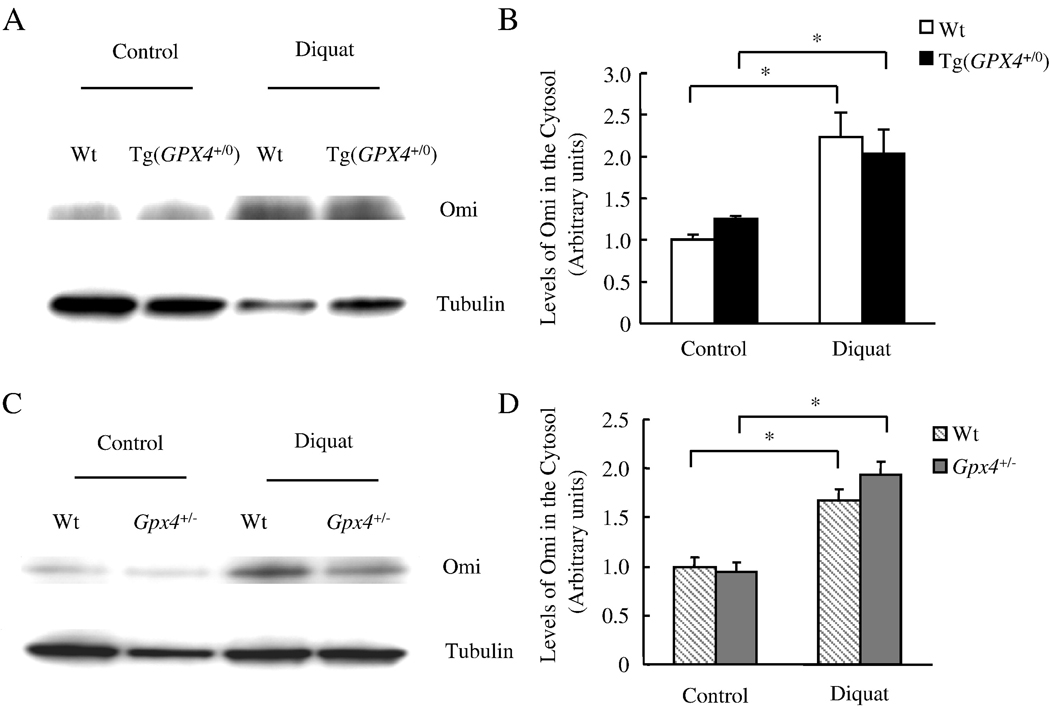
We measured the release of Smac/DIABLO and Omi/HtrA2 from mitochondria in response to diquat treatment. The data in Figure 4 show that diquat treatment significantly increased the levels of Smac/DIABLO in the cytosol from Wt, Tg(GPX4+/0), and Gpx4+/− mice; however, neither Tg(GPX4+/0) nor Gpx4+/− mice showed any alteration in level of Smac/DIABLO release by diquat. The data in Figure 5 show mitochondrial release of Omi/HtrA2 after diquat treatment. Similar to the release of Smac/DIABLO, the release of Omi/HtrA2 by diquat was unchanged in Tg(GPX4+/0) and Gpx4+/− mice when compared to Wt mice. These data indicate that Gpx4 does not affect diquat induced mitochondrial release of Smac/DIABLO and Omi/HtrA2.
Figure 4. Effect of Gpx4 on diquat-induced Smac/DIABLO release from liver mitochondria.
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat. Smac/DIABLO release was determined by measuring the levels of Smac/DIABLO in the cytosol using Western blotting as described in the Experimental Procedures. Panel A: Photograph of a representative Western blot showing Smac/DIABLO release into the cytosol from the Tg(GPX4+/0) mice and their Wt littermates. Panel B: Quantification of Smac/DIABLO release in the Tg(GPX4+/0) mice (solid bars) and their Wt littermates (open bars) as determined from the Western blotting. Panel C: Photograph of a representative Western blotting showing Smac/DIABLO release into the cytosol from the Gpx4+/− mice and their Wt littermates. Panel D: Quantification of Smac/DIABLO release in the Gpx4+/− mice (shaded bars) and their Wt littermates (hatched bars) as determined from the Western blotting. All values are mean ± SEM of data obtained from 3–4 mice. The asterisks indicate the differences that are significantly different.
Figure 5. Effect of Gpx4 on diquat-induced Omi/HtrA2 release from liver mitochondria.
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat. Omi/HtrA2 release was determined by measuring the levels of Omi/HtrA2 in the cytosol using Western blotting as described in the Experimental Procedures. Panel A: Photograph of a representative Western blot showing Omi/HtrA2 release into the cytosol from the Tg(GPX4+/0) mice and their Wt littermates. Panel B: Quantification of Omi/HtrA2 release in the Tg(GPX4+/0) mice (solid bars) and their Wt littermates (open bars) as determined from the Western blotting. Panel C: Photograph of a representative Western blotting showing Omi/HtrA2 release into the cytosol from the Gpx4+/− mice and their Wt littermates. Panel D: Quantification of Omi/HtrA2 release in the Gpx4+/− mice (shaded bars) and their Wt littermates (hatched bars) as determined from the Western blotting. All values are mean ± SEM of data obtained from 3–4 mice. The asterisks indicate the differences that are significantly different.
Mechanism Responsible for the Differential Regulation of Mitochondrial Apoptogenic Protein release by Gpx4
To understand why Gpx4 levels affected differently the release of apoptogenic proteins by mitochondria, we studied the submitochondrial localization of apoptogenic proteins and Gpx4 (Figure 6A). Liver mitochondria were subfractioned into outer membrane, intermembrane space, inner membrane, and matrix. The identity of the different submitochondrial fractions was demonstrated by the enrichment of marker proteins VDAC, ATPase β subunit, and MnSOD in the outer membrane, inner membrane, and matrix, respectively. Our fractionation scheme separated mitochondrial membranes from the soluble fractions as shown by the absence of membrane markers in the soluble fractions and the absence of soluble fraction markers in the membranes. Given that the outer membrane and inner membrane are closely apposed at various contact sites, the mitochondrial subfractionation did not allow a complete separation of the outer and inner mitochondrial membranes. This was indicated by the presence of small amounts of VDAC in the inner membrane and ATPase β subunit in the outer membrane. In addition, mitochondrial matrix marker MnSOD was found in the intermembrane space as well; however, considering matrix has more protein than intermembrane space, the majority of MnSOD would still exist in the matrix.
Figure 6. Submitochondrial localization of Gpx4 and apoptogenic proteins.
Submitochondrial fractions were obtained as described in the Experimental Procedures. Localizations of Gpx4 and apoptogenic proteins are shown by Western blotting. Panel A: Photograph of a representative Western blot showing the localization of Gpx4 and apoptogenic proteins in the mitochondria. Panel B: Photograph of a representative Western blotting showing the integral membrane association of Gpx4 with the inner membrane.
The data in Figure 6A showed that Smac/DIABLO was found in the intermembrane space with a small amount present in the matrix. Omi/HtrA2 was enriched in intermembrane space as well as in matrix. These data are consistent with the speculation that Smac/DIABLO and Omi/HtrA2 are freely soluble proteins [38]. In fact Rardin et al. [39], also reported the intermembrane space and matrix location of Smac/DIABLO in rat kidney mitochondria. In contrast, cyt. c was primarily found in the inner membrane of mitochondria. This is in good agreement with literature [40] because it is commonly accepted that cyt.c is associated with the lipids at the outer face of mitochondrial inner membrane through electrostatic force and hydrophobic interaction. It should be noted that in several studies, cyt. c has been found in the intermembrane space [41,42]; these studies mitochondria were obtained from neuronal and kidney cell lines whereas in our study liver mitochondria isolated from mice. In addition, submitochondrial fractionation protocols in the previous studies used buffer containing 5 mM MgCl2, which has been shown to dissociate cyt. c from the inner membrane by disrupting the ionic interaction between cyt. c and inner membrane [38]. In our study, we used an isolation buffer that had low ionic strength to prevent cyt. c dissociation.
Like cyt. c, Gpx4 was found in the membrane fractions and it was particularly enriched in the inner membrane. This is consistent with Gpx4 being a small hydrophobic protein. In addition, Gpx4 was an integral protein in the inner membrane but not peripherally bound because alkaline condition failed to elute Gpx4 from the inner membrane as shown in Figure 6B [43]. Because Gpx4 and cyt.c were both associated with inner mitochondrial membrane, Gpx4 could affect cyt.c release from mitochondria through its effect on lipid peroxidation.
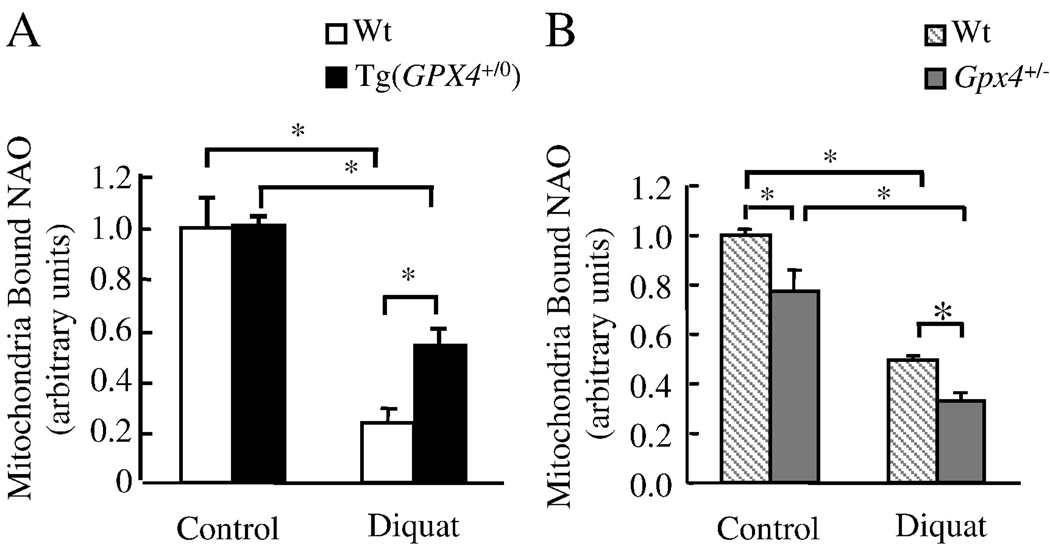
Studies with cells and isolated mitochondria suggest that CL peroxidation reduces cyt. c binding to the inner mitochondrial membrane and therefore promotes cyt. c release from the mitochondria [44,45]. We compared CL peroxidation in liver mitochondria isolated from Tg(GPX4+/0), Gpx4+/− and Wt mice before and after treatment with diquat by measuring NAO binding to the mitochondria. The lipophilic dye NAO binds to CL with high affinity and the fluorochrome has no affinity for CL hydroperoxide (CLOOH) [32,46]; however, NAO is able to bind to hydroxyl CL(CL-OH) [45]. Thus loss of mitochondrial bound NAO would suggest CL peroxidation under the assumption that CL content remains unchanged. Figure 7A shows that NAO binding to the mitochondria was reduced by 76% in Wt mice after diquat treatment, indicating increased CL peroxidation after diquat treatment. Mitochondria isolated from diquat-treated Tg(GPX4+/0) mice showed more (2.3-fold increase) NAO binding compared to Wt, indicating that overexpression of Gpx4 reduced the level of CL peroxidation induced by diquat. Figure 7B shows that NAO binding to the mitochondria was significantly lower in both untreated (23% lower) and treated (33% lower) Gpx4+/− mice when compared to Wt mice, indicating that Gpx4 deficiency increased endogenous and diquat induced CL peroxidation.
Figure 7. Effect of Gpx4 on CL peroxidation.
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat and CL peroxidation determined by measuring mitochondrial bound NAO as described in the Experimental Procedures. Panel A: Quantification of CL peroxidation in Tg(GPX4+/0) mice (solid bars) and their Wt littermates (open bars). Panel B: Quantification of CL peroxidation in Gpx4+/− mice (shaded bars) and their Wt littermates (hatched bars). All values are mean ± SEM of data obtained from 4 mice. The asterisks indicate the differences that are significantly different.
To exclude the possibility that altered CL content contributed to the differences in NAO binding, we measured the levels of CL (total CL including CL, CL-OOH, and CL-OH) in the liver mitochondria. The data in Table 1 show there was no difference in the CL content between the Wt and Tg(GPX4+/0) mice treated or untreated with diquat. When comparing Gpx4+/− and Wt mice, we found that Gpx4+/− mice tended to have less CL than Wt mice; however, this difference was significant only for the untreated mice. Therefore, the changes in NAO binding in the diquat-treated Tg(GPX4+/0) and Gpx4+/− mice observed in Figure 7 were not due to differences in CL content. We conclude that diquat induced CL peroxidation without altering CL content and that Gpx4 prevented diquat-induced CL peroxidation.
Table 1.
CL content of isolated mitochondria
| nmol CL/mg mitochondrial protein |
||||
|---|---|---|---|---|
| Wt | Tg(GPX4+/0) | Wt | Gpx4+/− | |
| Control | 32.7 ± 2.1 | 31.8 ± 2.1 | 40.4 ± 2.8 | 31.9 ± 0.9* |
| Diquat | 33.5 ± 1.5 | 33.4 ± 0.4 | 42.3 ± 0.8 | 37.5 ± 0.8 |
Tg(GPX4+/0) and Gpx4+/− mice and their corresponding Wt littermates were either untreated or treated with diquat for 6 hours. Liver mitochondria were isolated, and CL content in the isolated mitochondria determined by HPLC as described in the EXPERIMENTAL PROCEDURES. All values are mean ± SEM of data obtained from 3–4 mice.
significantly different from Wt control.
Discussions
Oxidative stress is known to induce apoptosis; however, the underling mechanism is not clear. In this study, we show for the first time that the levels of Gpx4 inversely correlate with the in vivo levels of apoptosis after an oxidative stress, e.g., Tg(GPX4+/0) mice are protected from diquat-induced apoptosis whereas Gpx4+/− mice are more sensitive to diquat-induced apoptosis. Our data are in agreement with previous studies showing that the overexpression of Gpx4 in cells inhibits the induction of apoptosis. Because the major function of Gpx4 is to repair membrane lipid peoxidation, lipid peroxidation appears to play a major role in oxidative stress induced apoptosis.
Cyt.c release from mitochondria in response to oxidative stress is a key step in apoptosis. Our data show that both cyt. c and Gpx4 were primarily located at the inner mitochondrial membrane, suggesting that Gpx4 is in a cellular location that allows it to affect directly cyt. c release. In fact, we found that Tg(GPX4+/0) mice, which were resistant to the induction of apoptosis, showed reduced cyt.c release. In contrast, Gpx4+/− mice, which were more sensitive to the induction of apoptosis, showed enhanced cyt.c release.
How does Gpx4 effectively protect against cyt. c release? At the inner mitochondrial membrane, cyt. c has been shown to bind to CL with high affinity through electrostatic and hydrophobic interactions [40]. CL is a unique phospholipid localized exclusively in mitochondria that is vulnerable to free radical oxidation because of its high content (80–90%) of linoleic acid [47]. Studies in vitro and with cell culture show that oxidation of CL reduces cyt. c binding to CL, allowing cyt.c to be released from the mitochondria and to initiate apoptosis [44,45,48]. Fore example, using monolyers of CL (and other phospholipids), Nomura et al. [45] showed that autoxidation of CL to CLOOH decreases cyt. c binding and cyt. c bound to CLOH with the same affinity as CL; autoxidation of other phospholipids had no effect on their binding of cyt. c. They also observed a similar phenomenon of cyt. c binding with liposomes containing CL, CLOOH and CLOH. Interestingly, they showed that CLOOH liposomes treated with Gpx4 were able to bind cyt. c as well as non-oxidized CL liposomes. Studies with isolated mitochondria [47] or cells [44,45,48] also showed the peroxidation of CL reduced cyt. c binding, and cells overexpressing Gpx4 showed reduced CLOOH levels and reduced release of cyt. c from mitochondria. Using oxidative lipidomics to study the role of lipid peroxidation in apoptosis, Kagan et al. [49] showed that CL was the only phospholipid in the mitochondria that underwent early oxidation during the induction of apoptosis in human leukemia HL-60 cells and mouse embryonic cells. Because CL peroxidation could play a pivotal role in cyt. c release, we measured the levels of CL peroxidation in Wt mice and mice with altered levels of Gpx4. We found that diquat-induced CL peroxidation was reduced in Tg(GPX4+/0) mice and increased in Gpx4+/− mice, compared to WT mice. In other words, our data show that diquat-induced CL peroxidation in vivo was correlated to cyt. c release from mitochondria.
It also has been shown that CL peroxidation promotes mitochondrial outer membrane permeabilization (MOMP) [49], another step involved in the release of cyt. c from mitochondria. Therefore, Gpx4 could alter cyt. c release in response to diquat-treatment at the level of MOMP. To test this possibility, we measured the release of the apoptogenic factors Smac/DIABLO and Omi/HtrA2 from mitochondria because they are found primarily in the intermembrane space and their release from mitochondria is dependent upon on the MOMP [38]. We found that Gpx4 expression had no effect on diquat-induced Smac/DIABLO and Omi/HtrA2 release from mitochondria, i.e., the alterations in cyt. c release in Tg(GPX4+/0) and Gpx4+/− mice do not appear to arise from alterations in the MOMP.
Our study would suggest that mitochondrial apoptogenic proteins are not equally important in diquat-induced liver apoptosis. Our data showed that the level of cyt.c release correlated with the level of apoptosis, suggesting that cyt.c release plays a critical role in diquat-induced liver apoptosis. In contrast, it appears that role of Smac/DIABLO and Omi/HtrA2 in diquat-induced liver apoptosis is limited. This is due to the fact that the level of Smac/DIABLO and Omi/HtrA2 release did not correlate with the level of apoptosis, e.g., Tg(GPX4+/0) mice showed significantly less apoptosis while they had the same levels of Smac/DIABLO and Omi/HtrA2 release compared to Wt mice; moreover, Gpx4+/− mice had significantly more apoptosis, but they still had the same levels of Smac/DIABLO and Omi/HtrA2 release comparable to Wt mice. It would be of great interest to study further the roles of various apoptogenic proteins as well as Gpx4 in diquat-induced apoptosis using isolated hepatocyte from Tg(GPX4+/0) and Gpx4+/− mice.
Oxidative stress and lipid peroxidation have been implicated in neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease [50]. Now emerging evidence shows that Gpx4 could play a critical role in neurodegeneration. Gpx4 mRNA has been found in neurons of cortex, hippocampus and cerebellum, whereas the expression was not detected in glial cells [51]. Primary culture cortical neurons derived from Tg(GPX4+/0) mice had increased cell survival and a reduced level of apoptosis after exposure to t-butyl hydroperoxide, hydrogen peroxide, and Abeta25–35 [52]. In contrast, knocking down Gpx4 in the neuronal cells by small, interfering RNA triggered apoptosis as evidenced by caspase-3 activation [51]. Recently, Seiler et al. reported the generation of neuronal-specific Gpx4 knockout mice [53]. It was found that neuronal-specific Gpx4 deletion caused massive neurodegeneration in hippocampus and cortex. The neuronal specific Gpx4 null pups developed seizures and had to be euthanized no later than postnatal day 13. Interestingly, it appeared that Gpx4 deletion resulted in an AIF-mediated neuronal cell death that was downstream of 12/15-lipoxygenase and was independent of caspase-3 activation. Understanding the mechanism by which Gpx4 inhibits apoptosis will be valuable for designing therapeutic approaches for treating neurodegenerative diseases, where neuronal cell death plays a critical role.
In summary, our study showed that Gpx4 plays a regulatory type role in oxidative stress induced apoptosis in vivo. Based on our data, we propose that Gpx4 prevents oxidative stress-induced release of cyt. c from the inner mitochondria membrane by reducing CL peroxidation because of its unique activity (ability to reduce complex lipid hydroperoxides) and its location in the inner mitochondrial membrane. Thus, alterations in Gpx4 levels/activity would lead to changes in the ability of cells/tissues to respond to oxidative stress.
Acknowledgments
This work was supported by NIH grants P01AG19316, P01AG020591, R37 AG026557 (Richardson); the San Antonio Nathan Shock Aging Center (1P30-AG13319); and VA Merit grants (Richardson and Ran) and a REAP from the Department of Veteran Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reed JC. Mechanisms of apoptosis. Am. J. Pathol. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–698. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 3.Spierings D, McStay G, Saleh M, Bender C, Chipuk J, Maurer U, Green DR. Connected to death: the (unexpurgated) mitochondrial pathway of apoptosis. Science. 2005;310:66–67. doi: 10.1126/science.1117105. [DOI] [PubMed] [Google Scholar]
- 4.Jabs T. Reactive oxygen intermediates as mediators of programmed cell death in plants and animals. Biochem. Pharmacol. 1999;57:231–245. doi: 10.1016/s0006-2952(98)00227-5. [DOI] [PubMed] [Google Scholar]
- 5.Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423:275–280. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht H, Tschopp J, Jongeneel CV. Bcl-2 protects from oxidative damage and apoptotic cell death without interfering with activation of NF-kappa B by TNF. FEBS Lett. 1994;351:45–48. doi: 10.1016/0014-5793(94)00817-5. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Ruiz C, Colell A, Mari M, Morales A, Fernandez-Checa JC. Direct effect of ceramide on the mitochondrial electron transport chain leads to generation of reactive oxygen species. Role of mitochondrial glutathione. J. Biol. Chem. 1997;272:11369–11377. doi: 10.1074/jbc.272.17.11369. [DOI] [PubMed] [Google Scholar]
- 8.Coyle JT, Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science. 1993;262:689–695. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- 9.Loo DT, Copani A, Pike CJ, Whittemore ER, Walencewicz AJ, Cotman CW. Apoptosis is induced by beta-amyloid in cultured central nervous system neurons. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7951–7955. doi: 10.1073/pnas.90.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majima HJ, Oberley TD, Furukawa K, Mattson MP, Yen HC, Szweda LI, St Clair DK. Prevention of mitochondrial injury by manganese superoxide dismutase reveals a primary mechanism for alkaline-induced cell death. J. Biol. Chem. 1998;273:8217–8224. doi: 10.1074/jbc.273.14.8217. [DOI] [PubMed] [Google Scholar]
- 11.Atabay C, Cagnoli CM, Kharlamov E, Ikonomovic MD, Manev H. Removal of serum from primary cultures of cerebellar granule neurons induces oxidative stress and DNA fragmentation: protection with antioxidants and glutamate receptor antagonists. J. Neurosci. Res. 1996;43:465–475. doi: 10.1002/(SICI)1097-4547(19960215)43:4<465::AID-JNR7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brigelius-Flohe R, Aumann KD, Blocker H, Gross G, Kiess M, Kloppel KD, Maiorino M, Roveri A, Schuckelt R, Usani F. Phospholipid-hydroperoxide glutathione peroxidase. Genomic DNA, cDNA, and deduced amino acid sequence. J. Biol. Chem. 1994;269:7342–7348. [PubMed] [Google Scholar]
- 14.Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic. Biol. Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 15.Antunes F, Salvador A, Pinto RE. PHGPx and phospholipase A2/GPx: comparative importance on the reduction of hydroperoxides in rat liver mitochondria. Free Radic. Biol. Med. 1995;19:669–677. doi: 10.1016/0891-5849(95)00040-5. [DOI] [PubMed] [Google Scholar]
- 16.Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 17.Arai M, Imai H, Koumura T, Yoshida M, Emoto K, Umeda M, Chiba N, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J. Biol. Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- 18.Imai H, Sumi D, Sakamoto H, Hanamoto A, Arai M, Chiba N, Nakagawa Y. Overexpression of phospholipid hydroperoxide glutathione peroxidase suppressed cell death due to oxidative damage in rat basophile leukemia cells (RBL-2H3) Biochem. Biophys. Res. Commun. 1996;222:432–438. doi: 10.1006/bbrc.1996.0762. [DOI] [PubMed] [Google Scholar]
- 19.Yagi K, Komura S, Kojima H, Sun Q, Nagata N, Ohishi N, Nishikimi M. Expression of human phospholipid hydroperoxide glutathione peroxidase gene for protection of host cells from lipid hydroperoxide-mediated injury. Biochem. Biophys. Res. Commun. 1996;219:486–491. doi: 10.1006/bbrc.1996.0260. [DOI] [PubMed] [Google Scholar]
- 20.Brigelius-Flohe R, Maurer S, Lotzer K, Bol G, Kallionpaa H, Lehtolainen P, Viita H, Yla-Herttuala S. Overexpression of PHGPx inhibits hydroperoxide-induced oxidation, NFkappaB activation and apoptosis and affects oxLDL-mediated proliferation of rabbit aortic smooth muscle cells. Atherosclerosis. 2000;152:307–316. doi: 10.1016/s0021-9150(99)00486-4. [DOI] [PubMed] [Google Scholar]
- 21.Wang HP, Qian SY, Schafer FQ, Domann FE, Oberley LW, Buettner GR. Phospholipid hydroperoxide glutathione peroxidase protects against singlet oxygen-induced cell damage of photodynamic therapy. Free Radic. Biol. Med. 2001;30:825–835. doi: 10.1016/s0891-5849(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 22.Hurst R, Korytowski W, Kriska T, Esworthy RS, Chu FF, Girotti AW. Hyperresistance to cholesterol hydroperoxide-induced peroxidative injury and apoptotic death in a tumor cell line that overexpresses glutathione peroxidase isotype-4. Free Radic. Biol. Med. 2001;31:1051–1065. doi: 10.1016/s0891-5849(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 23.Nomura K, Imai H, Koumura T, Arai M, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase suppresses apoptosis mediated by a mitochondrial death pathway. J. Biol. Chem. 1999;274:29294–29302. doi: 10.1074/jbc.274.41.29294. [DOI] [PubMed] [Google Scholar]
- 24.Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 25.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, Nakae S, Terry RD, Mokhtari GK, Gunawan F, Balsam LB, Kaneda H, Kofidis T, Tsao PS, Robbins RC. Cardiomyocyte-specific Bcl-2 overexpression attenuates ischemia-reperfusion injury, immune response during acute rejection, and graft coronary artery disease. Blood. 2004;104:3789–3796. doi: 10.1182/blood-2004-02-0666. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Fujimura M, Morita-Fujimura Y, Noshita N, Sugawara T, Kawase M, Chan PH. The cytosolic antioxidant copper/zinc-superoxide dismutase prevents the early release of mitochondrial cytochrome c in ischemic brain after transient focal cerebral ischemia in mice. J. Neurosci. 2000;20:2817–2824. doi: 10.1523/JNEUROSCI.20-08-02817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnaitman C, Greenawalt JW. Enzymatic properties of the inner and outer membranes of rat liver mitochondria. J. Cell Biol. 1968;38:158–175. doi: 10.1083/jcb.38.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 31.Petit JM, Maftah A, Ratinaud MH, Julien R. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 1992;209:267–273. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- 32.Kirkinezos IG, Bacman SR, Hernandez D, Oca-Cossio J, Arias LJ, Perez-Pinzon MA, Bradley WG, Moraes CT. Cytochrome c association with the inner mitochondrial membrane is impaired in the CNS of G93A-SOD1 mice. J. Neurosci. 2005;25:164–172. doi: 10.1523/JNEUROSCI.3829-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson J, Duchen MR, Heales SJ. Intracellular distribution of the fluorescent dye nonyl acridine orange responds to the mitochondrial membrane potential: implications for assays of cardiolipin and mitochondrial mass. J. Neurochem. 2002;82:224–233. doi: 10.1046/j.1471-4159.2002.00945.x. [DOI] [PubMed] [Google Scholar]
- 34.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 35.Nissen HP, Kreysel HW. Analysis of phospholipids in human semen by high-performance liquid chromatography. J. Chromatogr. 1983;276:29–35. doi: 10.1016/s0378-4347(00)85062-8. [DOI] [PubMed] [Google Scholar]
- 36.Mira D, Brunk U, Boveris A, Cadenas E. One-electron transfer reactions of diquat radical to different reduction intermediates of oxygen. Formation of hydroxyl radical and electronically excited states. Free Radic. Biol. Med. 1988;5:155–163. doi: 10.1016/0891-5849(88)90078-0. [DOI] [PubMed] [Google Scholar]
- 37.Smith CV. Evidence for participation of lipid peroxidation and iron in diquat-induced hepatic necrosis in vivo. Mol. Pharmacol. 1987;32:417–422. [PubMed] [Google Scholar]
- 38.Uren RT, Dewson G, Bonzon C, Lithgow T, Newmeyer DD, Kluck RM. Mitochondrial release of pro-apoptotic proteins: electrostatic interactions can hold cytochrome c but not Smac/DIABLO to mitochondrial membranes. J. Biol. Chem. 2005;280:2266–2274. doi: 10.1074/jbc.M411106200. [DOI] [PubMed] [Google Scholar]
- 39.Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE. Dual specificity phosphatases 18 and 21 target to opposing sides of the mitochondrial inner membrane. J. Biol. Chem. 2008;283:15440–15450. doi: 10.1074/jbc.M709547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berezhna S, Wohlrab H, Champion PM. Resonance Raman investigations of cytochrome c conformational change upon interaction with the membranes of intact and Ca2+-exposed mitochondria. Biochemistry. 2003;42:6149–6158. doi: 10.1021/bi027387y. [DOI] [PubMed] [Google Scholar]
- 41.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 42.Milon L, Meyer P, Chiadmi M, Munier A, Johansson M, Karlsson A, Lascu I, Capeau J, Janin J, Lacombe ML. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 43.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kriska T, Korytowski W, Girotti AW. Role of mitochondrial cardiolipin peroxidation in apoptotic photokilling of 5-aminolevulinate-treated tumor cells. Arch. Biochem. Biophys. 2005;433:435–446. doi: 10.1016/j.abb.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 45.Nomura K, Imai H, Koumura T, Kobayashi T, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem. J. 2000;351:183–193. doi: 10.1042/0264-6021:3510183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asumendi A, Morales MC, Alvarez A, Arechaga J, Perez-Yarza G. Implication of mitochondria-derived ROS and cardiolipin peroxidation in N-(4-hydroxyphenyl)retinamide-induced apoptosis. Br. J. Cancer. 2002;86:1951–1956. doi: 10.1038/sj.bjc.6600356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrosillo G, Ruggiero FM, Paradies G. Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria. FASEB J. 2003;17:2202–2208. doi: 10.1096/fj.03-0012com. [DOI] [PubMed] [Google Scholar]
- 48.Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 50.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat. Med. 2004;10 Suppl:S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 51.Savaskan NE, Borchert A, Brauer AU, Kuhn H. Role for glutathione peroxidase-4 in brain development and neuronal apoptosis: specific induction of enzyme expression in reactive astrocytes following brain injury. Free Radic. Biol. Med. 2007;43:191–201. doi: 10.1016/j.freeradbiomed.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 52.Ran Q, Gu M, Van Remmen H, Strong R, Roberts JL, Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J. Neurosci. Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- 53.Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]