Abstract
WD40 repeat proteins regulate biosynthesis of anthocyanins, proanthocyanidins (PAs), and mucilage in the seed and the development of trichomes and root hairs. We have cloned and characterized a WD40 repeat protein gene from Medicago truncatula (MtWD40-1) via a retrotransposon-tagging approach. Deficiency of MtWD40-1 expression blocks accumulation of mucilage and a range of phenolic compounds, including PAs, epicatechin, other flavonoids, and benzoic acids, in the seed, reduces epicatechin levels without corresponding effects on other flavonoids in flowers, reduces isoflavone levels in roots, but does not impair trichome or root hair development. MtWD40-1 is expressed constitutively, with highest expression in the seed coat, where its transcript profile temporally parallels those of PA biosynthetic genes. Transcript profile analysis revealed that many genes of flavonoid biosynthesis were down-regulated in a tissue-specific manner in M. truncatula lines harboring retrotransposon insertions in the MtWD40-1 gene. MtWD40-1 complemented the anthocyanin, PA, and trichome phenotypes of the Arabidopsis (Arabidopsis thaliana) transparent testa glabrous1 mutant. We discuss the function of MtWD40-1 in natural product formation in M. truncatula and the potential use of the gene for engineering PAs in the forage legume alfalfa (Medicago sativa).
Anthocyanins and proanthocyanidins (PAs; also called condensed tannins) are flavonoids that benefit both plant and human health. Anthocyanins attract pollinators, protect plant tissues from UV light damage, and defend plants against predators (Stapleton and Walbot, 1994; Sullivan, 1998). PAs are abundant in beverages such as tea, wine, and fruit juice and exhibit antioxidant activity and cardiovascular protective effects (Bagchi et al., 2000; Cos et al., 2004; Dixon et al., 2005). Moreover, a moderate PA level is an important quality trait in forage crops, because PAs can protect ruminant animals from lethal pasture bloat by binding proteins and thereby slowing down their fermentation in the rumen (Li et al., 1996; Aerts et al., 1999; Barry and McNabb, 1999; Dixon et al., 2005).
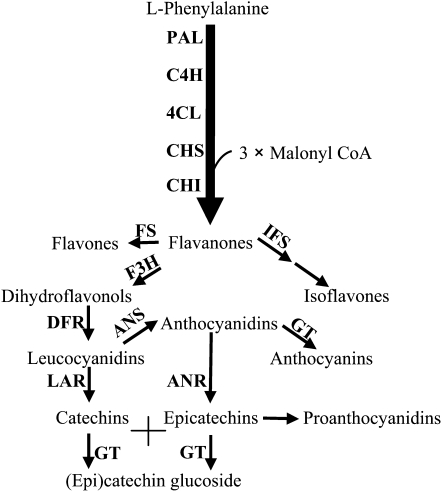
The PA biosynthetic pathway in Arabidopsis (Arabidopsis thaliana) has been studied primarily through the analysis of transparent testa (tt) or transparent testa glabrous (ttg) mutants, which exhibit seed coat (tt) or seed coat and trichome (ttg) phenotypes (Shirley et al., 1995; Lepiniec et al., 2006). The mutated genes have been found to encode either pathway enzymes or transcriptional regulators that function alone or in complexes to control the whole or branches of the pathway (Lepiniec et al., 2006). Anthocyanins and PAs share the same upstream phenylpropanoid/flavonoid pathway, and anthocyanidin is the immediate substrate for both glycosylation to anthocyanin or reduction to epicatechin in the biosynthesis of PAs in Arabidopsis (Fig. 1).
Figure 1.
The flavonoid pathway leading to anthocyanins and PAs. Enzymes are as follows: PAL, l-Phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; CHR, chalcone reductase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol reductase; FS, flavone synthase; IFS, isoflavone synthase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; GT, glucosyltransferase.
We are studying the formation of PAs in the model legume Medicago truncatula (Xie et al., 2003, 2006; Pang et al., 2007, 2008). Four structural genes encoding anthocyanidin synthase (ANS), leucoanthocyanidin reductase (LAR), anthocyanidin reductase (ANR), and epicatechin 3′-O-glucosyltransferase (UGT72L1) were characterized biochemically and/or genetically from this species (Xie et al., 2003; Pang et al., 2007, 2008). However, little is known about the regulatory network involved in anthocyanin/PA biosynthesis in M. truncatula.
A regulatory complex, comprising an R2R3-MYB transcription factor, a basic helix-loop-helix (bHLH) domain protein, and a WD40 repeat protein, regulates production of anthocyanins in foliar tissues and PAs and mucilage in seed coats; this complex also controls the formation of root hairs and trichomes on aerial tissues in some but not all plants (Baudry et al., 2004; Broun, 2005; Lepiniec et al., 2006; Morita et al., 2006; Serna and Martin, 2006; Gonzalez et al., 2008; Zhao et al., 2008). In Arabidopsis, these proteins are encoded by Transparent Testa2 (TT2, Myb), Transparent Testa8 (TT8, HLH), and Transparent Testa Glabrous1 (TTG1, WD40 repeat), which together regulate the late flavonoid pathway genes and the PA-specific pathway gene ANR (Baudry et al., 2004). Loss of function of either TT2 or TT8 leads to a lack of anthocyanin pigmentation in foliar tissue and a loss of PAs in the seed coat (Nesi et al., 2000, 2001). The presence of TTG1 is essential and irreplaceable in this complex for anthocyanin/PA biosynthesis, trichome formation, seed mucilage production, and root hair formation (Koornneef, 1981; Walker et al., 1999). Several other WD40 repeat proteins functionally orthologous to TTG1 have been described from other species such as petunia (Petunia hybrida), Perilla frutescens, cotton (Gossypium hirsutum), and maize (Zea mays); mutation of some affects both anthocyanin/PA and trichome phenotypes, whereas mutation of others only affects the anthocyanin/PA phenotype (Lloyd et al., 1992; de Vetten et al., 1997; Sompornpailin et al., 2002; Carey et al., 2004; Humphries et al., 2005).
In an attempt to identify genes involved in the regulation of anthocyanin and PA biosynthesis in M. truncatula, we have screened a Tnt1 retrotransposon insertion population for altered leaf (lack of red pigment) and seed (transparent testa) phenotypes. This led to the cloning and functional characterization of a gene, MtWD40-1, with high sequence identity to known WD40 repeat proteins. MtWD40-1 can complement the Arabidopsis ttg1 PA and trichome phenotypes, although the Medicago wd40-1 mutant retained normal trichomes. Loss of MtWD40-1 function has profound and differential effects on flavonoid biosynthesis in different plant organs. The potential of MtWD40-1 for engineering the PA pathway in alfalfa (Medicago sativa) was also investigated.
RESULTS
Phenotypic and Genotypic Characterization of M. truncatula Retrotransposon Insertion Mutants
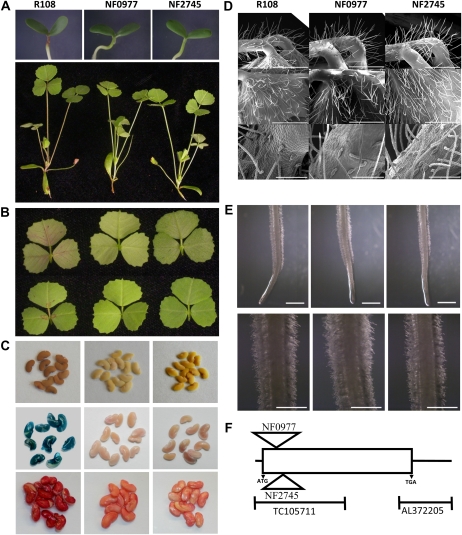
One mutant line (NF0977) drew our attention when screening the M. truncatula Tnt1 insertion population for visible anthocyanin phenotypes. This line lacked the typical red pigmentation in the stem, the anthocyanin-rich circle at the base of the axial side of the leaflet, and the small red spots on the adaxial side of the leaflet, all of which are seen in wild-type ecotype R108 (Fig. 2, A and B). The seed coat of this mutant line was transparent with a yellowish color that contrasted with the brown pigmentation of the wild type that arises from the presence of oxidized PAs (Fig. 2C). To further confirm the PA phenotype, seeds were stained with dimethylaminocinnamaldehyde (DMACA), a reagent that is specific for PAs and their flavan 3-ol precursors. Mature seeds from the mutant line did not exhibit the typical blue staining characteristic of the reaction of PAs with DMACA (Fig. 2C). The seeds from the mutant also produced less mucilage than those of the wild type, as seen by the reduced staining of the seed coat with ruthenium red (Fig. 2C). No other obvious phenotypes, such as altered density of glandular or nonglandular trichomes (Fig. 2D) or root hairs (Fig. 2E), were observed in the NF0977 mutant. Root hair density appeared to be unaffected on both young (4 d after germination; Fig. 2E) and mature (Supplemental Fig. S1) roots.
Figure 2.
Visible phenotypes resulting from insertional mutagenesis of MtWD40-1. A, Top, 4-d-old seedlings of M. truncatula R108 (wild type) and the two insertional mutant lines, showing pigmentation below the cotyledons. Bottom, aerial parts of older seedlings, shown in the same order. B, Axial side (bottom) and adaxial side (top) of leaves from a wild-type plant (left), NF0977 (center), and NF2745 (right). C, Mature seeds of the wild type (left), NF0977 (center), and NF2745 (right), either unstained (top), stained with DMACA reagent to detect PAs (center), or stained with ruthenium red to detect mucilage (bottom). D, Scanning electron microscopy analysis of trichomes on young petioles and leaves of the wild type (left), NF0977 (center), and NF2745 (right). The top panels show nonglandular petiole trichomes, the center panels show nonglandular leaf trichomes, and the bottom panels show glandular and nonglandular petiole trichomes. Bars = 1 mm in top and center panels and 200 μm in bottom panels. E, Root hair phenotypes of the wild type (left), NF0977 (center), and NF2745 (right). Bars = 2 mm in top panels and 1 mm in bottom panels (showing closeups of the hairs just behind the root tip). F, A diagram of the MtWD40-1 gene (1,364 bp) showing the positions of the independent Tnt1 insertions and the two probe sets on the Medicago Affymetrix GeneChip.
One of 12 plants from the NF0977 R2 generation exhibiting the lack of pigmentation phenotype was allowed to undergo self-pollination. All 29 plants from the R3 generation were homozygous, as confirmed by PCR with gene-specific primers and a primer for the Tnt1 insert, and retained the visible mutant phenotypes as characterized in Figure 2, A to C. Use of thermal asymmetric interlaced (TAIL)-PCR revealed that all individuals possessed a retrotransposon insertion in a WD40 gene with similarity to the TTG1 gene from Arabidopsis. After sequencing and alignment using the available M. truncatula genome database, this Tnt1 insertion was found to be between the first and second nucleotides of amino acid residue Ser-31 of the WD40 protein in the NF0977 mutant (Fig. 2F). A further 20 insertion sites in different regions of the genome were also recovered from NF0977 (Supplemental Table S1), typical for Tnt1 insertional mutagenesis in Medicago (Tadege et al., 2008). None of these insertions was in a gene that would be expected to affect flavonoid biosynthesis, although this does not rule out the possibility that the lack of pigmentation phenotype could have been the result of an insertion in one or more of these genes. Therefore, a reverse genetic approach was employed to screen the Tnt1 insertion mutant population for additional lines with insertions in the WD40 gene, and another mutant line, NF2745, was obtained. The insertion site in line NF2745 was between amino acid residues Ser-46 and Ile-47 (Fig. 2F). Homozygous NF2745 plants exhibited the same phenotype as NF0977 (Fig. 2, A–E), strongly suggesting that the loss of function of the WD40 gene is responsible for the pigmentation phenotypes in the two mutants.
Molecular Cloning and Characterization of MtWD40-1
BLASTX analysis of the partial WD40 sequence against the GenBank database showed that this gene was located on the M. truncatula bacterial artificial chromosome clone CR940305. Its full-length sequence was predicted to be 1,363 bp in length with a 49-bp 5′ untranslated region and a 285-bp 3′ untranslated region (designated as MtWD40-1; GenBank accession no. EU040206). MtWD40-1 is a single-copy gene lacking introns, as confirmed by DNA gel-blot analysis and amplification of the MtWD40-1 open reading frame (ORF) with genomic DNA as template (data not shown). MtWD40-1 encodes a predicted protein ORF of 343 amino acids, with a calculated pI of 4.99 and a molecular mass of 38 kD.
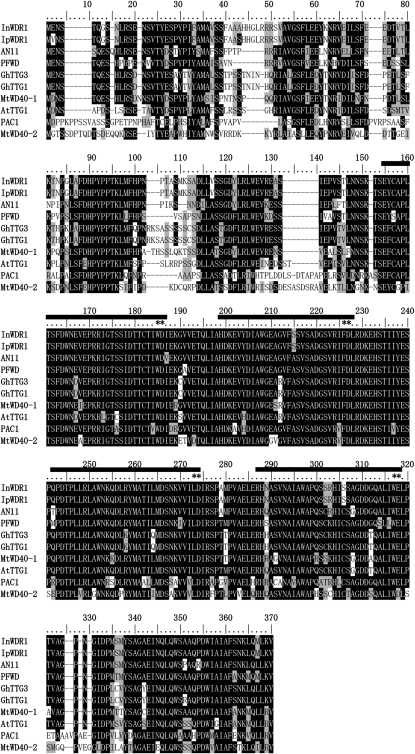
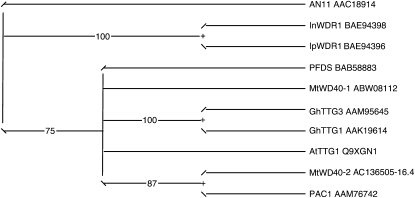
The deduced amino acid sequence of MtWD40-1 showed 77% to 79% identity to other known WD40 repeat proteins from different plant species, such as TTG1 from Arabidopsis and AN11 from petunia (Fig. 3). The four WD40 repeat domains are highly conserved among all the WD40 repeat proteins, including MtWD40-1, and the last two amino acids in each WD40 repeat are identical. Phylogenetic analysis (Fig. 4) showed that MtWD40-1 is most closely related to TTG1 from Arabidopsis. Another Medicago WD40-like protein, MtWD40-2, is less than 60% identical to MtWD40-1 at the amino acid level and somewhat closer to PAC1 from maize.
Figure 3.
Alignment of deduced amino acid sequences of plant WD40 repeat proteins. The WD40 repeat domains are marked with horizontal bars above the sequences, and the last two amino acids of each repeat domain are marked with stars. Identical residues are highlighted on a black background, and similar residues are highlighted on a gray background. The GenBank accession numbers are as follows: BAE94398, InWDR1 from Ipomoea nil; BAE94396, IpWDR1 from Ipomoea purpurea; AAC18914, AN11 from Petunia hybrida; BAB58883, PFDS from Perilla frutescens; AAM95645, GhTTG3 from Gossypium hirsutum; AAK19614, GhTTG1; ABW08112, MtWD40-1; Q9XGN1, AtTTG1; AAM76742, PAC1 from Zea mays; AC136505_16.4, MtWD40-2.
Figure 4.
Unrooted phylogram comparison of the amino acid sequences of MtWD40-1 and other functionally characterized plant WD40 repeat proteins. The sequences used are the same as in Figure 3. The phylogenetic tree was constructed by PAUP* 4.0b10, after alignment using MAFFT software. Node support was estimated using neighbor-joining bootstrap analysis (1,000 bootstrap replicates).
MtWD40-1 Complements the Arabidopsis ttg1 and Medicago NF0977 Mutants by Interacting with Glabrous3
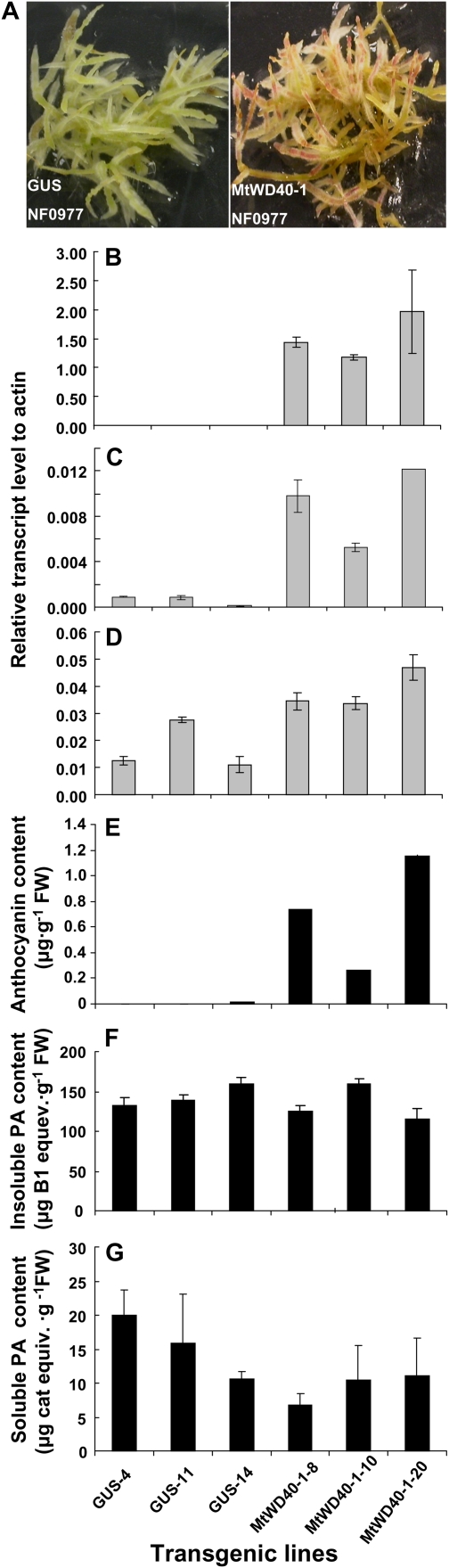
Hairy roots of M. truncatula R108 exhibit red anthocyanin pigmentation (Pang et al., 2008), but this was lacking in the NF0977 line. Hairy root transformation, therefore, was used as a rapid method to confirm that MtWD40-1 could complement the lack-of-pigment phenotype of the NF0977 Tnt1 insertion mutant. Red pigmentation was seen in all 101 phosphinothricin (ppt)-resistant hairy root lines transformed with MtWD40-1 but in none of the 30 ppt-resistant NF0977 lines transformed with the GUS gene (Fig. 5A). Quantitative reverse transcription (qRT)-PCR confirmed that MtWD40-1, ANS, and the anthocyanin-specific glucosyltransferase UGT78G1 (Modolo et al., 2007; Peel et al., 2009) were expressed at higher levels in hairy roots of the MtWD40-1-transformed lines than in the GUS transformants (Fig. 5, B–D), thus accounting for the high levels of extractable anthocyanins in the MtWD40-1-expressing lines (Fig. 5E). No significant differences were observed in the levels of insoluble PAs (PAs that bind to the cell wall and cannot be extracted by organic solvents such as 70% acetone) between the MtWD40-1-expressing NF0977 lines and the GUS control lines (Fig. 5G) or in the levels of transcripts encoding the PA pathway-specific genes ANR and UGT72L1 (data not shown). In contrast, soluble PA levels decreased slightly in the mutant line complemented with MtWD40-1 (Fig. 5F), possibly as a result of flux into soluble PAs being diverted back into the anthocyanin pathway.
Figure 5.
Genetic complementation of the anthocyanin and PA phenotypes of the NF0977 retrotransposon insertion line. A, Pigmentation of hairy roots of the NF0977 line expressing GUS (left) and MtWD40-1 (right). B, qRT-PCR analysis of MtWD40-1 transcript levels in hairy roots of NF0977 expressing GUS or MtWD40-1. C, As above, showing ANS transcript levels. D, As above, showing UGT78G1 transcript levels. E, Anthocyanin levels from NF0977 expressing GUS or MtWD40-1 (three independent lines of each). F, As above, showing insoluble PA levels. G, As above, showing soluble PA levels. FW, Fresh weight.
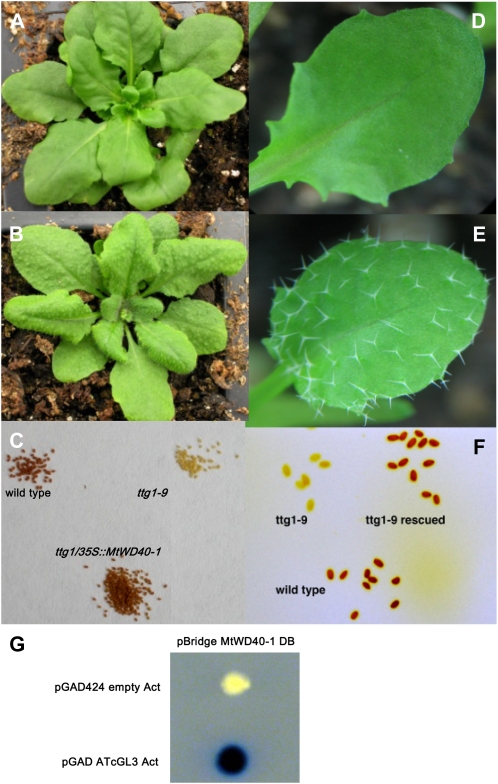
To determine whether MtWD40-1 is a functional ortholog of TTG1, the MtWD40-1 ORF under the control of the 35S promoter was transformed into the Arabidopsis ttg1-9 mutant, and expression of the foreign MtWD40-1 gene was confirmed by qRT-PCR (Supplemental Fig. S2). 35S:MtWD40-1 fully complemented the anthocyanin pigmentation, trichome deficiency, and seed coat PA phenotypes (Fig. 6, A–C). We also tested the ability of MtWD40-1 to complement the Arabidopsis ttg1-9 mutant when expressed under the control of the Arabidopsis Glabrous2 (GL2) promoter, which is active in the shoots of ttg1 mutants (Szymanski et al., 1998). Again, the phenotype was fully rescued (Fig. 6, D–F).
Figure 6.
Genetic complementation of the Arabidopsis ttg1-9 mutant. A, Leaves of the ttg1-9 mutant line. B, Leaves of the ttg1-9 mutant expressing 35S∷MtWD40-1. C, Seed coat pigmentation of the wild type, ttg1-9, and ttg1-9 expressing 35S∷MtWD40-1. D, A single leaf of ttg1-9 showing the glabrous phenotype. E, A single leaf of the ttg1-9 mutant expressing GL2∷MtWD40-1 showing the restoration of the trichome phenotype. F, Seed coat pigmentation of the wild type, ttg1-9, and ttg1-9 expressing GL2∷MtWD40-1. G, Yeast two-hybrid analysis of the interaction between MtWD40-1 and Arabidopsis GL3 (see text for details).
To further determine how MtWD40-1 might function to restore the trichome phenotype in ttg1-9 Arabidopsis, the yeast two-hybrid system was used to test the interaction of MtWD40-1 with GL3, a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1 (Payne et al., 2000). GL3 was fused to the activation domain (AD) of GAL4, and MtWD40-1 was fused to the binding domain (BD) of GAL4. Yeast containing empty pGAD424 (AD) and pBridge (BD) vectors in conjunction with MtWD40-1 did not exhibit β-galactosidase activity (Fig. 6G, top), whereas yeast containing GL3-AD and MtWD40-1BD exhibited strong activity (Fig. 6G, bottom), suggesting that MtWD40-1 can interact with GL3 for trichome formation in Arabidopsis, even though it is not necessary for trichome formation in M. truncatula.
Tissue- and Development-Specific Expression of MtWD40-1
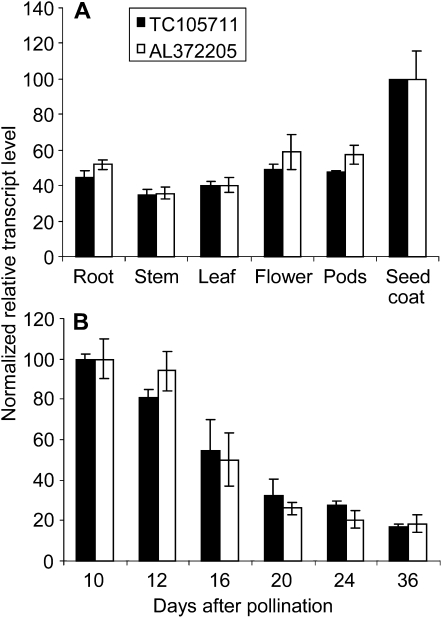
To determine the developmental expression pattern of MtWD40-1, normalized data were retrieved from the M. truncatula gene expression atlas (Benedito et al., 2008) together with seed coat microarray data (Pang et al., 2008). The expression patterns of two probe sets for MtWD40-1 (TC105711 and AL372205; probe set locations are shown in Fig. 2F) were essentially the same, confirming that, as is also the case for TTG1 in Arabidopsis (Walker et al., 1999), MtWD40-1 is expressed in all organs, with highest expression in the seed coat (Fig. 7A). During seed development, MtWD40-1 showed its highest expression level at or before 10 d after pollination (dap; Fig. 7B), with a subsequent decline toward seed maturity. This expression pattern parallels the expression of MtANR and UGT72L1 during seed development (Pang et al., 2008).
Figure 7.
MtWD40-1 transcript levels in M. truncatula ecotype Jemalong A17 as determined by microarray analysis. A, Tissue-specific expression. B, MtWD40-1 transcript levels during seed development. The data were retrieved from the M. truncatula gene expression atlas (Benedito et al., 2008) and the seed coat microarray data set (Pang et al., 2008).
We also analyzed the expression pattern of MtWD40-2 in the M. truncatula gene expression atlas (Benedito et al., 2008), where it is represented by probe set Mtr. 22605.1.S1_at (http://bioinfo.noble.org/gene-atlas/v2/). The highest expression level is in roots 24 h after salt stress and in developing root nodules, but the expression level in these tissues is nearly 2 orders of magnitude lower than the maximum expression level of MtWD40-1 (in developing seeds). MtWD40-2 is expressed around 15-fold lower than MtWD40-1 in trichome-containing leaf and petiole tissues and is only expressed very weakly if at all in isolated nonglandular trichomes from M. truncatula (only called present in one out of three Affymetrix data sets; M. David Marks, unpublished data). Furthermore, unlike MtWD40-1, MtWD40-2 is not induced in M. truncatula hairy roots expressing Arabidopsis TT2.
Tissue-Specific Effects of Loss of MtWD40-1 Function on Phenylpropanoid/Flavonoid Pathway Gene Transcripts and Metabolites
To determine the impacts of the loss of MtWD40-1 function on gene expression in seeds, we dissected seeds at 16 dap from both the NF0977 mutant line and the corresponding wild-type control (ecotype R108) for microarray analysis using the Affymetrix Medicago GeneChip. We have previously shown that phenylpropanoid/flavonoid biosynthetic pathway genes are highly expressed at 16 dap (Pang et al., 2007). The microarray data showed that 152 probe sets were down-regulated more than 2-fold in the MtWD40-1 mutant line; among these, three probe sets were down-regulated by more than 100-fold, 25 by more than 5-fold, with the remainder between 2- to 5-fold (Supplemental Table S2E). Classification using the GeneBins ontology tool (http://bioinfoserver.rsbs.anu.edu.au/utils/GeneBins/index.php) showed that a high percentage (43.5%) of the down-regulated genes were “unclassified with homology” followed by “biosynthesis of secondary metabolites” (25.9%; Supplemental Fig. S3). This latter class consisted primarily of phenylpropanoid/flavonoid pathway genes.
Among the 28 probe sets that exhibited a more than 5-fold reduction in expression level in the MtWD40-1 mutant (Table I), 17 were associated with the phenylpropanoid/flavonoid pathway and one had no homology to any known gene. The early phenylpropanoid pathway genes PAL, 4CL, CHS, F3′H, and F3′5′H were all down-regulated, almost 200-fold in the case of one CHS probe set (Table I; Supplemental Table S2). CHS is encoded by a large gene family in Medicago, and nine different CHS probe sets were down-regulated more than 5-fold (Supplemental Table S2). The two later anthocyanin pathway genes, DFR and ANS, were down-regulated by 2.6-fold and 9.2/10.0-fold, respectively (Table I; Supplemental Table S2), suggesting that MtWD40-1 regulates both early and later anthocyanin pathway genes in seeds. Three genes specific for the PA pathway, LAR, ANR, and UGT72L1, were down-regulated 3.9-, 34.6-, and 14.7-fold, respectively, highlighting the specific involvement of MtWD40-1 in the regulation of PA biosynthesis. MtWD40-1 might also regulate additional branches of the flavonoid pathway, as seen by the 40.6-fold and 2.1-fold reductions in expression of flavonol synthase and a putative isoflavone O-glycosyltransferase in the MtWD40-1 mutant.
Table I.
The gene probe sets that were down-regulated more than 5-fold in developing seed of the M. truncatula NF0977 mutant compared with the wild-type control
Expression values were obtained from RMA (Irizarry et al., 2003).
| Probe Sets | Target Description | Ratio (R108/NF0977) | Pa | Qb |
|---|---|---|---|---|
| Mtr.20567.1.S1_at | Type III polyketide synthase; naringenin-chalcone synthase (CHS) | 198.05 | 0.000017 | 0.060224 |
| Mtr.20185.1.S1_x_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 105.58 | 0.000147 | 0.089928 |
| Mtr.39897.1.S1_at | Similar to CPRD12 protein, partial (61%) | 104.58 | 0.000001 | 0.0219 |
| Mtr.20185.1.S1_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 95.97 | 0.000666 | 0.121225 |
| Mtr.36333.1.S1_at | Similar to flavonoid 3′-hydroxylase (fragment), partial (21%; F3′H) | 85.31 | 0.000002 | 0.032849 |
| Mtr.49421.1.S1_at | 2OG-Fe(II) oxygenase | 79.12 | 0.000005 | 0.043799 |
| Mtr.14017.1.S1_at | Flavonol synthase (based on similarity; FLS) | 40.62 | 0.000018 | 0.060224 |
| Mtr.6517.1.S1_at | Similar to gray pubescence flavonoid 3′-hydroxylase, partial (49%; F3′H) | 36.77 | 0.000264 | 0.105576 |
| Mtr.44985.1.S1_at | Anthocyanidin reductase, complete (ANR) | 34.55 | 0.000038 | 0.089283 |
| Mtr.14428.1.S1_x_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 26.53 | 0.000414 | 0.115755 |
| Mtr.51818.1.S1_at | Predicted protein | 23.29 | 0.00086 | 0.124286 |
| Mtr.16432.1.S1_at | Myb, DNA-binding; homeodomain-like | 23.04 | 0.003335 | 0.151484 |
| Mtr.14428.1.S1_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 22.59 | 0.000774 | 0.124286 |
| Mtr.20187.1.S1_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 16.74 | 0.000015 | 0.060224 |
| Mtr.20187.1.S1_x_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 15.91 | 0.000049 | 0.089283 |
| Mtr.21996.1.S1_x_at | Weakly similar to glucosyltransferase-13 (fragment; UGT72L1) | 14.72 | 0.000639 | 0.121225 |
| Mtr.49572.1.S1_s_at | Naringenin-chalcone synthase; type III polyketide synthase (CHS) | 14.19 | 0.000191 | 0.096689 |
| Mtr.47287.1.S1_at | Weakly similar to myosin heavy chain-related temporary automated functional | 11.91 | 0.000397 | 0.115149 |
| Mtr.3858.1.S1_at | Leucoanthocyanidin dioxygenase, anthocyanidin synthase, partial (24%; ANS) | 9.98 | 0.015726 | 0.183253 |
| Mtr.28774.1.S1_at | Anthocyanidin synthase, partial (53%; ANS) | 9.23 | 0.000069 | 0.089283 |
| Mtr.28714.1.S1_at | Homolog to chalcone synthase 3 (Sinapis alba), partial (12%; CHS) | 7.92 | 0.014403 | 0.182863 |
| Mtr.48474.1.S1_at | Weakly similar to nodulin N21 family protein integral membrane protein domain, partial (91%) | 7.65 | 0.002909 | 0.147343 |
| Mtr.6511.1.S1_at | Similar to GTP-binding protein, partial (47%) | 6.75 | 0.000189 | 0.096689 |
| Mtr.25305.1.S1_at | Weakly similar to The Arabidopsis Information Resource gene 2827885-GOpep 0.1 68409.m01848; expressed protein | 6.00 | 0.00022 | 0.10046 |
| Mtr.18797.1.S1_at | Proteinase inhibitor I3, Kunitz legume; Kunitz inhibitor ST1-like | 5.97 | 0.00559 | 0.164484 |
| Mtr.32965.1.S1_at | Similar to cytochrome b5 DIF-F, partial (36%) | 5.79 | 0.008992 | 0.176801 |
| Mtr.7095.1.S1_at | Similar to Na+/H+ antiporter NHX6, partial (28%) | 5.69 | 0.001118 | 0.127332 |
| Mtr.41031.1.S1_at |
Homolog to 4-coumarate-CoA ligase (4CL) |
5.26 |
0.000157 |
0.089928 |
The P values were obtained using associative analysis (Dozmorov and Centola, 2003).
The Q values were obtained using extraction of differential gene expression (Leek et al., 2006).
Another 271 probe sets were up-regulated in seeds of the mutant, most of them associated with primary metabolism or stress responses, but no phenylpropanoid/flavonoid pathway genes were up-regulated (data not shown).
The large number of changes observed in nonphenylpropanoid/flavonoid pathway genes in the above experiment could potentially occur as a result of the additional retrotransposon insertions in line NF0977. Therefore, we reexamined changes in key flavonoid pathway gene transcripts in seeds and other organs, in both NF0977 and the independent retrotransposon insertion line NF2745, using qRT-PCR (Table II). MtWD40-1 transcript levels were more strongly down-regulated in tissues of NF2745 than in NF0977 (Table II; Supplemental Tables S3 and S4). Compared with wild-type R108, PAL and CHI transcript levels were least affected in the two MtWD401 retrotransposon insertion mutants. The most consistent changes observed as a result of loss of MtWD40-1 function were strong reductions of CHS expression in flower (but only determined for one probe set corresponding to TC138581) and seed, DFR1 expression in leaf and flower, ANS expression in stem, leaf, and seed, LAR and ANR expression in flower and seed, and UGT72L1 expression in seed (Table II; Supplemental Tables S3 and S4). Thus, although MtWD40-1 is most strongly expressed in the seed (coat), its loss of function can affect flavonoid pathway gene expression in multiple tissues.
Table II.
Fold change (decrease) of flavonoid pathway gene transcripts in different tissues of the NF0977 and NF2745 retrotransposon insertion lines compared with wild-type R108
Transcript levels were determined by qRT-PCR with actin as an internal reference. Data represent average relative transcript levels to actin from biological triplicates, expressed as the ratio of transcript level in R108 to that in the mutants.
| Tissue | Ratio | PAL | CHS | CHI | F3H | DFR1 | DFR2 | ANS | LAR | ANR | UGT72L1 | MtWD40-1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Root | R108/NF0977 | 2.66 | 5.98 | 2.24 | 3.10 | 0.10 | 0.57 | 0.68 | 1.43 | 0.92 | 0.34 | 4.25 |
| R108/NF2745 | 0.56 | 1.62 | 1.20 | 4.30 | 1.11 | 0.35 | 1.01 | 0.95 | 1.49 | 0.54 | 2,132.75 | |
| Stem | R108/NF0977 | 0.98 | 0.52 | 0.58 | ∞a | 105.15 | 1.50 | 799.92 | 1.48 | 1.80 | 0.65 | 9.32 |
| R108/NF2745 | 0.87 | 2.02 | 1.02 | 4.48 | 1.54 | 0.82 | 76.42 | 1.39 | 1.01 | 0.62 | 2,454.75 | |
| Leaf | R108/NF0977 | 0.59 | 0.61 | 0.22 | ∞ | 81.06 | 1.00 | 76.22 | 0.17 | 0.16 | 1.55 | 5.14 |
| R108/NF2745 | 0.47 | 3.39 | 0.98 | ∞ | 11.07 | 6.13 | 33.21 | 0.17 | 0.64 | 1.68 | 1,568.11 | |
| Flower | R108/NF0977 | 0.89 | 67.38 | 0.96 | 0.91 | 5.48 | 1.55 | 2.71 | 20.62 | 216.01 | 7.97 | 17.10 |
| R108/NF2745 | 0.93 | 46.15 | 0.62 | 0.28 | 8.03 | 1.54 | 1.23 | 16.09 | 119.12 | 0.84 | 1,521.86 | |
| Seed (16 dap) | R108/NF0977 | 0.42 | 240.10 | 1.05 | ∞ | 1.73 | 3.11 | 12.25 | 6.47 | 58.83 | ∞ | 23.26 |
| R108/NF2745 |
2.12 |
80.22 |
0.72 |
∞ |
1.65 |
3.03 |
7.87 |
8.60 |
33.7 |
4.16 |
2,761.54 |
∞, Numerical ratio set to infinity due to the undetectable transcript level in the mutant line. See Supplemental Tables S3 and S4 for absolute values and sd values for each measurement.
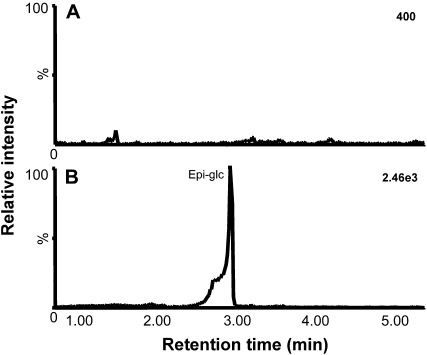
To further investigate the impact of loss of WD40-1 expression on flavonoid biosynthesis, levels of phenylpropanoid-derived secondary metabolites were measured by ultra-high-performance liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOF-MS) in various tissues of wild-type R108 and the two independent retrotransposon insertion lines (Table III). The greatest effects were seen in developing seed, where levels of epicatechin and its glucoside (Fig. 8) as well as cyanidin 3-O-glucoside, kaempferol 3-O-rutinoside, and two benzoic acid derivatives were reduced to undetectable levels in the insertion lines. In contrast, although epicatechin and its conjugate were likewise undetectable in flowers of the two mutant lines, levels of cyanidin 3-O-glucoside and other flavonoids were increased (Table III), in spite of the apparently strong reduction in CHS expression in these lines. Loss of function of MtWD40-1 had little effect on the levels of three flavonoids in leaves but resulted in reduced isoflavone (biochanin A) and aurone levels in roots (Table III). Flavonol (kaempferol 3-O-rutinoside) levels were reduced in developing seed of the mutant lines, consistent with the reduction in flavonol synthase expression (Table I). The less consistent results of MtWD40-1 down-regulation in nonseed tissue could either be because natural product levels are more variable as a result of environmental factors in nonseed tissues or because of effects of different additional retrotransposon inserts in the two mutant lines.
Table III.
Levels of selected flavonoid compounds in different tissues of wild-type and mutant M. truncatula determined by UPLC-MS analysis
The data represent the peak area corresponding to each compound divided by that of the internal standard and multiplied by 1,000. Results are presented as means ± sd from biological triplicates.
| Compound Name | MS Ion Used for Quantification | Retention Time | NF0977 | R108a | NF2745 | R108a |
|---|---|---|---|---|---|---|
| min | ||||||
| Root | ||||||
| Pelargonidin-3-O-glucoside | 449.11 | 2.45 | 28 ± 13 | 32 ± 3 | 1 ± 0 | 6 ± 9 |
| Formononetin-7-O-glucoside | 267.07 | 8.47 | 9 ± 4 | 14 ± 3 | 35 ± 8 | 139 ± 40 |
| 4,6-Dihydroxy-aurone | 253.04 | 10.06 | 6 ± 2 | 11 ± 2 | 3 ± 1 | 11 ± 2 |
| Biochanin A-7-O-glucoside | 283.06 | 10.68 | 2 ± 1 | 5 ± 0 | 2 ± 0 | 6 ± 2 |
| Leaf | ||||||
| Formononetin-7-O-glucoside | 267.07 | 8.47 | 12 ± 3 | 13 ± 2 | 21 ± 4 | 15 ± 6 |
| Apigenin | 269.04 | 10.19 | 73 ± 23 | 52 ± 6 | 3 ± 2 | 4 ± 1 |
| Flower | ||||||
| Epicatechin 3′-O-glucoside | 451.12 | 3.09 | NDb | 7 ± 3 | ND | 3 ± 0 |
| Epicatechin | 289.07 | 3.22 | ND | 4 ± 2 | ND | 1 ± 0 |
| Cyanidin 3-O-glucoside | 461.07 | 5.43 | 6 ± 1 | 2 ± 1 | 33 ± 2 | 14 ± 0 |
| Genistein-7-O-glucoside | 431.09 | 5.54 | 5 ± 2 | 3 ± 2 | 2 ± 1 | 4 ± 1 |
| Apigenin-7-O-glucoside | 431.09 | 7.18 | 4 ± 2 | 3 ± 2 | 2 ± 1 | 3 ± 1 |
| Luteolin-7-O-glucoside | 579.13 | 5.95 | 24 ± 2 | 19 ± 4 | 45 ± 5 | 32 ± 1 |
| Kaempferol | 285.04 | 10.34 | 24 ± 1 | 18 ± 3 | 20 ± 1 | 3 ± 0 |
| 16-d seed | ||||||
| 3,5-Dihydroxybenzoic acid | 153.02 | 1.17 | ND | 26 ± 3 | ND | 40 ± 2 |
| 2,4-Dihydroxybenzoic acid | 151.00 | 1.35 | ND | 8 ± 1 | ND | 10 ± 1 |
| Epicatechin 3′-O-glucoside | 451.12 | 3.09 | ND | 69 ± 7 | ND | 181 ± 22 |
| Epicatechin | 289.07 | 3.22 | ND | 34 ± 5 | ND | 59 ± 6 |
| Cyanidin 3-O-glucoside | 461.07 | 5.43 | ND | 6 ± 2 | ND | 19 ± 2 |
| Kaempferol 3-O-rutinoside | 593.15 | 6.25 | 1 ± 1 | 4 ± 1 | ND | 4 ± 1 |
| Mature seed | ||||||
| Apigenin |
269.04 |
10.19 |
18 ± 2 |
24 ± 2 |
40 ± 1 |
19 ± 2 |
R108 columns to the right of the mutant lines represent independent sets of plants grown in parallel with the corresponding mutants.
ND, Not detected.
Figure 8.
UPLC-ESI-QTOF-MS analysis with selected ion monitoring of epicatechin glucoside in developing M. truncatula seed. A, Extract of seed of mutant line NF0977. B, Extract of seed of wild-type R108. The peak in B corresponds to glucosylated epicatechin (Epi-glc; retention time of 3.06 min, mass of 451.125, 500 ppm). Seed were harvested at 16 dap. Numbers at right indicate absolute intensity of the peaks at retention time of 3.06 min.
Overexpression of MtWD40-1 in Medicago Hairy Roots
Ectopic expression of the Arabidopsis MYB transcription factor TT2 in M. truncatula hairy roots results in a massive induction of PAs accompanied by the up-regulation of several hundred genes, especially those of the anthocyanin/PA biosynthetic pathway (Pang et al., 2008), and TT2, at least in Arabidopsis, functions in a complex with TTG1 and TT8. Therefore, we introduced MtWD40-1 into hairy roots of wild-type M. truncatula to determine whether overexpression of this gene could modulate PA biosynthesis in the absence of TT2 overexpression. The MtWD40-1-overexpressing root lines did not exhibit obvious phenotypical differences compared with GUS control lines; both exhibited purple pigmentation, but neither stained blue with DMACA reagent (data not shown).
Three independent MtWD40-1-overexpressing hairy root lines were selected for high MtWD40-1 expression by qRT-PCR along with three GUS control lines (Supplemental Fig. S4A), and global transcript levels in these lines were compared by Affymetrix microarray analysis. Only 15 probe sets were up-regulated by at least 2-fold as a result of overexpression of MtWD40-1 in the hairy roots, and none of these, other than the 28.2-fold induced MtWD40-1 transcripts, appeared to be associated with the flavonoid pathway (Table IV). The lack of induction by MtWD40-1 of ANS and ANR was confirmed by qRT-PCR (data not shown). Consistent with the transcript levels, only a very small change in anthocyanin levels was observed in the MtWD40-1-overexpressing hairy roots (Supplemental Fig. S4B), and no significant changes in either soluble or insoluble PAs were recorded (Supplemental Fig. S4, C and D). Quantitative and qualitative flavonoid profiles, as detected by HPLC, also remained unchanged (data not shown).
Table IV.
The 15 gene probe sets that were up-regulated by expression of MtWD40-1 in M. truncatula hairy roots
| Probe Sets | Target Description | Ratio (WD40/GUS) | P | Q |
|---|---|---|---|---|
| Mtr.39774.1.S1_at | TC105711 Ttg1-like protein, partial (46%) | 28.19 | 0 | 0.05 |
| Mtr.11660.1.S1_at | TC110633 /FEA = mRNA /DEF= | 4.69 | 3.6E-22 | 0.998392 |
| Mtr.40780.1.S1_at | TC108029 /FEA = mRNA /DEF= | 4.35 | 6.4E-22 | 0.998392 |
| Mtr.20158.1.S1_s_at | Zinc finger, CCHC type; peptidase aspartic | 4.10 | 6.6E-15 | 0.998392 |
| Mtr.42612.1.S1_s_at | Similar to UP|Q6BGZ6 (Q6BGZ6); similarity, partial (9%) | 3.90 | 5.9E-98 | 0.998392 |
| Mtr.5918.1.S1_at | Weakly similar to GB|AAP21357.1|30102878|BT006549 At1g56300 (Arabidopsis), partial (60%) | 3.01 | 1.4E-40 | 0.998392 |
| Mtr.50164.1.S1_at | Heat shock protein Hsp20; HSP20-like chaperone | 2.86 | 1.4E-07 | 0.998392 |
| Mtr.51122.1.S1_at | Hypothetical protein AC126009.22.141 47552 46950 mth2-15c20 01/13/05 | 2.70 | 6.7E-08 | 0.998392 |
| Mtr.18796.1.S1_s_at | T26F17.17-related | 2.69 | 2.4E-06 | 0.998392 |
| Mtr.40781.1.S1_s_at | Similar to UP|Q6BE36 (Q6BE36) protein 7, partial (23%) | 2.62 | 4E-10 | 0.998392 |
| Mtr.37337.1.S1_at | Homolog to UP|HS12_MEDSA (P27880) 18.2-kD class I heat shock protein, complete | 2.53 | 6.1E-08 | 0.998392 |
| Mtr.40779.1.S1_at | Similar to UP|Q25783 (Q25783) Plasmodium falciparum parasite antigen DNA, partial coding sequence (fragment), partial (10%) | 2.52 | 2.9E-09 | 0.998392 |
| Mtr.20165.1.S1_s_at | Hypothetical protein | 2.48 | 1E-69 | 0.998392 |
| Mtr.45232.1.S1_at | Similar to UP|DR2A_ARATH (O82132) dehydration-responsive element-binding protein 2A, partial (23%) | 2.44 | 4.7E-25 | 0.998392 |
| Mtr.4076.1.S1_s_at |
Weakly similar to UP|O24249 (O24249) methyltransferase, partial (10%) |
2.01 |
6.5E-12 |
0.998392 |
Expression of MtWD40-1 in Alfalfa
The MtWD40-1 gene driven by the 35S promoter was introduced into alfalfa by Agrobacterium tumefaciens-mediated stable transformation. Fourteen out of 20 independent ppt-positive transgenic lines were further confirmed by qRT-PCR, and the three lines with the highest MtWD40-1 gene transcript levels (Supplemental Fig. S5A) were selected for global transcript level analysis using the Affymetrix Medicago GeneChip. Two hundred sixty probe sets were up-regulated in leaf tissue from MtWD40-1-overexpressing alfalfa by at least 2-fold, the top 30 of which are listed in Supplemental Table S5. The two probe sets for MtWD40-1 itself were up-regulated by 8.2/7.7-fold, respectively. More than half of the probe sets were grouped into the unclassified category when analyzed for gene function classification (Supplemental Fig. S6). No genes up-regulated more than 2-fold appeared to be associated with flavonoid biosynthesis. Eleven of the probe sets that were up-regulated in alfalfa expressing MtWD40-1 were also up-regulated in M. truncatula hairy roots expressing AtTT2 (Pang et al., 2008; Supplemental Table S6; Supplemental Fig. S7); these include a 51-kD seed maturation protein precursor that is seed coat preferentially expressed and down-regulated in the NF0977 mutant and a glucosyltransferase with yet uncharacterized function.
Anthocyanin levels almost doubled in leaf tissue of the MtWD40-1-overexpressing lines (Supplemental Fig. S5B), although the plants showed no visible increase in pigmentation. Only very small changes in soluble and insoluble PAs were detected in leaves of the MtWD40-1-overexpressing lines compared with the GUS control lines (Supplemental Fig. S5, C and D).
DISCUSSION
The Role of MtWD40-1 in Anthocyanin/PA Biosynthesis in M. truncatula
In this study, a M. truncatula gene encoding a WD40 repeat protein necessary for the biosynthesis of anthocyanins/PAs was identified by forward genetic screening of a Medicago Tnt1 insertional mutant population.
In Arabidopsis leaf tissue, anthocyanin/PA biosynthesis is blocked at the DFR step in the ttg1 mutant (Shirley et al., 1995; Pelletier et al., 1997), with expression of upstream genes such as CHS, CHI, and F3H being unaffected (Shirley et al., 1995). However, the steps at which the pathways were blocked in other tissues were not determined. MtWD40-1 is expressed in both pigmented (leaf and stem) and nonpigmented (root, flower, and seed) tissues, and its expression level is similar in all tissues except the seed coat, where it exhibits the highest expression. However, transcript and metabolite analyses revealed different effects of its down-regulation on pathway genes and/or pathway products in different tissues. For example, the anthocyanin biosynthetic pathway is blocked at the DFR step in pigmented leaf and stem tissue of the NF0977 mutant. In contrast, the pathway was more strongly blocked at the CHS step in flowers (based on qRT-PCR, but targeting only one CHS probe set) and seed (based on both qRT-PCR and microarray). In particular, expression of the PA-specific pathway genes LAR and ANR was very strongly reduced in flower and seed, which in turn led to a deficiency of epicatechin and its glucoside in flower and of PAs in seed. The expression pattern of MtWD40-1 during seed development was similar to that of ANR, which encodes the enzyme that catalyzes the first committed step of PA biosynthesis in M. truncatula (Pang et al., 2007).
It is interesting that loss of function of MtWD40-1 expression results in a large reduction in the levels of multiple phenylpropanoid classes (benzoic acids, flavonols, flavan-3-ols, anthocyanins) in seed, whereas only flavan-3-ols were strongly down-regulated in flower (where anthocyanin levels were actually increased). Although the qRT-PCR data indicated strong down-regulation of one specific CHS family member in flower, it is likely that other members of the CHS gene family remain expressed. Additional anthocyanin accumulation would be predicted in flowers in which ANR is strongly down-regulated but ANS remains unaffected, since cyanidin is the immediate precursor of epicatechin (Xie et al., 2003).
Although WD40 proteins are known to regulate anthocyanin and PA biosynthesis, their potential involvement in other areas of phenylpropanoid biosynthesis is less clear. Our data indicate that loss of function of MtWD40-1 also results in reduction in the levels of an aurone and an isoflavone glycoside in roots and complete loss of benzoic acids in seeds. Levels of the latter compounds are likely directly regulated through the action of MtWD40-1, whereas the smaller change in isoflavone levels in roots might be an indirect effect of altered metabolic flux. We did, however, record a 2-fold decrease in isoflavone synthase transcripts in two independent mtwd40-1 alleles by qRT-PCR (data not shown). Together, these data suggest a critical role for MtWD40-1 in the control of seed PA biosynthesis, with additional but less precise (and possibly indirect) effects on the formation of other flavonoid compounds in other tissues.
The Role of MtWD40-1 in Trichome Formation
WD40 repeat proteins are critical for trichome formation in Arabidopsis, but not in all plant species (Serna and Martin, 2006). In Arabidopsis, a regulatory complex consisting of GL1-GL3/EGL3 (for Enhance Glabrous3)-WD40 triggers expression of the downstream GL2 gene by binding to its promoter region to regulate trichome formation in the epidermal cell layer (Oppenheimer et al., 1991; Payne et al., 2000; Zhang et al., 2003). Lack of TTG1 expression in Arabidopsis leads to the loss of trichomes on aerial tissues (Walker et al., 1999). M. truncatula stems and leaves harbor two types of trichomes: nonglandular hairs and, at a lower density, small glandular structures that generally lie flat against the epidermal surface (Damerval, 1983). MtWD40-1 mutations do not qualitatively affect trichome distribution on young leaves and petioles, even though MtWD40-1 can apparently interact with GL3 to activate GL2 expression and therefore restore (nonglandular) trichome formation in the Arabidopsis ttg1 mutant; however, subtle changes due to either the direct loss of MtWD40-1 or secondary effects caused by changes in metabolite production have not been assessed in M. truncatula. Similar observations with WD40 repeat proteins have been reported in other plant species. For example, neither mutation nor ectopic expression of the single-copy AN11 gene caused any obvious change in trichome phenotype in petunia (de Vetten et al., 1997). In maize, the WD40 protein encoded by the pale aleurone colors1 (PAC1) locus is required for anthocyanin production in the aleurone and scutellum of the seed and can complement the ttg1 trichome phenotype, although loss of PAC1 expression does not affect trichomes in maize (Carey et al., 2004). PFWD, a WD40 repeat protein from P. frutescens, controls both anthocyanin production and trichome initiation in gain-of-function tests (Sompornpailin et al., 2002), and the WD40 repeat genes GhTTG1 and GhTTG3 from cotton can rescue the trichome phenotype of Arabidopsis ttg1 and the anthocyanin deficiency phenotype of the Matthiola incana ttg1 mutant (Humphries et al., 2005). However, it was not shown whether PFWD and GhTTG1/2 control trichome initiation in their host species.
Like AN11 from petunia and PAC1 from maize, MtWD40-1 is also a single-copy gene, as determined by DNA gel-blot analysis under high stringency (data not shown). BLASTN analysis of the M. truncatula genome databases with the MtWD40-1 nucleotide sequence as query recovered no other WD40 repeat protein genes. Furthermore, when the deduced amino acid sequence was queried (by BLASTP), no other WD40 repeat protein with more then 30% identity was recovered. MtWD40-2, which is only represented as an EST in the Medicago sequence available to date, is less than 60% identical to MtWD40-1 at the amino acid level. MtWD40-1 is related to maize PAC1, which can complement the Arabidopsis ttg1 mutant. It is possible, therefore, that the absence of a trichome phenotype in the MtWD40-1 mutant is due to genetic redundancy, although the expression level and pattern of MtWD40-2 based on microarray data are not obviously supportive of a primary role in trichome development.
Biotechnological Applications of MtWD40-1
Transcription factors have already been employed for bioengineering of the anthocyanin/PA pathway. Successful examples of engineering anthocyanin production include ectopic expression of the Myb transcription factors Production of Anthocyanin Pigment1 (PAP1) in tobacco (Nicotiana tabacum) and Arabidopsis (Borevitz et al., 2000; Xie et al., 2006), Legume Anthocyanin Production1 in alfalfa and white clover (Trifolium repens; Peel et al., 2009), and the maize bHLH transcriptional regulators Lc and Sn in alfalfa and Lotus corniculatus, respectively (Ray et al., 2003; Robbins et al., 2003). Coexpression of PAP1 and TT2 led to the accumulation of detectable PA levels in Arabidopsis, although the plants did not survive (Sharma and Dixon, 2006). Coexpression of ANR with PAP1 led to accumulation of PAs in tobacco leaves (Xie et al., 2006). However, none of the components of the TT2-TT8-WD40 transcription complex has been tested for engineering PAs in foliage of forage legumes.
In a previous study, we introduced the TT2 gene from Arabidopsis into M. truncatula hairy roots, and this alone led to massive accumulation of PAs (Pang et al., 2008). Increased transcript levels of both MtWD40-1 and a TT8 homolog were observed in these lines, associated with strong induction of flavonoid pathway enzymes, which included an over 400-fold increase in ANR expression (Pang et al., 2008). In contrast, overexpression of MtWD40-1 alone did not induce PA formation or increased expression of flavonoid biosynthetic pathway genes. One explanation could be that basal levels of MtWD40-1 and MtTT8 are sufficient to support PA biosynthesis in the hairy roots and that TT2 expression is the limiting factor in this tissue. Therefore, MtWD40-1 is necessary, but not sufficient, for PA biosynthesis. Expression of MtWD40-1 did not induce either PA-specific genes or PA accumulation in alfalfa foliage. Similarly, expression of AtTT2 alone does not induce PA biosynthesis in alfalfa foliage (Peel et al., 2009). There are three possible explanations for these observations. First, even though sufficient MtWD40-1 or AtTT2 may be present in the foliar tissue, ANR expression will not be triggered if there is insufficient partner protein present to form the TT2-TT8-WD40 complex. Coexpression of all three genes would address this possibility. Second, low levels of anthocyanidin substrate might limit PA monomer formation in foliar tissues. Finally, we cannot rule out the potential existence of suppressors of the anthocyanin/PA biosynthetic pathway in leaves. A protein with a single MYB domain has recently been shown to act as a negative regulator of anthocyanin biosynthesis in Arabidopsis (Matsui et al., 2008), and CAPRICE (CPC), TRIPTYCHON, and ENHANCER OF TRY AND CPC1 (ETC1) and ETC2 function as suppressors of the GL1-GL3/EGL3-WD40 complex to repress trichome formation (Schnittger et al., 1999; Schellmann et al., 2002; Kirik et al., 2004a, 2004b) and possibly the anthocyanin/PA-promoting function of the complex. It is clear that the successful bioengineering of PAs in forage crops will depend largely on our gaining a better understanding of the endogenous regulatory controls for PA biosynthesis.
MATERIALS AND METHODS
Insertion Mutant Screening and Molecular Confirmation by TAIL-PCR
Generation of the Medicago truncatula Tnt1 insertional mutant population and growth of R1 seed were as described previously (Tadege et al., 2005). The mutant line NF0977 was selected due to its lack of anthocyanins in the aerial tissues. Genomic DNA from the mutant was isolated using the method of Dellaporta et al. (1983). Tnt1 flanking sequences were recovered using TAIL-PCR (Liu et al., 1995, 2005). PCR fragments were purified using a PCR Purification Kit (Qiagen) and then cloned into pGEM-T Easy vector (Promega), followed by sequencing with the Tnt1-specific primer Tnt1-F2 (Supplemental Table S7). The sequenced fragments were then analyzed by BLASTN against the M. truncatula genome at the National Center for Biotechnology Information.
Seeds from the identified Tnt1 insertion lines were scarified with concentrated sulfuric acid, cold treated for 3 d at 4°C on filter paper, and grown in Metro-Mix 350 (Scott) with an 18-h-light/25°C and 6-h-dark/22°C photoperiod in the greenhouse. Genomic DNA from the R2 and R3 progeny was extracted and analyzed as above, using the Tnt1-R1 and MtWD40-1F1 primers (Supplemental Table S7) to confirm the Tnt1 insertion and the MtWD40-1F1 and MtWD40-1R1 primers to check if an individual plant is homozygous or heterozygous with respect to the mutated MtWD40-1 gene.
Reverse Genetic Screening for Tnt1 Retrotransposon Insertions in MtWD40-1
DNA samples used for mutant screening were 10 superpools of pooled DNA samples from 5,000 Tnt1 insertional mutant lines of M. truncatula (Tadege et al., 2005, 2008). A PCR approach was taken for reverse genetic screening to uncover MtWD40-1 mutants. Briefly, two rounds of PCR were used to screen the superpools; the primers used for the primary PCR were Tnt1 reverse primer Tnt1-R and gene-specific primer MtWD40-1F. For nested PCR, Tnt1-R1 and MtWD40-1F1 were used (Supplemental Table S7). The PCR products from the final target plants were then purified with the QIAquick PCR Purification Kit (Qiagen) and sequenced with the primer Tnt1-R2.
Sequence Alignment and Phylogeny Analysis
A multiple alignment of the deduced amino acid sequences of MtWD40-1 and other WD40 repeat domain proteins was constructed using ClustalX 1.81 (Thompson et al., 1997). For phylogeny analysis, the alignment was performed using MAFFT (Katoh et al., 2005). The resulting alignment was further edited manually using Mesquite (Maddison and Maddison, 2009). The unrooted consensus tree was constructed using PAUP* 4.0b10 with 1,000 bootstrap replicates (Swofford, 2003).
Sample Collection, RNA Extraction, qRT-PCR, and Microarray Analysis
Root, stem, leaf, flower, and seed samples from three independent homozygous NF0977 and NF2745 R3 generation and wild-type R108 plants were collected 1 month after planting in soil. Additional flowers were labeled individually according to pollination date, and seed pods were harvested at 16 dap; the seeds were collected and stored at −80°C. RNA was extracted from triplicate biological replicates of the above samples using the cetyl-trimethyl-ammonium bromide method (Jaakola et al., 2001) followed by treatment with Turbo DNase I (Ambion) and reverse transcription of 3 μg of RNA from each sample. The cDNA samples were used for qRT-PCR with technical duplicates. The 10-μL reaction included 2 μL of primers (0.5 μm of each primer), 5 μL of Power Sybr (Applied Biosystems), 2 μL of 1:20 diluted cDNA from the RT step, and 1 μL of water. The gene-specific primers used for qRT-PCR are listed in Supplemental Table S7. RNA samples from seed collected at 16 dap were further purified with a Qiagen RNeasy MinElute Cleanup Kit, and 10 μg-samples were subjected to microarray analysis. RNA from transgenic hairy roots and alfalfa (Medicago sativa) leaf tissue were extracted with Tri-reagent (Gibco-BRL Life Technologies) for qRT-PCR, and 10 μg of purified RNA samples was used for microarray analysis.
qRT-PCR data were analyzed using SDS 2.2.1 software (Applied Biosystems). PCR efficiency (E) was estimated using the LinRegPCR software (Ramakers et al., 2003), and the transcript levels were determined by relative quantification (Pfaffl, 2001) using the M. truncatula actin gene (tentative consensus no. 107326) as a reference.
Probe labeling, hybridization, and scanning for microarray analysis were conducted according to the manufacturer's instructions (Affymetrix). For each sample, the .CEL file was exported from the GeneChip Operating System program (Affymetrix). All .CEL files were imported into RMA (for Robust Multi-Chip Average) and normalized as described by Irizarry et al. (2003). The presence/absence call for each probe set was obtained from dCHIP (Li and Wong, 2001). Differentially expressed genes between wild-type R108 versus NF0977 seed coat sample and MtWD40-1-overexpressing hairy roots versus GUS controls were selected using associative analysis as described (Dozmorov and Centola, 2003). Type I family-wise error rate was reduced using a Bonferroni-corrected P value threshold of 0.05/N, where N represents the number of genes present on the chip. The false discovery rate was monitored and controlled by calculating the Q value (false discovery rate) using extraction of differential gene expression (http://www.biostat.washington.edu/software/jstorey/edge/; Storey and Tibshirani, 2003; Leek et al., 2006).
All microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/array express). Accession numbers are as follows: E-MEXP-1757, experiment name “Medicago truncatula MtTTG1 gene mutant seed transcript profiling”; E-MEXP-1758, experiment name “Medicago truncatula TTG1 over-expressing hairy root”; E-MEXP-1759, experiment name “MtTTG1 over-expression transgenic alfalfa gene profiling.”
Staining Seeds for PA and Mucilage
To determine the presence of PAs in the seed coat, seeds were soaked in DMACA reagent (0.1% [w/v] DMACA in methanol-3 n HCl) for 1 h and then destained with ethanol:acetate acid (75:25). To stain for mucilage, seeds were imbibed in sterilized deionized water for 1 h, transferred to 0.01% ruthenium red solution for 10 min, and then washed twice with water.
Scanning Electron Microscopy
Young developing leaves with attached petioles were mounted on copper stubs, frozen in liquid nitrogen, sputter coated with gold using an Emitech K1150 cryopreparation system, and imaged with a Hitachi S3500N scanning electron microscope as described by Ahlstrand (1996).
Analysis of Anthocyanins, PAs, and Total Flavonoids
For extraction of anthocyanins, 2 to 3 mL of 0.1% HCl/methanol was added to 0.1 g of ground fresh samples, followed by sonication for 30 min and standing overnight at 4°C. Following centrifugation at 2,500g for 10 min, the extraction was repeated once and the supernatants were pooled. An equal volume of water and chloroform was added to remove chlorophyll, and the absorption of the aqueous phase was recorded at 530 nm. Total anthocyanin content was calculated based on the molar absorbance of cyanidin-3-O-glucoside.
For PA analysis, 0.5 to 0.75 g of ground samples was extracted with 5 mL of 70% acetone/0.5% acetic acid (extraction solution) by vortexing and then sonicated at room temperature for 1 h. Following centrifugation at 2,500g for 10 min, the residues were reextracted twice as above. The pooled supernatants were then extracted three times with chloroform and once with hexane, and the supernatants (containing soluble PAs) and residues (containing insoluble PAs) from each sample were freeze dried separately. The dried soluble PAs were suspended in extraction solution to a concentration of 3 mg mL−1.
Total soluble PA content was determined spectrophotometrically after reaction with DMACA reagent (0.2% [w/v] DMACA in methanol-3 n HCl) at 640 nm, with (+)-catechin as standard. For quantification of insoluble PAs, 2 mL of butanol-HCl (95:5, v/v) was added to the dried residues and the mixtures were sonicated at room temperature for 1 h, followed by centrifugation at 2,500g for 10 min. The absorption of the supernatants was measured at 550 nm; the samples were then boiled for 1 h and cooled to room temperature, and the A550 was measured again, with the first value being subtracted from the second. Absorbance values were converted into PA equivalents using a standard curve generated with procyanidin B1 (Indofine).
For determination of total flavonoids, 0.1-g batches of ground samples were extracted with 2 mL of 80% methanol, sonicated for 1 h, and then kept at 4°C overnight. The extract was centrifuged to remove tissue debris and the supernatant was dried under nitrogen gas, followed by hydrolysis in 2 mL of 5 mg mL−1 β-glucosidase (34 units) from almond (Prunus dulcis; Sigma). After extracting twice with 2 mL of ethyl acetate, the supernatants were pooled, dried under nitrogen, and resuspended in 200 μL of methanol. Fifty microliters of the methanolic solution was used for reverse-phase HPLC analysis on an Agilent HP1100 system using the following gradient with solvent A (1% phosphoric acid) and solvent B (acetonitrile) at 1 mL min−1 flow rate: 0 to 5 min, 5% B; 5 to 10 min, 5% to 10% B; 10 to 25 min, 10% to 17% B; 25 to 30 min, 17% to 23% B; 30 to 65 min, 23% to 50% B; 65 to 79 min, 50% to 100% B; 79 to 80 min, 100% to 5% B. Data were collected at 254 nm for flavonoid compounds. Identifications were based on chromatographic behavior, and UV spectra were compared with those of authentic standards.
Extraction and UPLC-ESI-QTOF-MS Analysis of Flavonoids
Dried tissues (10.0 ± 0.06 mg) were weighed into a 1-g glass vial. The samples (biological triplicates) were extracted in 2 mL of 80% methanol containing 2 μg mL−1 puerarin and 18 μg mL−1 umbelliferone (internal standards) for 2 h at room temperature with constant agitation. Samples were centrifuged at 2,900g for 30 min, and the supernatants were transferred to liquid chromatography vials and analyzed with a Waters Acquit UPLC system fitted with a hybrid quadrupole time-of-flight (QTOF) Premier mass spectrometer (Waters). A reverse-phase, 1.7-μm UPLC BEH C18, 2.1 × 150 mm column (Waters) was used for separations. The mobile phase consisted of eluent A (0.1% [v/v] acetic acid/water) and eluent B (acetonitrile), and separations were achieved using a linear gradient of 95% to 30% A over 30 min, 30% to 5% A over 3.0 min, and 5% to 95% A over 3.0 min. The flow rate was 0.56 mL min−1, and the column temperature was maintained at 60°C. Masses of the eluted compounds were detected in the negative ESI mode from 50 to 2,000 mass-to-charge ratio. The QTOF Premier was operated under the following instrument parameters: desolvation temperature of 400°C; desolvation nitrogen gas flow of 850 L h−1; capillary voltage of 2.9 kV; cone voltage of 48 eV; and collision energy of 10 eV. The MS system was calibrated using sodium formate, and raffinose was used as the lockmass. Metabolites were identified based on accurate masses and retention times relative to authentic standards. Mass Lynx version 4.1, Data Bridge, was used to convert the raw data files to NetCDF. Relative abundances were calculated using MET-IDEA (Broeckling et al., 2006), and the peak areas were normalized by dividing each peak area by the value of the internal standard peak area.
Construction of Binary Vectors for MtWD40-1 Expression in Plants
The ORF of the MtWD40-1 gene was amplified from cDNA produced from total RNA isolated from M. truncatula seed coats, using the primers MtWD40-1CF and MtWD40-1R1 and DNA polymerase with proofreading activity. The PCR product was purified and cloned into the Gateway Entry vector pENTR/D-TOPO (Invitrogen), and the MtWD40-1 ORF in the resulting vector pENTR-MtWD40-1 was confirmed by sequencing.
The primers MtWD40-1NF (with an NcoI site) and MtWD40-1BR (with a BstEII site; Supplemental Table S7) were used to amplify the ORF region (with added NcoI and BstEII restriction sites) from pENTR-MtWD40-1 template with proofreading DNA polymerase. The resulting fragment was digested, purified, and ligated into plasmid pCAMBIA3301-HP (Xiao et al., 2005) digested with NcoI and BstEII to produce a new construct, p3301-MtWD40-1. This construct, as well as a control construct containing the GUS ORF in place of WD40-1, was transformed into Agrobacterium rhizogenes strain Arqua1 (Quandt et al., 1993) by electroporation. Single colonies were confirmed by PCR and used for M. truncatula transformation. Both wild-type M. truncatula Jemalong A17 and the mutant line NF0977 (Genotype R108 as background) were transformed using the protocol of Chabaud et al. (2006) with 2.5 mg L−1 ppt as selection. The generated hairy roots were maintained on B5 agar medium in petri dishes supplied with 2.5 mg L−1 ppt under fluorescent light (140 μE m−2 s−1) with a photoperiod of 16 h and were subcultured every month onto fresh medium.
For stable transformation by Agrobacterium tumefaciens, the MtWD40-1 ORF was first transferred into the Gateway plant transformation destination vector pB2GW7 (Karimi et al., 2002) using Gateway LR Clonase enzyme mix with pENTR-MtWD40-1 according to the manufacturer's instructions (Invitrogen). The reading frame of the resulting vector, pB2GW7-MtWD40-1, was confirmed by sequencing. pB2GW7-MtWD40-1 was transformed into A. tumefaciens strain AGL1 by electroporation. A single colony containing the target construct was confirmed by PCR and used for genetic transformation of Arabidopsis (Arabidopsis thaliana) and alfalfa. The protocol of Austin et al. (1995) was used for alfalfa transformation with minor modifications and 10 mg L−1 ppt selection.
Rescue of the Arabidopsis ttg1-9 Mutant
The construct used to generate Arabidopsis expressing GL2∷MtWD40-1 was derived from pGL2∷GUS (Szymanski et al., 1998). This plasmid was modified by removal of the GUS coding sequence by SmaI/SacI digestion followed by blunt ending with Klenow. The RFA Gateway recombination fragment RFA from Invitrogen was inserted into this site. The coding region of MtWD40-1 was derived from cDNA using total RNA isolated from M. truncatula (Jemalong A17) shoots as a template. Primers flanking the MtWD40-1 coding region were used to generate a double-stranded DNA product via PCR that was first subcloned into pCR8 (Invitrogen) before being moved into the Gateway GL2 promoter vector.
The Arabidopsis ttg1-9 mutant (Walker et al., 1999) was transformed by the floral dip infiltration method (Clough and Bent, 1998). Selection of transformants was conducted on 0.5× Murashige and Skoog medium supplied with 7.5 mg L−1 ppt. The ppt-resistant seedlings were then transferred into soil to set seed. Progeny from self-fertilized primary transformants were grown in soil for observation of trichome phenotype.
Yeast Two-Hybrid Assay
For the yeast two-hybrid assays, PCR was used generate a copy of the MtWD40-1 coding region with leading and tailing EcoRI and BamHI restriction enzyme sites. The coding region was then moved into the corresponding sites of pBridge (Clontech) to create pMtWD40-1DB. The empty vector pGAD424 was from Clontech, and pGL3-AD was as described previously (Esch et al., 2003). β-Galactosidase activity was detected as adapted from Duttweiler (1996) and further described at http://www.fccc.edu/research/labs/golemis/betagal/plates_vs_overlay.html
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU040206 (MtWD40-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root hairs on mature, greenhouse-grown M. truncatula plants (wild type and two lines harboring transposon insertions in MtWD40-1).
Supplemental Figure S2. MtWD40-1 transcript levels in the Arabidopsis ttg1-9 mutant and two lines complemented with MtWD40-1.
Supplemental Figure S3. Gene functional categories of probe sets that were down-regulated by more than 2-fold in the NF0977 mutant compared with wild-type R108.
Supplemental Figure S4. Anthocyanin and PA levels in hairy roots of M. truncatula A17 overexpressing MtWD40-1.
Supplemental Figure S5. Anthocyanin and PA levels in leaves of alfalfa R2336 lines overexpressing MtWD40-1.
Supplemental Figure S6. Gene functional categories of probe sets that were up-regulated by more than 2-fold in leaves of alfalfa expressing MtWD40-1 compared with a GUS-expressing control line.
Supplemental Figure S7. Venn diagram showing overlap between probe sets induced by MtWD40-1 and AtTT2 in M. truncatula hairy roots and by MtWD40-1 in alfalfa leaves.
Supplemental Table S1. BLASTN analysis of all Tnt1 flanking sequences retrieved from the NF0977 mutant.
Supplemental Table S2. All gene probe sets that were down-regulated by more than 2-fold in developing seed of the M. truncatula NF0977 mutant.
Supplemental Table S3. Changes of flavonoid pathway gene transcripts in different tissues of M. truncatula R108 and the NF0977 retrotransposon insertion mutant as determined by qRT-PCR.
Supplemental Table S4. Changes of flavonoid pathway gene transcripts in different tissues of M. truncatula R108 and the NF2745 retrotransposon insertion mutant as determined by qRT-PCR.
Supplemental Table S5. The 30 gene probe sets that were most up-regulated by expression of MtWD40-1 in alfalfa leaf tissue.
Supplemental Table S6. The gene probe sets that were up-regulated by MtWD40-1 in alfalfa leaf and by AtTT2 in hairy roots of M. truncatula.
Supplemental Table S7. The primer sequences used in the present study.
Supplementary Material
Acknowledgments
We thank Dr. Ji He for BLAST analysis, Ms. Darla Boydston for assistance with artwork, and Dr. Elison Blancaflor and Alan Sparks for help with root hair analysis.
This work was supported by the National Science Foundation Plant Genome Program (grant nos. DBI–0605033 and DBI–0703285 to R.A.D. and K.S.M., respectively), by Forage Genetics International, and by the Samuel Roberts Noble Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard A. Dixon (radixon@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aerts RJ, Barry TN, McNabb WC (1999) Polyphenols and agriculture: beneficial effects of proanthocyanidins in forages. Agric Ecosyst Environ 75 1–12 [Google Scholar]
- Ahlstrand G (1996) Low-temperature low-voltage scanning microscopy (LTLVSEM) of uncoated frozen biological materials: a simple alternative. In G Bailey, J Corbett, R Dimlich, J Michael, N Zaluzec, eds, Proceedings of Microscopy Microanalysis. San Francisco Press, San Francisco, p 918
- Austin S, Bingham ET, Mathews DE, Shahan MN, Will J, Burgess RR (1995) Production and field performance of transgenic alfalfa (Medicago sativa L.) expressing alpha-amylase and manganese-dependent lignin peroxidase. Euphytica 85 381–393 [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148 187–197 [DOI] [PubMed] [Google Scholar]
- Barry TN, McNabb WC (1999) The implications of condensed tannins on the nutritive value of temperate forages fed to ruminants. Br J Nutr 81 263–272 [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray J, Andriankaja A, Allen S, Kakar K, Wandrey M, Thomson R, Ott T, Moreau S, et al (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55 504–513 [DOI] [PubMed] [Google Scholar]
- Borevitz J, Xia Y, Blount JW, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW (2006) MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78 4334–4341 [DOI] [PubMed] [Google Scholar]
- Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8 272–279 [DOI] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL (2004) Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Boisson-Dernier A, Zhang J, Taylor CG, Yu O, Barker DG (2006) Agrobacterium rhizogenes-mediated root transformation. In U Mathesius, EP Journer, LW Sumner, eds, The Medicago truncatula Handbook, Version November 2006. The Samuel Roberts Noble Foundation, Ardmore, OK, http://www.noble.org/MedicagoHandbook
- Clough SJ, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ (2004) Proanthocyanidins in health care: current and new trends. Curr Med Chem 11 1345–1359 [DOI] [PubMed] [Google Scholar]
- Damerval C (1983) Micromorphologie des epidermes foliares chez quelques especes de Medicago. Can J Bot 61 3461–3470 [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1 19–21 [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11 1422–1434 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Sharma SB, Xie D (2005) Proanthocyanidins: a final frontier in flavonoid research? New Phytol 165 9–28 [DOI] [PubMed] [Google Scholar]
- Dozmorov I, Centola M (2003) An associative analysis of gene expression array data. Bioinformatics 19 204–211 [DOI] [PubMed] [Google Scholar]
- Duttweiler HM (1996) A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet 12 340–341 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD (2003) A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53 814–827 [DOI] [PubMed] [Google Scholar]
- Humphries JA, Walker AR, Timmis JN, Orford SJ (2005) Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol Biol 57 67–81 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L, Pirttila AM, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19 201–203 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004. a) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M (2004. b) ENHANCER of TRY and CPC2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55 389–398 [DOI] [PubMed] [Google Scholar]
- Koornneef M (1981) The complex syndrome of ttg mutants. Arabidopsis Inf Serv 18 45–51 [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD (2006) EDGE: extraction and analysis of differential gene expression. Bioinformatics 22 507–508 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57 405–430 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Tanner G, Larkin P (1996) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70 89–101 [Google Scholar]
- Liu YG, Chen Y, Zhang Q (2005) Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. Methods Mol Biol 286 341–348 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production in dicots activated by maize anthocyanin-specific regulators R and C1. Science 258 1773–1775 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (2009) Mesquite: a modular system for evolutionary analysis. Version 2.6. http://mesquiteproject.org (April 6, 2009)
- Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55 945–967 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang X, Dixon RA (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64 499–518 [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47 457–470 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493 [DOI] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105 14210–14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang ZY, Dixon RA (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel GJ, Pang Y, Modolo LV, Dixon RA (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59 136–149 [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Murrell JR, Shirley BW (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Plant Physiol 113 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Puhler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6 699–706 [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 13 62–66 [DOI] [PubMed] [Google Scholar]
- Ray H, Yu M, Auser P, Blahut-Beatty L, McKersie B, Bowley S, Westcott N, Coulman B, Lloyd A, Gruber MY (2003) Expression of anthocyanins and proanthocyanidins after transformation of alfalfa with maize Lc. Plant Physiol 132 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins MP, Paolocci F, Hughes JW, Turchetti V, Allison G, Arcioni S, Morris P, Damiani F (2003) Sn, a maize bHLH gene, modulates anthocyanin and condensed tannin pathways in Lotus corniculatus. J Exp Bot 54 239–248 [DOI] [PubMed] [Google Scholar]
- Schellmann TS, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jurgens G, Hulskamp M (1999) Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11 274–280 [DOI] [PubMed] [Google Scholar]
- Sharma SB, Dixon RA (2006) Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J 44 62–75 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8 659–671 [DOI] [PubMed] [Google Scholar]
- Sompornpailin K, Makita Y, Yamazak M, Saito K (2002) A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol Biol 50 485–495 [DOI] [PubMed] [Google Scholar]
- Stapleton AE, Walbot V (1994) Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol 105 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genome wide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J (1998) Anthocyanin. Carnivorous Plant Newsletter 27 86–88 [Google Scholar]
- Swofford DL (2003). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4 Beta 10. Sinauer Associates, Sunderland, MA
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171 [DOI] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS (2005) Insertional mutagenesis: a Swiss army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10 229–235 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al (2008) Large scale insertional mutagenesis using Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 45 335–347 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Zhang C, Harrison M, Wang ZY (2005) Isolation and characterization of a novel plant promoter that directs strong constitutive expression of transgenes in plants. Mol Breed 15 221–231 [Google Scholar]
- Xie D, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299 396–399 [DOI] [PubMed] [Google Scholar]
- Xie D, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45 895–907 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135 1991–1999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.