Abstract
Plant α-dioxygenases initiate the synthesis of oxylipins by catalyzing the incorporation of molecular oxygen at the α-methylene carbon atom of fatty acids. Previously, α-DOX1 has been shown to display α-dioxygenase activity and to be implicated in plant defense. In this study, we investigated the function of a second α-dioxygenase isoform, α-DOX2, in tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana). Recombinant Slα-DOX2 and Atα-DOX2 proteins catalyzed the conversion of a wide range of fatty acids into 2(R)-hydroperoxy derivatives. Expression of Slα-DOX2 and Atα-DOX2 was found in seedlings and increased during senescence induced by detachment of leaves. In contrast, microbial infection, earlier known to increase the expression of α-DOX1, did not alter the expression of Slα-DOX2 or Atα-DOX2. The tomato mutant divaricata, characterized by early dwarfing and anthocyanin accumulation, carries a mutation at the Slα-DOX2 locus and was chosen for functional studies of α-DOX2. Transcriptional changes in such mutants showed the up-regulation of genes playing roles in lipid and phenylpropanoid metabolism, the latter being in consonance with the anthocyanin accumulation. Transgenic expression of Atα-DOX2 and Slα-DOX2 in divaricata partially complemented the compromised phenotype in mature plants and fully complemented it in seedlings, thus indicating the functional exchangeability between α-DOX2 from tomato and Arabidopsis. However, deletion of Atα-DOX2 in Arabidopsis plants did not provoke any visible phenotypic alteration indicating that the relative importance of α-DOX2 in plant physiology is species specific.
Plants have evolved elaborate signaling systems to regulate a variety of physiological responses to the environment and to facilitate intercellular cross talk in development and reproduction. Oxylipins comprise a large class of oxygenated fatty acid-derived lipid mediators that contribute to such signaling circuits (Weber, 2002; Farmer et al., 2003). A variety of functions have been ascribed to plant oxylipins, including critical roles in plant defense against microbial pathogens, as well as in reproduction and tissue development (Howe and Schilmiller, 2002; Browse, 2005; Kachroo and Kachroo 2009).
The biosynthesis of oxylipins is initiated by hydroperoxide formation catalyzed by fatty acid oxygenases, among which the 9- and 13-lipoxygenases have been studied most intensively (Shibata and Axelrod, 1995; Feussner and Wasternack, 2002). α-Dioxygenase, first encountered about 10 years ago, also catalyzes primary fatty acid oxygenation. This enzyme was first identified in Nicotiana tabacum plants as a pathogen-induced protein showing homology to mammalian prostaglandin endoperoxide synthases (Sanz et al., 1998). Studies of the catalytic function of the recombinant tobacco protein and of a homologous protein from Arabidopsis (Arabidopsis thaliana) revealed that these plant enzymes, designated as α-dioxygenase-1 or α-DOX1, catalyze the incorporation of molecular oxygen at the α-methylene carbon atom of fatty acids. The products are chemically unstable 2(R)-hydroperoxy fatty acids, which are either reduced to 2R-hydroxy fatty acid or spontaneous decarboxylated to the corresponding lower fatty aldehyde (Hamberg et al., 1999).
Expression of α-DOX1 in tobacco and Arabidopsis leaves is activated in response to bacterial inoculation and by agents that generate oxidative stress (Sanz et al., 1998; Ponce de León et al., 2002; Hamberg et al., 2003). In these responses, α-dioxygenase was proposed to play a tissue-protective role as concluded from results showing a negative correlation between the extent of α-dioxygenase activity and the level of tissue damage (Ponce de León et al., 2002). In addition, α-dioxygenase expression has been shown to be up-regulated in response to herbivore attack (Hermsmeier et al., 2001), and a variety of different types of abiotic stresses, such as salt stress, cold, drought, and heavy metal exposure (Wei et al., 2000; Seki et al., 2002; Koeduka et al., 2005); however, the contribution of the α-DOX activity during these responses remains unknown.
Sequence analyses of reported α-dioxygenases as well as database searches reveal the presence of plant proteins that show high level of homology to the first identified α-dioxygenases, α-DOX1, in plant species such as rice (Oryza sativa), tomato (Solanum lycopersicum), N. tabacum, Nicotiana attenuata, Pisum sativum, Capsicum annuum, Vitis vinifera, Ricinus communis, and Populus trichocarpa (Supplemental Fig. S1). Additionally, these analyses identify a second group of predicted α-dioxygenases, termed α-DOX2, which cluster together as a phylogenetic group distinct from the first identified α-dioxygenases (Hamberg et al., 2005; Supplemental Fig. S1). In tomato, mutations of α-DOX2, also known as FEEBLY, and divaricata (div) result in defects in plant development as well as accumulation of anthocyanins, pointing to a role of Slα-DOX2 in plant development (Stevens and Rick, 1986; van der Biezen et al., 1996). However, no information is available on the enzymatic activity of Slα-DOX2 or on any other putative α-DOX2 reported to date.
This study concerns the biochemical characterization and function of the α-DOX2 proteins from tomato (Slα-DOX2) and Arabidopsis (Atα-DOX2). We show that these two α-DOX2 enzymes are functionally interchangeable. However, in contrast to tomato in which mutation of α-DOX2 provokes large phenotypic effects, mutation of Atα-DOX2 does not have any visible phenotypic consequence.
RESULTS
α-Dioxygenase Activity of Tomato and Arabidopsis α-DOX2
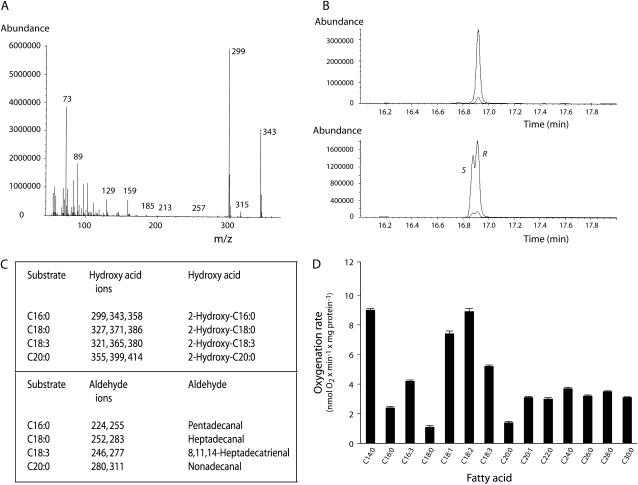
Infection of High Five insect cells with recombinant baculovirus containing Slα-DOX2 pFastBac or Atα-DOX2 pFastBac constructs, respectively, resulted in the expression of Slα-DOX2 and Atα-DOX2 proteins with molecular masses in accordance with the predicted size (72.8 and 72.5 kD, respectively; Supplemental Fig. S2). No α-dioxygenase was detected when insect cells were infected with baculovirus prepared from empty pFastBac vector. Incubation of palmitic acid (C16:0) with Slα-DOX2-expressing cell homogenates led to the generation of the corresponding 2-hydroperoxy fatty acid as shown by the identification of the decarboxylation product pentadecanal and the reduction product 2-hydroxypalmitic acid by gas chromatography-mass spectrometry (GC-MS; Fig. 1A). In a similar fashion, Slα-DOX2 catalyzed the formation of pairs of aldehydes and 2-hydroxy acids when incubated with stearic acid (C18:0), linolenic acid (C18:3), or arachidic acid (C20:0; data not shown). Likewise, incubations of the mentioned fatty acids with Atα-DOX2-expressing cells led to the formation of 2-hydroperoxides as shown by the identification of corresponding aldehydes and 2-hydroxy acids (Fig. 2, A and C). Steric analysis of the (−)-menthoxycarbonyl derivative of 2-hydroxylinolenic acid isolated after incubation of linolenic acid with Slα-DOX2 and of 2-hydroxypalmitic acid isolated after incubation of palmitic acid with Atα-DOX2 demonstrated exclusive formation of the 2(R) enantiomers (Figs. 1B and 2B). These results demonstrated that both Slα-DOX2 and Atα-DOX2 catalyze stereospecific introduction of molecular oxygen at the α-carbon to produce fatty acid 2(R)-hydroperoxides (Supplemental Fig. S3).
Figure 1.
Determination of α-dioxygenase activity of Slα-DOX2. A, GC-MS identification of products formed by incubation of palmitic acid with Slα-DOX2-containing insect cells. Top: Peaks due to pentadecanal (O-methyloxime syn/anti isomers), palmitic acid (methyl ester; corresponding to substrate remaining not converted), and 2-hydroxypalmitic acid (methyl ester/trimethylsilyl ether derivative) were observed. The reaction products observed arouse by decarboxylation or reduction of 2-hydroperoxypalmitic acid, the primary α-DOX product. Bottom: Mass spectrum of 2-hydroxypalmitic acid (methyl ester/trimethylsilyl ether derivative). B, Steric analysis of 2-hydroxylinolenic acid as its (−)-menthoxycarbonyl/methyl ester derivative. Top: 2-Hydroxylinolenic acid prepared from an incubation of linolenic acid with Slα-DOX2. Bottom: Synthetic 2(R,S)-hydroxylinolenic acid elution order 2(S) followed by 2(R). C, Fatty acid substrate specificity of oxygenation by Slα-DOX2 (mean ± se of n = 3–4 measurements). Enzymatic oxygenation rates were determined at 23°C after addition of approximately 100 μg total protein to 1.5 mL 0.1 m Tris, pH 7.4, containing 100 μm fatty acid substrate and 100 μm tert-butylhydroperoxide.
Figure 2.
Determination of α-dioxygenase activity of Atα-DOX2. A, MS identification of 2-hydroxypalmitic acid formed by incubation of palmitic acid with a homogenate of Atα-DOX2-expressing insect cells. The methyl ester/trimethylsilyl ether derivative was used. B, Steric analysis of 2-hydroxypalmitic acid as its (−)-menthoxycarbonyl/methyl ester derivative. Top: 2-Hydroxypalmitic acid prepared from an incubation of palmitic acid with Atα-DOX2. Bottom: Synthetic 2(R,S)-hydroxypalmitic acid elution order 2(S) followed by 2(R). C, Mass-spectral ions (m/z) recorded on Atα-DOX2-derived 2-hydroxy fatty acids (methyl ester/trimethylsilyl ether derivatives) and fatty aldehydes (O-methyloxime derivatives). D, Fatty acid substrate specificity of oxygenation by Atα-DOX2 (mean ± se of n = 3–4 measurements).
In order to determine the substrate specificity of Slα-DOX2 and Atα-DOX2, the oxygenation rates of a wide range of long chain (C14-22) and very-long-chain (C24-30) fatty acids (VLCFAs) were determined. Both enzymes oxygenated all of these fatty acids. Differences in oxygenation rates observed with Slα-DOX2 were not large (Fig. 1C). Thus, linoleic acid was oxygenated most efficiently, but only 4 times faster than its least efficient substrate, i.e. arachidic acid. Atα-DOX2 displayed a substrate preference for unsaturated fatty acids of 18 carbon chain length, i.e. oleic, linoleic, and linolenic acids, and unexpectedly also showed high activity with myristic acid (C14:0; Fig. 2D). VLCFAs ranging in chain length from 20 to 30 carbons were also efficiently oxygenated by both enzymes. These results demonstrate that the gene products of Slα-DOX2 and Atα-DOX2 are authentic α-dioxygenases, which can oxygenate a wide range of fatty acids.
Slα-DOX2 Activity Is Required for Normal Vegetative Growth and Fruit Development
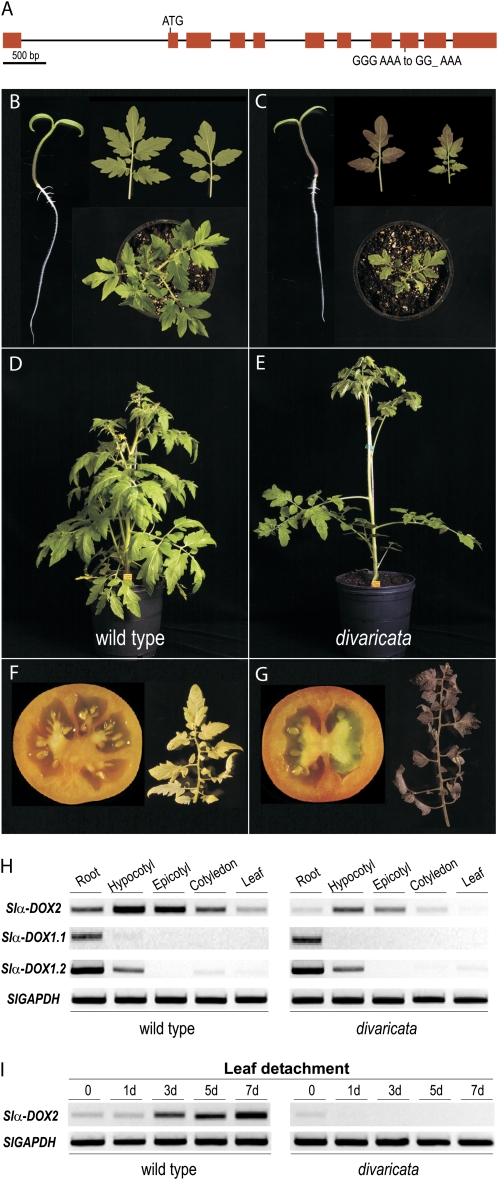
Previously, based on allele complementation tests and on phenotypic similarities of the mutation, the α-DOX2 was shown to correspond to DIV (van der Biezen et al., 1996). Here, sequence comparison of the Slα-DOX2 gene from both wild-type and div plants revealed a single nucleotide deletion in exon 9 of Slα-DOX2 creating a stop codon at amino acid 433 of the predicted protein, thus confirming the identity of α-DOX2 and DIV (Fig. 3A). As a result of this mutation, the div mutant displayed significant phenotypic alterations and was selected here to further examine the function of the Slα-DOX2 protein. To this end, expression and phenotypic analyses were performed in wild-type and div plants. Analyses of Slα-DOX2 expression revealed the presence of Slα-DOX2 RNA in seedlings of wild-type plants (Fig. 3H). Expression was found preferentially in aerial tissues with the highest levels of transcript accumulation in the hypocotyls. Slα-DOX2 expression is lower in leaves and decreased markedly as plants matured. Analyses of the two α-DOX1 genes found in tomato, designated as Slα-DOX1.1 and Slα-DOX1.2 (Supplemental Fig. S1), revealed their expression mainly in roots, although a low level of Slα-DOX1.2 transcript was found also in hypocotyls (Fig. 3H). The expression of Slα-DOX2 was strongly reduced in div plants compared to wild-type plants.
Figure 3.
Vegetative growth and fruit development in wild-type and div tomato plants. A, Genomic structure of Slα-DOX2 gene (GenBank accession no. FN428743), indicating the div mutation. B, Seedlings, young leaves, and top view of wild-type plants. C, Seedlings, young leaves, and top view of div plants. D, Lateral view of adult wild-type tomato plant (5 weeks old). E, Lateral view of adult div tomato plant (5 weeks old). F, Cross section of ripe fruit and senescent leaf of wild-type tomato plants. G, Cross section of ripe fruit and senescent leaf of div mutant. H, Gene expression levels of the three tomato α-dioxygenase genes Slα-DOX2, Slα-DOX1.1, and Slα-DOX1.2 in roots, hypocotyls, epicotyls, cotyledons, and leaves of seedlings of tomato wild-type or div plants. I, Gene expression levels of Slα-DOX2 during a 1-week period after detachment of young leaves of wild-type and div plants.
As α-DOX1 from different plant species has previously been shown to be induced in response to biotic stress (Sanz et al., 1998; Ponce de León et al., 2002), the expression of Slα-DOX2 was examined in leaves from wild-type tomato infected with Pseudomonas syringae pv tomato or with the necrotrophic fungus Botrytis cinerea; however, these treatments did not lead to Slα-DOX2 expression (data not shown). Only one stimulus was found to consistently activate Slα-DOX2 expression, namely, the detachment of mature leaves, a treatment that is frequently used to induce senescence (Gepstein et al., 2003; Guo and Gan, 2005). Slα-DOX2 mRNA expression increased in leaves 3 d after detachment and was maintained elevated up to at least 1 week after treatment (Fig. 3I). In contrast, no marked increase of Slα-DOX2 mRNA levels was found after detachment in div plants (Fig. 3I).
The expression of Slα-DOX2 in growing seedlings is in accordance with the phenotypic alterations of div, i.e. delayed development accompanied by strong anthocyanin accumulation in cotyledons and leaves of young plants (Fig. 3, B and C). Also, in line with the activation of Slα-DOX2 during senescence induced by leaf detachment, a marked anthocyanin production was found in senescent leaves of div (Fig. 3, F and G). Inspection of mature div plants revealed further phenotypic alterations, including increased internodal length and reduced number of lateral shoots (Fig. 3, D and E), as well as a reduction in the number of locules from 3 to 10 in fruits from wild-type plants to a single symmetrical division into two locules (Fig. 3, F and G). However, no significant variations between control and mutant plants were detected in traits such as weight and size of the fruits or seed yield. Taken together, the morphological changes recapitulated in Figure 3 are consistent with a role of Slα-DOX2 at different stages of plant development. Moreover, the alterations found in mature plants suggested that Slα-DOX2 might be expressed in additional tissues to those identified here, in which the defect of Slα-DOX2 could account for the observed phenotypic alterations.
Identification of Gene Expression Changes in div Plants
To gain further understanding of the function of Slα-DOX2, gene expression profiling was performed on div and wild-type plants. Three individual microarrays were hybridized using RNA extracted from three independent biological replicates. Complete microarrays data sets were analyzed, and a list of differentially expressed genes is shown in Supplemental Table S1. Details of computational methods to process gene expression are described in “Materials and Methods.” Of over 9,200 tomato transcripts represented on the Affymetrix tomato array, 42 showed altered expression according to a statistical false discovery rate (FDR) value of 0.05. Most changes correspond to genes induced in div plants (69% of genes shown in red in Supplemental Table S1), while a lower proportion (31% of genes shown in green in Supplemental Table S1), including Slα-DOX2, decreased their expression compared to wild-type plants. In accordance with the accumulation of anthocyanins, we found that genes related to the flavonoid-phenylpropanoid pathway were among the up-regulated genes in div seedlings. From these, the activation of DFR encoding dihydroflavonol 4-reductase, an enzyme that directs anthocyanin biosynthesis by catalyzing the conversion of dihydroflavonols to leucoanthocyanidins (Lepiniec et al., 2006), was confirmed by reverse transcription (RT)-PCR (Fig. 4). In addition, a substantial percentage of the genes with increased expression in div (28%) encoded proteins predicted to play a role in lipid deacylation, lipid transfer, and lipid metabolism. Among these, the activation of two GDSL-motif lipase/hydrolases, one class 3 lipase, three lipid transfer proteins, and two proteins mediating the synthesis of VLCFAs (CER1 and KCS6), was confirmed by semiquantitative RT-PCR (Fig. 4). Additional genes related to lipid modifications with a higher FDR value (between 0.05 and 0.21) were also selected for RT-PCR examination. These included genes encoding a GDSL-motif lipase/hydrolase, three lipid transfer proteins, a lipoxygenase, and a cytochrome P450 CYP94B1 that has been implicated in the biosynthesis of suberin (Kandel et al., 2006). As shown in Figure 4, the expression of the former five first genes was confirmed by RT-PCR and found to be increased in div, whereas no significant change was observed for CYP94B1. Finally, a high percentage of genes with decreased expression in div plants with respect to controls were chloroplastic (54%). Within the down-regulated group of genes, the expression of a nuclear gene (cDNA sequence with GenBank accession no. AK247303) encoding a noncoding RNA was confirmed by RT-PCR (Fig. 4).
Figure 4.
Gene expression analysis in the div mutant compared to wild-type tomato plants. Differentially expressed transcripts obtained from microarray analyses were examined by RT-PCR. Fold change and statistical value FDR are indicated for each probe from Affymetrix GeneChip Tomato Genome Array. Tomato GAPDH was used to normalize transcript levels in each sample. Gene-specific primer sets used for the evaluation of RNA are shown in Supplemental Table S2.
Taken together, the results from these studies showed that the mutation of Slα-DOX2 leads to increased anthocyanin production by a pretranslational control mechanism. Moreover, the additional identified transcriptional changes suggest that the div mutation would give rise to important changes in lipid composition of the plant.
Functional Analyses of Arabidopsis α-DOX2
Given that Slα-DOX2 and Atα-DOX2 cluster together within a phylogenetic group distinct from that of the first identified α-DOX1 α-dioxygenases (see Supplemental Fig. S1), we speculated that Atα-DOX2 might be a functional homolog of Slα-DOX2. To examine this possibility, we assessed whether Arabidopsis mutants defective in Atα-DOX2 display developmental alterations as observed for Slα-DOX2 in tomato. To this end, two Arabidopsis T-DNA insertion mutants, α-dox2-1 and α-dox2-2, lacking Atα-DOX2 function (see Supplemental Fig. S4 for a scheme of genome structure and transcript levels detection) were examined for seed formation, seed yield, germination, and growth. In these studies, we found, in contrast to the results in tomato, that the lack of Atα-DOX2 function did not provoke any visible phenotypic alteration or premature senescence. This finding could indicate that Atα-DOX2 might differ from Slα-DOX2 with regard to its expression characteristics. Alternatively, these results may reflect species-specific differences distinguishing tomato and Arabidopsis. In this respect, a functional redundancy of Atα-DOX2 could be implied. A candidate to substitute for the Atα-DOX2 defect was Atα-DOX1, which is the closest gene homolog to Atα-DOX2 in Arabidopsis. This possibility was tested by generating a double Arabidopsis mutant lacking both enzymatic activities: α-DOX1 and α-DOX2 (see Supplemental Fig. S4 for the genome structure and transcript levels detection). As found in the single α-DOX mutants, the examined double mutants did not show any visible phenotypic alteration compared to wild-type plants (data not shown), which demonstrated that the lack of phenotype in the α-dox2 mutants was not due to a compensatory effect of Atα-DOX1.
Atα-DOX2 Is Expressed in Different Plant Tissues during Plant Development
In order to compare the expression patterns of Atα-DOX2 and Slα-DOX2, the expression of Atα-DOX2 was analyzed using plants expressing an Atα-DOX2 promoter GUS (Atα-DOX2∷GUS) construct and by northern analysis. Atα-DOX2 was expressed in cotyledons, young leaves, and hypocotyls of transgenic seedlings (Fig. 5A). Expression decreased as plants matured and was only occasionally detected in leaves of flowering plants (Fig. 5A). In contrast to leaves, no significant levels of GUS activity were observed in roots at any developmental stage. Detailed examination by GUS staining of flowers and siliques revealed that Atα-DOX2 was expressed in anthers and ovules prior to fertilization as well as in developing seeds (Fig. 5A). The GUS activity found in the examined transgenic lines correlated with the accumulation of Atα-DOX2 transcripts detected by northern blot in RNA samples prepared from different tissues of wild-type plants. Thus, examination of untreated wild-type plants revealed Atα-DOX2 expression in seedlings, young leaves, stems, flowers, and siliques (Fig. 5B). Further examination of gene expression after stress treatments revealed that, as found for Slα-DOX2, the expression of Atα-DOX2 was increased after leaf detachment. Atα-DOX2 expression was observed 3 d after detachment and maintained up to at least 1 week (Fig. 5C). In summary, the results from these studies revealed significant similarities in the expression pattern of Atα-DOX2 and Slα-DOX2.
Figure 5.
Expression of Atα-DOX2. A, Histochemical localization of GUS gene expression in transgenic plants containing an Atα-DOX2∷GUS chimeric construct. Bright-field micrographs reveal in blue the presence of GUS enzyme activity in seedlings and mature tissues of transgenic plants. B, RNA was extracted from different plant organs of healthy untreated plants. Blots were hybridized with riboprobes derived from an Atα-DOX2 cDNA. C, Atα-DOX2 expression during detachment of mature Arabidopsis leaves. Histochemical localization of GUS gene expression in adult leaves of Atα-DOX2∷GUS transgenic plants during a 1-week period after detachment. RNA blots were hybridized with riboprobes derived from an Atα-DOX2 cDNA. Loading control was analyzed by ethidium bromide staining followed by hybridization against an 18S rRNA radioactive probe.
Complementation of the Tomato div Mutant by Arabidopsis or Tomato α-DOX2
To obtain further insight into the functionality of the α-DOX2 α-dioxygenases, we tested whether Arabidopsis Atα-DOX2 could substitute for the function of the tomato enzyme in vivo. This was assessed by complementation studies of the tomato div mutant with the wild-type Arabidopsis Atα-DOX2 cDNA. Transformation with a 35S∷Slα-DOX2 construct was performed and used as a control in these experiments. Two transgenic lines with constitutive expression of Atα-DOX2 or Slα-DOX2 were selected (see Supplemental Fig. S5 for characterization of transgene expression). The phenotype of a transgenic line for each construct is shown in Figure 6. The phenotypic alterations that characterize div seedlings (delayed development and anthocyanin accumulation in seedlings; Fig. 3C) were reversed by stable transformation of tomato div mutant plants with a 35S∷Atα-DOX2 construct. Complementation was also obtained by transformation with a 35S∷Slα-DOX2 construct (Fig. 6, A and B). However, in mature plants, clear differences between transgenic and wild-type plants were detected in traits such as the morphology of the leaves, the distance between internodes, the number of lateral shoots, and the number of fruit locules. Thus, phenotypic alterations of div were only partially complemented in adult plants (Fig. 6, C–H), possibly due to a dose-dependent effect of α-DOX2-complementation. Independently of this, these results proved that Atα-DOX2 can substitute for the function of the α-DOX2 tomato gene in vivo, demonstrating the functional similarity of the two enzymes and indicating that the different phenotypic effect of mutation of Slα-DOX2 and Atα-DOX2 is not due to intrinsic differences between these proteins.
Figure 6.
Complementation of tomato div mutant plants with Slα-DOX2 or Atα-DOX2 rescues the phenotypic defects of div plants. A, Young leaves and top view of complemented div 35S∷Slα-DOX2 plants. B, Young leaves and top view of complemented div 35S∷Atα-DOX2 plants. C, Lateral view of adult complemented div 35S∷Slα-DOX2 plants (5 weeks old). D, Lateral view of adult complemented div 35S∷Atα-DOX2 plants (5 weeks old). E, Adult leaf from the second node below the first inflorescence of a tomato wild-type plant (5 weeks old). F, Adult leaf from the second node below the first inflorescence of tomato div plant (5 weeks old). G, Adult leaf from the second node below the first inflorescence of tomato div 35S∷Slα-DOX2 plant (5 weeks old). H, Adult leaf from the second node below the first inflorescence of a tomato div 35S∷Atα-DOX2 plant (5 weeks old). See Figure 3 for phenotype of wild-type tomato and div seedlings and 5-week-old adult plants.
DISCUSSION
α-DOX2 from Tomato and Arabidopsis Are Authentic Fatty Acid Oxygenases with Broad Substrate Specificity
Fatty acid oxygenases initiate the synthesis of a family of lipid mediators playing critical roles in physiological and pathological processes in plants and vertebrates. Up to now, two types of fatty acid oxygenases, lipoxygenases (9- and 13-lipoxygenases) and α-dioxygenases, have been identified in plants (Hamberg et al., 2005; Liavonchanka and Feussner, 2006). Previous characterization of the α-DOX1 α-dioxygenases from tobacco and Arabidopsis revealed their activity catalyzing the incorporation of molecular oxygen at the α-methylene carbon atom of fatty acids to generate 2R-hydroperoxy fatty acids derivatives (Hamberg et al., 1999). In this study, we focused our interest on the characterization of a second group of predicted α-dioxygenases, α-DOX2, that cluster as a phylogenetic group distinct from the first identified α-DOX1 (Supplemental Fig. S1). Tomato (Slα-DOX2) and Arabidopsis (Atα-DOX2) α-DOX2 were selected here to examine their catalytic activity and determine the nature of their enzymatic products. Biochemical characterization of recombinant proteins from heterologously expressed Slα-DOX2 and Atα-DOX2 genes, demonstrated the α-dioxygenase activity of Slα-DOX2 and Atα-DOX2. The proteins characterized catalyzed the oxygenation of fatty acids to form the same products as α-DOX1, namely, the 2R-hydroxy fatty acid and the corresponding one-carbon atom chain shortened aldehyde (Hamberg et al., 1999). The substrate specificity of the two examined recombinant α-DOX2 proteins appeared to be broad, as all tested fatty acids possessing chain lengths from C14 to C30, which included fully saturated fatty acids to fatty acids possessing up to three carbon double bonds, were all substrates for oxygenation. In contrast, the characterized type-1 α-dioxygenases from rice and Arabidopsis have been shown to possess a much more pronounced substrate preference for α-linolenic acid, linoleic acid, and oleic acid (Liu et al., 2006; Koszelak-Rosenblum et al., 2008). A number of conserved amino acid residues that are essential for catalytic activity in type-1 α-dioxygenases and cyclooxygenases, namely, the distal and proximal His residues involved in binding of heme, and the hydrogen-abstracting Tyr (His-163, His-389, and Tyr-389 in Atα-DOX1; Sanz et al., 1998; Liu et al., 2004) are conserved in type-2 α-dioxygenases as well (His-157, His-381, and Tyr-378 in AtDOX2 and His-157, Tyr-379, and His-382 in Slα-DOX2; Hamberg et al., 2002). In summary, our results indicate that the two type-2 α-dioxygenases characterized in this study are authentic α-dioxygenases that likely function in an enzymatically similar manner as type-1 α-dioxygenases.
Tomato and Arabidopsis α-DOX2 Genes Are Expressed during Development and Share a Common Expression Pattern
In addition to the catalytic activity of Slα-DOX2 and Atα-DOX2, studies on the expression of Slα-DOX2 and Atα-DOX2 revealed significant similarities. Thus, both genes were expressed during early development, preferentially in the aerial part of the seedlings, with expression waning as the leaf matured. Whereas a typical stimulus activating the transcription of tobacco and Arabidopsis α-DOX1 genes, such as infection of leaves with strains of P. syringae (Sanz et al., 1998; Ponce de León et al., 2002) did not stimulate expression of the Slα-DOX2 or Atα-DOX2 genes, marked expression of both genes was induced by leaf detachment. These results revealed important similarities between Slα-DOX2 and Atα-DOX2. On the other hand, the expression of Slα-DOX2 and Atα-DOX2 differs significantly from that previously characterized Atα-DOX1 (Sanz et al., 1998; Ponce de León et al., 2002) and shown here for Slα-DOX1, pointing out to a different role of the two types of α-dioxygenase isoforms.
Further characterization of Atα-DOX2 expression in wild-type and Atα-DOX2∷GUS transgenic plants revealed GUS activity in stems, stamens, ovules, and siliques. Whereas the expression of Slα-DOX2 in these organs has not been directly addressed, the fact that the anatomy of mature plants and fruits in the div mutant differed from that of wild-type tomato plants suggested that as found for Atα-DOX2, the Slα-DOX2 gene might also be expressed in specific cells of stems and floral organs in which the activity of the encoded Slα-DOX2 protein is required for normal development. Taken together, these findings indicated that the two characterized α-DOX2 α-dioxygenases share a similar expression pattern consistent with a common function in plant development. Moreover, the characterized pattern of expression suggests that the role of α-DOX2 is not restricted to a particular phase of development but rather exerts a specific function in different plant organs throughout plant development.
Mutation of Slα-DOX2 May Alter Plant Development by Causing Alterations in Lipid Homeostasis
That Slα-DOX2 plays a role in plant development is concluded from the phenotypic abnormalities found in div mutants. Further support for the role of Slα-DOX2 in plant development derived from the results showing that the div phenotype could be reversed by transformation with a wild-type version of the Slα-DOX2 gene. The phenotypic differences distinguishing wild-type tomato plants from div mutants encompass young plants, morphology of vegetative organs in adult and senescent plants, and development of fruits, indicating that wild-type levels of Slα-DOX2 activity are required for normal growth throughout the plant life cycle.
As concluded from the characterized transcriptional changes, the defect in Slα-DOX2 activity of the div mutation might alter the lipid composition of the plant. Thus, a high percentage of genes changing their expression are related to lipid release, transport, and metabolism. Among these, the α-DOX2 defect modifies the expression of genes that encode enzymes mediating the synthesis and posterior modification of VLCFAs, such as KCS6 (encoding a ketoacyl-CoA synthase catalyzing the first rate limiting step in the synthesis of VLCFAs) and CER1 (encoding an aldehyde decarbonylase; Aarts et al., 1995; Lai et al., 2007). Attempts to pinpoint lipid biosynthetic defect(s) in div plants by profiling cutin monomers and surface lipids have been initiated but so far produced no unequivocal result. For example, no obvious differences in the content of VLCFAs or 2-hydroxy fatty acids in cutin from leaves of wild-type or div plants were observed. In addition to its lipidic constituents, cutin and suberin contains low amounts of phenylpropanoids (Molina et al., 2006; Mintz-Oron et al., 2008), which might have altered content as concluded from the strong accumulation of anthocyanins (a class of phenylpropanoids) in div plants. Further biochemical examination will be required to examine these possibilities and to determine the nature of the lipidic changes of the div plants. Based on the overall alteration of the plant morphology, it is likely that changes in hormonal levels or transcription factors regulating development may also contribute to the div phenotype. Potential candidates to be examined in this respect are several transcription factors found to be induced in our microarray results when examined with a more relaxed stringency (0.1 > FDR > 0.05) and a gene encoding a noncoding RNA that is down-regulated in div plants (Supplemental Table S1).
Significant Differences Regarding the Effect of Inactivating α-DOX2 Function Distinguish Tomato and Arabidopsis Plants
That Atα-DOX2 knockout plants did not display visible defects in growth and development stands in clear contrast to the phenotype observed in tomato plants. However, results showing that constitutive expression of Atα-DOX2 reverses the phenotypic alterations of the div mutants demonstrated the functional similarity of Slα-DOX2 and Atα-DOX2. The different phenotypes associated with the absence of α-DOX2 in tomato and Arabidopsis suggest that additional differences related to the function of α-DOX2 distinguish these two plant species. As a possibility, the α-DOX2 defect could be more easily substituted in Arabidopsis than in tomato plants by other enzymatic systems. A candidate for this could be the α-DOX1 protein. However, results shown here revealed that this is not the case as a double α-dox1 α-dox2 Arabidopsis mutant does not show any visible phenotypic modification or senescence alteration. Alternatively, the differences between tomato and Arabidopsis could be due to intrinsic changes in the composition of the lipid structures influenced by the α-DOX2 function or in the importance of such lipidic components in the development of these two types of plants. As reported (Franke et al., 2005; Nawrath, 2006; Mintz-Oron et al., 2008), the amount and the composition of cutin and likely of other lipid structures that may be affected in the div mutant could differ between tomato and Arabidopsis plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tomato (Solanum lycopersicum) wild-type cultivar Condine Red and α-DOX2 div mutant were provided by the Tomato Genetics Resource Center (TGRC; University of California, Davis). Sterilized tomato seeds were grown in vertically oriented square petri dishes, containing 0.5× Murashige and Skoog (MS), pH 6.0, 2% Suc (w/v), and 1.5% w/v agar (Bacto Agar; Becton-Dickinson). Growth conditions were 16 h of light, 8 h of dark, and 22°C. Seedlings were transplanted to individual pots and grown in the greenhouse. Arabidopsis (Arabidopsis thaliana) wild-type and transgenic Atα-DOX2∷GUS plants used in this study were derived from Arabidopsis plant ecotype Columbia (Col-0). Insertion mutants used were identified using the SIGnAL T-DNA Express Arabidopsis gene mapping tool (http://signal.salk.edu/). SALK lines SALK_005633 (α-dox1-1), SALK_029547 (α-dox2-1), and SALK_089649 (α-dox2-2) were distributed by the Nottingham Arabidopsis Stock Centre (http://Arabidopsis.info). Homozygous insertion mutants were identified by PCR using T-DNA and gene-specific primer sets as described on the T-DNA Express homepage. Sterilized Arabidopsis seeds were vernalized for 3 d at 4°C and grown at 16 h of light, 8 h of dark, and 22°C in petri dishes containing 0.5× MS, pH 6.0, 1.5% Suc (w/v), and 0.8% (w/v) agar (Bacto Agar). For assays with mature plants, seeds were sown on soil and vernalized for 3 d at 4°C and then grown in chamber (22°C, 70% relative humidity, 250 μE m−2 s−1 fluorescent lighting) under a 14 h light/10 h dark photoperiod.
Sequence Alignment and Phylogenetic Relationship Analysis of Plant α-Dioxygenases
Twenty-one plant α-dioxygenases were aligned using the ClustalW2 program (Larkin et al., 2007) and subjected to phylogenetic analysis by the neighbor-joining and maximum parsimony methods using the PHYLIP package (Felsenstein, 1989) through the facilities of the Mobyle platform from the Institut Pasteur server (http://mobyle.pasteur.fr/cgi-bin/portal.py). To maximize the statistical significance of the phylogenetic trees generated by the distance and parsimony methods, 1,000 bootstrap replicates were obtained by both methods.
Cloning and Expression of Tomato and Arabidopsis α-DOX2
Recombinant baculoviruses expressing tomato (Slα-DOX2) and Arabidopsis (Atα-DOX2) α-DOX2 were generated using the Bac-to-Bac baculovirus expression system (Invitrogen). Slα-DOX2 cDNA was isolated from a tomato cDNA library. Clone R16142 containing full-length Atα-DOX2 cDNA was obtained from the Arabidopsis Biological Resource Center (Ohio State University). Slα-DOX2 and Atα-DOX2-encoding cDNAs (GenBank accession nos. AJ850958 and AY081283, respectively) were excised from their host plasmids and ligated into the pFastBac vector. Correct cloning of the insert was verified by sequence analysis. The recombinant plasmids were transferred into DH10Bac Escherichia coli cells containing the baculovirus shuttle vector bMON14272 and the helper plasmid pMON7124. Recombinant bacmid DNAs were prepared from positive bacterial clones, and recombinant baculovirus was obtained by transfection of the bacmids into High Five insect cells according to the manufacturer's instructions. In contrast to the enzymatic characterization of Atα-DOX1 (Sanz et al., 1998) the heterologous expression of α-DOX2 from Arabidopsis and tomato proved to be intricate. Baculovirus-mediated expression in insect cells led to a protein lacking enzymatic activity and was to a large extent present as aggregated protein. Satisfactory enzyme expression with enzyme activity was obtained when the cells were supplemented with hemin, added concomitantly to infection of the cells with baculovirus.
Slα-DOX2 and Atα-DOX2 were expressed by infecting High Five insect cell cultures, grown at 28°C in Tc-100 medium supplemented with 10% fetal calf serum, 10 μm hemin, and the recombinant baculovirus. At 48 h after infection, cells were collected by centrifugation (5 min, 3,000g), washed twice with Dulbecco's phosphate-buffered saline (pH 7.4), divided in aliquots, and pelleted by centrifugation (5 min, 3,000g). Cell pellets were snap-frozen in liquid nitrogen and stored at −80°C. In order to limit enzymatic inactivation, broken cell preparations from recently thawed frozen cell pellets were used to determine enzyme activity in this study. Total protein lysates of α-dioxygenase-expressing High Five insect cells were prepared in sample buffer (30 mm Tris, pH 6.8, 0.5% SDS, 0.5% β-mercaptoethanol, 5% glycerol, 1 mm EDTA, 1× protease inhibitor cocktail [Sigma-Aldrich P-2714], and 0.005% bromphenol blue), and separated by SDS-PAGE (9% cross-linked gels) at 100 V for 3 h in a Bio-Rad gel electrophoresis system (Lowry et al., 1951). Proteins were stained with Coomassie Brilliant Blue. The apparent Mr of the recombinant proteins was determined using Mr marker proteins (Precision Plus protein standards; Bio-Rad). Total protein content was determined by the method of Bradford using cell homogenates prepared in 0.1 m Tris buffer, pH 7.4, with 0.1% Triton X-100 (Bradford, 1976).
α-Dioxygenase Activity
α-Dioxygenase activity was measured by using a Clark-type oxygen electrode (Hansatech Instruments). High Five insect cell pellets containing Slα-DOX2 or Atα-DOX2 (approximately 100 μg total protein) were thawed in 50 μL 0.1 m Tris, pH 7.4, passed five times through a 100-μL Hamilton syringe, and rapidly brought to room temperature. The broken cell preparations were added to the measuring cell containing 1.5 mL 0.1 m Tris, pH 7.4, 100 μm fatty acid substrate, and 100 μm tert-butyl-hydroperoxide. Oxygen consumption was recorded at room temperature, and the rate of enzyme activity calculated as nmol oxygen consumed during the first minute per mg protein. The oxygenase activities were determined using saturated fatty acids ranging in chain length from 14 to 30 carbons and the following unsaturated fatty acids: 7(Z),10(Z),13(Z)-hexadecatrienoic (C16:3), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), and 11(Z)-eicosenoic acid (C20:1). Fatty acids with carbon chain lengths ranging from 14 to 18 were added from ethanol stocks (final assay concentration of ethanol 0.05%), whereas stock solutions of the less soluble C20 to C30 fatty acids were prepared in 1% Tween 20 containing 0.1 n NaOH (final assay concentration of Tween 20 was 0.01%).
Enzyme Incubations and Product Identification
For identification of reaction products, homogenates of High Five cells (approximately 6 × 106 cells) containing Atα-DOX2 or Slα-DOX2 were incubated with palmitic, stearic, linolenic, or arachidic acids (100 μm) in 8 mL of 0.1 m potassium phosphate buffer, pH 7.0, at 23°C under oxygen atmosphere. After 30 min, 20 mL of 30 mm methoxyamine hydrochloride solution in methanol was added to derivatize aldehydes. After 1 h at 23°C, the products were extracted with diethyl ether, derivatized with diazomethane and trimethylchlorosilane, and analyzed by GC-MS. GC-MS analysis was carried out with a Hewlett-Packard model 5970B mass selective detector connected to a Hewlett-Packard model 5890 gas chromatograph equipped with a phenylmethylsiloxane capillary column (12 m, film thickness 0.33 μm). Helium at a flow rate of 25 cm/s was used as the carrier gas.
Steric analysis of 2-hydroperoxide derivatives generated following incubation of palmitic and linolenic acids with Atα-DOX2 or Slα-DOX2 was carried out following reduction with sodium borohydride, methyl-esterification, and isolation of the 2-hydroxyesters by thin-layer chromatography. (−)-Menthoxycarbonyl derivatives were prepared by treatment of the 2-hydroxyesters with (−)-menthylchloroformate/toluene/pyridine (10:10:3, v/v/v) at 23°C for 15 h and purified by thin-layer chromatography. Separation of the (−)-menthoxycarbonyl derivatives of 2(R)- and 2(S)-hydroxyesters was achieved by GC-MS under the conditions described above. The corresponding derivatives of 2(R)- and 2(R,S)-hydroxypalmitic acids and of 2(R)- and 2(R,S)-hydroxylinolenic acids (Lipidox) were used as references.
Plant Treatments and RNA Isolation
For microarray analyses, hypocotyls and cotyledons of 7-d-old tomato seedlings grown on MS medium were excised and used to compare gene expression between the wild-type control and the div mutant. For detachment assays, mature leaves were detached at the petiole from the stem using a forceps and placed onto a Whatman paper filter in water in a 14-cm-diameter petri dish. The dish was placed in a growth chamber in normal lighting conditions, and leaves were collected at days 0, 1, 3, 5, and 7 after detachment. In all cases, collected tissues were frozen in liquid nitrogen and stored at −80°C until analysis. Tomato total RNA was isolated by using the RNeasy plant mini kit (Qiagen), whereas Arabidopsis total RNA was isolated according to Logemann et al. (1987).
Microarray Hybridization and Analysis
Gene expression of wild-type tomato seedling aerial parts grown on MS medium versus α-DOX2 div mutant was compared using the Affymetrix GeneChip Tomato Genome Array. This array consists of over 10,000 tomato probe sets to interrogate over 9,200 tomato transcripts. More information can be found at the Affymetrix homepage (http://www.affymetrix.com/products_services/arrays/specific/tomato.affx). Total RNA was isolated from three independent biological replicates. RNA samples from the wild type and div mutant were quantified using a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technology) and assessed using an Agilent 2100 bioanalyzer (Agilent Technologies). cDNA was synthesized from 4 μg of total RNA using one-cycle target labeling and control reagents (Affymetrix) to produce biotin-labeled cRNA. The cRNA preparation (15 μg) was fragmented at 94°C for 35 min into 35 to 200 bases in length. If the quality control was correct, 5 μg of fragmented cRNA were hybridized to the Tomato Genome Array. Each sample was added to a hybridization solution containing 100 mm MES, 1 m Na+, and 200 mm of EDTA in the presence of 0.01% of Tween 20 to a final cRNA concentration of 0.05 μg/mL. Hybridization was performed for 16 h at 45°C. Each microarray was washed and stained with streptavidin-phycoerythrin in a Fluidics station 450 (Affymetrix) and scanned at 1.56-μm resolution in a GeneChipScanner 3000 7G System (Affymetrix). Data analyses were performed using GeneChip Operating Software. Arrays were hybridized, stained, washed, and screened for quality at the Genomics Service of the Centro Nacional de Biotecnología (Consejo Superior de Investigaciones Científicas). The robust multiarray analysis algorithm was used for background correction, normalization, and expression level summarization (Irizarry et al., 2003). Raw data and normalized data were deposited at ArrayExpress data library (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MEXP-2265. Differentially expressed transcripts were determined using the rank products method (Breitling et al., 2004). The multiple testing problem inherent to microarray experiments was corrected using the FDR method (Benjamin and Hochberg, 1995; Reiner et al., 2003). An FDR of 5% means that only 5% or less of the transcripts up to this position is expected to be observed by chance (false positives), with the remaining 95% being transcripts that are indeed significantly affected (true positives). Significantly up-regulated and down-regulated transcripts obtained in seedlings aerial parts of div mutant compared with the wild type (at FDR of 5%), represented in red and green, respectively, are listed in Supplemental Table S1 online in ascending order of FDR. Additionally, fold change representing differential expression ratio is listed for each probe. In order to improve the annotation of transcripts (at FDR of 21%), the most closely related Arabidopsis homologous loci found using the BLAST algorithm (Zhang et al., 2000) against the National Center for Biotechnology Information nonredundant database of 2009-07-09 (lower E-value) are listed for each probe. Statistical analysis and graphical visualization of data were performed with the interactive tool FIESTA (http://bioinfogp.cnb.csic.es/tools/FIESTA).
Analysis of Gene Expression
RT-PCR analyses were performed with a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems) using the Titan One Tube RT-PCR system (Roche Applied Science) as specified by the manufacturer. Total RNA was treated with DNase TURBO DNA-free (Ambion) to remove contaminating DNA. A quantity (100 ng) of this RNA was used in each one-step RT-PCR procedure. Primers used and the lengths of tomato and Arabidopsis amplification products are described in Supplemental Tables S2 and S3, respectively, online. Tomato gene GAPDH encoding glyceraldehyde 3-phosphate dehydrogenase and Arabidopsis cytoplasmic ribosomal protein L3A were used as internal standards, respectively. For northern blots, RNA (5 μg per lane) was analyzed in agarose-formaldehyde gels, transferred to Hybond N membranes, and hybridized to single-stranded riboprobes following standard procedures (Sambrook et al., 1989). Radioactive probes were prepared for Arabidopsis α-DOX2 from clone R16142 containing full-length cDNA. The amount of loaded RNA was verified by addition of ethidium bromide to the samples and photography under UV light after electrophoresis, followed by hybridization to 18S rRNA (Ruiz-García et al., 1997). Blots shown are representative examples of the results obtained in three independent experiments.
Construction of Transgenic Lines and Analyses of GUS Activity
Genomic sequence extending to approximately 1 kb from the translational start site of the Arabidopsis Atα-DOX2 gene was PCR amplified from wild-type Col-0 using Expand High Fidelity polymerase (Roche). The forward and reverse primers used were 5′-GCTAATAATTCCGAGGGACAGAA-3′ and 5′-CTGTTTTACATATCATTTCTTTTACGG-3′, respectively. The resulting PCR fragment was inserted into the plasmid pGEM-T Easy vector system I (Promega) and sequenced to ensure correct amplification. The promoter sequence was fused to the coding region of the GUS gene present in the plasmid pBI101.2, which confers resistance to kanamycin in planta, introduced into Agrobacterium tumefaciens, and transferred into Col-0 wild-type plants. Homozygous transgenic lines were selected for these studies, and examination of GUS activity was performed as described (Malamy and Benfey, 1997).
Generation of Tomato Transgenic Lines
Slα-DOX2 and Atα-DOX2 genes were excised from their host plasmids and cloned into pGSJ780A and pROK binary vectors harboring a cauliflower mosaic virus 35S promoter resulting in 35S∷Slα-DOX2 and 35S∷Atα-DOX2 constructs, respectively. These constructs were introduced into A. tumefaciens and transferred into tomato div mutant plants according to McCormick (1991). Two different homozygous transgenic lines were selected in each case. Genotype of transgenic plants was verified by PCR amplification and sequencing. Transgene overexpression was confirmed by RT-PCR (Supplemental Fig. S5). Amplification of Atα-DOX2 was performed with oligonucleotides 5′-ACACCAATCTTGTGGCGCATT-3′ and 5′-CTTCATCATCTGTCAACTCTTCC-3′ generating a 221-bp amplicon.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ850958 (tomato α-DOX2 cDNA) and FN428743 (tomato α-DOX2 genomic sequence).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of available α-dioxygenases.
Supplemental Figure S2. Heterologous expression of Slα-DOX2 and Atα-DOX2 in insect cells.
Supplemental Figure S3. Schematic representation of the α-dioxygenation reaction catalyzed by Slα-DOX2 and Atα-DOX2.
Supplemental Figure S4. Scheme of Atα-DOX1 and Atα-DOX2 genomic structures and T-DNA insertion mutants.
Supplemental Figure S5. α-DOX2 expression in tomato wild-type, div mutant, and div transgenic lines.
Supplemental Table S1. Differentially expressed genes in seedlings of tomato div mutant versus wild type.
Supplemental Table S2. Sets of primers used to examine tomato gene expression.
Supplemental Table S3. Sets of primers used to examine Arabidopsis gene expression.
Supplementary Material
Acknowledgments
We thank Michael Bartsch, Pilar Cubas, and Javier Paz-Arés for critical reading of the manuscript; M. Irigoyen for help with phenotypic analyses; R. Piqueras and M. Peinado for help with in vitro plant growth; I. Poveda for expert photography; and Gunvor Hamberg and Tomas Cascón for excellent technical assistance. We also thank G. Martin for the tomato cDNA library. The tomato wild-type cultivar Condine Red and div mutant were provided by the Tomato Genetics Resource Center. The Arabidopsis α-DOX2 cDNA was from the Arabidopsis Biological Resource Center (Ohio State University). The T-DNA insertion lines used in these studies were from the SALK collection and were obtained from the Nottingham Arabidopsis Stock Centre. Microarray analyses were performed at the genomic facilities of the Centro Nacional de Biotecnología (http://www.cnb.csic.es/∼genomica).
This work was supported by the Ministry of Education and Science (Spain; grant no. BIO2006–08581 to C.C.). G.B. is a Ramón y Cajal fellow supported by the Spanish Ministry of Education and Science and the Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas, Spain.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Carmen Castresana (ccastresana@cnb.csic.es).
The online version of this article contains Web-only data.
References
- Aarts MGM, Keijzer CJ, Stiekema WJ, Pereira A (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B 57 289–300 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573 83–92 [DOI] [PubMed] [Google Scholar]
- Browse J (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72 431–456 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Almeras E, Krishnamurthy V (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6 372–378 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP-Phylogeny inference package (version 3.2). Cladistics 5 164–166 [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53 275–297 [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues. A typical suberin and a particular cutin. Phytochemistry 66 2643–2658 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36 629–642 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2005) Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol 71 83–112 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Ponce de León I, Rodríguez MJ, Castresana C (2005) α-Dioxygenases. Biochem Biophys Res Commun 338 169–174 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Ponce de León I, Sanz A, Castresana C (2002) Fatty acid α-dioxygenases. Prostaglandins Other Lipid Mediat 68-69 363–374 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C (1999) α-Oxidation of fatty acids in higher plants. Identification of a pathogen-inducible oxygenase (piox) as an α-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem 274 24503–24513 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Rodríguez MJ, Calvo AP, Castresana C (2003) Activation of the fatty acid α-dioxygenase pathway during bacterial infection of tobacco leaves. Formation of oxylipins protecting against cell death. J Biol Chem 278 51796–51805 [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol 125 683–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5 230–236 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Colllin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Kachroo P (2009) Fatty acid-derived signals in plant defense. Annu Rev Phytopathol 47 153–176 [DOI] [PubMed] [Google Scholar]
- Kandel S, Sauveplane V, Olry A, Diss L, Benviste I, Pinot F (2006) Cytochrome P450-dependent fatty acid hydroxylases in plants. Phytochem Rev 5 359–372 [Google Scholar]
- Koeduka T, Matsui K, Hasegawa M, Akakabe Y, Kajiwara T (2005) Rice fatty acid α-dioxygenase is induced by pathogen attack and heavy metal stress: activation through jasmonate signaling. J Plant Physiol 162 912–920 [DOI] [PubMed] [Google Scholar]
- Koszelak-Rosenblum M, Krol AC, Simmons DM, Goulah CC, Wroblewski L, Malkowski MG (2008) His-311 and Arg-559 are key residues involved in fatty acid oxygenation in pathogen-inducible oxygenase. J Biol Chem 283 24962–24971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Kunst L, Jetter R (2007) Composition of alkyl esters in the cuticular wax on inflorescence stems of Arabidopsis thaliana cer mutants. Plant J 50 189–196 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, López R, et al (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57 405–430 [DOI] [PubMed] [Google Scholar]
- Liavonchanka A, Feussner I (2006) Lipoxygenases: occurrence, functions and catalysis. J Plant Physiol 163 348–357 [DOI] [PubMed] [Google Scholar]
- Liu W, Rogge CE, Bambai B, Palmer G, Tsai AL, Kulmacz RJ (2004) Characterization of the heme environment in Arabidopsis thaliana fatty acid α-dioxygenase-1. J Biol Chem 279 29805–29815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang LH, Fabian P, Hayashi Y, McGinley CM, van der Donk WA, Kulmacz RJ (2006) Arabidopsis thaliana fatty acid alpha-dioxygenase-1: evaluation of substrates, inhibitors and amino-terminal function. Plant Physiol Biochem 44 284–293 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willlmitzer L (1987) Improved method for isolation of RNA from plant tissues. Anal Biochem 163 16–20 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Roseborough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin phenol reagent. J Biol Chem 193 265–273 [PubMed] [Google Scholar]
- Malamy J, Benfey PN (1997) Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J 12 957–963 [DOI] [PubMed] [Google Scholar]
- McCormick S (1991) Transformation of Tomato with Agrobacterium tumefaciens, Vol B. Kluwer, Amsterdam
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang ZY, Jetter R, Venger I, Adato A, et al (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M (2006) The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 67 2597–2610 [DOI] [PubMed] [Google Scholar]
- Nawrath C (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9 281–287 [DOI] [PubMed] [Google Scholar]
- Ponce de León I, Sanz A, Hamberg M, Castresana C (2002) Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J 29 61–62 [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamin Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 368–375 [DOI] [PubMed] [Google Scholar]
- Ruiz-García L, Madueño F, Wilkinson M, Haughn G, Salinas J, Martínez-Zapater JM (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9 1921–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanz A, Moreno JI, Castresana C (1998) PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell 10 1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Shibata D, Axelrod B (1995) Plant lipoxygenases. J Lipid Mediat Cell Signal 12 213–228 [DOI] [PubMed] [Google Scholar]
- Stevens MA, Rick CM (1986) Genetics and breeding. In JG Atherton, J Rudich, eds, The Tomato Crop, A Scientific Basis for Crop Improvement. Chapman and Hall, New York, pp 35–100
- van der Biezen EA, Brandwagt BF, van Leeuwen W, John J, Nijkamp J, Hille J (1996) Identification and isolation of the FEEBLY gene from tomato by transposon tagging. Mol Genet Genomics 251 267–280 [DOI] [PubMed] [Google Scholar]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7 217–224 [DOI] [PubMed] [Google Scholar]
- Wei J, Tirajoh A, Effendy J, Plant AL (2000) Characterization of salt-induced changes in gene expression in tomato (Lycopersicon esculentum) roots and the role played by abscisic acid. Plant Sci 159 135–148 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7 203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.