Abstract
A family of generation one, two and three triazine dendrimers differing in their core flexibility was prepared and evaluated for their ability to accomplish gene transfection. Dendrimers and dendriplexes were analyzed by their physicochemical and biological properties such as condensation of DNA, size, surface charge, morphology of dendriplexes, toxic and hemolytic effects and ultimately transfection efficiency in L929 and MeWo cells. Flexibility of the backbone was found to play an important role with generation 2 dendrimer displaying higher transfection efficiencies than 25 kDa poly(ethyleneimine) or SuperFect™ at a lower cytotoxicity level. This result is surprising as PAMAM dendrimers require generations 4 or 5 to become effective transfection reagents. The ability to delineate effects of molecular structure and generation of triazine dendrimers with biological properties provides valuable clues for further modifying this promising class of non-viral delivery systems.
Keywords: Triazine dendrimers, PAMAM, transfection efficiency, structure function relationship, cytotoxicity, physico-chemical characterization
Introduction
Gene therapy has gained importance in recent years due to its potential for targeting genetic diseases and inhibiting tumor growth. However, clinical application is limited by numerous biological barriers to delivering plasmid DNA (1). Both viral and non-viral vector systems have been developed to overcome these hurdles. Viral delivery systems, which include retroviruses, lentiviruses, and adenoviruses, have encountered potential safety problems such as recombination, insertional mutagenesis, and immunogenicity as well as scale-up difficulties (2). Therefore, synthesis and evaluation of non-viral vector systems becomes increasingly important in spite of their comparatively lower transfection efficiency (3).
Non-viral vectors such as cationic liposomes, linear and branched polycations and dendrimers have been shown to be effective transfection agents and offer advantages over viral vectors regarding safety and manufacturing. Several non-viral gene delivery systems based on poly(amidoamine) (PAMAM) (4–6), poly(propylene imine) (PPI) (7, 8), and poly(ethylene imine) (PEI) (9, 10) have yielded efficient transfection in vivo with relatively low toxicity. The potential of triazine dendrimers as gene delivery systems is largely unexplored, but has been described by us in a study involving generation 2 rigid structures (11), which reveal an optimized peripheral group for further study. These dendrimers may be attractive candidates as they can be synthesized in high yields via a divergent strategy that creates multiple surface functionalities in a completely monodisperse manner (12). In addition, triazine dendrimers show low toxicity, both in vitro and in vivo: mice tolerate i.p. doses up to 40 mg/kg (13). This level of toxicity is comparable to PAMAM (14) suggesting that triazine dendrimers could be considered as a platform for transfection studies. Here, the physicochemical properties, cytotoxicities and transfection efficiencies of a panel of triazine dendrimers were evaluated to establish structure function relationships. The effect of dendrimer size (generations) as well as core structure is probed in an attempt to maximize transfection efficiency. Increasing generation increases size and the number of available cations. Changing the composition of the core affords dendrimers that differ in the flexibility, a characteristic that is thought to influence interactions with p-DNA (6). The dendrimers used in this study are shown in Figure 1. Here we present results from the synthesis, physicochemical and biological in vitro evaluation of a panel of 5 triazine dendrimers and compare them to commercially available PAMAM dendrimers of different generations to define structural parameters such as generation and core flexibility for the design of triazine dendrimers as non-viral gene delivery systems.
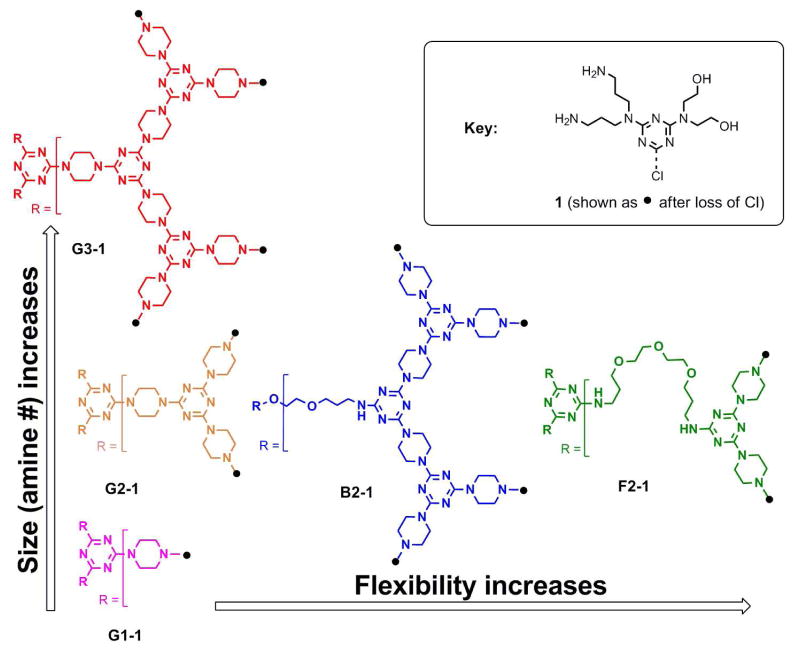
Figure 1.
The dendrimer panel is given as chemical structures and sketches in a color code used throughout the study. Structure activity relationships of the dendrimers in Table 1 were investigated in a two-dimensional approach: By variation of the generation (vertical arrows) and the core structure (horizontal arrows), influence of both parameters were investigated. 123×101mm (300 × 300 DPI)
Experimental Section
Materials
Poly(ethylene imine) (Polymin™, 25 kDa) was a gift from BASF (Ludwigshafen, Germany), PAMAM (ethylenediamine core, amino surface) 2nd and 3rd generation was bought from Sigma-Aldrich Laborchemikalien GmbH, Seelze, Germany and SuperFect™ from Qiagen, Hilden, Germany. All chemicals used for synthesis were obtained from Sigma-Aldrich (St. Louis, MO).
Synthesis
The synthesis of all dendrimers used in this study proceeded via a divergent strategy originating from cyanuric chloride. The rigid, first generation dendrimer was synthesized by reacting a tris(piperazyl) triazine core with MCT1, followed by deprotection. The rigid, second generation dendrimer, G2-1, was synthesized as previously described (11). To form the third generation rigid dendrimer, G3-1, the rigid, deprotected G1-dendrimer, G1-C was reacted with di(Boc-piperazyl)monochlorotriazine. This second generation structure was deprotected and reacted with MCT1 to form the protected third generation compound, which was deprotected using HCl (Supplementary Materials). A detailed description of the synthesis is provided in the supplementary material section.
The flexible core dendrimer, F2-1, was synthesized by reacting cyanuric chloride with mono-Boc-protected O,O′-bis(3-aminopropyl)diethylene glycol. This core was deprotected with HCl, reacted with Boc-piperazyl-dichlorotriazine, and capped with Boc-piperazine. After removal of the protecting groups with HCl, the compounds were reacted with MCT1 and deprotected to form the flexible, second generation dendrimer, F2-1 (Supplementary Materials). A detailed description of the synthesis is provided in the supplementary material section.
Finally, the second generation bow-tie structure, B2-1, was synthesized by reacting di(Boc-piperazyl)monochlorotriazine with O,O′-bis(3-aminopropyl)diethylene glycol. This core structure was deprotected with HCl and reacted with di(Boc-piperazyl)monochlorotriazine to form a first generation bow-tie structure. This structure was deprotected, reacted with MCT1, and deprotected again to afford the dendrimer B2-1 (Supplementary Materials). A detailed description of the synthesis is provided in the supplementary material section.
Cell Culture
L929 murine fibroblasts and MeWo human melanoma cells were purchased from LG Promochem, Wesel, Germany, and maintained in DMEM low or high glucose (PAA Laboratories, Cölbe, Germany), respectively, and supplemented with 10% fetal calf serum (Cytogen, Sinn, Germany) in humidified atmosphere with 5% CO2 at 37°C.
Preparation of Dendriplexes
Dendriplexes were formed by adding 25 μl of a calculated concentration depending on “protonable unit” and N/P ratio) of dendrimer to an equal volume of the DNA solution (pCMV-Luc, Plasmid Factory, Bielefeld, Germany) and then mixing the two components before letting them mature for 20 minutes. The final concentration of pDNA in the dendriplex solution was 0.02 μg/μl. Both pDNA and dendrimers were diluted with 10 mM HEPES buffer, pH 7.4, and the appropriate amount of dendrimer was calculated by considering the desired N/P ratio and the “protonable unit” of each dendrimer, which represents the mass of dendrimer per protonable nitrogen atom. For comparison, standard transfection reagents, including poly(ethylene imine) (PEI 25 kDa), second and third generation PAMAM (ethylenediamine core, amino surface), and SuperFect (Qiagen, Hilden, Germany) were used. Polyplexes with PEI 25 kDa and dendriplexes with PAMAMs of 2nd and 3rd generation were formed as described above, whereas dendriplexes with SuperFect were prepared as recommended by the manufacturer.
Ethidium Bromide Quenching Assay
To quantify condensation efficiency of the dendrimers, an indirect dye-quenching assay was performed as previously reported (15). Briefly, 4 μg of herring testes DNA (Sigma-Aldrich Chemie GmbH, Schnelldorf, Germany) was complexed with increasing amounts of dendrimer in a final volume of 280 μl 10 mM HEPES buffer. After incubation for 20 minutes at room temperature, 20 μl of a 0.1 mg/ml ethidium bromide solution (Carl Roth GmbH, Karlsruhe, Germany) was added and incubated for 20 minutes in the dark following intensive mixing. Intercalation-caused fluorescence was quantified using a fluorescence plate reader (LS 50 B, Perkin-Elmer, Rodgau-Jügesheim, Germany) at 518 nm excitation and 605 nm emission wavelengths. Results are given as relative fluorescence intensity values where intercalation of free DNA represents 100% fluorescence and non-intercalating ethidium bromide in buffer represents 0% remaining fluorescence. Results are given as means of triplicate measurements ± standard deviation (SD).
Dynamic Light Scattering and Zeta Potential Analysis
Polyplex size and zeta potential were determined as previously reported (15). Briefly, dendriplexes of 0.6 μg pCMV-Luc and the corresponding amount of dendrimer were prepared as described above. Complexes were incubated for 20 minutes and then diluted to a total volume of 800 μl with 10 mM HEPES buffer, pH 7.4, to be measured using a Zetasizer Nano ZS, Malvern, Herrenberg, Germany. Position and attenuator were optimized by the device. The accuracy of the size measurements was routinely checked using reference polymer particles (Nanosphere Size Standards, 50, 100 and 200 nm, Duke Scientific Corp., Palo Alto, CA, USA), and the zeta potential values were calibrated with the Zeta Potential Transfer Standard (Malvern Instruments, Herrenberg, Germany). The data was analyzed using a High Resolution Mulitmodal algorithm. All measurements were conducted in triplicates. For better characterization of the size distributions, in addition to Z Average, the PDI of the distribution is given as the mean value of three independent measurements ± SD.
Atomic Force Microscopy
The size and morphology of dried dendriplexes were analyzed by atomic force microscopy as previously described for polyplexes (16). Briefly, dendriplexes were prepared by mixing 1 μg pCMV-Luc and a dendrimer at an N/P ratio of 5 as described above, incubating for 20 minutes, and drying on silicon chips. Microscopy was performed on a NanoWizard (JPK instruments, Berlin, Germany). Commercial NSC16 AlBS-tips (Micromasch, Estonia) on an I-type cantilever with a length of 230 μm and a nominal force constant of 40 N/m were used. All measurements were performed in tapping mode to avoid damage to the sample surface. The scan frequency was 0.6 Hz and scan size was 5×5 μm. Multiple images were recorded, of which one representative topograph is presented.
Erythrocyte Aggregation and Hemolysis
Unspecific cell interaction was visualized by erythrocyte aggregation and quantified by the measurement of hemolysis according to previously described procedures (17). Briefly, human erythrocytes were isolated from fresh citrated blood from healthy volunteers by centrifugation, washed with PBS buffer, and diluted to 500.000.000 cells/ml. Dendriplexes of 0.1 μg pDNA per ml were prepared at N/P 5, and 50 μl aliquots of dendriplex solutions, 1 % Triton-X 100 (100 % lysis) or PBS (0 % lysis) were mixed with 50 μl erythrocyte suspension. After incubation for 30 minutes at 37 °C, samples were centrifuged at 850 g. Hemolysis was measured by VIS-absorption of supernatants at 541 nm and the data is given as a mean percentage (n = 3) of the released hemoglobin ± SD, normalized to total hemolysis induced by 1% Triton-X 100. Pelleted erythrocytes were resuspended in 50 μl PBS and microscopically examined to determine erythrocyte aggregation.
Cell viability
The cytotoxicity of the dendrimers was investigated using an MTT assay (15). Murine L929 fibroblasts were seeded on 96-well plates at a density of 8000 cells per well approximately 24 hours before they were treated with dendrimer solutions of increasing concentrations in the range of 0.001 to 1 mg/ml. Another 24 hrs later, the medium was changed. MTT solution was added into fresh serum-free medium and incubated for 4 hours. The remaining mitochondrial enzyme activity as compared to untreated cells was determined by measuring the absorbance of enzymatically-formed formazane at 580 nm with 690 nm background corrections after cell lysis in 200 μl DMSO. The results are given as mean values of a replicate of 4 ± SD.
Transfection Experiments
To compare transfection efficiency in different cell lines, murine 3T3 fibroblasts (L929) and human melanoma cells (MeWo) were seeded in 96-well plates at a seeding density of 8000 cells per well. Approximately 24 hours later, the cells were transfected with dendriplexes consisting of 0.25 μg pCMV-Luc per well and the amount of dendrimer calculated to be N/P 2.5, 5 or 7.5. Branched PEI of 25 kDa, second and third generation PAMAM, and 4th generation SuperFect were used as standards and free plasmid was used as a negative control.
Polyplexes were added to full serum-containing medium, and cells were incubated for 4 hours before the medium was changed. Another 44 hours later, the cells were washed with PBS, lysed with CCLR (Promega, Mannheim, Germany), and assayed for luciferase expression using a commercial luciferase assay kit (Promega, Mannheim, Germany) on a BMG LUMIstar OPTIMA luminometer plate reader (BMG Labtech, Offenburg, Germany). Transfection assays were performed in triplicates and the mean values ± SD are given.
Results and Discussion
Nomenclature adopted
Throughout the manuscript, the targets are named to reflect common and disparate features in structure with the core/generation-surface groups identified. The cores increase in flexibility from G (standard) to B (bowtie) to F (flexible), and the effect of size is determined over three (1, 2, 3) generations using the standard core structure. Each dendrimer contains a common surface group (1) that was previously reported to avoid obtaining variations in transfection efficiency that result from the peripheral functionalities (11).
Synthesis and design criteria
As the products of multistep organic syntheses, triazine dendrimers allow us access to well-defined structures to investigate structure-activity relationships such as the influence of size and backbone structure on transfection efficiency and cytotoxicity. For this purpose, a preliminary panel of dendrimers was synthesized consisting of different generations (G1, G2, G3) with a rigid backbone and a common surface group (1), yielding 6, 12 or 24 amino-groups in the periphery. With each increase in generation number, the number of primary amino groups presented in the periphery doubles.
In addition to generation number, the flexibility of the core could hypothetically affect transfection efficiency by influencing the distribution and accessibility of the peripheral groups for interacting with p-DNA (18). The role of flexibility has been supported experimentally by Szoka who showed enhanced transfection efficiency resulting from fragmentation of PAMAM dendrimers (6). In this study, this effect was investigated with second generation triazine dendrimers, each having differing core flexibilities (G2-1, F2-1, B2-1). Synthetic yields and NMR data are provided within the Supporting Information.
The composition of the panel is summarized in Figure 1 and Table 1. Branched poly(ethylene imine) of 25 kDa (PEI 25kDa) was used as a control. The transfection efficiencies of the triazine dendriplexes were additionally compared to SuperFect™, a commercially available 4th generation fractured PAMAM dendrimer and to second and third generation PAMAM as dendrimer standards.
Table 1.
Summary of properties of the screened dendrimers concerning in vitro parameters 162×88mm (300 × 300 DPI)
| Generation-1 rigid | Generation-2 rigid | Generation-3 rigid | Generation-2 flexible | Generation-2 bow-tie | |
|---|---|---|---|---|---|
| Primary amines | 6 | 12 | 24 | 12 | 16 |
| Amine density [Da/N atom] | 211.27 | 245.30 | 262.20 | 262.32 | 262.20 |
| DNA condensation at plateau [%] | 48.0 | 30.2 | 4.2 | 36.4 | 5.9 |
| Size [nm] | 338.3 | 154.8 | 230.2 | 426.3 | 882.0 |
| Zeta potential [mV] | −0.05 | 22.6 | 17.7 | −7.5 | 3.1 |
| Hemolysis [%] | 12.6 | 10.9 | 12.9 | 6.11 | 13.0 |
| Toxicity (IC50) [μg/ml] | 501.2 | 64.3 | 20.1 | 22.8 | 115.3 |
| Transfection efficiency (N/P 5, MeWo) [RLU/s] | 2480.0 | 4948.3 | 7087.3 | 44184.0 | 4487.0 |
Condensation properties of triazine dendrimers
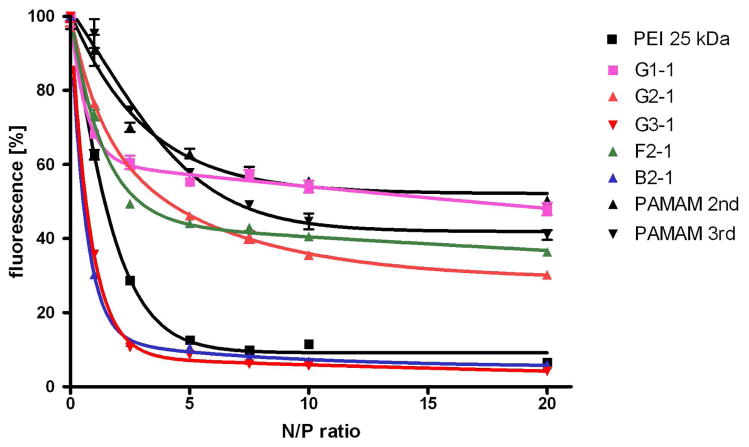
The formation of dendriplexes results from electrostatic interactions between polycationic dendrimers and negatively charged DNA. The quantification of condensation properties was performed using an ethidium bromide quenching assay (15) which reveals the fraction of uncomplexed DNA that is accessible for intercalation with ethidium bromide at a given N/P ratio. PAMAM dendrimers of generation 2 and 3 showed significant residual fluorescence leading to a plateau around N/P 7.5 (compare Figure 2). A similar trend was seen for several triazine dendrimers, including G1-1, G2-1 and F2-1. The third generation triazine dendrimer, G3-1, yielded higher condensation than PEI, even at low N/P ratios. Interestingly, the bow-tie dendrimer B2-1 showed better DNA condensation than PEI, with a condensation profile comparable to the third generation rigid dendrimer G3-1.
Figure 2.
DNA condensation efficiency was quantified by ethidium bromide quenching assays and compared to PAMAMs of the same generations and PEI 25 kDa. 169×105mm (300 × 300 DPI)
Similar to PAMAM, triazine dendrimers showed an increase in condensation efficiency with increasing generation. The first generation dendrimer G1-1 condensed DNA less effectively than the second generation analogue, G2-1, which in turn was less efficient than the third generation dendrimer, G3-1. However, the effect of the core structure on condensation efficiency seems minimal. This may result from the type of amines present in the triazine core. The reason that more flexible dendrimers are thought to complex with DNA more effectively than rigid analogues has been attributed to the increased capacity of flexible dendrimers to interact with DNA, as all amine groups, both inside the core and at the surface, contributed to the electrostatic interactions for binding (19). This increased availability of internalized amines in flexible structures results in the formation of more compact complexes (7, 20, 21). However, in the case of triazine dendrimers, the nitrogen atoms in the core are part of a delocalized electron system and hence not available for protonation under physiologic conditions (pH 7.4). Therefore, only the primary amino functional groups at the dendrimer periphery contribute to DNA condensation due to their accessibility to protonation (22). As the perhipheral groups are identical and the triazine core amines remain unprotonated at physiological pH, the trend of increased condensation efficiency for more flexible structures is not apparent; the semi-flexible bow-tie dendrimer B2-1 more effectively condenses DNA than either the rigid G2-1 or flexible F2-1 analogs.
Dendriplex sizes and surface charges
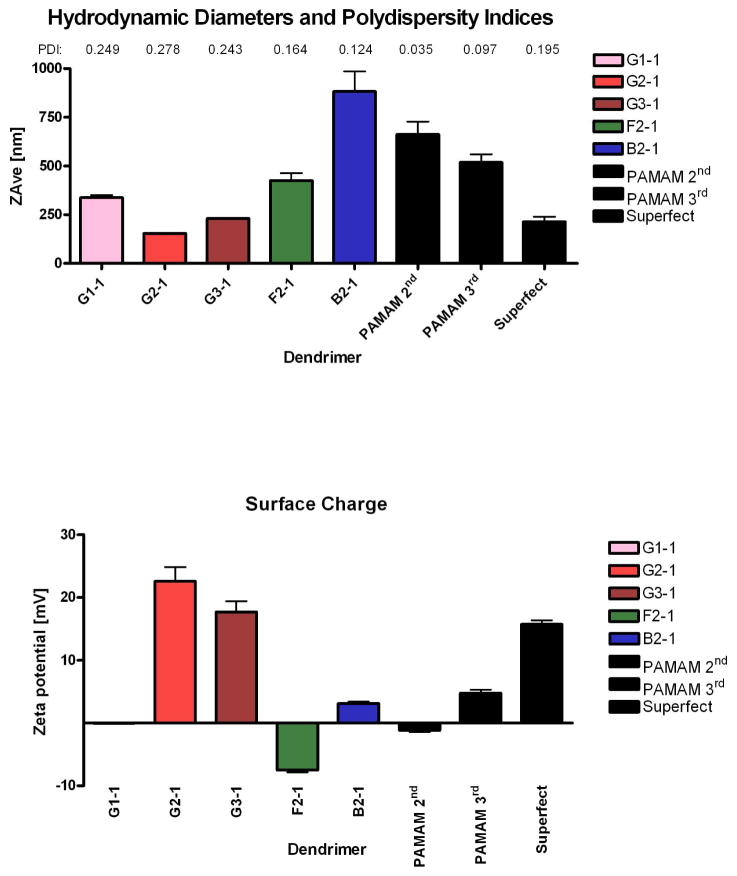
Polyplexes with diameters < 200 nm in size are internalized by most cell lines (23), while larger particles can sediment faster leading to enhanced transfection efficiency under in vitro conditions (24). A positive surface charge generally facilitates endocytosis, whereas negatively charged particles are often repelled by the negatively charged cell surface. Previous studies have shown that the complexation of DNA with PEI decreases polyplex sizes for increasing N/P ratios up to 20 (25). This trend could not be observed for the triazine dendrimers (data not shown). In the case of triazine dendriplexes, there seems to be an optimal N/P ratio at which minimal sizes can be achieved. When this N/P ratio is exceeded, larger dendriplexes (and possibly aggregates) form. By comparing rigid G1-1, G2-1 and G3-1 dendrimers (Figure 3A), no clear trend between dendriplex sizes and the generation number emerges. Generally, dendrimers with a higher number of primary amines on the periphery tend to form smaller polyplexes with DNA, as shown for PAMAM in this study. This behavior is not observed for the rigid triazine dendrimers, G2-1, G2-1, and G3-1. Our findings indicated that the rigid second generation dendrimer, G2-1, forms the smallest complexes. This may be due to the “nitrogen density” (26) or “charge density” (27, 28), a parameter that is known to influence the interaction of polycations with pDNA. This parameter is defined as the density of protonable amines in the polycation per molecular weight and can also be called the “protonable unit”. The protonable unit can therefore be measured in g/mol N and helps to characterize polycations. The charge density of PEI, where every third atom is a nitrogen atom, is 43.1 g/mol N (29), which is a comparably high charge density considering that the charge density in our triazine dendrimers is about 200 g/mol N: G1-1 has a density of primary amines of 211.27 Da/N atom, G2-1 has a density of 245.3 Da/N atom, and G3-1 has a density of 262.2 Da/N atom. Therefore, G2-1 may form smaller complexes as compared to G3-1 because of the difference in charge density. However, despite the lower charge density of G2-1, G2-2 formed smaller complexes, which can be explained by the twofold increase in primary amines per molecule. The formation of large dendriplexes using the flexible dendrimer, F2-1 and the bow-tie dendrimer, B2-1, may result partially from the low charge densities of these dendrimers (F2-1: 262.32 Da/N atom; B2-1: 262.2 Da/N atom). These values may be too low for the formation of very small complexes. Additionally, the size of dendriplexes formed with the flexible dendrimer, F2-1, were much larger than those of G2-1, suggesting that loose structures were obtained, in accordance with data from ethidium bromide quenching assay. The bow-tie dendrimer B2-1 formed the largest dendriplexes despite its DNA condensation efficiency, possibly caused by the formation of inter-dendriplex aggregates (7). The dendriplex size distributions were monomodal but showed polydispersity indices around 0.2 (Figure 3A) as reported previously for dendriplexes (30–32).
Figure 3.
Hydrodynamic diameters and polydispersity indices (PDI) (A) and zeta potentials (B) of various dendriplexes at an N/P of 5 were determined in 10 mM HEPES buffer, pH 7.4. 180×213mm (300 × 300 DPI)
Zeta potentials of the PAMAM dendriplexes generally increased as particle size decreased. For example, the PAMAM 2nd generation dendrimer did not condense DNA efficiently as illustrated by a negative zeta potential and large complexes sizes. As the number of primary amines on the periphery (generation) increases, the PAMAM dendrimers condensed DNA more efficiently (Figure 2) due to their very high number of primary amines (PAMAM G4 = SuperFect™: 64 surface amines) and charge density (PAMAM G4: 222.10 Da/N atom). The rigid triazine dendrimers (G1-1, G2-1, and G3-1) did not show a clear generation-dependent trend for surface charge. G1-1 exhibited a zeta potential of −0.0513 mV. The zeta potential of G2-1 dendriplexes, which had proven to form the smallest particles among the rigid dendrimers, was observed to be strongly positive. By further increasing the generation of the rigid dendrimer, not only sizes increased (Figure 3A), but also the zeta potential decreased (Figure 3B). Taken together, third generation dendrimers condense and protect DNA (Figure 2) efficiently, but may result in larger aggregates in solution. It is possible that, due to their rather low charge density at the same N/P ratio and molecular composition, G3-1 dendrimers do not shield the negative charge of DNA effectively, which results in lower electrostatic repulsion and stronger aggregation of dendriplexes. According to the data acquired in the ethidium bromide assay (Figure 2B), the flexible dendrimer F2-1 only loosely bound DNA and was not able to fully protect the complex. Therefore, it seems reasonable to assume that the net negative charge measured for F2-1 dendriplexes was caused by segments of free DNA which were not incorporated into any particle but were only bound to the surface of the flexible polycation. The dendriplexes formed with the bow-tie dendrimer B2-1 exhibited a low but positive surface charge, which can again be explained by inter-dendriplex interaction.
Atomic Force Microscopy
Although dynamic light scattering is an effective method for measuring particle size, the method is limited because it assumes spherically shaped particles. Therefore atomic force microscopy (AFM) was used to characterize the morphology and heterogeneity of dendriplexes (33). The broad polydispersities observed with the dynamic light scattering experiments was also reflected in the AFM micrographs (Figure 4). The flexible core dendrimer F2-1, produced two populations or particles: very small complexes (<50 nm) as well as larger ones (>100 nm). The bow-tie dendrimer, B2-1, formed clusters of aggregates, which supports the assumption of inter-dendriplex interactions (7). The third generation dendrimer G3-1 showed the most uniform distribution.
Figure 4.
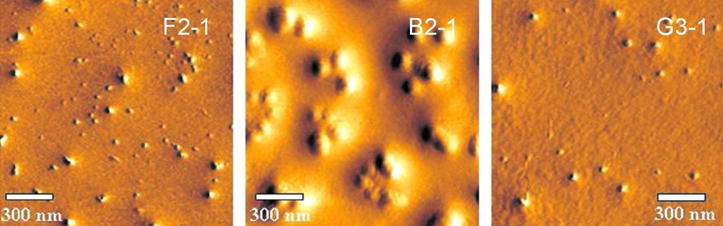
Morphology and density of dendriplexes were investigated by AFM revealing spherically shaped nanoplexes. Differing polyplex sizes as measured by dynamic light scattering are probably due to sample preparation (drying). 159×54mm (300 × 300 DPI)
Both the formation of clusters and the discrepancy between size measurement of dried particles and complexes in solution are not surprising: Similar observations have been made for PAMAM dendriplexes (21). The sizes measured for F2-1 and G3-1 dendriplexes by DLS are confirmed by AFM. In AFM, a distribution of 20–100 nm (F2-1) and narrow size distribution of particles of 100 nm (G3-1) could be observed. The larger sizes measured by DLS can be explained by the high number of short ethylene glycol chains that might be hydrated in solution but carry no weight after drying the sample for AFM. The picture of the B2-1 dendriplex-clusters confirmed the DLS determined size of ~900.
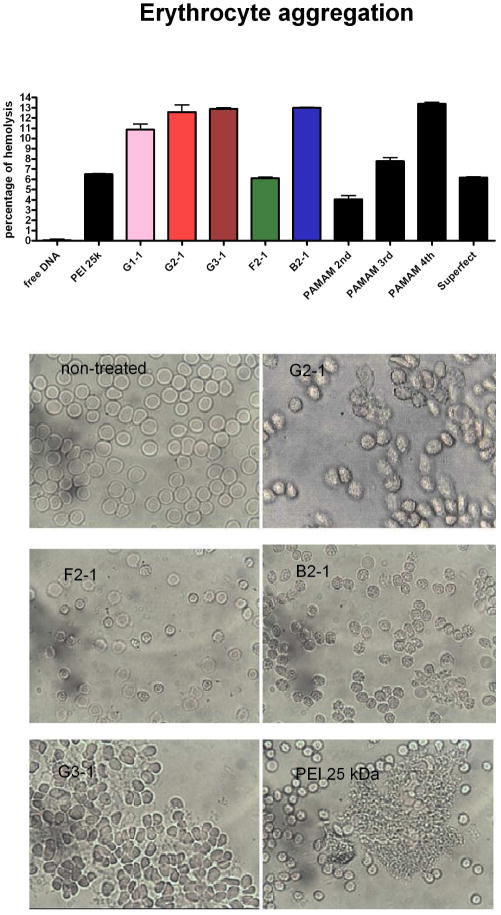
Erythrocyte Aggregation and Hemolysis
Red blood cells can be used to visualize and quantify the membrane interactions of dendriplexes by determining erythrocyte aggregation or hemolysis. Positive surface charges can lead to unspecific binding, membrane interactions and the cytotoxicity associated with amino groups (34). The concentration dependent hemolysis and perturbation of red blood cells after incubation with PAMAM, diaminobutane (DAB), diamonoethane (DAE) and poly(ethylene oxide) (PEO) grafted carbosilane (CSi–PEO) was reported previously (35). Generation-dependent hemolysis similar to PAMAM was seen with triazine dendrimers as well, though not as pronounced (Figure 5A). The hemolytic activity of triazine dendrimers seems to depend on their core structure, as hemolysis of the flexible dendrimer, F2-1, was markedly decreased compared to the rigid analogue, G2-1. However, dendriplexes formed from the bow-tie dendrimer, B2-1, caused hemolytic effects comparable to G3-1 dendriplexes. A possible explanation is the strong aggregation of B2-1 dendriplexes, which could facilitate electrostatic interactions with cells. The extent of hemolytic activity of a dendriplex was clearly shown to be influenced by the zeta potential.
Figure 5.
(A) Hemolysis after incubation with dendriplexes at an N/P of 5 was UV-metrically quantified and (B) morphologic changes of erythrocytes were microscopically captured. Hemolysis depended strongly on the core structure, whereas erythrocyte aggregation increased by generation. 157×274mm (300 × 300 DPI)
Cytotoxicity
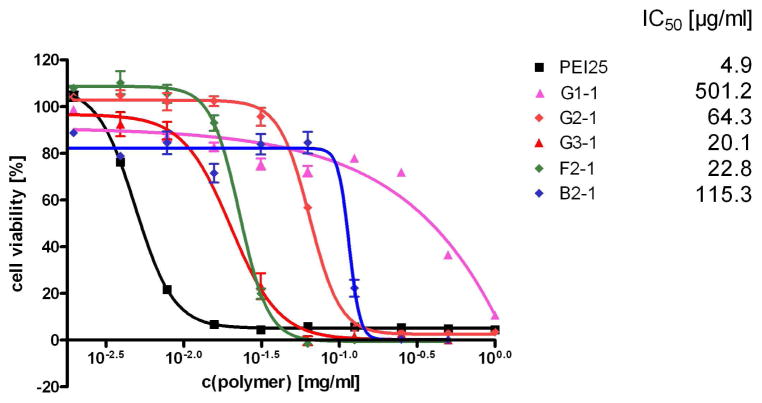
Cell viability or cytotoxicity can routinely be measured by quantification of mitochondrial enzyme activity using an MTT assay. In order to mimic the “worst case scenario”, murine fibroblasts (L929) were incubated with various concentrations of free dendrimer, ranging from 0.001 to 1 mg/ml (Figure 6). The half enzyme-inhibitory concentrations (IC50) were determined. While PEI 25 kDa exhibited the strongest toxic effects for the panel investigated, there was a clear trend between cytotoxicity and generation number for the rigid triazine dendrimers. For G1-1, we determined an IC50 value of 501.2 μg/ml, which is about 100 fold higher than the IC50 value of PEI 25 kDa (4.9 μg/ml, compare Figure 6). The IC50 values decreased with increased generation among the rigid dendrimers, which can be explained by the increasing number of peripheral primary amino groups. Nontheless, G3-1, the dendrimer that contains the highest number of primary amines per molecule, was approximately 5 fold less toxic than PEI 25 kDa and showed a 2.5 fold lower IC50 value than the third generation PAMAM (35). The F2-1 dendrimer showed a higher toxicity profile than G2-1, which can be explained as resulting from an increased accessibility of both the core and the terminal amine groups, although the hemolytic activity which we measured for F2-1/DNA complexes was comparably low. The effect on remaining cell viability, on the other hand, which is expressed in the IC50 values, can have various reasons. Highly surface active substances are expected to strongly impede the phospholipids double layer, leading to decreased cell viability and cell death. But substances that do not exhibit strong membrane toxicity can still alter cellular mechanisms, leading to decreased cell viability. It is therefore not surprising that F2-1, which shows comparably low hemolysis, is more toxic than G2-1 and B2-1, which have higher hemolytic potentials.
Figure 6.
Toxicity profiles of all the dendrimers according to the MTT assays were compared to that of PEI 25 kDa, and for clarity IC50 values are given. 184×99mm (300 × 300 DPI)
The bow-tie dendrimer B2-1, on the other hand, exhibited lower toxicity than the rigid dendrimer G2-1, despite causing comparable hemolytic effects. For all of the triazine dendrimers, our cytotoxicity results are in line with an earlier report (13) revealing an IC50 value of 0.1 mg/ml for a related melamine structure. Triazine dendrimers seem to be less toxic than PEI and therefore more suitable for possible in vivo studies. Moreover, cytotoxicity strongly depends on the core structure and can be significantly reduced by dendriplex formation.
Transfection Efficiency
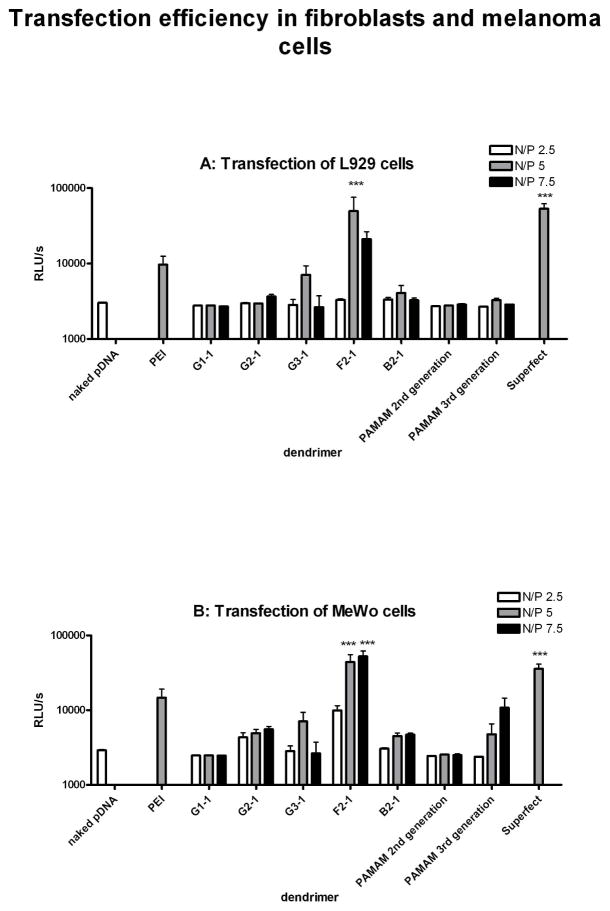
The primary interest for non-viral vectors is the efficiency of transfection. This property was studied using a luciferase-expressing plasmid in two different cell lines, namely murine L929 fibroblasts (Figure 7A) and human MeWo (Figure 7B) melanoma cells. The transfection efficiency of dendriplexes at different N/P ratios was quantified using a commercial luciferase assay and was compared to the transfection efficiencies of PEI 25 kDa, SuperFect™, and second and third generation PAMAM.
Figure 7.
Transfection efficiency of the dendrimers at various N/P ratios in (A) L929 and (B) MeWo cells are given as relative light units detected by luciferase assay. F2-1 and Superfect exhibited significantly (***p < 0.01) higher transgene expression than PEI 25 kDa and all other dendrimers. 185×279mm (300 × 300 DPI)
PAMAM (2nd and 3rd generation) dendrimers showed little transfection efficiency with increasing N/P ratios, as reported in the literature (5). An increase in transfection efficiency with increasing generation was revealed for rigid triazine dendrimers (G1-1, G2-1, and G3-1). The lowest generation dendrimer, G1-1, did not induce transgene expression in L929 or MeWo cells, while G2-1 successfully transfected MeWo and L929 at N/P ratio > 7.5. Interestingly, G3-1 yielded a maximum luciferase expression at N/P 5 in both cell lines. The reduced transfection efficiency at a higher N/P ratio for the higher generation dendrimer may be caused by cytotoxic effects. In both cell lines, F2-1 achieved the highest transgene expression, even higher than the expression of PEI 25 or SuperFect. The bow-tie dendrimer, B2-1, exhibited moderate transfection efficiency at N/P ratio of 5 in L929 and at N/P 5 and 7.5 in MeWo cells.
These results demonstrate that triazine dendrimers could provide a promising platform for gene delivery systems as they induce transfection at generation two, while established transfection reagents such as PAMAM are ineffective at that size. Similarly, the phosphorous containing dendrimers of Majoral show a transfection optimum at generation 5 (36). Higher generation, flexible triazine dendrimers are therefore under investigation with the goal of providing more efficient transfection reagents.
Because the flexible dendrimer, F2-1, formed larger complexes than the third generation dendrimer, G3-1, it presumably sedimented faster (24). This could have contributed to the higher transfection efficiency seen for this dendriplex. However, this route of endocytosis may not prove viable for in vivo applications. But also a higher availability to RNA polymerase inside the nucleus of DNA in these less compact dendriplexes, as reported by Yamagata et al. (37), might be an explanation. Hence the colloidal stability and the systematic variation of the triazine dendrimer surface are further goals for synthetic modifications.
Conclusion
Efforts to optimize the triazine dendrimers will require consideration of a number of lessons learned from these studies. The first generation rigid dendrimer, G1-1, yielded very low condensation efficiency, and although the structure had a good biocompatibility profile it did not show any transfection efficiency. The second generation rigid dendrimer G2-1 showed improved DNA condensation, formed small dendriplexes, and was less toxic but more hemolytic than PEI. However, it showed intermediate transfection efficiency. The G3-1 dendrimer condensed DNA well, formed small particles within a narrow size distribution, but exhibited erythrocyte aggregation. Transfection efficiency of this dendrimer was possibly limited by toxic effects at higher N/P ratios.
The change in core structure had the most profound impact on physicochemical as well as biologic properties of the triazine dendrimers. While the rigid, second generation dendrimer, G2-1, formed comparably small dendriplexes, both B2-1 and F2-1 assembled DNA into larger complexes with lower or even negative surface charge. DNA condensation was affected differently: DNA condensation with F2-1 and G2-1 were similar, whereas B2-1 protected DNA even better than PEI 25 kDa. The flexible dendrimer F2-1 was most effective in transfection experiments. The low density particles exhibited the lowest hemolytic effect and almost no erythrocyte aggregation.
The bow-tie dendrimer, B2-1, caused the most pronounced hemolytic effects which are attributed to the formation of aggregates and clusters as observed with DLS and AFM. Due to their large size, B2-1 dendriplexes were not effectively taken up by L929 or MeWo cells, leading to low transfection efficiency. We remain optimistic that triazine dendrimers demonstrate a promising platform for non-viral delivery systems, meriting further investigations, while our most important result is that flexible triazine dendrimers that are well biocompatible may be highly efficient vectors for non-viral gene delivery at comparably low generations. This is especially true for the flexible dendrimer F2-1, which exhibits higher transfection efficacy than SuperFect.
Supplementary Material
Acknowledgments
We thank Eva Mohr and Sandra Engel (DPB, Marburg, Germany) for their support in the cell culture lab and Michael Dutescu (Dept. of Clinical Chemistry, Marburg, Germany) for providing us with fresh blood samples. EES acknowledges support of the NIH (NIGMS R01 45640).
Footnotes
Supporting Information: Detailed descriptions of the syntheses of all dendrimers and precursors as well as yields, 1H NMR and MS (MALDI) data are provided as supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Anderson WF. Human gene therapy. Nature. 1998;392:25–30. doi: 10.1038/32058. [DOI] [PubMed] [Google Scholar]
- 2.Verma IM, Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389:239–42. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 3.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 4.Haensler J, Szoka FC., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug Chem. 1993;4:372–9. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 5.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR., Jr Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proc Natl Acad Sci U S A. 1996;93:4897–902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang MX, Redemann CT, Szoka FC., Jr In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjug Chem. 1996;7:703–14. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 7.Zinselmeyer BH, Mackay SP, Schatzlein AG, Uchegbu IF. The lower-generation polypropylenimine dendrimers are effective gene-transfer agents. Pharm Res. 2002;19:960–7. doi: 10.1023/a:1016458104359. [DOI] [PubMed] [Google Scholar]
- 8.Hollins AJ, Benboubetra M, Omidi Y, Zinselmeyer BH, Schatzlein AG, Uchegbu IF, Akhtar S. Evaluation of generation 2 and 3 poly(propylenimine) dendrimers for the potential cellular delivery of antisense oligonucleotides targeting the epidermal growth factor receptor. Pharm Res. 2004;21:458–66. doi: 10.1023/B:PHAM.0000019300.04836.51. [DOI] [PubMed] [Google Scholar]
- 9.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–9. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 10.Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, Kissel T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–25. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 11.Mintzer MA, Merkel OM, Kissel T, Simanek E. Polycationic triazine-based dendrimers: Effect of peripheral groups on transfection efficiency. New Journal of Chemistry. 2009 doi: 10.1039/b908735d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim J, Guo Y, Rostollan CL, Stanfield J, Hsieh JT, Sun X, Simanek EE. The role of the size and number of polyethylene glycol chains in the biodistribution and tumor localization of triazine dendrimers. Mol Pharm. 2008;5:540–7. doi: 10.1021/mp8000292. [DOI] [PubMed] [Google Scholar]
- 13.Neerman MF, Zhang W, Parrish AR, Simanek EE. In vitro and in vivo evaluation of a melamine dendrimer as a vehicle for drug delivery. Int J Pharm. 2004;281:129–32. doi: 10.1016/j.ijpharm.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Roberts JC, Bhalgat MK, Zera RT. Preliminary biological evaluation of polyamidoamine (PAMAM) Starburst dendrimers. J Biomed Mater Res. 1996;30:53–65. doi: 10.1002/(SICI)1097-4636(199601)30:1<53::AID-JBM8>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Germershaus O, Mao S, Sitterberg J, Bakowsky U, Kissel T. Gene delivery using chitosan, trimethyl chitosan or polyethylenglycol-graft-trimethyl chitosan block copolymers: establishment of structure-activity relationships in vitro. J Control Release. 2008;125:145–54. doi: 10.1016/j.jconrel.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Germershaus O, Merdan T, Bakowsky U, Behe M, Kissel T. Trastuzumab-polyethylenimine-polyethylene glycol conjugates for targeting Her2-expressing tumors. Bioconjug Chem. 2006;17:1190–9. doi: 10.1021/bc0601119. [DOI] [PubMed] [Google Scholar]
- 17.Germershaus O, Neu M, Behe M, Kissel T. HER2 targeted polyplexes: the effect of polyplex composition and conjugation chemistry on in vitro and in vivo characteristics. Bioconjug Chem. 2008;19:244–53. doi: 10.1021/bc700311n. [DOI] [PubMed] [Google Scholar]
- 18.Han M, Chen P, Yang X. Molecular dynamics simulation of PAMAM dendrimer in aqueous solution. Polymer. 2005;46:3481–3488. [Google Scholar]
- 19.Kabanov VA, Sergeyev VG, Pyshkina OA, Zinchenko AA, Zezin AB, Joosten JGH, Brackman J, Yoshikawa K. Interpolyelectrolyte Complexes Formed by DNA and Astramol Poly(propylene imine) Dendrimers. Macromolecules. 2000;33:9587–9593. [Google Scholar]
- 20.Gebhart CL, Kabanov AV. Evaluation of polyplexes as gene transfer agents. J Control Release. 2001;73:401–16. doi: 10.1016/s0168-3659(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 21.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 1997;4:823–32. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 22.Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, Strohalm J, Ulbrich K. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- 23.Wood KC, Little SR, Langer R, Hammond PT. A family of hierarchically self-assembling linear-dendritic hybrid polymers for highly efficient targeted gene delivery. Angew Chem Int Ed Engl. 2005;44:6704–8. doi: 10.1002/anie.200502152. [DOI] [PubMed] [Google Scholar]
- 24.Mahato RI, Rolland A, Tomlinson E. Cationic lipid-based gene delivery systems: pharmaceutical perspectives. Pharm Res. 1997;14:853–9. doi: 10.1023/a:1012187414126. [DOI] [PubMed] [Google Scholar]
- 25.Erbacher P, Bettinger T, Belguise-Valladier P, Zou S, Coll JL, Behr JP, Remy JS. Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI) J Gene Med. 1999;1:210–22. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<210::AID-JGM30>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Kim TI, Seo HJ, Choi JS, Jang HS, Baek JU, Kim K, Park JS. PAMAM-PEG-PAMAM: novel triblock copolymer as a biocompatible and efficient gene delivery carrier. Biomacromolecules. 2004;5:2487–92. doi: 10.1021/bm049563j. [DOI] [PubMed] [Google Scholar]
- 27.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 28.Wittmar M, Ellis JS, Morell F, Unger F, Schumacher JC, Roberts CJ, Tendler SJB, Davies MC, Kissel T. Biophysical and Transfection Studies of an Amine-Modified Poly(vinyl alcohol) for Gene Delivery. Bioconjugate Chemistry. 2005;16:1390–1398. doi: 10.1021/bc0500995. [DOI] [PubMed] [Google Scholar]
- 29.Kunath K, von Harpe A, Petersen H, Fischer D, Voigt K, Kissel T, Bickel U. The structure of PEG-modified poly(ethylene imines) influences biodistribution and pharmacokinetics of their complexes with NF-kappaB decoy in mice. Pharm Res. 2002;19:810–7. doi: 10.1023/a:1016152831963. [DOI] [PubMed] [Google Scholar]
- 30.Khandare JJ, Jayant S, Singh A, Chandna P, Wang Y, Vorsa N, Minko T. Dendrimer versus linear conjugate: Influence of polymeric architecture on the delivery and anticancer effect of paclitaxel. Bioconjug Chem. 2006;17:1464–72. doi: 10.1021/bc060240p. [DOI] [PubMed] [Google Scholar]
- 31.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19:1396–403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Camb) 2006:2362–4. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 33.Manunta M, Tan PH, Sagoo P, Kashefi K, George AJ. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucleic Acids Res. 2004;32:2730–9. doi: 10.1093/nar/gkh595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun CS, Vetro JA, Tomalia DA, Koe GS, Koe JG, Middaugh CR. Structure/function relationships of polyamidoamine/DNA dendrimers as gene delivery vehicles. J Pharm Sci. 2005;94:423–36. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- 35.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, Meijer EW, Paulus W, Duncan R. Dendrimers: relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo. J Control Release. 2000;65:133–48. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 36.Padié C, Maszewska M, Majchrzak K, Nawrot B, Caminade AM, Majoral Jean-Pierre. Polycationic phosphorus dendrimers: synthesis, characterization, study of cytotoxicity, complexation of DNA, and transfection experiments. New Journal of Chemistry. 2009;33:318. [Google Scholar]
- 37.Yamagata M, Kawano T, Shiba K, Mori T, Katayama Y, Niidome T. Structural advantage of dendritic poly(L-lysine) for gene delivery into cells. Bioorg Med Chem. 2007;15:526–32. doi: 10.1016/j.bmc.2006.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.