Expanding legume research beyond the model members of the subfamily Papilionoideae (papilionoids) is necessary if we wish to capture more of the diversity of the enormous, economically important legume family. Chamaecrista fasciculata is emerging as a nonpapilionoid model, belonging to the paraphyletic subfamily Caesalpinioideae within the mimosoid clade. Mimosoids diverged from the common ancestor of soybean (Glycine max), Medicago truncatula, and Lotus japonicus nearly 60 million years ago—nearly contemporaneously with the origin of legumes. There is growing interest within the legume community in C. fasciculata as a complementary legume model for a number of reasons, including phylogenetic position, nodulation within a clade of limited nodulating species, nonpapilionoid floral morphology, herbaceous growth habit, and tractability in laboratory and field settings. Whole-transcriptome sequencing (WTS) of C. fasciculata shoots, roots, and nodules, along with gene expression and single nucleotide polymorphism (SNP) profiling, provides community resources to address fundamental questions about legume evolution. A range of ecotypes, development of functional genomics tools, and an integration of research and undergraduate education leverage these genomic resources.
WHY CHAMAECRISTA?
Legumes are the third-largest family of flowering plants, constituting about 8% of all angiosperms. Analysis of genomes of three model legumes, M. trucatula, L. japonicus, and soybean are advancing our understanding of this economically important family, yet the models all fall within one large clade of subfamily Papilionoideae (papilionoids) that encompasses less than half of the species diversity of the legumes (Fig. 1). In 2005, the Cross-Legume Advances through Genomics Conference was held in Santa Fe, New Mexico, to set priorities for future research in this enormous and tremendously diverse family. Among these priorities was the expansion beyond model and crop legumes by developing new legume experimental systems. To quote from the conference report (Gepts et al., 2005, p. 1232):
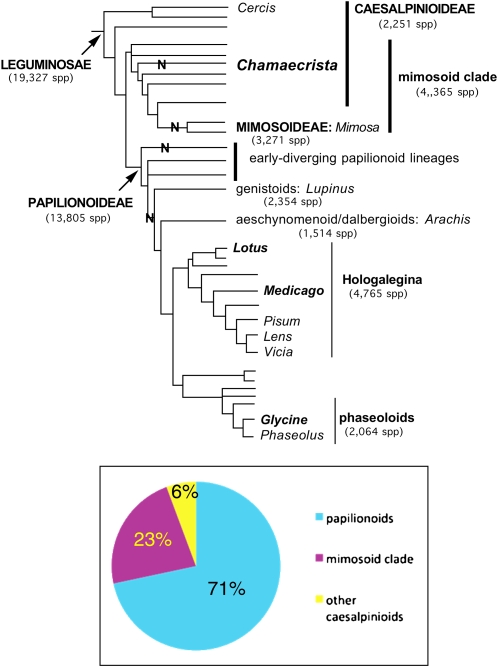
Figure 1.
Relationship of Chamaecrista to other legumes. Top: legume phylogeny (modified from Lavin et al., 2005; species numbers from Lewis et al., 2005). Chamaecrista is a member of the mimosoid clade, which includes the monophyletic subfamily Mimosoideae and over 800 species of the paraphyletic subfamily Caesalpinioideae. “N” along a branch indicates a group in which nodulation is known to occur in some or all members (Sprent, 2001). Bottom: The mimosoid clade comprises over 20% of all legume species.
“Some of the fundamental biological questions, such as the origin of legume-characteristic traits, require an evolutionary approach that encompasses the entire legume family. Such traits include reproductive development (especially floral and pod development), the origin of nodulation, and the evolution and importance of polyploidy. The two foci, while providing coverage for most economic legumes involved in food and feed, do not come close to covering the biodiversity included in the Fabaceae. Hence, to address the issue of legume-characteristic traits, other species may have to be considered, including species in the basal clades of the Papilionoideae, the Caesalpinioideae (e.g. Chamaecrista sp.), and the Mimosoideae. In these species, ad hoc genomic resources targeted toward evolutionary genomics questions of interest will have to be developed to allow comparisons with reference and other legume species.”
It was no accident that Chamaecrista (Fig. 2) was specifically mentioned as a complementary model for the legumes in this recommendation. A primary reason is its phylogenetic position in the family. Chamaecrista belongs to an evolutionary lineage (the mimosoid clade) that is the sister group to the papilionoid clade (e.g. Arachis, Glycine, Medicago, and Lotus), from which it diverged nearly 60 million years ago—nearly contemporaneous with the origin of legumes (Lavin et al., 2005; Fig. 1). The mimosoid clade comprises 141 genera and 4,365 species (Lewis et al., 2005), making it the second-largest lineage in the family after the papilionoids (478 genera, 13,805 species). This currently modelless 23% of the legume family is a very diverse group in its own right, as reflected in the historical, morphologically based taxonomy of its members. The majority of its genera and species (82 genera, 3,271 species) are a distinctive group long recognized as the subfamily Mimosoideae (e.g. acacia and sensitive plant mimosa), a natural group typically with distinctive inflorescences of small flowers with showy anthers and mostly doubly compound leaves. The remaining genera of the mimosoid clade have traditionally been classified as members of the paraphyletic subfamily Caesalpinioideae, a group with often showy and relatively radially symmetric flowers, like Chamaecrista (Fig. 2). Papilionoid flowers have strong bilateral symmetry, a derived condition within the angiosperms (Tucker, 1989). Comparisons of the floral transcriptomes of Chamaecrista and papilionoid legumes may offer clues to the origins of bilateral symmetry within the legumes.
Figure 2.
C. fasciculata is a prairie plant found throughout the northeastern, southeastern, north-central, and south-central United States. A, Inflorescence. B, Flower. C, Root with nodules.
The mimosoid clade in general, and Chamaecrista in particular, is useful in obtaining a more complete picture of legume evolution, including: (1) understanding the role of polyploidy in legume genome evolution; (2) elucidating origins of nodulation in the family; and (3) characterizing the evolution of floral development in legumes. Beyond the intrinsic interest in understanding the biology and evolution of the mimosoid clade, the group is important in that it serves as an outgroup for the papilionoids. In addition, Chamaecrista has utility for more applied studies.
GENOME EVOLUTION
The common ancestor of the three papilionoid legume models experienced a whole-genome duplication event (Schlueter et al., 2004; Pfeil et al., 2005) that is shared with many other papilionoid legumes (e.g. Arachis: Bertioli et al., 2009) but not shared with Arabidopsis (Arabidopsis thaliana) or poplar (Populus trichocarpa; Cannon et al., 2006). It is not known whether this event took place in the most recent ancestor of the three models, in an earlier papilionoid ancestor, somewhere deeper within the legumes, or perhaps even in an ancestor shared with members of the nitrogen-fixing clade of rosid families. The phylogenetic position of Chamaecrista (Fig. 1) makes it an excellent taxon for understanding where, in the evolution of legumes, this polyploid event occurred. If Chamaecrista, despite its low chromosome number and relatively small genome, shares the event identified in papilionoids it will mean that the vast majority of legumes share a common polyploid origin, suggesting that the radiation of the third-largest flowering plant family could have been affected by genome duplication. Alternatively, the polyploid event may be confined within the papilionoid clade—the largest species radiation within the family. Polyploidy is a potent generator of evolutionary novelty, leading to morphological innovation (De Bodt et al., 2005; Freeling and Thomas, 2006). Thus it is possible that polyploidy could be associated with major species radiations and with the origin of novel structures such as the nodule.
NODULATION
Like the three legume models and the vast majority of their papilionoid relatives, Chamaecrista participates in nitrogen-fixing symbiosis with rhizobial bacteria and forms nodules. This, however, is nearly unique among caesalpinioid legumes, though the core Mimosoideae also nodulate (Sprent, 2001, 2007, 2009). Phylogenetic considerations suggest that nodulation may have evolved in Chamaecrista independently from papilionoids, either along with or independently from Mimosoideae (Doyle, 1998; Fig. 1). Chamaecrista species all have indeterminate nodules of the caesalpinoid type common also to all mimosoids and many papilionoids, though some Chamaecrista species have persistent infection threads, a trait that has been considered primitive by some workers (Naisbitt et al., 1992). Even if Chamaecrista nodules are homologous with those of papilionoids, they represent around 60 million years of independent evolution relative to any of the better-characterized nodules of the three models, and thus could afford insights into the origin of the nodule and such phenomena as gene recruitment in generating evolutionarily novel structures.
FLOWERING
Analysis of the regulation of flowering time, inflorescence architecture, and floral morphology of Chamaecrista affords the opportunity to sort out patterns of gene expression that are unique to the legumes, patterns that are conserved within the legumes, and patterns that diverge in the papilionoids.
Understanding the genetics of flowering time in Chamaecrista could also further our understanding of evolutionary and ecological genomics, specifically the interplay between ecotypic differentiation and vulnerability to climate change. For example, Etterson and Shaw (2001) analyzed three ecotypes of C. fasciculata, a northern American prairie plant, which vary in a number of traits, including time to flowering. Predicted rates of evolutionary response in the ecotypes to a warmer, more arid climate were much slower than the predicted rate of climate change. Identification of allelic variation in genes that regulate the time to flowering in region-specific ecotypes could be used to create models for regulation of flowering time. These models could be applied to breeding programs to enhance survival of species threatened by climate change.
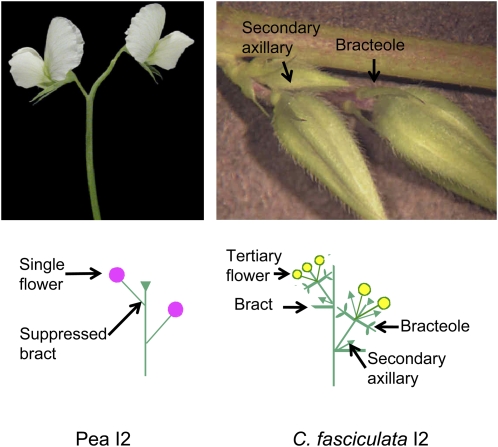
Chamaecrista's phylogenetic position also informs our understanding of inflorescence architecture. Unlike Arabidopsis, legumes have compound rather than simple racemes. That is, the axillary meristem that forms in a node after the floral transition has occurred produces an inflorescence meristem (I2 for second-order inflorescence) rather than a floral meristem (Fig. 3). C. fasciulata produces floral meristems from the developing I2, accompanied by the outgrowth of other organs including bracts, bracteoles, additional axillary flowers, and vegetative shoots. In contrast, papilionoids, specifically the well-studied garden pea (Pisum sativum), produce flowers and suppress the outgrowth of additional organs (Tucker, 1989; Singer et al., 1990; Fig. 4).
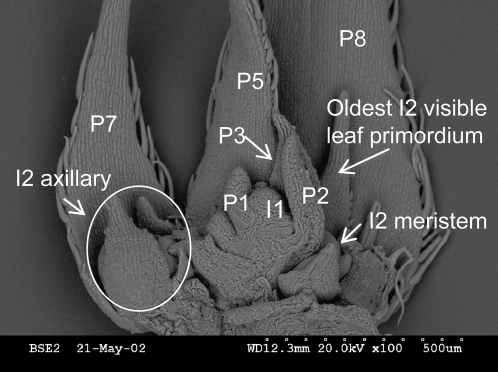
Figure 3.
Scanning electron micrograph (100×) of a C. fasciculata shoot tip. The I1 is the first-order meristem that has initiated the eight-leaf primordia (P4 and P6 were removed for visual clarity). Each leaf primordium is at a node containing, in this case, an I2 inflorescence meristem, but only the older nodes have substantial organ development visible. Note that none of the leaf primordia have matured to the point where leaflets are developing. In this one shoot tip, eight different stages of leaflet and I2 meristem/I2 axillary development are represented.
Figure 4.
Comparison of pea and C. fasciculata I2 inflorescences (axillary inflorescences). Garden peas exhibit extensive suppression of organ outgrowth from bracts to second flowers and/or vegetative shoots in a node, to bracteoles, which subtend flowers.
At the level of the flower, the three genomic models are all characterized by the derived, bilaterally symmetric, papilionoid floral morphology most people associate with legumes. Papilionoid flowers are closed with petals covering stamens and carpels. Chamaecrista has an open, relatively radial floral morphology with helical organ initiation that is more ancestral in form (Tucker, 1996). We are fortunate to have an unusually rich set of morphological analyses of floral and inflorescence development in the basal legumes, including Chamaecrista, from Shirley Tucker's research (for review, see Tucker, 2003). Coupling her work with the extensive characterization of floral and inflorescence mutants in pea, many of which have been cloned, provides a framework for a meaningful, comparative genomic analysis of flowering in Chamaecrista and the sequenced papilionoid models. From an evolutionary perspective, analysis of floral development in Chamaecrista will permit the discrimination of traits unique to papilionoids from characteristics shared with the mimosoid clade. Insights will thus be obtained into how a once common gene repertoire has been modified over roughly 60 million years to produce the very different morphologies in these two groups.
ECOLOGICAL, AGRONOMIC, AND PRACTICAL UTILITY
In addition to the insights Chamaecrista can provide into these basic research questions it also is ideal for some more practical applications. From an applied perspective, Chamaecrista, with its nitrogen-fixing capacity and rapid growth, is an ideal legume for early establishment of mixed prairie for synfuel production. As demonstrated by Tilman et al. (2006), mixed prairie provides significantly higher net-energy gain than crop-based biofuels and allows productive, carbon-negative use of marginal farmland and other low-fertility soils without significant nutrient input.
Among the 330 species of Chamaecrista, practical considerations make C. fasciculata well suited to be a model species. It includes both annual and perennial genotypes, is small and herbaceous, making it tractable in laboratory and field settings. Related Chamaecrista species have small genomes, approximately 650 Mbp.
DEVELOPING C. FASCICULATA AS A MODEL NONPAPILIONOID LEGUME
The Chamaecrista community held its inaugural workshop in December, 2008 at the International Conference on Legume Genomics and Genetics (http://www.ccg.unam.mx/iclgg4/program.php) in Puerto Vallarta, Mexico. We focused on resource and tool development to address biological questions raised earlier in this update, with an emphasis on integrating research and undergraduate education. Community resources, including sequence data, are available at the National Center for Biotechnology Information Trace Archive, the Legume Information System (http://www.comparative-legumes.org), and http://serc.carleton.edu/chamaecrista/index.html.
GENOMIC RESOURCES
A C. fasciculata WTS effort at Carleton College, Cornell and Iowa State Universities, and the National Center for Genome Resources demonstrated the feasibility of using next-generation WTS to provide useful information about the gene space of an organism without closely related reference species. Chamaecrista plants were raised and developmentally staged in soil or hydroponic units (for nodule development) in controlled growth chambers. RNA was isolated from a Minnesota ecotype from: shoot tips of plants at seeds 4 d postimbibition and with two, four, six, eight, 12 to 16, and 18 to 21 expanded leaves, root tips, branched and unbranched roots, roots with nodules, whole nodules, and senescent and nonsenescent portions of nodules. Scanning electron microscopy was used to identify stages of shoot development (Fig. 5). RNA was also isolated from floral shoot tips of Oklahoma and Kansas ecotypes. Seed was provided by J. Etterson, University of Minnesota, Duluth, and then inbred for five generations.
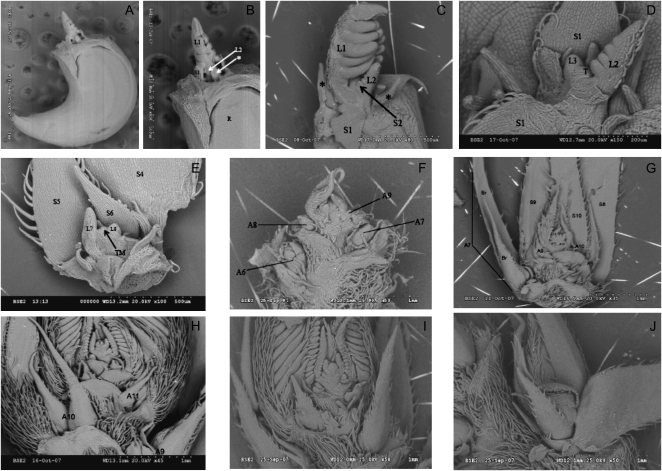
Figure 5.
Shoot development in C. fasciculata. Scanning electron micrographs of shoot tips isolated at different developmental stages. A to C, Twenty-four hours after imbibition. There are two leaf primordia at this stage. D, Four days after imbibition. Three leaf primordial have been initiated. E, Twenty-four days after imbibition. Eight leaf primorida have been initiated. There is very little axillary bud development. F, Thirty-three days after imbibition. Twelve leaves have been initiated. Axillary buds in nodes 6 and above are floral. G, Forty-nine days after imbibition. Fourteen leaf primordial have been initiated. Axillary buds are floral in nodes 5 and above. H, Fifty-four days after imbibition. Eighteen leaves have been initiated. Flower development is advanced in lower inflorescence axillaries. I and J, Ninety-two days after imbibition. Twenty-six leaves have been initiated. J, Floral axillary bud with the bract removed to expose the developing flowers. L, Leaf blade; S, stipule; R, radical; *, possible scalar leaf; TM or T, terminal meristem; T, terminal meristem; A, axillary bud; Br, bract (numbers following symbol indicate the node number).
Libraries were sequenced using two methods: with Roche 454 titanium on pooled RNA from all Minnesota tissues to generate long reads to assemble as a reference to serve as alignment templates, and with the Illumina genome analyzer to generate deep sequence coverage and transcript counts. The 454 and Illumina data are available through the National Center for Biotechnology Information Trace Archive. The Illumina sequence data were used for identification of genetic variants (SNPs) and gene expression profiling of individual libraries.
Illumina sequencing generated more than 143 million passing 46-bp DNA sequence reads, that yielded a total greater than 6.5 Gbp of sequence. The 454 titanium run yielded approximately one million reads, each with average length 370 bp, for a total of 370 Mbp of sequence. Assembly proceeded by contigging the 454 reads, followed by several rounds of contig extensions. Sequences were aligned in frame to the most similar soybean peptide sequence from the Glyma1.01 annotation. Our final assembly consists of 60,015 contigs, with average length 571 bp. These were also translated and trimmed to longest open reading frames, then recontigged, producing 21,781 contig sequences, with average length of 463 bp.
GENOME EVOLUTION
Comparison of phylogenetic patterns involving duplicated genes from several species permits the discrimination of shared versus independent events. For example, phylogenetic analyses of 39 gene families provided strong evidence that the whole-genome duplications detected in Medicago and Glycine were the same event, shared by their common ancestor (Pfeil et al., 2005). The same approach is being used with Chamaecrista, comparing duplicated contigs with homologous genes in Glycine and Medicago.
NODULATION
Though limited, research on C. fasciculata nodulation provides clues about nodule evolution. Nodulation in C. fasciculata has been described by Naisbitt et al. (1992). Hirsch et al. (2009) propose that Rhizobium infection threads in the nodule enter secondarily through crack entry since rhizobial infection threads abort in root hairs. Within nodules, a hemoglobin with reduced O2 affinity has been characterized that is intermediate between nonsymbiotic hemoglobins and leghemoglobin, resembling a putative ancestral leghemoglobin (Gopalasubramaniam et al., 2008). Rhizobial strains selected for C. fasciulata were collected by the late Peter Graham and will become available from the U.S. Department of Agriculture Agricultural Research Service National Rhizobium Germplasm Resource Collection curated by Peter van Berkum.
Thousands of genes were identified in our transcriptome analysis that are highly up-regulated in nodules versus roots or shoots. Similarly, thousands of genes differ in expression between active and senescent portions of nodules.
GENOTYPE DIVERSITY
C. fasciulata is distributed from southern New England to Florida and westward to New Mexico in the United States (Fenster et al., 2003; distribution map available at the U.S. Department of Agriculture Natural Resources Conservation Service Plant Profiles Web site: http://plants.usda.gov/java/profile?symbol=chfa2). MN, KS, and OK ecotypes have distinct flowering times, nodes of floral initiation (NFI), and nodes of first open flower in controlled environments that are affected by temperature and light intensity (Etterson and Shaw, 2001). Additional C. fasciculata accessions are available from the National Plant Germplasm System/Germplasm Resources Information Network collection. Plants from 24 accessions showed a range of flowering times and NFI with some populations having greater variation than others under controlled growth conditions. NFI correlates with latitude of accession with NFI being the lowest in the most northern populations.
A high level of sequence diversity among C. fasciculata accessions justifies efforts to develop mapping populations. An amplified fragment length polymorphism (AFLP) analysis on the 24 U.S. Department of Agriculture accession plants and MN, KS, and OK ecotypes used for the flowering time analysis identified 317 loci from the 12 AFLP primers (T. Kisha, Washington State University, personal communication). Geographically proximal accessions were most similar in terms of the loci identified. R.V. Penmetsa (University of California, Davis) evaluated sequences from ortholog-targeting markers in the MN, KS, and OK reference plants used for the WTS analysis. In agreement with AFLP analysis, polymorphism levels were proportional to geographic distance, and sufficiently high to allow the genetic mapping of several hundred orthologs in the MN98 × OK37 genotype pair for purposes of comparative structural genomics (R.V. Penmetsa and D. Cook, personal communication).
Comparative analysis of transcriptome data for the MN, KS, and OK ecotypes yielded 297,410 SNPs, 3,069 insertions, and 3,385 deletions. Interpretation of these data is limited as shoot apices from 15 plants were pooled for each ecotype for the WTS. The pattern of SNP distribution is consistent with greater similarity among geographically proximal ecotypes (KS and OK versus MN).
FUNCTIONAL GENOMICS TOOLS
Tools for functional analysis are in development. Fluorescence in situ hybridization markers for cytogenetics are being developed in the laboratory of G. Stacy (University of Missouri). Early results with viral-induced gene silencing using a vector based on Pea early browning virus developed by Constantin et al. (2004) show promise in the laboratory of S. Singer. The laboratory of A. Hirsch (University of California, Los Angeles) has established a hairy root transformation system for implementing RNAi mutagenesis in the caesalpinioid legumes, Gleditisia and Gymnocladus, which, like Chamaecrista, are members of the mimosoid clade.
UNDERGRADUATE EDUCATION
The National Science Foundation grant that funded our WTS project was titled “Big Science at Small Schools Collaboration: Genomics of Chamaecrista fasciculata, a native prairie plant with potential for mixed prairie biomass.” An overarching goal was to engage students in authentic genomic research, in a formal teaching setting, where the questions they ask are both original and of value to the research community. WTS can provide undergraduates with large data sets forming a foundation for authentic research experiences in the context of a teaching laboratory, while the nonmodel system keeps the biological questions at the fore. C. fasciculata is an exemplar for all nonmodel plants and forms a context to keep students focused on the integration of the genomics with organismal biology. The concept of integrating genomics and organismal biology in a pedagogically sound way is at the core of a broader “Teaching Big Science at Small Colleges” curricular development consortium (http://serc.carleton.edu/genomics/). The advent of 454 and Illumina sequencing at increasingly affordable prices has the tantalizing potential for more fully integrating both genomics and organismal biology and research and education.
We developed a laboratory experience that integrates in silico and wet laboratories and supports students who generate and explore original research questions focused on C. fasciculata (http://serc.carleton.edu/genomics/units/33519.html). Students frame questions after reading selected literature and working with different plant ecotypes, address the questions using transcriptome analysis scaffolded with a web-based genomics explorer, design and conduct experiments using PCR approaches to address questions arising from the in silico analysis, and prepare final papers and presentations. The Chamaecrista activity takes place over 8 to 9 weeks of laboratory sessions in an undergraduate genetics course. The first 2 weeks are focused on working with whole plants and framing questions and hypotheses that can be addressed with in silico data during the following 2 weeks. The in silico analysis is used to develop questions and hypotheses that can be experimentally tested during the following 3 weeks. Students then give presentations and use feedback from peers and instructors to prepare a final group paper on all three aspects of their work. It is possible to modify any of the modules for use as stand-alone activities. Assessment of student learning shows an increase in critical thinking skills as a result of participating in the Chamaecrista research (data not shown).
SUMMARY
WTS from shoots, roots, and nodules provides the opportunity to explore the gene space of C. fasciculata and begin addressing questions about genome evolution in legumes, the origins of nodulation, and floral development in nonpapilionoid legumes. Emerging functional genomics tools, a range of C. fasciculata populations, and venues for undergraduates to participate in genomics research will leverage the value of the WTS resource. Our approach to developing C. fasciculata community resources models a culture shift where the big science of genomics provides potential opportunities to integrate research and education at big and small schools, enabled by outsourcing of increasingly affordable high-tech, high-throughput science.
Acknowledgments
We thank Heidi Mullen for her excellent scanning electron microscopy work and plant care. Anna Newman and Fang yu Lee thoughtfully critiqued our manuscript. Carleton's Science Education Resource Center participated in the development and assessment of the Chamaecrista genomics laboratories. We thank the Carleton genetics laboratory students for their wonderful sense of adventure in exploring the Chamaecrista gene space with us.
This work was supported by the National Science Foundation (grant nos. DEB–0746571 and DUE–0837375).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Susan R. Singer (ssinger@carleton.edu).
References
- Bertioli DJ, Moretzsohn MC, Madsen LH, Sandal N, Leal-Bertioli SC, Guimarães PM, Hougaard BK, Fredslund J, Schauser L, Nielsen AM, et al (2009) An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics 10 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy JP, Wang X, Mudge J, Vasdewani J, Scheix T, et al (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103 14959–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin GD, Krath BN, MacFarlane SA, Nicolaisen M, Johansen IE, Lund OS (2004) Virus induced gene silencing as a tool for functional genomics in a legume species. Plant J 40 622–631 [DOI] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20 591–597 [DOI] [PubMed] [Google Scholar]
- Doyle JJ (1998) Phylogenetic perspective on nodulation: evolving views of plants and symbiotic bacteria. Trends Plant Sci 3 473–478 [Google Scholar]
- Etterson JR, Shaw RG (2001) Contsraint to adaptive evolution in response to global warming. Science 294 151–154 [DOI] [PubMed] [Google Scholar]
- Fenster CB, Vekemans X, Hardy OJ (2003) Quantifying gene flow from spatial genetic structure data in a metapopulation of Chamaecrista fasciculata (Leguminosae). Evolution 57 995–1007 [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16 805–814 [DOI] [PubMed] [Google Scholar]
- Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND (2005) Legumes as a model plant family: genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol 137 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalasubramaniam SK, Kovacs F, Violante-Mota F, Twigg P, Arredondo-Peter R, Sarath G (2008) Cloning and characterization of a caesalpinoid (Chamaecrista fasciculata) hemoglobin: the structural transition from a nonsymbiotic hemoglobin to a leghemoglobin. Proteins 72 252–260 [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Yu N, Ma N, Schwartz AR, De Hoff PL (2009) The Chamaecrista fasciculata nitrogen-fixing symbiosis. Botany and Mycology 2009 Abstract 446. http://www.botanyconference.org/engine/search/index.php?func=detail&aid=446 (September 30, 2009)
- Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54 530–549 [DOI] [PubMed] [Google Scholar]
- Lewis G, Schrire B, Mackinder B, Lock M, editors (2005) Legumes of the World. Royal Botanic Gardens, Kew, UK, pp 21–54
- Naisbitt T, James EK, Sprent JI (1992) The evolutionary significance of the legume genus Chamaecrista, as determined by nodule structure. New Phytol 122 487–492 [DOI] [PubMed] [Google Scholar]
- Pfeil B, Schlueter J, Shoemaker RC, Doyle J (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54 441–454 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Schoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47 868–876 [DOI] [PubMed] [Google Scholar]
- Singer SR, Hsiung L, Huber SC (1990) The determinate (det) mutant of Pisum sativum L. (Leguminosae: Papilionoidae) exhibits an indeterminate growth pattern. Am J Bot 77 1330–1335 [Google Scholar]
- Sprent JI (2001) Nodulation in Legumes. Royal Botanic Gardens, Kew, UK, pp 1–54
- Sprent JI (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174 11–25 [DOI] [PubMed] [Google Scholar]
- Sprent JI (2009) Legume Nodulation—A Global Perspective. Wiley-Blackwell, West Sussex, UK, pp 3–7
- Tilman D, Hill J, Lehman C (2006) Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 314 1598–1600 [DOI] [PubMed] [Google Scholar]
- Tucker SC (1989) Overlapping organ initiation and common primordia in flowers of Pisum sativum (Leguminosae: Papilionoideae). Am J Bot 76 714–729 [Google Scholar]
- Tucker SC (1996) Trends in evolution of floral ontogeny in Cassia senus stricto, Senna, and Chamaecrista (Leguminosae: Caesalpiniodeae: Cassiaea: Cassiinae); a study in convergence. Am J Bot 83 687–711 [Google Scholar]
- Tucker SC (2003) Floral development in legumes. Plant Physiol 131 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]