Abstract
In the N-end rule pathway of protein degradation, the destabilizing activity of N-terminal Asp, Glu or (oxidized) Cys residues requires their conjugation to Arg, which is recognized directly by pathway's ubiquitin ligases. N-terminal arginylation is mediated by the Ate1 arginyltransferase, whose physiological substrates include the Rgs4, Rgs5 and Rgs16 regulators of G proteins. Here, we employed the Cre-lox technique to uncover new physiological functions of N-terminal arginylation in adult mice. We show that postnatal deletion of mouse Ate1 (its unconditional deletion is embryonic lethal) causes a rapid decrease of body weight and results in early death of ∼15% of Ate1-deficient mice. Despite being hyperphagic, the surviving Ate1-deficient mice contain little visceral fat. They also exhibit an increased metabolic rate, ectopic induction of the Ucp1 uncoupling protein in white fat, and are resistant to diet-induced obesity. In addition, Ate1-deficient mice have enlarged brains, an enhanced startle response, are strikingly hyperkinetic, and are prone to seizures and kyphosis. Ate1-deficient males are also infertile, owing to defects in Ate1−/− spermatocytes. The remarkably broad range of specific biological processes that are shown here to be perturbed by the loss of N-terminal arginylation will make possible the dissection of regulatory circuits that involve Ate1 and either its known substrates, such as Rgs4, Rgs5 and Rgs16, or those currently unknown.
Introduction
N-terminal arginylation of intracellular proteins by Arg-tRNA-protein transferase (R-transferase) is a part of the N-end rule pathway of protein degradation (Fig. 1A). In eukaryotes, this pathway is a part of the ubiquitin (Ub)-proteasome system. The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue (reviewed in [1], [2], [3], [4]). Degradation signals (degrons) that can be targeted by the N-end rule pathway are of two distinct kinds: N-terminal degrons, called N-degrons, and internal (non-N-terminal) degrons [1], [5]. The main determinant of an N-degron is a destabilizing N-terminal residue of a substrate protein (Fig. 1A). The other determinants of N-degron are a substrate's internal Lys residue (the site of formation of a poly-Ub chain) and a nearby unstructured region [6], [7]. An N-degron is produced from a precursor, called a pre-N-degron, through a protease-mediated cleavage of a substrate that exposes a destabilizing N-terminal residue.
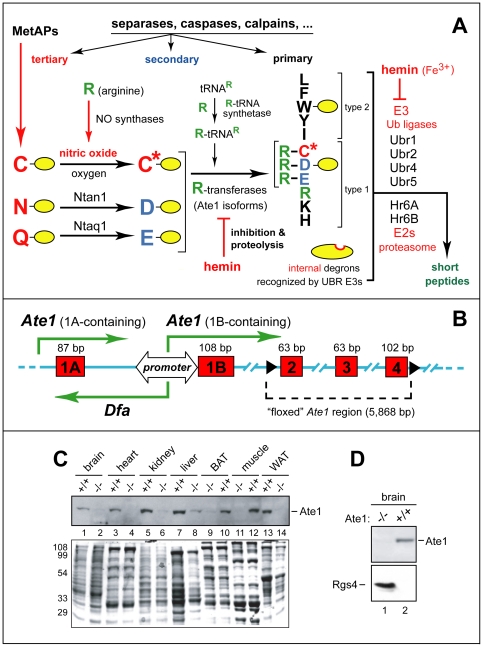
Figure 1. Postnatal ablation of the mouse Ate1 R-transferase, a component of the N-end rule pathway.
(A) The mammalian N-end rule pathway. N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. “Primary”, “secondary” and “tertiary” denote mechanistically distinct subsets of destabilizing N-terminal residues (see Introduction). C* denotes oxidized Cys, either Cys-sulfinate or Cys-sulfonate. MetAPs, Met-aminopeptidases. (B) Bidirectional promoter between the mouse Ate1 exons 1A and 1B [14]. Green arrows indicate transcriptional units, including a previously uncharacterized gene, termed Dfa (“divergent of Ate1), that is transcribed from the bidirectional promoter. (C) Immunoblotting-based comparisons of Ate1 levels in the indicated mouse tissues from Ate1+/+ and Ate1flox/−;CaggCreER mice 76 days after the tamoxifen (TM)-induced, Cre-mediated Ate1flox→Ate1− conversion that yielded Ate1-deficient mice. The band of 60-kDa Ate1, detected by antibody to mouse Ate1, is indicated on the right. Total (Ponceau-stained) protein patterns are shown below, with positions of molecular-mass markers on the left. (D) IB assays for the levels of Ate1 and Rgs4 (25 kDa) in brain extracts from Ate1+/+ and Ate1-deficient mice (Ate1flox/−;CaggCreER mice 30 days after TM treatment).
The N-end rule has a hierarchic structure (Fig. 1A). N-terminal Asn and Gln are tertiary destabilizing residues in that they function through their enzymatic deamidation, to yield the secondary destabilizing N-terminal residues Asp and Glu [8]. Destabilizing activity of N-terminal Asp and Glu requires their conjugation to Arg, one of the primary destabilizing residues, by the Ate1-encoded R-transferase [9], [10], [11], [12]. In eukaryotes that produce nitric oxide (NO), R-transferase arginylates not only N-terminal Asp and Glu but also Cys, after its conversion to Cys-sulfinate or Cys-sulfonate, in reactions that require NO and oxygen (Fig. 1A) [11], [13]. Alternative splicing of the mammalian Ate1 pre-mRNA produces isoforms of R-transferase, a metabolically unstable protein whose enzymatic activity and the in vivo half-life are down-regulated by heme [10], [12], [14]. E3 Ub ligases of the N-end rule pathway are called N-recognins. An N-recognin is an E3 that can recognize (target for polyubiquitylation) at least a subset of N-degrons (Fig. 1A) [1], [4]. Some of substrate-binding sites of an N-recognin target N-degrons, while other sites of the same N-recognin are specific for structurally unrelated internal (non-N-terminal) degrons [15], [16]. At least four N-recognins, Ubr1, Ubr2, Ubr4 and Ubr5, mediate the mammalian N-end rule pathway (Fig. 1A) [4], [17].
The functions of the N-end rule pathway in eukaryotes include selective degradation of misfolded proteins; the sensing of heme, oxygen, nitric oxide (NO), and short peptides; the regulation of DNA repair and peptide import; the signaling by transmembrane receptors, through the NO/O2-controlled degradation of G-protein regulators Rgs4, Rgs5 and Rgs16; the fidelity of chromosome segregation; regulation of apoptosis, meiosis, spermatogenesis, neurogenesis, and cardiovascular development; the functioning of specific organs, in particular the brain and the pancreas; and regulation of leaf senescence, seed germination, and other processes in plants ([2], [12], [16], [18], [19], [20], and refs. therein). A partial N-terminal arginylation of the apparently long-lived mammalian α-actin [21] suggests that arginylation of some proteins may not alter their in vivo half-lives.
Although there are many putative intracellular substrates of the Ate1 R-transferase, for example, among C-terminal fragments of proteins that are cleaved in vivo by proteases such as MetAPs, caspases, calpains or secretases, the set of definitively identified Ate1 substrates is still small. It includes the Drosophila antiapoptotic Ub ligase DIAP1 [22]; the mammalian G-protein regulators Rgs4, Rgs5 and Rgs16 [11], [13]; and the separase-produced fragment of the mammalian Rad21/Scc1 cohesin subunit that bears N-terminal Glu, a secondary destabilizing residue (Fig. 1A) (J. Zhou, D. Pati and A.V., unpublished data) [23]. Heterozygous Ate1+/− mice appear indistinguishable from their wild-type counterparts, whereas Ate1−/− mice die around embryonic day 15 (E15) with abnormalities that include cardiovascular defects [10].
To bypass the embryonic lethality of nonconditional Ate1−/− mice, we employed the Cre-lox technique [24]. As shown below, a systemic postnatal deletion of the sole active Ate1flox allele in juvenile Ate1flox/− mice causes a rapid decrease of body weight and results in early death of ∼15% of Ate1-deficient mice, with surviving mice attaining only ∼70% of normal weight. This failure to thrive occurs despite higher than normal food intake by Ate1-deficient mice. These mice contain little or no visceral fat, exhibit an increased metabolic rate, a decreased fasting blood glucose level, and an increased intestinal import and retention of amino acids and/or peptides. Ate1-deficient mice are also resistant to diet-induced obesity and exhibit ectopic induction of the Ucp1 uncoupling protein in white adipose tissue (WAT). In addition, Ate1-deficient mice have enlarged brains, an enhanced startle response, and are strikingly hyperkinetic. They often suffer from kyphosis, i.e., an excessive curvature of the upper back, and from frequent seizures as well. Ate1-deficient males are also infertile, owing to defects in meiotic Ate1−/− spermatocytes. The remarkably broad range of specific biological processes that are shown here to be perturbed by the loss of N-terminal arginylation will facilitate the dissection of regulatory circuits that involve Ate1 and either its known substrates, such as Rgs4, Rgs5 and Rgs16 [11], [13], or those currently unknown.
Results
Ate1flox/− Mouse Strains and Production of Ate1−/− Mice
Standard methods were employed to produce, initially, ATEflox/+ mouse strains in which a specific segment of Ate1 was “floxed”, i.e., flanked by 34-bp loxP repeats (Fig. 2C–E). The targeting vector contained ∼14 kb of Ate1, including the exon 1A-exon 4 segment that encodes an essential part of R-transferase [9] (Fig. 2A). Our previous work has shown that the Ate1 promoter (PAte1) is bidirectional, expressing both Ate1 and an oppositely oriented gene termed Dfa (divergent from Ate1), which overlaps with exon 1A of Ate1 (Fig. 1B) ([14]; C.S.B. and A.V., unpublished data). To minimize the possibility of perturbing the expression of Dfa, the “floxed” region of Ate1 encompassed exons 2–4, away from exon 1A (Fig. 1B). Our aim was to produce ATEflox/− mouse strains that were “poised” to lose their remaining active ATEflox allele through the expression of Cre recombinase. To do so, heterozygous matings were carried out among the above ATEflox/+ mice, the previously constructed ATE+/− mice [10], and a mouse strain that contained the CaggCreER gene, expressed from the ubiquitously active chimeric Cagg promoter [25]. CaggCreER encoded CreER, a fusion between Cre and a derivative of the mouse estrogen receptor ligand binding domain. CreER was functionally inactive (sequestered in the cytosol) but could be activated by intraperitoneal (IP) injections of tamoxifen (TM) [25]. Depending on configurations of their Ate1 alleles, the resulting mice, poised for the loss of Ate1, were termed Ate1flox/−;CaggCreER or Ate1flox/flox;CaggCreER.
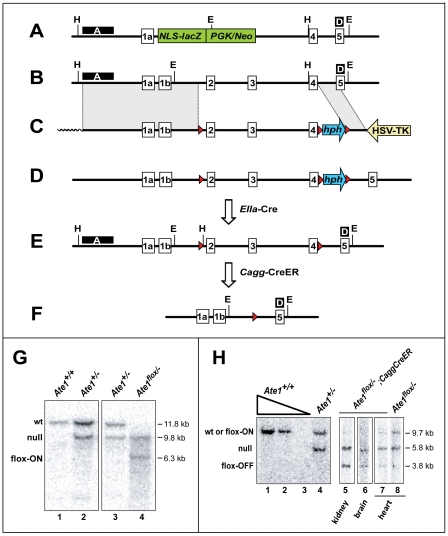
Figure 2. Genomic configurations at the Ate1 locus of Cre-lox-based mouse strains constructed in the present work.
(A) The 5′ end of the previously produced unconditional Ate1− allele [10], in which the Ate1 exons 1b through 3 were replaced by a cassette encoding a promoter-lacking, NLS-containing LacZ (NLS-βgal) (it was expressed from the endogenous PAte1 promoter) and the Neo selection marker expressed from the phosphoglycerate kinase PPGK promoter (green rectangles). (B) A diagram of the 5′ end of wild-type (wt) mouse Ate1, indicating approximate locations of exons 1a through 5. (C) The ∼22.5 kb targeting construct containing a ∼6 kb long-arm region of Ate1 homology (shown as a shaded rectangle on the left); a single loxP site (red triangle) upstream of Ate1 exon 2, a “floxed”-hygromycin-resistance (hph) cassette, expressed from the PPGK promoter (blue arrow between two red triangles) downstream of Ate1 exon 4; a ∼2 kb short-arm region of homology (an inclined shaded rectangle), and the HSV thymidine kinase (tk) negative-selection cassette expressed from the PHSV promoter (yellow arrow). Wavy line indicates an abutting sequence of the pBR322 plasmid DNA. (D) The tri-lox Ate1 allele obtained after a correctly targeted double crossover event. (E) In the notations here and elsewhere in the paper, “flox-on” indicates a configuration depicted in this panel (the functionally active Ate1flox allele), whereas “flox-off” indicates a configuration depicted in panel F (the null Ate1− allele). The functionally active, “flox-on” (Ate1flox) allele, obtained by the removal of the hph cassette, using the in vivo expression of Cre-recombinase driven by the PEIIA promoter, which is active only in pre-implantation blastocysts. (F) The null “flox-off” (Ate1−) allele obtained by the inducible expression of CreER recombinase from the PCagg promoter and posttranslationally induced by tamoxifen (TM) treatment (see the main text and Materials and Methods). H, approximate locations of HindIII sites used in Southern analyses with DNA probe A (see panel G); E, approximate locations of EcoRI sites used in Southern analyses with DNA probe D (see panel H); black boxes marked “A” and “D” indicate the regions specific for DNA probes A and D, respectively. (G) Southern hybridization analysis using DNA probe A and HindIII-digested genomic DNA. The wt Ate1 allele (panel B) yields the 11.8 kb HindIII fragment. The previously constructed [10] unconditionally null Ate1− allele (panel A), denoted as “null” on this panel, yields the 9.8 kb HindIII fragment. The functionally active flox-on (Ate1flox) allele (panel E) yields the 6.3 kb HindIII fragment. Lane 1, Ate1+/+; lane 2, Ate1+/−; lane 3, Ate1+/−; lane 4, Ate1flox/−. (H) Southern hybridization analysis using DNA probe D (external to targeting vector) and EcoRI-digested genomic DNA. The previously constructed [10] unconditionally null Ate1− allele (denoted as “null”) yields the 5.8 kb fragment. Both the wild-type Ate1 allele and the flox-on (Ate1flox) allele yield the 9.7 kB fragment, whereas the null flox-off (Ate1−) allele yields the characteristic 3.8 kb fragment. The use of DNA probe D and EcoRI-digested DNA from specific tissues of tamoxifen (TM)-treated Ate1flox/−;CaggCreER mice allowed approximate estimates of the levels of Cre-mediated recombination that produced the flox-off (Ate1−) allele. For example, whereas no flox-on (Ate1flox) allele could be detected in the kidney and brain of Ate1flox/−;CaggCreER mice after TM treatment (lanes 5, 6), approximately equal amounts of flox-on (Ate1flox) and flox-off (Ate1−) alleles were present in the heart of TM-treated Ate1flox/−; CaggCreER mice. Lanes 1–3, 1,000, 250, and 25 ng of EcoRI-digested wt mouse genomic DNA (from a tail biopsy), respectively. Lane 4, EcoRI-digested genomic DNA from the tail of a previously constructed [10] Ate1+/− mouse. Lanes 5–7, EcoRI-digested genomic DNA from the indicated tissues of TM-treated Ate1flox/−;CaggCreER mice. Lane 8, same as lane 7, but from a TM-treated Ate1flox/− mouse (lacking the CaggCreER transgene).
Using standard methods, we could demonstrate the presence of Ate1flox/−;CaggCreER mice, at expected (Mendelian) frequencies, in the progeny of above matings. These mice expressed TM-inducible CreER recombinase and contained a single copy of Ate1flox, the active Ate1 allele (Figs. 2 and 3A). The functional intactness of Ate1flox was inferred from the fact that Ate1flox/−;CaggCreER mice survived embryogenesis (in contrast to Ate1−/− mice [10]) and were phenotypically similar (in the absence of TM treatment) to Ate1+/− and Ate1+/+ mice. To induce the Ate1flox→Ate1− conversion, ∼1 month old Ate1flox/−;CaggCreER mice and their Ate1flox/+;CaggCreER (as well as Ate1+/−;CaggCreER) littermates, used as controls, were treated with TM (see Materials and Methods for details, including the ages of TM-treated mice). Southern hybridization and PCR-based analyses of DNA from tissues of the resulting mice (sampled ∼1 month after TM treatment) confirmed the TM-induced, Cre-mediated excision of the Ate1flox allele in Ate1flox/−;CaggCreER mice. The frequency of Ate1flox→Ate1− conversion was nearly 100% in the brain and kidney of these mice, but significantly lower in several other tissues (Figs. 2G, H and 3B).
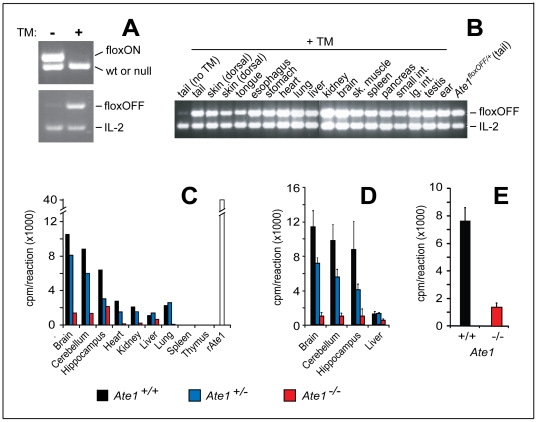
Figure 3. Cre-mediated conversion to Ate1-null genotype in different mouse tissues.
(A) PCR-based genotyping of tail DNA to detect the Cre-mediated Ate1flox→Ate1− conversion of the functionally active flox-on (Ate1flox) allele to the null Ate1− allele in a 27-day old Ate1flox/−;CaggCreER mouse immediately after the fourth (daily) intraperitoneal (IP) injection of tamoxifen (TM+), or in the absence of TM treatment (TM-). Upper panel: the 512 bp DNA fragment characteristic of the flox-on (Ate1flox) allele and the 472 bp DNA fragment characteristic of either wild-type or the previously constructed [10] unconditionally null Ate1− allele, using primers CB156 and CB157 (Table 4). Lower panel: the 470 bp DNA fragment characteristic of the Cre-produced flox-off (Ate1−) allele, with primers CB110 and CB157 (Table 4); and the 324 bp DNA fragment (control), amplified from the IL-2 gene using primers IMR42 and IMR43, in the same PCR reaction. (B) The Cre-mediated Ate1flox→Ate1− conversion, detected by PCR (as described in panel A) in genomic DNA isolated from the indicated tissues immediately after the fourth (daily) IP injection of tamoxifen in a 24-day old Ate1flox/−;CaggCreER mouse. (C) Relative in vitro arginylation activity (cpm/reaction) in extracts of the indicated tissues from a wild type mouse (Ate1+/+) (black bar), a heterozygous mouse (Ate1+/−) (blue bar), and an Ate1−/− mouse (the latter mouse was initially Ate1flox/−;CaggCreER) (red bar) from the same litter 76 days after TM treatment. A white bar on the right indicates the relative arginylation activity obtained with purified recombinant mouse Ate1 (denoted as “rAte1”) that had been expressed in S. cerevisiae. Shown here are “cpm/reaction” after subtracting “cpm/reaction” in the null-control (“buffer alone”) sample. The control incorporation was approximately equal to that observed in extracts from spleen and thymus. In other words, the assay configured as described in this panel and in Materials and Methods was not sensitive enough to robustly detect the arginylation activity in extracts from spleen and thymus. (D) Relative in vitro arginylation activity (cpm/reaction) in the whole brain, cerebellum, and hippocampus harvested from wild type mice (Ate1+/+; n = 3), heterozygous mice (Ate1+/−; n = 3), and Ate1−/− mice (specifically, Ate1flox/−;CaggCreER mice; n = 3) mice 40 days after TM treatment. Standard deviations are indicated. (E) Relative in vitro arginylation activity (cpm/reaction) in testis extracts from Ate1+/+ mice (n = 3) and Ate1−/− mice (specifically, Ate1flox/−;CaggCreER mice; n = 3) ∼130 days after TM treatment. Standard deviations are indicated.
We also used an affinity-purified antibody to mouse Ate1 [11] to carry out immunoblotting (IB) with extracts from brain, heart, kidney, liver, muscle, brown adipose tissue (BAT) and white adipose tissue (WAT) that were harvested up to 8 months after TM treatment of Ate1flox/−;CaggCreER mice, versus identically TM-treated control littermates. No Ate1 could be detected by IB in several tissues of TM-treated Ate1flox/−;CaggCreER mice, in contrast to readily detectable Ate1 in TM-treated control mice (Fig. 1C). The only significant exception was liver (Fig. 1C, lanes 7, 8; cf. lanes 5, 6 or lanes 11–14; see also below). One effect of Ate1 depletion in the mouse brain was a striking increase of Rgs4, a physiological Ate1 substrate (see Introduction) that down-regulates specific G proteins by acting as a GTPase-activating protein (GAP) (Fig. 1D). Whereas no Rgs4 could be detected in the Ate1-containing brain (owing to degradation of Rgs4 by the N-end rule pathway [11]), an intense band of Rgs4 was present in the Ate1-deficient brain, illustrating high penetrance of Ate1 deletion in the brain (Fig. 1D).
We also performed in vitro arginylation assays with extracts from several tissues of Ate1flox/−;CaggCreER mice 21 days after TM treatment, versus extracts from identically treated Ate1 +/+ or ATE1 +/− mice. The TM-induced decrease of arginylation activity in specific organs of Ate1−/−;CaggCreER mice ranged from ∼90% in the brain and kidney to ∼60% in the liver (Fig. 3C–E). Although heterozygous Ate1 +/− mice were phenotypically similar to their wild-type (Ate1 +/+) counterparts, we found that Ate1 +/− mice grew slightly but consistently slower than Ate1 +/+ mice, and reached a lower average weight (Fig. 4B). In agreement with this mild but detectable haploinsufficiency of Ate1, the arginylation activity in extracts from, e.g., brains or hearts of Ate1 +/− mice was significantly below its wild-type (Ate1 +/+) levels (Fig. 3C), implying the absence of a compensatory (e.g., autoregulated) increase of Ate1 expression upon a decrease of Ate1 gene dosage.
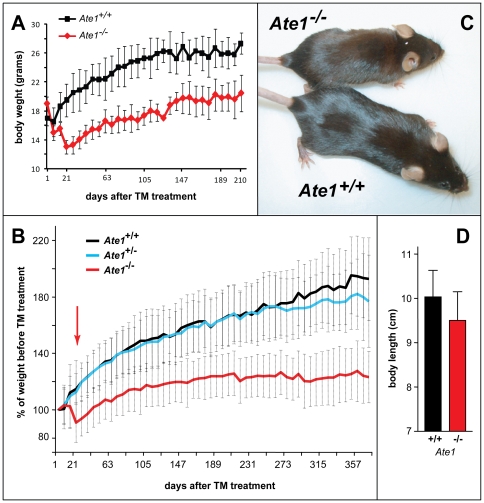
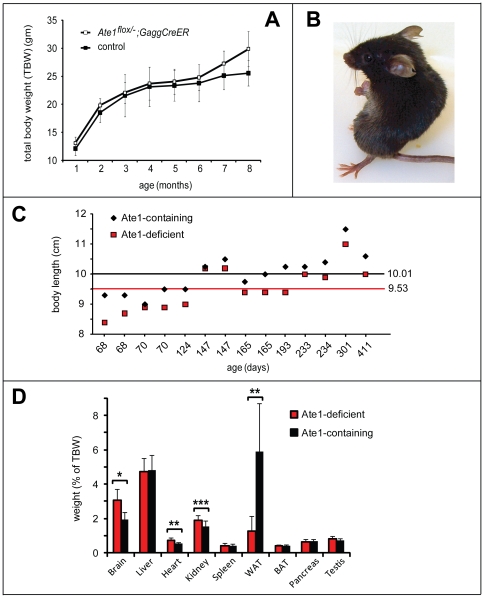
Figure 4. Growth rate consequences of postnatal ablation of Ate1.
(A) Weights of Ate1-containing (n = 4; black curve) and Ate1-deficient (n = 2; red curve) mice from the same litter as a function of time after tamoxifen (TM) treatment. Weights were measured at weekly intervals. Vertical bars indicate the ranges of measured weights. (B) Averaged growth curves for the indicated numbers of mice after TM treatment, plotted as a percentage of their weight immediately before TM treatment. Red, black and blue curves: Ate1−/− (n = 87), Ate1+/+ (n = 55), and Ate1+/− (n = 66) mice. Red arrow indicates the time (∼21 days) after TM treatment by which ∼15% of Ate1-deficient mice have died while the rest of them began to gain weight. Note a slightly but clearly decreased weight of heterozygous (Ate1+/−) mice (blue curve), in comparison to Ate1+/+ mice (black curve) ∼1 year after TM treatment. Error bars indicate standard deviations (SD). (C) Typical appearance of Ate1−/− versus wt mice (a smaller, leaner Ate1−/− mouse) ∼1 year after TM-mediated ablation of Ate1. (D) Mean body lengths (± SD) (from tip-of-nose to base-of-tail) between pairs of Ate1−/− (red bar) and Ate1+/+ (black bar) mice. This comparison was derived from the data in Fig. 5C. Statistical analysis was performed using an unpaired t-test (p<0.08).
Retarded Growth, Kyphosis, and a Transient Increase in Lethality of Ate1-Deficient Mice
TM treatment produced abnormal phenotypes within 1 week in Ate1flox/−;CaggCreER mice, in comparison to identically TM-treated controls. Specifically, ∼1 month old and previously growing Ate1flox/−;CaggCreER mice failed to thrive (in comparison to control mice) after their TM-induced conversion to Ate1−/−;CaggCreER mice (Figs. 4A–C and 5A). During the first ∼3 weeks after becoming Ate1−/−, these mice experienced a rapid loss of weight and decreased growth (Fig. 4A, B), despite no decrease in their consumption of food (see below). The average body length (measured from tip-of-nose to base-of-tail) of Ate1−/−;CaggCreER mice was 5% smaller (p<0.08) than that of their Ate1-containing, identically TM-treated counterparts (Ate1+/+;CaggCreER, Ate1+/−;CaggCreER, or Ate1flox/+;CaggCreER mice) (Figs. 4D and 5C).
Figure 5. Comparison of organ sizes and other parameters of Ate1−/− versus Ate1+/+ mice.
(A) Averaged growth curves (total body weight (TBW)) for Ate1flox/−;CaggCreER mice versus control mice in the absence of TM treatments. A cohort of “control” mice contained Ate1flox/+ mice (n = 2); Ate1flox/− mice (n = 2); Ate1+/+ mice (n = 1) and Ate1+/+;CaggCreER mice (n = 1) from 1 month of age through 8 months. None of the mice were treated with TM. Vertical bars indicate standard deviations. (B) Typical “kyphoid” posture of an Ate1-deficient mouse (see also the main text). (C) A plot of body lengths (in cm from tip-of-nose to base-of tail) in individual sets of Ate1-containing (black diamonds) and Ate1-deficient (red boxes) siblings at the indicated ages. Each pair of symbols, at a given age, represents a single pair of siblings. The black horizontal line indicates the averaged body length of all Ate1-containing mice (n = 14). The red horizontal line indicates the averaged body length of all Ate1-deficient mice (n = 14). (D) Comparison of tissue weights (as a percentage of total body weight (TBW)). Numbers in parentheses indicate the numbers of mice sampled and averaged for each tissue (Ate1-containing and Ate1-deficient). Brain (n = 43), liver (n = 28), heart (n = 17), kidney (n = 17), spleen (n = 16), white adipose tissue (WAT; n = 10), brown adipose tissue (BAT; n = 10), pancreas (n = 6), and testis (n = 8) from Ate1-containing (black bars) and Ate1-deficient mice (red bars). * = p<8×10−15; ** = p<5×10−5; and *** = p<0.003. Statistical analysis was performed using an unpaired t-test. Standard deviations are indicated.
In the entire cohort of TM-treated post-natal Ate1flox/−;CaggCreER mice, 15% of them (18 of 119 mice) died over 42 days after TM treatment. Crucially, none of identically TM-treated control mice (Ate1+/+;CaggCreER, Ate1+/−;CaggCreER, or Ate1flox/+;CaggCreER) died in the same time interval. The frequency of Ate1−/−;CaggCreER mice succumbing upon the acquisition of Ate1−/− genotype was age-dependent. Specifically, 46% of Ate1−/−;CaggCreER mice younger than 30 days at the beginning of TM treatment died within 42 days after TM treatment. In contrast, only 12% of Ate1−/−;CaggCreER mice died if they were older than 30 days (by up to 56 days) at the beginning of TM treatment. Those among Ate1−/−;CaggCreER mice that survived for at least 42 days after TM treatment eventually resumed growth, but the rate of growth and their maximum weight were significantly below those parameters for identically TM-treated control mice (Fig. 4A–C).
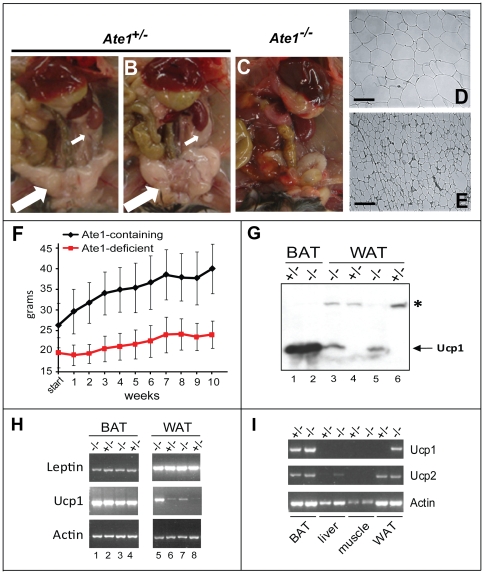
In addition to their retarded growth (despite a higher than normal food intake; see below), 53% of Ate1−/−;CaggCreER mice (95 of 180 mice) appeared “scruffy”, and 66% of them (109 of 180) had a kyphotic posture, i.e., an excessive curvature of the upper back (Fig. 5B). In contrast, only 3% of Ate1-containing mice (8 of 244) were scruffy, and only 2% (5 of 244) exhibited kyphosis. Among surviving Ate1−/−;CaggCreER mice, 10% (8 of 80) developed patches of red hair among their normally black hair, in contrast to identically TM-treated Ate1-containing mice (data not shown), suggesting a misregulation of melanocytes in Ate1-deficient mice. The liver, spleen, intrascapular brown adipose tissue (BAT), pancreas, and testis of Ate1−/−;CaggCreER mice appeared normal and were of appropriate sizes (if the smaller size of these mice (Fig. 4C) was taken into account), whereas the brains, hearts and kidneys of these Ate1-deficient mice were disproportionately large, in comparison to those of Ate1-containing siblings (Fig. 5D). Intact brains of Ate1-deficient mice appeared swollen, in comparison to brains harvested, in parallel, from identically treated Ate1-containing siblings (Fig. 6A). In addition, Ate1-deficient males were infertile, in agreement with defects in their testes (Fig. 6F–I). Yet another abnormality of Ate1-deficient mice was their strikingly lower content of the peritoneal white adipose tissue (WAT), on average only 16% of WAT in Ate1-containing mice (Figs. 5D and 7A–C). These phenotypes are discussed below.
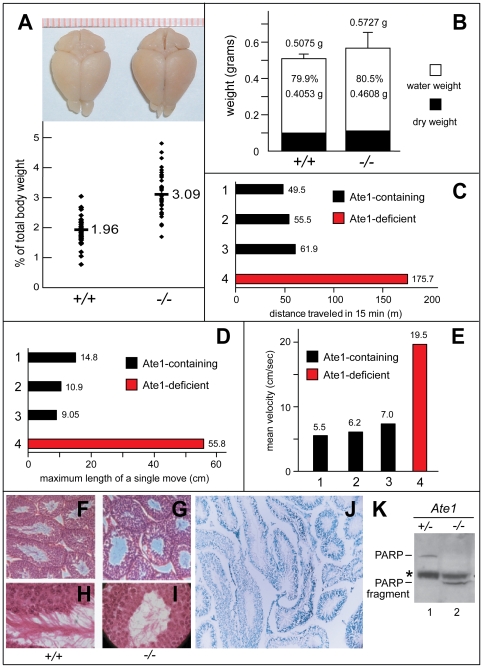
Figure 6. Brain, behavioral, and testis abnormalities of Ate1-deficient mice.
(A) Enlarged brains of Ate1-deficient mice. Upper panel: comparison of representative brains harvested from an Ate1+/+ and an Ate1−/− mouse, respectively, 134 days after tamoxifen (TM) treatment. Lower panel: brain weights expressed as percentages of total body weights in Ate1+/+ (n = 41) and Ate1−/− (n = 40) mice. Horizontal bars and numbers indicate mean values. (B) Wet (0.4053 g versus 0.4608 g) and dry (0.1022 g versus 0.1119 g) weight components of the total mean brain weights (±SD) in Ate1+/+ and Ate1−/− mice. (C) Total distance traveled (in meters), over 15 min, in an open field test among mice of different genotypes belonging to the same litter, 44 days after TM-treatment. Bar 1, Ate1flox/+;CaggCreER mouse. Bar 2, Ate1+/+;CaggCreER mouse. Bar 3, Ate1+/+ mouse. Bar 4, Ate1flox/−;CaggCreER mouse that was converted to Ate1−/− by TM treatment. Blue and red bars denote Ate1-containing and Ate1-deficient mice, respectively. (D) Same as in C but maximum lengths of single movements (in centimeters). (E) Same as in C but mean velocities (in cm/second) over 15 min. (F) Paraffin sections (4 µm) of testis showing cross-sections of seminiferous tubules in Ate1+/+ testis stained with hematoxylin and eosin (150× magnification). (G) Same as in F but Ate1−/− testis. Note that sperm tails in the lumens of Ate1−/− tubules are sparse in comparison to those in Ate1+/+ testis. (H) Same as in F but at 600× magnification. (I) Same as in G but at 600× magnification. (J) XGal staining for βgal activity in a 10-µm section of Ate1+/− testis in which one copy of Ate1 was replaced by an ORF encoding NLS-β-galactosidase and expressed from the PAte1 promoter (100× magnification). (K) Immunoblotting analysis, using antibody to poly (ADP-ribose) polymerase (PARP), of testis extracts from an Ate1-containing (Ate1flox/− (+/−)) and an Ate1-deficient (Ate1flox/−;CaggCreER (−/−)) mouse 16 days after TM treatment. Note the loss of the full-length length 116 kDa PARP and the presence of the 85 kDa PARP fragment (lane2). An asterisk denotes a protein crossreacting with anti-PARP antibody.
Figure 7. Loss of white adipose tissue (WAT), resistance to high fat diet-induced obesity, and ectopic Ucp1 in WAT of Ate1-deficient mice.
(A–C) Visceral fat content of Ate1-containing mice. Shown here are representative examples of Ate1-containing (Ate1flox/+;CaggCreER) (A) and Ate1flox/− (B)) and Ate1-deficient (Ate1flox/−;CaggCreER (C)) mice 37 days after TM-treatment. Note the loss of both visceral fat (large white arrow in A and B) and fat surrounding the kidney (small white arrows in A and B) in an Ate1-deficient mouse (C). (D) Hematoxylin/eosin staining of a 10-µm section of white adipose tissue (WAT) harvested from an Ate1-containing mouse (TM-treated Ate1flox/+;CaggCreER). The bar denotes 100 µm. (E) Same as in D except that WAT was from an Ate1-deficient mouse (TM-treated Ate1flox/−;CaggCreER). (F) Average weights of TM-treated Ate1-containing (n = 12; black curve) and Ate1-deficient (n = 11; red curve) mice as a function of time after the beginning of ad libitum high-fat diet. Weights were measured at weekly intervals for 10 weeks. Error bars indicate ±SD. (G) Comparisons, by immunoblotting, of Ucp1 protein levels in extracts from brown adipose tissue (BAT) (lanes 1 and 2) and WAT (lanes 3 through 6) from Ate1+/− and Ate1−/− mice 46 days (lanes 1 and 2) or ∼1 year (lanes 3–6) after TM treatment. Specific genotypes were as follows (genotypes after TM treatment are indicated in parentheses here, and also on top of the gel): lane 1, Ate1flox/− (+/−); lane 2, Ate1flox/−;CaggCreER (−/−); lane 3, Ate1flox/−;CaggCreER (−/−); lane 4, Ate1flox/− (+/−); lane 5, Ate1flox/−;CaggCreER (−/−); lane 6, Ate1flox/+;CaggCreER (+/−). Note abnormally high expression of Ucp1 in WAT of Ate1-deficient mice (lanes 3 and 5). An asterisk denotes a protein in WAT that cross-reacts with anti-Ucp1 antibody. (H) RT-PCR analyses of leptin and Ucp1 mRNA levels in BAT (lanes 1–4) and WAT (lanes 5–8) of Ate1-containing (denoted as “+/−”; lanes 2, 4, 6, and 8) and Ate1-deficient (denoted as “−/−”; lanes 1, 3, 5, and 7) mice ∼1 year after TM treatment. Specific genotypes: lanes 1 and 5, Ate1flox/flox;CaggCreER (−/−); lanes 2 and 6, Ate1flox/+;CaggCreER (+/−); lanes 3 and 5, Ate1flox/flox;CaggCreER (−/−); lanes 4 and 8, Ate1flox/− (+/−). (I) RT-PCR analyses of Ucp1 and Ucp2 mRNA levels in BAT, liver, muscle, and WAT of an Ate1flox/+ mouse (denoted as “+/−”) and an Ate1flox/−;CaggCreER mouse (denoted as “−/−”)∼1 year after TM treatment.
Spermatogenesis Defects and Infertility of Ate1-Deficient Male Mice
The marking of Ate1− allele with NLS-β-galactosidase (βgal) expressed from the PAte1 promoter revealed high levels of Ate1 expression in the neural tube and other specific, often sharply delineated, regions of Ate1+/− embryos [10]. An earlier study detected high levels of Ate1 expression in spermatogonia (stem cells, located at the periphery of testis' seminiferous tubules), and possibly also in early meiotic spermatocytes of adult mice [26]. Male Ate1−/−;CaggCreER mice that were produced by TM treatment (Fig. 4C) were found to be infertile in matings with Ate1-containing females, in contrast to identically TM-treated Ate1-containing males (data not shown). XGal staining of testis sections of NLS-βgal-marked Ate1 +/− mice in the present work (Fig. 6J) confirmed and extended the earlier evidence [26] for the pattern of Ate1 expression in testis. Whereas the lumens of seminiferous tubules in Ate1-containing testis were filled with inward-pointing sperm tails, the lumens of tubules in Ate1-deficient testis contained few sperm cells, in a disorganized arrangement (Fig. 6F–I), in agreement with the observed infertility of Ate1-deficient males.
To address the timing of requirement for Ate1 during spermatogenesis, we mated wild-type females with Ate1flox/− males that contained (instead of the CaggCreER gene) the PrpCreER gene (line 28.8) [27] or the PrmCre gene [28]. PrpCreER expresses TM-inducible CreER from the Prp promoter, whose activity in testis is confined to spermatogonia and meiotic spermatocytes [27]. In contrast, PrmCre expresses the (unconditionally active) Cre recombinase from the protamine promoter, which is active at later stages of spermatogenesis, in (haploid) round and elongating spermatids [28]. Three breeding pairs for each of two kinds of Ate1flox/− males (PrpCreER-based and PrmCre-based) and wild-type females were set up. 33% fewer litters and 50% fewer pups were produced with Ate1flox/−;PrpCreER males, in comparison to Ate1flox/−;PrmCre males (Table 1). (This substantial difference is expected to be even larger in a setting where an expressed Cre does not require a second, TM-mediated step for activation, as is the case with TM-independent Cre expressed from the Prm promoter, but not with TM-inducible CreER, expressed from the Prp promoter.) Nearly equal numbers of the Ate1flox (active) and Ate1− (inactive) alleles were present in the heterozygous progeny of matings that involved Ate1flox/−;PrmCre males (13 versus 14 pups containing Ate1flox versus Ate1− alleles, respectively). In contrast and most revealingly, only one Ate1− (inactive) allele but 12 Ate1flox (active) alleles were present in the progeny of matings that involved Ate1flox/−;PrpCreER males (Table 1). These findings suggest that the PrmCre-mediated inactivation of the Ate1flox allele, which occurs at a post-meiotic stage of spermatogenesis [28], takes place at a time when Ate1 is no longer essential for production of viable sperm cells, thus accounting for high frequency of the Ate1− allele in the progeny of matings that involve Ate1flox/−;PrmCre males. In contrast, the PrpCreER-mediated inactivation of the Ate1flox allele, which takes place in meiotic spermatocytes [27], clearly discriminated against the transmission of the Ate1− allele, in comparison to the Ate1flox (active) allele, most likely because spermatocytes that became Ate1-deficient before they became haploid were sufficiently perturbed by the absence of arginylation to either undergo apoptosis or differentiate abnormally, yielding defective sperm cells.
Table 1. Genotypes of mice from matings of Ate1+/+ females with Ate1flox/− males containing testis-specific Cre transgenes.
| Ate1flox/−;PrmCre ♂ x wild type ♀ | Ate1flox/−;Prp28.8Cre ♂ x wild type ♀ | |
| # breeding pairs | 3 | 3 |
| # litters | 6 | 4 |
| Average litter size | 7 | 5.25 |
| total pups | 42 | 21 |
| # floxOFF | 14 | 1 |
| % floxOFF | 33 | 4.7 |
| # floxON | 13 | 12 |
| % floxON | 31 | 57 |
Matings involving the Prp28.8 Cre strain occurred ∼1 month following TM treatment.
Previous work demonstrated a defective assembly of synaptonemal complexes and massive apoptosis of spermatocytes in Ubr2−/− mice [26]. The Ate1 R-transferase acts upstream of Ubr2 and other Ub ligases of the N-end rule pathway (Fig. 1A). Given a role of Ate1 in spermatogenesis demonstrated in the present study, it is possible that the currently unknown N-end rule substrate(s) whose degradation is in down-regulated in Ubr2−/− spermatocytes is an Ate1 substrate. To assess the extent of apoptosis in Ate1-deficient spermatocytes, we employed immunoblotting with antibody to poly(ADP-ribose)-polymerase (PARP), which is cleaved by caspases late in apoptosis. Anti-PARP antibody detected the (expected) 116 kDa full-length PARP in extracts from Ate1-containing mouse testis, but no 85-kDa PARP fragment, a marker of apoptosis (Fig. 6K, lane 1) [29]. In contrast, Ate1-deficient testis contained the 85-kDa fragment of PARP but virtually no full-length PARP, indicating extensive apoptosis in the absence of Ate1 (Fig. 6K, lane 2; cf. lane 1), in agreement with cytological and Ate1−/− male-infertility data (Fig. 6F–I). The 85-kDa PARP fragment is expected to bear N-terminal Gly [29], which is not a substrate of the Ate1 R-transferase (Fig. 1A). Thus the absence of the 85-kDa PARP fragment in Ate1-containing testis (Fig. 6K, lane 1) signifies the lack of production of this fragment by caspases, rather than its degradation by the arginylation branch of the N-end rule pathway. Proteins that require N-terminal arginylation for their degradation and that are likely to be relevant to meiotic functions of Ate1 include Rec8 [30], [31], a subunit of meiotic cohesin whose cleavage by separase is expected to produce an Ate1 substrate, similarly to the cleavage of Scc1/Rad21, the somatic counterpart of Rec8 (see Introduction).
Hyperkinesia, Seizures, and Enlarged Brains of Ate1-Deficient Mice
Most of Ate1−/−;CaggCreER mice (96 of 180) were strikingly hyperactive (hyperkinetic) (Figs. 6C–E and 8A). Intact brains harvested from Ate1-deficient mice appeared swollen, in comparison to brains harvested, in parallel, from Ate1-containing siblings (Fig. 6A). While the average brain weight, as a percentage of total body weight (TBW), of Ate1-containing mice was 1.96%, that of Ate1−/−;CaggCreER mice was 3.09% (Fig. 6A). In addition, there was a larger scatter of relative brain weights for Ate1-deficient mice, in comparison to identically TM-treated Ate1-containing controls. In particular, the brains of some Ate1-deficient mice reached 5% of TBW (Fig. 6A). Histological patterns of NLS-βgal [10] expressed from the PAte1 promoter in the brains of Ate1+/− mice (data not shown) were in agreement with in situ hybridization data in the Allen Brain Atlas (http://www.brain-map.org/), in that Ate1 was expressed at varying but significant levels throughout the mouse brain, particularly in the hippocampus, dorsal thalamus, and cerebellum. No Ate1 protein could be detected in brain extracts of Ate1−/−;CaggCreER mice, in contrast to extracts from wild-type or Ate1+/− brains (Fig. 1C, D). The virtually null Ate1 state of the brain in Ate1−/−;CaggCreER mice was also indicated by a strong accumulation of Rgs4, a physiological substrate of Ate1 (see Introduction) (Fig. 1D).
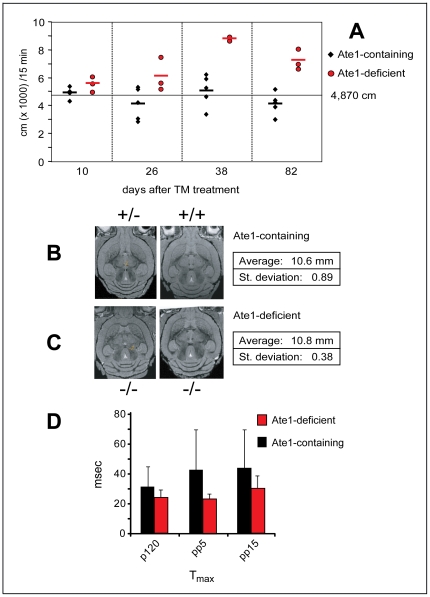
Figure 8. Brain abnormalities and behavioral phenotypes of Ate1-deficient mice.
(A) Ate1-deficient mice become hyperactive as a function of time after TM-mediated ablation of Ate1. Total distance (in cm) traveled over 15 min in the open field test box (2500 cm2). This test was repeated every ∼2 weeks after the end of TM treatment. The data for Ate1-containing mice (n = 5; their genotypes were Ate1flox/+, Ate1flox/−, and Ate1flox/+;CaggCreER) and Ate1-deficient mice (n = 3; Ate1flox/−;CaggCreER) are indicated by black diamonds and red circles, respectively. The horizontal bars indicate mean values. The average total distance traveled over 15 min for all Ate1-containing mice (n = 37) was 4,870 cm. (B) Representative magnetic resonance images showing equivalent horizontal planes of Ate1-containing (Ate1flox/+;CaggCreER on the left, Ate1flox/+ on the right) brains ∼3 months after TM treatment. The indicated average width of the skull (measured at the widest point from left to right in the same plane) of four Ate1-containing mice was 10.6 mm (±0.89 mm). (C) Same as in B except with brains from Ate1-deficient (Ate1flox/−;CaggCreER) mice ∼3 months after TM-treatment. The average width of the skull (measured as in B) of four Ate1-deficient mice was 10.8 mm (±0.38 mm). (D) Comparison of the response latency (Tmax; recorded in msec) between Ate1-containing (n = 3; black bars) and Ate1-deficient mice (n = 3; red bars) to a 40-msec pulse of 120 dB (p120; p<0.3), a 40-msec pulse of 120 dB preceded by a pre-pulse of 5 dB (pp5; p<0.09), or a 40-msec pulse of 120 dB preceded by a pre-pulse of 15 dB (pp15; p<0.01). Statistical analysis was performed using an unpaired t-test.
We carried out cell proliferation assays with Ate1−/−;CaggCreER mice (and controls), using 5-ethynyl–2′-deoxyuridine (EdU). In examinations of EdU-labeled brain sections, we paid particular attention to regions such as the hippocampus and the periventricular zone of the lateral ventricles, where neurogenesis is known to occur. However, no differences in EdU incorporation between Ate1-deficient and Ate1-containing brains were observed (data not shown), consistent with a brain edema (fluid accumulation) being a significant cause of brain enlargement in Ate1-deficient mice. We also determined the water content of freshly isolated brains, by subtracting their “dry weights” (after freeze-drying) from their total weights. The average water content and dry weight of control (Ate1-containing) brains was 79.9% and 20.1%, respectively, versus 80.5% and 19.5%, respectively (p<0.03), for Ate1-deficient brains (Fig. 6B). Thus cerebral edema at least contributes to the observed differences in brain weight between Ate1-deficient and Ate1-containing mice. It remains to be determined whether an edema (owing, e.g., to an osmotic imbalance or inflammation) suffices to account for consistently observed Ate1-dependent differences in brain weights (Fig. 6A, B).
There was also a 10-fold higher propensity for seizures among Ate1-deficient mice. For example, during routine cage changes and handling of mice, ∼3.1% of Ate1-deficient mice (38 of 1,232) versus ∼0.3% of identically TM-treated Ate1-containing mice had tonic-clonic seizures. The skulls of Ate1-deficient mice appeared to be thinner, “softer” than the sculls of Ate1-containing mice. Although MRI analyses did not reveal statistically significant abnormalities in the shape or size of skulls in Ate1-deficient mice (Fig. 8B, C), the MRI data did not preclude the possibility that bone structure may be perturbed in the absence of Ate1. These issues remain to be addressed.
The neurological/behavioral abnormalities of Ate1-deficient mice included an enhanced startle response, a marker for increased anxiety in rodents. Specifically, the latency between stimulus and response (Tmax) for Ate1-deficient mice was between 54% and 76% of the average latency for Ate1-containing controls, i.e., Ate1-deficient mice reacted significantly faster (Fig. 8D), thus exhibiting an enhanced startle response. The open field test is used to assess locomotor, exploratory and anxiety-like behavior in rodents. This test revealed a remarkably hyperkinetic behavior of Ate1-deficient mice (Figs. 6C–E and 8A), consistent with their enhanced startled response (Fig. 8D). The initial test involved a 15-min comparison of movements of Ate1-deficient mice versus Ate1-containing siblings of the same litter. An Ate1-deficient mouse traveled, during the test, a 3-fold greater distance than their (identically TM-treated) Ate1-containing counterpart (175.71 m versus 55.63 m, respectively) (Fig. 6C). The mean velocity of an Ate1−/−;CaggCreER mouse was 19.5 cm/sec, in comparison to 7.0 cm/sec for a wild-type (Ate1 +/+) mouse, 6.2 cm/sec for an Ate1+/+;CaggCreER mouse, and 5.5 cm/sec for an Ate1flox/+;CaggCreER mouse (Fig. 6E).
To assess generality of this striking phenotype, we repeated the open field test with three Ate1-deficient mice at 10, 26, 38, and 82 days after TM treatment, in parallel with TM-treated Ate1-containing (control) mice. At 10 days after TM treatment, i.e., soon after the acquisition of the Ate1−/− genotype, the differences between distances travelled by Ate1-deficient versus Ate1-containing mice were small (Fig. 8A). However, by 26 days after TM treatment, there was a statistically significant difference between Ate1-deficient and Ate1-containing mice in regard to their locomotor activity (Fig. 8A). By 82 days after TM treatment, the locomotor activity of Ate1-deficient mice, in conjunction with their elevated overall anxiety, increased so much that the device in which the open field tests were performed became nearly impractical, as Ate1-deficient mice (in contrast to Ate-containing ones) kept jumping out of the testing box.
Depletion of White Adipose Tissue in Ate1-Deficient Mice, and Their Resistance to Diet-Induced Obesity
To address the cause of a strikingly lower content of the peritoneal white adipose tissue (WAT) in Ate1-deficient mice, on average only 16% of WAT in Ate1-containing mice (Figs. 5D and 7A–C), we examined sections of intraperitoneal WAT. The average diameter of WAT adipocytes from Ate1-deficient mice was ∼30% of the average diameter of such cells in identically TM-treated Ate1-containing mice (25.5±7.4 µm versus 76.2±16.2 µm, respectively) (Fig. 7D, E). Thus, at least the bulk of WAT decrease in Ate1-deficient mice resulted from a decreased lipid content of individual adipocytes, rather from an extensive loss of adipocytes. Similar results were obtained with intrascapular brown adipose tissue (BAT) (Fig. 9A, B). The leanness of Ate1-deficient mice was particularly striking in view of their hyperphagy (see below).
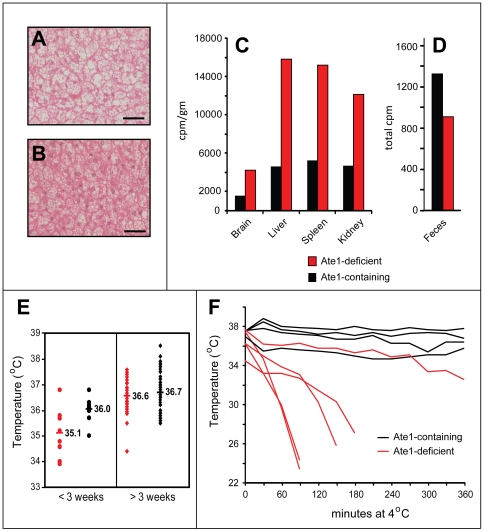
Figure 9. Body temperature, amino acid utilization and other properties of Ate1−/− versus Ate1+/+ mice.
(A) Hematoxylin/eosin staining (200× magnification) of a 10-µm section of brown adipose tissue (BAT) harvested from an Ate1-containing mouse (TM-treated Ate1flox/+;CaggCreER). The bar denotes 100 µm. (B) Same as in A except that BAT was from an Ate1-deficient mouse (TM-treated Ate1flox/−;CaggCreER). (C) Relative efficiencies of the import of 14C-amino acids and/or peptides from gastrointestinal tract in an Ate1-containing mouse (black bars; Ate1flox/+;CaggCreER) versus an Ate1-deficient mouse (red bars; Ate1flox/−;CaggCreER) 26 days after TM treatment. Shown here are representative comparisons of the retention of 14C (in cpm/gm) in the brains, livers, spleens, and kidneys 48 hr after gavage with a single bolus of 14C-labeled proteins (see Materials and Methods). (D) Total 14C (cpm) in the feces produced by mice in C within the first 48 hr after gavage with a bolus of 14C-labeled proteins. (E) Average core body temperatures of Ate1-containing (n = 8; black circles) versus Ate1-deficient (n = 11; red circles) mice during the first 3 weeks after TM treatment, in comparison to average core body temperatures of Ate1-containing (n = 54; black diamonds) versus Ate1-deficient (n = 36; red diamonds) mice beyond the first 3 weeks after TM treatment. (F) Core body temperature of individual Ate1-containing (black curves) and Ate1-deficient (red curves) mice, recorded at 30-min intervals after placing mice in a room at 4°C. Mice were removed from the cold room after 6 hr or when their core body temperature fell below 28°C.
We also asked whether the consistent difference in weight between Ate1-deficient and Ate1-containing mice on a standard ad libitum diet (Fig. 4A–C) could be reduced by an energy-rich, high-fat diet (HFD). At the end of the resulting 10-week test, the average weight of HFD-treated Ate1-containing mice was 152% of their starting weight (40.0 g versus 26.3 g). In contrast, the average weight of identically HFD-treated Ate1-deficient mice was only 122% (24.0 g versus 19.7 g) of their starting weight (Fig. 7F), indicating their relative resistance to diet-induced obesity. Yet another phenotype of Ate1-deficient mice, observed during their initial loss of weight after TM treatment (Fig. 4A, B), was their lower core body temperature, on average 35.1°C, in comparison to identically TM-treated Ate1-containing control mice, whose average core body temperature was 36.0°C during the same time, in the absence of weight loss (Fig. 9E). After the early deaths of ∼15% of Ate1−/−;CaggCreER mice (Fig. 4A, B), the average temperature of surviving mice (36.6°C) was not significantly different from that of Ate1-containing control mice (36.7°C) (Fig. 9E). As one would expect from their depletion of WAT (Fig. 7A–C), Ate1-deficient mice were strongly hypersensitive to cold (Fig. 9F).
Ucp1 is a proton carrier in the mitochondrial inner membrane that mediates a partial uncoupling of oxidative phosphorylation from ATP synthesis, an alteration that can increase heat production and thereby regulate body temperature and energy homeostasis. Although Ucp1 is normally expressed in BAT but not in WAT, several mouse mutants other than Ate1−/− that are resistant to diet-induced obesity have been shown to ectopically express Ucp1 in WAT [32], [33]. Using RT-PCR and immunoblotting with anti-Ucp1 antibody, we found that the levels of Ucp1 mRNA and Ucp1 protein in BAT did not change significantly between Ate1-deficient and Ate1-containing mice (Fig. 7G–I). Remarkably, however, the levels of both Ucp1 mRNA and Ucp1 were strongly increased in WAT of Ate1-deficient mice (Fig. 7G–I). A Ucp1-Ate1 connection revealed by these findings adds a new dimension to the understanding of Ucp1 regulation ([32] and refs. therein), and may also provide an experimental route to identifying a relevant circuit that involves Ate1.
Increased Metabolic Rate in Ate1-Deficient Mice
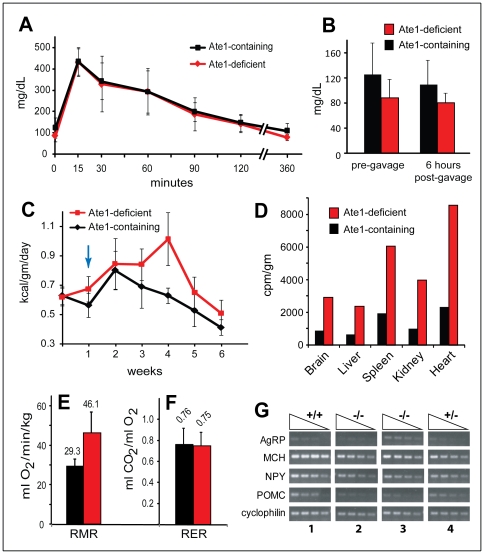
During the week prior to TM treatment, Ate1flox/−;CaggCreER and control (Ate1flox/+;CaggCreER) mice (at that point, both strains contained Ate1) consumed 0.63 and 0.62 kcal of standard chow per gram of body weight per day, respectively (Fig. 10C). Within a week after TM treatment the now Ate1-deficient Ate1−/−;CaggCreER mice increased their food consumption on average to 125% of identically TM-treated Ate1-containing mice (Fig. 10C). This pattern of significant hyperphagia of Ate1-deficient mice continued for the duration of this study, i.e., up to ∼8 months, with regular measurements for 6 weeks following TM treatment and intermittent comparisons afterwards (Fig. 10C). Thus, despite their initial decline of weight shortly after TM treatment and the early death of ∼15% of Ate1-deficient mice, and despite their subsequent failure to gain, on average, more than ∼63% and ∼69% of the weights of Ate1 +/+ and Ate1 +/− mice, respectively, the Ate1-deficient mice consumed significantly more food than their Ate1-containing counterparts (Fig. 10C). To address their patterns of glucose utilization, we fasted these mice for 16 hr and measured blood glucose before after administering a 50-mg (0.2 ml) bolus of glucose by gavage. The kinetics of rise and fall of blood glucose levels under these conditions was similar for Ate1-deficient and Ate1-containing mice (Fig. 10A). Ate1-deficient mice had lower fasting glucose levels than Ate1-containing mice (88.6 mg/dl versus 125.3 mg/dl, respectively; p<0.04), and also lower glucose levels 6 hr after administration of glucose (80.9 mg/dl versus 109.7 mg/dl, respectively; p<0.04), consistent with the (expected) higher energy expenditure of Ate1-deficient mice, and suggesting normal sensitivity of these mice to insulin (Fig. 10B). There were no other significant differences in blood composition (as well as urine composition) between Ate1-containing and Ate1-deficient mice (Tables 2 and 3).
Figure 10. Energy balance and metabolic rate in Ate1-deficient mice.
(A) Glucose tolerance test. Glucose concentration (mg/dL) in whole blood of Ate1-containing mice (n = 15; black curve) and Ate1-deficient mice (n = 11; red curve), at different times after a bolus of glucose by gavage, following a 16-hr fast. Glucose was administered at time zero. Error bars indicate ±SD. (B) Fasting blood glucose levels. Average blood glucose levels (mg/dL) in Ate1-containing mice (n = 15; black bar) and Ate1-deficient mice (n = 11; red bar), with measurements shortly before glucose gavage (after a 16-hr fast) and 6 hr after the gavage in A. Standard deviations are indicated. Statistical analysis was performed using an unpaired t-test (p<0.04). (C) Average daily energy consumption (kcal/gm of body weight) for Ate1-containing mice (n = 5; black curve) and Ate1-deficient mice (n = 3; red curve), with measurements from 1 week prior to tamoxifen (TM) treatment. Vertical arrow indicates the beginning of a 5-day TM treatment. Error bars indicate ±SD. (D) Relative efficiencies of the import of 14C-amino acids and 14C-peptides from gastrointestinal tract in Ate1-containing mice (black bars) versus Ate1-deficient mice (red bars). Shown here are representative comparisons of the retention of 14C (in cpm/gm) in the brains, livers, spleens, kidneys, and hearts of indicated mice 16 days after gavage with a single bolus of 14C-labeled proteins (see Materials and Methods). Mice were gavaged 26 days after TM treatment. (E) Comparison of resting metabolic rate (RMR) (measured in O2 (ml) consumed per kg of body weight per min) for Ate1-containing mice (n = 6; black bar) versus Ate1-deficient mice (n = 6; red bar). Standard deviations are indicated in E and F. Statistical analysis was performed using an unpaired t-test (p<0.008). (F) Comparison of the respiratory exchange ratio (RER), measured as CO2 (in ml) per ml of O2, for Ate1-containing mice (n = 6; black bar) and Ate1-deficient mice (n = 6; red bar) mice. No statistically significant difference in RER was observed. (G) RT-PCR analyses of AgRP, MCH, HPY, and POMC mRNA levels in the hypothalami of TM-treated Ate1-containing mice (Sets 1 and 4) versus Ate1-deficient mice (Sets 2 and 3). Set 1, Ate1flox/flox (+/+); Set 2, Ate1flox/flox;CaggCreER (−/−); Set 3, Ate1flox/flox;CaggCreER (−/−); Set 4, Ate1flox/+;CaggCreER (+/−). In sets 1 and 2, hypothalami were isolated 93 days after TM treatment. In sets 3 and 4 hypothalami were isolated ∼1 year after TM treatment. Sloping triangles indicate decreasing inputs (by 2-fold) of total RNA.
Table 2. Serum analyses.
| Ate1-deficient | Ate1-containing | |||||
| Mean | StDev | Mean | StDev | Units | normal range | |
| Glucose | 153.3 | 41.9 | 182.6 | 73.3 | mg/dl | 62–175 |
| BUN | 26.0 | 3.8 | 22.0 | 8.0 | mg/dl | 12–28 |
| Creatinine | 0.4 | 0.1 | 0.4 | 0.1 | mg/dl | 0.3–1.0 |
| Sodium | 158.0 | 2.8 | 129.8 | 62.3 | ||
| Potassium | 8.5 | 1.2 | 6.7 | 2.8 | ||
| NA/K ratio | 18.8 | 2.4 | 17.1 | 7.2 | ||
| Chloride | 112.8 | 3.4 | 91.9 | 43.4 | ||
| CO2 | 21.5 | 2.8 | 20.8 | 9.1 | ||
| Anion gap | 32.3 | 1.9 | 24.3 | 11.6 | ||
| Calcium | 9.3 | 0.4 | 8.1 | 3.4 | mg/dl | 3.2–8.5 |
| Phosphorus | 10.5 | 2.8 | 8.6 | 3.8 | mg/dl | 2.3–9.2 |
| Osm, Calc | 328.0 | 6.6 | 270.9 | 129.6 | ||
| TP | 5.2 | 0.4 | 4.5 | 1.8 | g/dl | 3.5–7.2 |
| Albumin | 3.1 | 0.3 | 2.7 | 1.1 | g/dl | 2.5–4.8 |
| Globulin | 2.0 | 0.1 | 1.8 | 0.7 | g/dl | 0.6 |
| Albumin/Globulin | 1.5 | 0.1 | 1.3 | 0.5 | 4.1–8 | |
| Bilirubin | 0.2 | 0.1 | 0.1 | 0.0 | mg/dl | 0.1–0.9 |
| AP | 102.3 | 41.6 | 113.7 | 48.0 | U/L | ∼70 |
| gamma gt | 0.0 | 0.0 | 0.0 | 0.0 | U/L | |
| ALT | 57.0 | 14.4 | 39.5 | 11.5 | U/L | ∼60 |
| AST | 409* | 286.5 | 199.2 | 83.9 | U/L | ∼100 |
| Cholesterol | 70.1 | 4.0 | 71.5 | 31.8 | mg/dl | 26–82 (∼1.5 mmol/L) |
| T4 | 2.7 | 0.9 | 2.4 | 0.8 | ng/dl | |
| T3 | 54.3 | 7.4 | 50.9 | 19.7 | µg/dl | |
Ate1-deficient mice with more severe phenotypes tended to have higher AST levels.
Table 3. Urinalysis.
| - | Ate1-deficient | Ate1-containing | Units |
| Glucose | neg | neg | mg/dL |
| Bilirubin | neg/small | neg/small | |
| Ketone | neg | neg | mg/dL |
| Specific Gravity | 1.03 | 1.02 | |
| Blood | neg | neg | |
| pH | 6.2* | 7.1 | |
| Protein | 100 | 100 | mg/dL |
| Urobilinogen | 0.2 | 0.2 | mg/dL |
| Nitrite | neg | neg | |
| Leukocytes | neg | neg |
Ate1-deficient mice had a significantly lower urine pH.
To measure metabolic rate, we employed indirect calorimetry (see Materials and Methods), determining O2 consumption and CO2 production by mice under resting conditions. The metabolic rate (resting metabolic rate, RMR) of Ate1-deficient mice was indeed higher than normal: they consumed on average 46.12 ml of O2 per kg per min, versus 29.3 ml of O2 per kg per min for Ate1-containing mice (Fig. 10E). In contrast, the respiratory exchange ratio, RER (the ratio of CO2 eliminated from the lungs to O2 taken into the lungs), a parameter that depends on a preferred source of fuel (e.g., carbohydrates versus fat), was similar for Ate1-deficient and Ate1-containing mice: 0.75 and 0.76, respectively (Fig. 10F).
The S. cerevisiae N-end rule pathway regulates the import of short peptides through the conditional degradation of Cup9, the import's repressor [34]. It is likely (but remains to be verified) that the N-end rule pathway regulates the transmembrane traffic of peptides in mammals as well. To address the possibility that significantly lower weights (despite hyperphagia) of Ate1-deficient mice might stem, at least in part, from an impaired ability to import peptides and/or amino acids from their gastrointestinal (GI) tract, we labeled E. coli with a mixture of 14C-amino acids and isolated a 14C-protein fraction that was essentially free of nucleic acids, fatty acids, lipids and carbohydrates (see Materials and Methods). Ate1-deficient and Ate1-containing mice were gavaged with a bolus of this 14C-protein preparation, followed by measurements of 14C in several organs of these mice (and in their feces) as a function of time post-gavage. Ate1-deficient mice passed less 14C in feces than Ate1-containing mice (Fig. 9D). Moreover, Ate1-deficient mice accumulated more of 14C in their brains, livers, spleens, kidneys and hearts than Ate1-containing mice (Figs. 9C and 10D). Irrespective of mechanistic causes involved (they remain to be understood), higher than wild-type levels of protein-derived 14C delivered to tissues of Ate1-deficient mice indicated the absence of significant defects in their transport of peptides and/or amino acids from GI tract.
Given the metabolic and behavioral abnormalities of Ate1-deficient mice (Figs. 6C–E, 8A, D and 10E, F), we also examined them for expression of neuropeptides. As we would be interested, at present, only in strong differences, a semiquantitative RT-PCR was employed. Using total RNA from hypothalami of Ate1-deficient versus Ate1-containing mice, we found no consistent differences between these mice in regard to the levels of hypothalamic mRNAs that encoded the agouti-related protein (AgRP) and the neural peptide Y (NPY) (Fig. 10G). Strikingly, however, there was a consistent and strong decrease of expression, in Ate1-deficient mice, of mRNA encoding proopiomelanocortin (POMC) (Fig. 10G). POMC is a precursor of several neurohormones with broad systemic and brain-specific functions ([35] and refs. therein). These functions include a role in melanocyte regulation (a process that is likely to be perturbed in Ate1-deficient mice; see above) and a down-regulation of food intake (the observed deficiency in POMC is consistent with hyperphagia of Ate1-deficient mice (Fig. 10C, G)). Similarly to a connection between Ate1 and the Ucp1 uncoupling protein (Fig. 7G–I), our finding of a link between N-terminal arginylation and the expression of POMC is likely to provide an experimental route to identifying the relevant Ate1-dependent circuit.
Discussion
A cell is alive owing to a cell-wide dynamic network of structurally or functionally interacting biopolymers. Some parts of this network can be sufficiently insulated, through their design, to be considered, in the first approximation, as distinct circuits. The N-end rule pathway is one such circuit. Its enzymes receive as their input specific degron-bearing proteins and convert them, through deamidation, arginylation, polyubiquitylation and processive degradation, into an output of proteolysis-derived short peptides (Fig. 1A). The rate and selectivity of the proteasome-mediated protein degradation by the N-end rule pathway are modulated by physiological effectors, including specific phosphokinases, short peptides, redox, heme and nitric oxide (see Introduction). Some of N-end rule substrates are produced by proteases that include MetAPs, separases, caspases and calpains. These and other nonprocessive proteases, which function as upstream components of the N-end rule pathway, have in common their ability to convert, through a cleavage, a pro-N-degron into an N-degron.
The present study expanded the earlier understanding of the Ate1 R-transferase (Fig. 1A) by making possible a postnatal inactivation of mouse Ate1. (Unconditional deletion of Ate1 results in embryonic lethality [10].) Described and discussed in Results is a large set of defects, some of them quite striking, in juvenile and adult mice upon the postnatal inactivation of Ate1 and the resulting loss of N-terminal arginylation (Figs. 1C, D). The initial abnormality is a rapid decrease of body weight and early death of ∼15% of Ate1-deficient mice, with surviving mice attaining, gradually, only ∼70% of the weight of wild-type mice identically treated with tamoxifen (TM) (Fig. 4A–C). Both “partial” lethality and the transiency of acute crisis, over ∼3 weeks after TM treatment (red arrow in Fig. 4B), remain to be understood in molecular terms. This crisis and subsequent failure to thrive occur despite higher than normal food intake by Ate1-deficient mice (Fig. 10C). These mice contain little or no visceral fat (Figs. 5D and 7A–E), and exhibit an increased metabolic rate (Fig. 10E), resistance to diet-induced obesity (Fig. 7F), enlarged brains (Figs. 5D and 6A), kyphosis (Fig. 5B), a striking hyperkinesia (Figs. 6C–E and 8A), and male sterility (Fig. 6F–K).
Owing to current constraints of the Cre-lox technology ([25] and refs. therein), the extent of Ate1 inactivation, while nearly 100% in some mouse tissues, was variable in others. The TM-induced Ate1−/−;CaggCreER mouse strains are thus mosaics of Ate1flox/− and Ate1−/− cells, where Ate1−/− cells are a great majority in most organs, such as the brain, but even there do not reach 100% of all cells (see Results). The initial weight loss upon the TM-induced conversion of Ate1flox/− mice to Ate1−/− mice was accompanied by death of ∼15% of Ate1−/− mice (Fig. 4A, B). Such a “partially” lethal phenotype suggests that an adult-onset Ate1−/− genotype in all cells (as distinguished from most cells) of a mouse might be incompatible with viability, similarly to the embryonic lethality of unconditional Ate1−/− mice. Thus, paradoxically, the discovery, in the present study, of specific Ate1-linked defects in adult mice might have been made possible by incomplete penetrance of the Cre-induced conversion of Ate1flox to the Ate1− allele.
The set of definitively identified mammalian N-end rule substrates that involve N-terminal arginylation consists, at present, of fewer than 10 proteins. They subserve different functions, from chromosome segregation to control of apoptosis and regulation of G proteins (see Introduction). This set is the tip of the iceberg, as several considerations [2], in addition to our findings above, strongly suggest a larger number of physiological Ate1 substrates. Given this complexity, the specific and often striking phenotypes of Ate1-deficient mice that were discovered in the present work will be of major assistance in deciphering the underlying Ate1 circuits.
Methods
Animal Care and Treatments
All animal care and procedures in the present study were conducted according to the relevant NIH guidelines, and were approved (Protocol #1328) by the Institutional Animal Care and Use Committee, the Office of Laboratory Animal Research (OLAR) at the California Institute of Technology, where the entire present study was carried out. Mice were housed at ∼22°C, at a pathogen-free (barrier) facility, using a 12 hr light/12 hr dark cycle, with Laboratory Rodent Diet 5001 (PMI International, Richmond, IN) ad libitum. Mice aged between 3 and 8 weeks were treated with tamoxifen (TM) (Sigma) (2 mg in 0.2 ml sesame oil) by daily intraperitoneal (IP) injections over 5 days. Mice were weighed weekly, starting 3 days before the first TM treatment. For a high-fat diet (HFD) study, mice were fed ad libitum a diet containing 35.5% fat (BioServe, Frenchtown, NJ), and were weighed weekly.
Construction of Ate1flox/−;CaggCreER Mouse Strains
Mouse genomic DNA encoding Ate1 was isolated from a BAC library [10]. Two pBluescript-based plasmids were used to construct the targeting vector. In one insert, a ∼12 kb HindIII fragment contained Ate1 exons 1a, 1b, 2, and 3 as well as ∼1.9 kb of DNA 3′ of exon 3 (ending just before exon 4). In the other insert, a ∼2.9 kb fragment contained Ate1 exons 4 and 5. The entire ∼12 kb HindIII fragment and a part of the ∼2.9 kb fragment were modified as described below and assembled into a final ∼22.5 kb targeting vector consisting of the following parts (Fig. 2C): (i) pBR322 backbone (New England Biolabs, Ipswich, MA); (ii) a ∼6.3 kb “long arm” of Ate1 homology containing the Ate1 exon 1a, the bidirectional PAte1 promoter [14], and exon 1b; (iii) A single loxP site ∼300 bp upstream of Ate1 exon 2; (iv) a ∼2 kb fragment that contains, 50 bp downstream of Ate1 exon 4, a “floxed” Hph (hygromycin) antibiotic-resistance marker, expressed from the PPGK promoter [36]; (v) a ∼1.2 kb “short arm” of Ate1 homology that spans most of the intron between exons 4 and 5; (vi) a gene encoding HSV-TK (herpes simplex virus thymidine kinase), expressed from the PPGK promoter. The targeting vector was linearized with BamHI and electroporated into CJ7 embryonic stem (ES) cells (a gift from Dr. Thomas Gridley, formerly of Jackson Laboratories, Bar Harbor, ME). ES cells were grown in DMEM supplemented with 15% fetal bovine serum (FBS), 0.1 mM non-essential amino acids, 0.1 mM β-mercaptoethanol, 2 mM glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 1 mM pyruvate, and 1,000 U/ml leukemia inhibitory factor (LIF) [37], using a feeder layer of hygromycin-resistant mouse primary fibroblasts that had been treated with 10 µg/ml mitomycin C for 3 hr at 37°C. Selection with hygromycin (at 0.2 mg/ml) and 1-(2′-deoxy, 2′-fluoro-β-D-arabinofuranosyl)-5-iodouracil (FIAU; at 0.4 µM) was started 24 hr after electroporation. Correctly targeted ES cell clones that contained “tri-loxed” Ate1 allele (Figs. 2C, D) were identified using Southern hybridization and PCR. Southern DNA probes and positions of primers for PCR are indicated in Fig. 2.
Two correctly targeted, independently produced ES cell lines that had apparently normal karyotypes were injected into 3.5-days-postcoitum C57BL/6J blastocysts and implanted into pseudopregnant females. The resulting male chimeric offspring were mated with C57BL/6J females. In some of the progeny, “floxed” ES cells became a part of germ line. Standard mating techniques [36], [38] were then used to produce, initially, mouse strains that contained a “tri-lox” Ate1 configuration, in that they also contained the floxed positive-selection PPGK-hph cassette (Fig. 2D). This DNA segment was removed by mating Ate1-tri-lox heterozygotes with EIIa-Cre mice that expressed Cre recombinase only in early, pre-implantation blastocysts [39], [40], [41]. Owing to the presence of three loxP sites at the initial floxed Ate1 locus, F1 progeny from this cross were mosaic, i.e., their tissues, including germ line, contained varying configurations of retained loxP sites, depending on specific patterns of Cre-mediated recombination (Fig. 2A–F). To isolate a mouse strain with the desired configuration of (retained) loxP sites (Fig. 2E), the above F1 mosaic mice were mated to wild-type C57BL/6 mice. This produced, among other progeny, a strain that lacked the PPGK-Hph cassette and had the desired Ate1flox/+ genotype, in the (mixed) C75BL/6J-129SvEv background.
Through the use of appropriate mating pairs, with genotyping of resulting progeny, we produced Ate1flox/−;CaggCreER mice as well as Ate1flox/flox;CaggCreER mice (Fig. 2). The former strain harbored one unconditionally null Ate1− allele (derived, through matings, from the previously constructed unconditional heterozygous Ate1+/− mice [10]) and one “floxed”, conditionally active Ate1flox allele that could be made null in the presence of active Cre recombinase. In the latter strain (Ate1flox/flox;CaggCreER), both copies of Ate1 were Ate1flox. These mouse strains also contained the CaggCreER gene, expressed from the ubiquitously active chimeric Cagg promoter (Fig. 2E) [25]. CaggCreER encoded CreER, a fusion between the phage P1 Cre recombinase and a derivative of the mouse estrogen receptor ligand binding domain. CreER was functionally inactive (sequestered in the cytosol) but could be activated by intraperitoneal (IP) injection of tamoxifen (TM) [25]. To produce Ate1flox/−;CaggCreER mice, we mated Ate1flox/+ mice with Ate1+/−;CaggCreER mice (the latter were generated by mating Ate1+/− with Ate1+/+;CaggCreER mice). To produce Ate1flox/flox;CaggCreER mice, we mated Ate1flox/+ mice with Ate1flox/+;CaggCreER mice (the latter were generated by mating Ate1flox/+ mice with Ate1+/+;CaggCreER mice). In the notations here and elsewhere in the paper, “flox-on” indicates a configuration depicted in Fig. 2E (the functionally active Ate1flox allele), whereas “flox-off” indicates a configuration depicted in Fig. 2F (the null Ate1− allele).
Southern Hybridization and PCR
Total genomic DNA was isolated from ES cells by washing them twice with phosphate-buffered saline PBS, followed by an overnight incubation at 50°C in 10 mM EDTA, 10 mM NaCl, 0.5% Sarcosyl,10 mM Tris-HCl (pH 7.5) containing Proteinase K at 0.2 mg/ml. Thereafter an equal volume of 75 mM NaCl in 100% ethanol was added. Precipitated genomic DNA was then gently washed twice with 70% ethanol and resuspended in T10E0.1 buffer (10 mM Tris (pH 8.0), 0.1 mM EDTA). Total genomic DNA was isolated from mouse tails or other tissues by overnight incubation at 55°C, with constant rotation, in 5 mM EDTA, 0.2 M NaCl, 0.3% SDS, 0.1 M Tris (pH 8.5) containing Proteinase K at 0.4 mg/ml. Thereafter an equal volume of isopropanol was added, and the mixture was gently inverted several times. Genomic DNA was then precipitated, and washed twice, with 70% ethanol, followed by a gentle resuspension in T10E0.1 buffer.
Southern hybridization was performed as described [42], with a 32P-labeled mouse DNA probes that was produced by PCR using the following primers: CB108F and CB107R (Table 2) to amplify a 219 bp genomic fragment containing Ate1 exon 5 (probe D, external probe); CB23 and CB24 (Table 4) to amplify a 929 bp genomic fragment that was a part of the long arm of the targeting vector (Probe A, internal probe) (Fig. 2). DNA probes were labeled with 32P-labeled using the Rediprime-II Random Prime Labeling System (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's protocol. Hybridization with Probe A was carried out overnight at 57°C in ExpressHyb solution (Clontech). The membrane was then washed once for 10 min at room temperature (RT) in 2xSSC/0.1% SDS, once for 30 min at 55°C in 2xSSC/0.1% SDS, once for 30 minutes at 58°C in 0.5xSSC/0.1% SDS, and once for 30 minutes at 65°C in 0.1xSSC/0.1% SDS, followed by autoradiography. (1xSSC is 0.15 M NaCl, 15 mM Na-citrate, pH 7.4.) Hybridization with Probe D was carried out overnight at 55°C in ExpressHyb solution (Clontech). The membrane was then washed once for 10 min at RT in 2xSSC/0.1% SDS, once for 30 min at 55°C in 2xSSC/0.1% SDS, and once for 30 min at 58°C in 1xSSC/0.1% SDS, followed by autoradiography.
Table 4. PCR primers used in the present study.
| Name | Nucleotide sequence (5′ to 3′) | Use |
| CB23 | ACTTTACAGTTGCTAGATAAGC | for PCR of Southern Probe A |
| CB24 | AGCAGGTTACTTGTCCAGTC | for PCR of Southern Probe A |
| CB107R | AATTCTTTAGACCCTTCTTTGTTT | for PCR of Southern Probe D |
| CB108F | TGTCAATAATGCAGCTGATGATGGGCTTTCATTGTCTTCTCATTCTTAGATGAGCCCATGGATTCTAC | for PCR of Southern Probe D |
| CB156F | CAA GCAG GGG AAG GAG GC | PCR detection ATE1-floxON |
| CB157R | TTC AGG AGT TAG CCA TTG CC | PCR detection ATE1-floxON and ATE1-floxOFF |
| AK49 | GGT ATT TGC TGC CGT CCT TTG GTG GT | PCR detection of ATE1-null |
| YT641 | CTG TTC CAC ATA CAC TTC ATT CTC AG | PCR detection of ATE1-null |
| AK83-Cbfix | CTG GAG ACA AAG CCC CAG CCA GAC | PCR detection of ATE1-null |
| Cre-1 | GTT CGC AAG AAC CTG ATG GAC A | PCR detection of Cre gene |
| Cre-2 | CTA GAG CCT GTT TTG CAC GTT C | PCR detection of Cre gene |
| CB159R | AC TGT AGA ATC CAT GGG CTC | PCR detection wild type ATE1 |
| CB160F | ACA GCA TAA GTG AGA CAC TCA | PCR detection wild type ATE1 |
| CB110F | GTT TGT GTC ACC ACT CCT ACC | PCR detection ATE1-floxOFF |
| oIMR0042 | CTA GGC CAC AGA ATT GAA AGA TCT | PCR detection IL-2 control |
| oIMR0043 | GTA GGT GGA AAT TCT AGC ATC ATC C | PCR detection IL-2 control |
| AgRP-for | GCGGAGGTGCTAGATCCA | RT-PCR |
| AgRP-rev | AGGACTCGTGCAGCCTTA | RT-PCR |
| NPY-for | CTCCGCTCTGCGACACTAC | RT-PCR |
| NPY-rev | AATCAGTGTCTCAGGGCT | RT-PCR |
| POMC-for | ACCTCACCACGGAGAGCA | RT-PCR |
| POMC-rev | GCGAGAGGTCGAGTTTGC | RT-PCR |
| MCH-for | ATTCAAAGAACACAGGCTCCAAAC | RT-PCR |
| MCH-rev | CGGATCCTTTCAGAGCAAGGTA | RT-PCR |
| cyclophilin-for | GGTGGAGAGCACCAAGACAGA | RT-PCR |
| Cyclophilin-rev | GCCGGAGTCGACAATGATG | RT-PCR |
| Ucp2-5′ | GGGGCGGCCGCATGGTTGGTTTCAAGGCCAC | RT-PCR |
| Ucp2-3′ | GGGGCGGCCGCTCAGAAAGGTGCCTCCCGAG | RT-PCR |
| Actb-5′ | ATGGATGACGATATCGCTGCG | RT-PCR |
| Actb-3′ | GAAGCTGTAGCCACGCTCGG | RT-PCR |
| Leptin-5′ | GGGGCGGCCGCATGTGCTGGAGACCCCTGTG | RT-PCR |
| Leptin-3′ | GGGGCGGCCGCTCAGCATTCAGGGCTAACAT | RT-PCR |
| Ucp1-5′ | GGGGCGGCCGCATGGTGAACCCGACAACTTC | RT-PCR |
| Ucp1-3′ | GGGGCGGCCGCTTATGTGGTACAATCCACTG | RT-PCR |
Genotyping of Mouse Strains
PCR-based genotyping was carried out with total genomic DNA isolated from various mouse tissues. Routine genotyping was performed using DNA from mouse tails. Specific Ate1 alleles and transgenes encoding specific derivatives of Cre were identified as follows. A two-primer PCR using the CB156F and CB157R primers (Table 4) was employed to produce and detect a 512 bp fragment of the Ate1flox (“floxON”, active) allele as well as a 472 bp fragment of the wild-type Ate1+ allele. A four-primer PCR using the CB110F, CB157R, OIMR0042, and OIM0043 primers Z (Table 4) was employed to produce and detect a 470 bp fragment of the Ate1flox-derived Ate1− allele (“floxOFF”) as well as a (control) 324 bp fragment of the Il-2 gene. A three-primer PCR using the AK49, YT641, and AK83-CBfix (Table 4) was employed to detect both a 300 bp fragment of the unconditional Ate1− allele [10] and a 560-bp fragment of the wild-type Ate1+ allele. A four-primer PCR using the Cre-1, Cre-2, CB159R, and CB160F primers (Table 4) was employed to detect both a 320 bp fragment of the CaggCreER transgene as well as a 1,060 bp fragment of the wild-type Ate1+ allele. All PCR reactions except for those to detect the CaggCreER transgene were carried out using HotStar Taq DNA polymerase, standard buffer conditions (Qiagen, Valencia, CA), 35 cycles of template denaturation for 30 seconds at 95°C, followed by primer annealing for 30 seconds at 60°C and primer extension for 1 minute at 72°C. PCR reactions for detecting CaggCreER were carried out using 30 cycles of template denaturation for 30 seconds at 95°C, followed by primer annealing for 30 seconds at 58°C and primer extension for 45 seconds at 72°C.
Northern and RT-PCR Analyses of RNA
Total RNA was isolated from various mouse tissues using the RNeasy Protect Mini Kit (Qiagen). Tissue disruption and homogenization were done in Buffer RLT and the MP FastPrep-24 instrument with Lysing Matrix D (MP Biomedicals, Solon, OH) for 2 runs at 6.5 m/s. First-strand cDNA was primed with oligo-dT using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) and PCR was carried out using primers cited in legends to the corresponding figures and in Table 4.
Tissue Extracts and Immunoblotting
Various mouse tissues were harvested and lysed in “Tissue Lysis Buffer” (10% glycerol, 0.05% NP40, 0.15 M NaCl, 2 mM EDTA, 1 mM dithiothreitol (DTT) 1 mM phenylmethylsulfonyl fluoride PMSF 50 mM HEPES, pH 7.5) plus freshly dissolved “Complete EDTA-Free Protease Inhibitors” (Roche), using the MP FastPrep-24 instrument and Lysing Matrix D (MP Biomedicals, with 2 or 3 runs at 6.5 m/s for 25 sec each, and with 5-min incubations on ice between the runs. The lysates were centrifuged at 10,000g for 20 min at 4°C. The supernatants were fractionated by SDS-12.5% PAGE, transferred to Immobilon-P PVDF membranes (Millipore, Billerica, MA), and analyzed by immunoblotting (IB) with antibodies indicated in specific figures. Immunoblots were visualized using SuperSignal West Pico or SuperSignal West Dura reagents (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions.
Other Analyses of Mouse Tissues
Specific mouse tissues were dissected immediately after euthanasia by CO2 inhalation. The tissues washed with PBS, blotted dry on Kimwipes, and weighed (wet). For dry-weight measurements, mouse brains were dissected intact, washed in PBS, blotted dry on Kimwipes, weighed wet, then incubated overnight in acetone. After acetone incubation, individual brains were lyophilized until their (dry) weight no longer decreased.
For routine histological examinations, tissues or organs were fixed in Bouin's solution or in 4% formaldehyde, using standard procedures [37]. Fixed samples were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. To stain for LacZ (NLS-βgal), dissected tissues or organs were fixed in LacZfix (0.2% glutaraldehyde, 5 mM EGTA, 0.1 M MgCl2 in PBS (pH 7.3)) for 4 hr, rinsed twice with PBS, dehydrated overnight at 4°C in 30% sucrose, 2 mM MgCl2 in PBS, and embedded and frozen in Tissue-Tek O.C.T. Compound (Sakura Finetek USA, Inc. Torrance, CA). Cryosections (prepared using a Tissue Tek Microtome/Cryostat model 4553) were mounted onto glass slides, fixed in LacZfix for 10 min at RT, washed 3 times in LacZWash (0.02% NP40, 01% Na-deoxycholate, 2 mM MgCl2 in PBS), and stained overnight at 37°C with LacZ stain (LacZWash containing 1 mg/ml XGal, 5 mM K4Fe(CN)6 and 5 mM K3Fe(CN)6). Stained sections were washed with PBS and mounted with Permount for light microscopy. Apoptosis was assessed by TUNEL, a nuclear DNA fragmentation assay, using a TUNEL kit (Roche, Indianapolis, IN), fluorescein-dUTP, and manufacturer's instructions. Cell proliferation was assayed using the Click-It Edu Cell Proliferation kit (Invitrogen).
In Vitro Arginylation Assay
The arginyl-transferase (R-transferase) reaction (50 µl) contained extracts for a specific mouse tissue (2.5 mg of total protein per ml), α-lactalbumin (arginylation reporter [14]) (0.5 mg/ml), total E. coli tRNA (0.6 mg/ml) (Sigma), total E. coli aminoacyl-tRNA synthetases (800 U/ml) (Sigma), 5 mM MG132 (proteasome inhibitor) (Sigma), 1 mM ATP, 30 mM KCl, 2 mM MgCl2, 2 mM β-mercaptoethanol, 10 mM Tris-HCl (pH 8.0) and 0.3 mM 3H-arginine (PerkinElmer, NEN Radiochemicals, Waltham, MA). The reaction mixture was incubated for 30 min at 37°C and deposited onto GF/C filter disks (GE-Healthcare, Pittsburg, PA). The filters were thereafter incubated for 10 min in 10% cold CCl3COOH, followed by 10 min in 5% CCl3COOH at 95°C. The filters were then washed in 5% CCl3COOH 3 times at RT, followed by a single ether∶ethanol (1∶1) wash, two ether washes, and measurements of 3H retained on a filter using a scintillation spectrometer.
Blood and Urine Analyses
Blood (∼0.6 ml per mouse) was withdrawn by cardiac puncture and transferred into BD Microtainer SST tubes (BD, Franklin Lakes, NJ). The serum fraction was prepared by centrifugation in a microcentrifuge after clotting occurred, immediately frozen in liquid N2 and stored at −80°C. The levels of glucose, cholesterol, sodium, potassium, chloride, calcium, phosphorus, blood urea nitrogen, creatine, total protein, albumin, total bilirubin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase, γ-glutamyltransferase (gamma gt), as well as the T3 and T4 hormones were determined by Phoenix Central Laboratories (Everett, WA).
Urine was obtained by placing the external urethra over a test tube. Urine samples collected from Ate1-deficient mice were pooled and compared with pooled urine from Ate1-containing mice. The levels of glucose, bilirubin, ketones, blood, protein, urobilinogen, nitrite, leukocytes, as well as the pH and specific gravity were determined using the Multistix 10 SG Reagent Strips (Bayer, Tarry Town, NY).
Measurements of Body Temperature and Cold Sensitivity
Mice (housed at one mouse per cage, without bedding but with food and water) were exposed to a 4°C environment for up to 6 hr Their core body temperature was monitored every 30 min via a rectal probe digital thermometer (Thermalert TH-8; Physitemp Instruments., Clifton, NJ).
Measurements of 14C-Protein Uptake from Gastrointestinal Tract
E. coli DH5α cells were grown in minimal M9 media supplemented with 0.5% glucose and 50 µCi of 14C-amino acids (derived from 14C-protein hydrolysate (Amersham)) until the incorporation of ∼70% of the added 14C amino acids. Cells were lysed by one freeze/thaw cycle in PBS containing 1 mg/ml lysozyme and incubated with RNase H and DNase I for 45 min at 37°C. A crude protein fraction was isolated by precipitation with cold 10% TCA (CCl3COOH), and the pellet was washed with ice-cold acetone. The pellet was redissolved in PBS, with a brief sonication to facilitate solubilization. 0.2 ml of the resulting sample, containing 260,000 cpm of 14C-labeled E. coli proteins was fed to a mouse by oral gavage. Urine and feces was collected at various times post-gavage. Total 14C was measured, using a scintillation spectrometer, in feces, urine, and (eventually) in mouse tissue samples that were collected either 48 hr or 15 days post-gavage.
Measurements of Glucose Uptake
For glucose analyses, mice were fasted for 24 hr, then gavaged with 50 mg glucose in 0.2 ml of water. Blood was collected through the lateral tail vein at 15, 30, 60, 90, 120, and 360 min post-gavage. Blood glucose levels were determined using the OneTouch UltraMini Blood Glucose Monitoring System (LifeScan, Johnson and Johnson, Milpitas, CA).
Metabolic Analyses
The resting metabolic rate was determined at the Mouse Physiology Laboratory in the Department of Physiology at the Geffen School of Medicine, UCLA using indirect calorimetry as previously described [43], [44]. Single mice were placed into a custom-made enclosed plexiglas chamber (25 cm×12 cm×7.5 cm, with 4 room air intake vents and one outflow port) and allowed to come to rest over a period of 30 min to 2 hr. Outflow of expired gases was sampled by the gas analyzer and recorded using a computerized acquisition system during a 30-min resting interval.
MRI Analyses of Mouse Brains
The procedures used were essentially the same as previously described [45]. Mice were given an IP injection of 40 mg/kg of Na-pentobarbital (Nembutal, Hospira, Inc., Lake Forest, IL). Once fully anesthetized, mice were transcardially perfused with 4% formaldehyde in PBS. MRI analysis was performed by the Caltech Brain Imaging Center. Briefly, mouse heads were excised and postfixed in 4% formaldehyde/PBS overnight. Hair and skin were removed from fixed heads, which were then soaked in 5 mM Gadolinium-based MR contrast agent (Prohance Bracco Diagnostics, Durham, NC) for 10 days, to decrease the intrinsic tissue relaxation rates and improve the MR acquisition efficiency. A gradient echo sequence (TE/TR = 8 msec/50 msec, 16 averages) was used to acquire 3D data sets of the mice heads, using a Bruker 7T Biospec animal magnet system. Images were reconstructed with an isotropic resolution of ∼90 µm and analyzed using Brainsuite 2 software [46].
Open-Field and Startle Response Tests
For the open-field activity measurements, individual mice were placed into a square chamber (50 by 50 cm). Movements along the x and y axes were tracked and analyzed using Ethovision software (Noldus, Leesburg, VA) over 15-min intervals. Startle response tests were carried out essentially as described previously [47].
Acknowledgments
We are grateful to current and former members of the Varshavsky laboratory for their advice and help. We thank R.-G. (Cory) Hu for antibody to Ate1, K. I. Piatkov for helpful discussions, N. V. Malkova for advice and assistance with behavioral tests, S. Pease for advice regarding ES cell manipulation, D. Procissi and K. P. Roos for help with MRI and metabolic rate tests, respectively, and E. Udartseva for assistance with genotyping mouse strains.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grants to A.V. from the National Institutes of Health (GM31530 and DK39520), the American Asthma Foundation, and the March of Dimes Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283:34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogk A, Schmidt R, Bukau B. The N-end rule pathway of regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–689. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 7.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Piatkov KI, Brower CS, Varshavsky A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol Cell. 2009;34:686–695. doi: 10.1016/j.molcel.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon YT, Kashina AS, Davydov IV, Hu R-G, An JY, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 11.Hu R-G, Sheng J, Xin Q, Xu Z, Takahashi TT, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 12.Hu R-G, Wang H, Xia Z, Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci USA. 2008;105:76–81. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu R-G, Brower CS, Wang H, Davydov IV, Sheng J, et al. Arginyl-transferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J Biol Chem. 2006;281:32559–32573. doi: 10.1074/jbc.M604355200. [DOI] [PubMed] [Google Scholar]
- 15.Xia Z, Webster A, Du F, Piatkov K, Ghislain M, et al. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang C-S, Shemorry A, Varshavsky A. Two proteolytic pathways regulate DNA repair by co-targeting the Mgt1 alkyguanine transferase. Proc Natl Acad Sci USA. 2009;106:2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasaki T, Zakrzewska A, Dudgeon D, Jiang Y, Lazo JS, et al. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284:1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holman TJ, Jones PD, Russell L, Medhurst A, Ubeda TS, et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4549–4554. doi: 10.1073/pnas.0810280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisele F, Wolf DH. Degradation of misfolded proteins in the cytoplasm by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Hwang C-S, Varshavsky A. Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc Natl Acad Sci USA. 2008;105:19188–19193. doi: 10.1073/pnas.0808891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakozova M, Kozak M, Wong CC, Bailey A O, Yates JRr, et al. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- 22.Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 23.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–960. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 24.Rajewsky K, Gu H, Kühn R, Betz UA, Müller W, et al. Conditional gene targeting. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi S, McMahon A. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 26.Kwon YT, Xia ZX, An JY, Tasaki T, Davydov IV, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber P, Schuler M, Gerard C, Mark M, Metzger D, et al. Temporally controlled site-specific mutagenesis in the germ cell lineage of the mouse testis. Biol Reprod. 2003;68:553–559. doi: 10.1095/biolreprod.102.005801. [DOI] [PubMed] [Google Scholar]
- 28.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulares HA, Yakovlev AG, Ivanova V, Stoica BA, Wang G, et al. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 30.Eijpe M, Offenberg H, Jessberger R, Revenkova E, Heyting C. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1-beta and SMC3. J Cell Biol. 2003;160:657–670. doi: 10.1083/jcb.200212080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, et al. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiskinis E, Hallberg M, Christian M, Olofsson M, Dilworth SM, et al. RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J. 2007;26:4831–4840. doi: 10.1038/sj.emboj.7601908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narvaez CJ, Matthews D, Broun E, Chan M, Welsh J. Lean phenotype and resistance to diet-induced obesity in vitamin D receptor knockout mice correlates with induction of uncoupling protein-1 in white adipose tissue. Endocrinology. 2009;150:651–661. doi: 10.1210/en.2008-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau K, Kauser S, Pritchard LE, Warhurst A, Oliver RL, et al. Proopiomelanocortin (POMC), the ACTH/melanocortin precursor, is secreted by human epidermal keratinocytes and melanocytes and stimulates melanogenesis. FASEB J. 2007;21:1–12. doi: 10.1096/fj.06-7398com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyner AL. Oxford, UK: Oxford University Press; 2000. Gene targeting: A practical approach.293 [Google Scholar]
- 37.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. Manipulating the Mouse Embryo: A Laboratory Manual.800 [Google Scholar]
- 38.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leneuve P, Colnot S, Hamard G, Francis F, Niwa-Kawakita M, et al. Cre-mediated germline mosaicism: a new transgenic mouse for the selective removal of residual markers from tri-lox conditional alleles. Nucl Acids Res. 2003;31:e21. doi: 10.1093/nar/gng021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holzenberger M, Lenzner C, Leneuve P, Zaouri R, Hamard G, et al. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucl Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, et al. New York: Wiley-Interscience; 2006. Current Protocols in Molecular Biology. [Google Scholar]
- 43.Wasserman K, Hansen JE, Sue DY, Stringer WW, Whipp BJ. Philadephia, PA: Lippincott, Williams & Wilkins; 2005. Principles of Exercise Testing and Interpretation: Including Pathophysiology and Clinical Applications, 4th edition.568 [Google Scholar]
- 44.Dauger S, Nsegbe E, Vardon G, Gaultier C, Gallego J. The effects of restraint on ventilatory responses to hypercapnia and hypoxia in adult mice. Respir Physiol. 1998;112:215–225. doi: 10.1016/s0034-5687(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 45.Redwine JM, Kosofsky B, Jacobs RI, Games D, Reilly JF, et al. Dentate gyrus volume is reduced before onset of plaque formation in PDAPP mice: A magnetic resonance microscopy and stereologic analysis. Proc Natl Acad Sci USA. 2003;100:1381–1386. doi: 10.1073/pnas.242746599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Medical Image Analysis. 2002;8:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 47.Geyer MA, Swerdlow NR. Measurement of startle response, prepulse inhibition, and habituation. Curr Protoc Neurosci. 1998;8.7:8.7.1–8.7.15. doi: 10.1002/0471142301.ns0807s03. [DOI] [PubMed] [Google Scholar]