Abstract
Relatively little is known about the small subset of peroxisomal proteins with predicted protease activity. Here, we report that the peroxisomal LON2 (At5g47040) protease facilitates matrix protein import into Arabidopsis (Arabidopsis thaliana) peroxisomes. We identified T-DNA insertion alleles disrupted in five of the nine confirmed or predicted peroxisomal proteases and found only two—lon2 and deg15, a mutant defective in the previously described PTS2-processing protease (DEG15/At1g28320)—with phenotypes suggestive of peroxisome metabolism defects. Both lon2 and deg15 mutants were mildly resistant to the inhibitory effects of indole-3-butyric acid (IBA) on root elongation, but only lon2 mutants were resistant to the stimulatory effects of IBA on lateral root production or displayed Suc dependence during seedling growth. lon2 mutants displayed defects in removing the type 2 peroxisome targeting signal (PTS2) from peroxisomal malate dehydrogenase and reduced accumulation of 3-ketoacyl-CoA thiolase, another PTS2-containing protein; both defects were not apparent upon germination but appeared in 5- to 8-d-old seedlings. In lon2 cotyledon cells, matrix proteins were localized to peroxisomes in 4-d-old seedlings but mislocalized to the cytosol in 8-d-old seedlings. Moreover, a PTS2-GFP reporter sorted to peroxisomes in lon2 root tip cells but was largely cytosolic in more mature root cells. Our results indicate that LON2 is needed for sustained matrix protein import into peroxisomes. The delayed onset of matrix protein sorting defects may account for the relatively weak Suc dependence following germination, moderate IBA-resistant primary root elongation, and severe defects in IBA-induced lateral root formation observed in lon2 mutants.
Peroxisomes are single-membrane-bound organelles found in most eukaryotes. Peroxin (PEX) proteins are necessary for various aspects of peroxisome biogenesis, including matrix protein import (for review, see Distel et al., 1996; Schrader and Fahimi, 2008). Most matrix proteins are imported into peroxisomes from the cytosol using one of two targeting signals, a C-terminal type 1 peroxisome-targeting signal (PTS1) or a cleavable N-terminal type 2 peroxisome-targeting signal (PTS2) (Reumann, 2004). PTS1- and PTS2-containing proteins are bound in the cytosol by soluble matrix protein receptors, escorted to the peroxisome membrane docking complex, and translocated into the peroxisome matrix (for review, see Platta and Erdmann, 2007). Once in the peroxisome, many matrix proteins participate in metabolic pathways, such as β-oxidation, hydrogen peroxide decomposition, and photorespiration (for review, see Gabaldon et al., 2006; Poirier et al., 2006).
In addition to metabolic enzymes, several proteases are found in the peroxisome matrix. Only one protease, DEG15/Tysnd1, has a well-defined role in peroxisome biology. The rat Tysnd1 protease removes the targeting signal after PTS2-containing proteins enter the peroxisome and also processes certain PTS1-containing β-oxidation enzymes (Kurochkin et al., 2007). Similarly, the Arabidopsis (Arabidopsis thaliana) Tysnd1 homolog DEG15 (At1g28320) is a peroxisomal Ser protease that removes PTS2 targeting signals (Helm et al., 2007; Schuhmann et al., 2008).
In contrast with DEG15, little is known about the other eight Arabidopsis proteins that are annotated as proteases in the AraPerox database of putative peroxisomal proteins (Reumann et al., 2004; Table I). Two of these proteins, At4g14570 and At4g36195, have been identified in vacuolar proteomics studies (Carter et al., 2004; Shimaoka et al., 2004), which, in combination with the minor PTS found in both of these predicted proteases (Reumann, 2004), suggests that these enzymes may not be peroxisomal. Along with DEG15, only two of the predicted peroxisomal proteases, an M16 metalloprotease (At2g41790), which we have named PXM16 for peroxisomal M16 protease, and a Lon-related protease (At5g47040/LON2; Ostersetzer et al., 2007), are found in the proteome of peroxisomes purified from Arabidopsis suspension cells (Eubel et al., 2008). DEG15 and LON2 also have been validated as peroxisomally targeted using GFP fusions (Ostersetzer et al., 2007; Schuhmann et al., 2008).
Table I.
Putative Arabidopsis proteases predicted or demonstrated to be peroxisomal
| AGI Identifier | Alias | Protein Class | T-DNA Insertion Alleles | PTS | Localization Evidence | Localization References |
|---|---|---|---|---|---|---|
| At1g28320 | DEG15 | PTS2-processing protease | SALK_007184 (deg15-1) | SKL>a | GFP | Reumann et al., 2004; Helm et al., 2007; Eubel et al., 2008; Schuhmann et al., 2008) |
| Proteomics | ||||||
| Bioinformatics | ||||||
| At2g41790 | PXM16 | Peptidase M16 family protein | SALK_019128 (pxm16-1) | PKL>b | Proteomics | Reumann et al., 2004, 2009; Eubel et al., 2008) |
| SALK_023917 (pxm16-2) | Bioinformatics | |||||
| At5g47040 | LON2 | Lon protease homolog | SALK_128438 (lon2-1) | SKL>a | GFP | Reumann et al., 2004, 2009; Ostersetzer et al., 2007; Eubel et al., 2008) |
| SALK_043857 (lon2-2) | Proteomics | |||||
| Bioinformatics | ||||||
| At2g18080 | Ser-type peptidase | SALK_020628 | SSI>c | Bioinformatics | (Reumann et al., 2004) | |
| SALK_102239 | ||||||
| At2g35615 | Aspartyl protease | SALK_090795 | ANL>b | Bioinformatics | (Reumann et al., 2004) | |
| SALK_036333 | ||||||
| At3g57810 | Ovarian tumor-like Cys protease | SKL>a | Bioinformatics | (Reumann et al., 2004) | ||
| At4g14570 | Acylaminoacyl-peptidase protein | CKL>b | Bioinformatics (peroxisome) | (Reumann et al., 2004; Shimaoka et al., 2004) | ||
| Proteomics (vacuole) | ||||||
| At4g20310 | Peptidase M50 family protein | RMx5HLd | Bioinformatics | (Reumann et al., 2004) | ||
| At4g36195 | Ser carboxypeptidase S28 family | SSM>b | Bioinformatics (peroxisome) | (Carter et al., 2004; Reumann et al., 2004) | ||
| Proteomics (vacuole) |
Major PTS1 (Reumann, 2004).
Minor PTS1 (Reumann, 2004).
Validated PTS1 (Reumann et al., 2007).
Minor PTS2 (Reumann, 2004).
PXM16 is the only one of the nine Arabidopsis M16 (pitrilysin family) metalloproteases (García-Lorenzo et al., 2006; Rawlings et al., 2008) containing a predicted PTS. M16 subfamilies B and C contain the plastid and mitochondrial processing peptidases (for review, see Schaller, 2004), whereas PXM16 belongs to M16 subfamily A, which includes insulin-degrading peptidases (Schaller, 2004). A tomato (Solanum lycopersicum) M16 subfamily A protease similar to insulin-degrading enzymes with a putative PTS1 was identified in a screen for proteases that cleave the wound response peptide hormone systemin (Strassner et al., 2002), but the role of Arabidopsis PXM16 is unknown.
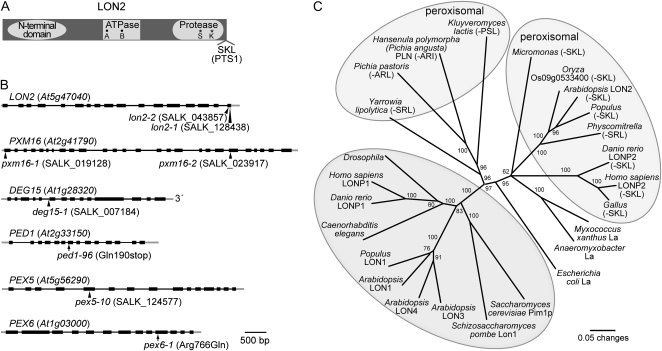
Arabidopsis LON2 is a typical Lon protease with three conserved domains: an N-terminal domain, a central ATPase domain in the AAA family, and a C-terminal protease domain with a Ser-Lys catalytic dyad (Fig. 1A; Lee and Suzuki, 2008). Lon proteases are found in prokaryotes and in some eukaryotic organelles (Fig. 1C) and participate in protein quality control by cleaving unfolded proteins and can regulate metabolism by controlling levels of enzymes from many pathways, including cell cycle, metabolism, and stress responses (for review, see Tsilibaris et al., 2006). Four Lon homologs are encoded in the Arabidopsis genome; isoforms have been identified in mitochondria, plastids, and peroxisomes (Ostersetzer et al., 2007; Eubel et al., 2008; Rawlings et al., 2008). Mitochondrial Lon protesases are found in a variety of eukaryotes (Fig. 1A) and function both as ATP-dependent proteases and as chaperones promoting protein complex assemblies (Lee and Suzuki, 2008). LON2 is the only Arabidopsis Lon isoform with a canonical C-terminal PTS1 (SKL-COOH; Ostersetzer et al., 2007) or found in the peroxisome proteome (Eubel et al., 2008; Reumann et al., 2009). Functional studies have been conducted with peroxisomal Lon isoforms found in the proteome of peroxisomes purified from rat hepatic cells (pLon; Kikuchi et al., 2004) and the methylotrophic yeast Hansenula polymorpha (Pln; Aksam et al., 2007). Rat pLon interacts with β-oxidation enzymes, and a cell line expressing a dominant negative pLon variant has decreased β-oxidation activity, displays defects in the activation processing of PTS1-containing acyl-CoA oxidase, and missorts catalase to the cytosol (Omi et al., 2008). H. polymorpha Pln is necessary for degradation of a misfolded, peroxisome-targeted version of dihydrofolate reductase and for degradation of in vitro-synthesized alcohol oxidase in peroxisomal matrix extracts, but does not contribute to degradation of peroxisomally targeted GFP (Aksam et al., 2007).
Figure 1.
Diagram of LON2 protein domains, gene models for LON2, PXM16, DEG15, PED1, PEX5, and PEX6, and phylogenetic relationships of LON family members. A, Organization of the 888-amino acid LON2 protein. Locations of the N-terminal domain conserved among Lon proteins, predicted ATP-binding Walker A and B domains (black circles), active site Ser (S) and Lys (K) residues (asterisks), and the C-terminal Ser-Lys-Leu (SKL) peroxisomal targeting signal (PTS1) are shown (Lee and Suzuki, 2008). B, Gene models for LON2, PXM16, DEG15, PED1, PEX5, and PEX6 and locations of T-DNA insertions (triangles) or missense alleles (arrows) used in this study. Exons are depicted by black boxes, introns by black lines, and untranslated regions by gray lines. C, Phylogenetic relationships among LON homologs. Sequences were aligned using MegAlign (DNAStar) and the ClustalW method. The PAUP 4.0b10 program (Swofford, 2001) was used to generate an unrooted phylogram from a trimmed alignment corresponding to Arabidopsis LON2 residues 400 to 888 (from the beginning of the ATPase domain to the end of the protein). The bootstrap method was performed for 500 replicates with distance as the optimality criterion. Bootstrap values are indicated at the nodes. Predicted peroxisomal proteins have C-terminal PTS1 signals in parentheses and are in light-gray ovals. Proteins in the darker gray oval have N-terminal extensions and include mitochondrial and chloroplastic proteins. Sequence identifiers are listed in Supplemental Table S2.
In this work, we examined the roles of several putative peroxisomal proteases in Arabidopsis. We found that lon2 mutants displayed peroxisome-deficient phenotypes, including resistance to the protoauxin indole-3-butyric acid (IBA) and age-dependent defects in peroxisomal import of PTS1- and PTS2-targeted matrix proteins. Our results indicate that LON2 contributes to matrix protein import into Arabidopsis peroxisomes.
RESULTS
LON2 Is Necessary for IBA-Stimulated Lateral Root Formation
We analyzed mutants defective in predicted peroxisomal proteases to assess contributions of the proteases to peroxisome functions. pxm16-1 and pxm16-2 have T-DNA insertions in exons 1 and 21 of PXM16, respectively; lon2-1 and lon2-2 carry independent insertions in the final LON2 exon (Fig. 1B). The lon2-1 and lon2-2 T-DNAs may allow LON2 expression; however, the lon2-1 and lon2-2 products would lack the C-terminal PTS1 and likely mislocalize to the cytosol. The previously described T-DNA insertion in the fifth intron of DEG15 (deg15-1; Helm et al., 2007; Schuhmann et al., 2008) was included for comparison (Fig. 1B).
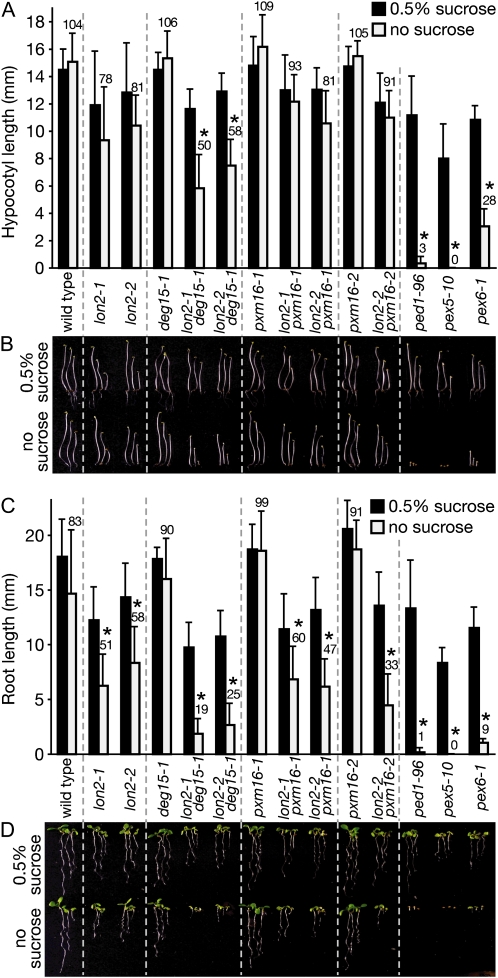
To determine whether the protease mutations impaired peroxisomal fatty acid β-oxidation, we assessed seedling hypocotyl elongation in the dark and root elongation in the light with and without supplemental Suc. Peroxisome biogenesis mutants often display defects in peroxisomal β-oxidation of fatty acids stored in seeds and consequently require Suc during early seedling development (Hayashi et al., 1998; Zolman et al., 2000). We compared the protease mutants to pex5-10 and pex6-1, Suc-dependent peroxin mutants (Zolman and Bartel, 2004; Zolman et al., 2005), and ped1-96, a likely null allele that we identified (our unpublished data) in the gene encoding the predominant seedling 3-ketoacyl-CoA thiolase isozyme (Germain et al., 2001). Like other ped1/kat2 alleles in different accessions (Hayashi et al., 1998; Germain et al., 2001), the ped1-96 mutant exhibited severe hypocotyl elongation defects in the dark (Fig. 2, A and B) and root elongation defects in the light (Fig. 2, C and D) in the absence of Suc supplementation.
Figure 2.
The weak Suc dependence of lon2 mutants is enhanced when combined with deg15-1. A and B, Suc-dependent hypocotyl elongation in the dark. Seedlings were grown for 36 h in light and 4.5 d in darkness on media with or without 0.5% Suc. C and D, Suc-dependent root elongation in the light. Seedlings were grown for 7 d under white light on media with or without 0.5% Suc. Seeds were stratified for 1 d prior to plating. In A and C, error bars represent sds of the means (n ≥ 12), numbers above light-gray bars indicate the mean percentages of elongation on media without Suc, and asterisks denote no Suc values significantly different than Suc values (Student's t test, P < 0.001). B and D show three seedlings representing the range of observed phenotypes. [See online article for color version of this figure.]
As previously reported (Schuhmann et al., 2008), we found that the deg15-1 mutant, like the wild type, had long hypocotyls and roots regardless of presence or absence of Suc (Fig. 2). Similarly, pxm16-1 and pxm16-2 displayed Suc-independent hypocotyl and root elongation (Fig. 2). By contrast, although the lon2 mutant hypocotyls were not significantly Suc dependent in the dark, lon2-1 and lon2-2 displayed root elongation defects in the light that were partially restored by Suc (Fig. 2).
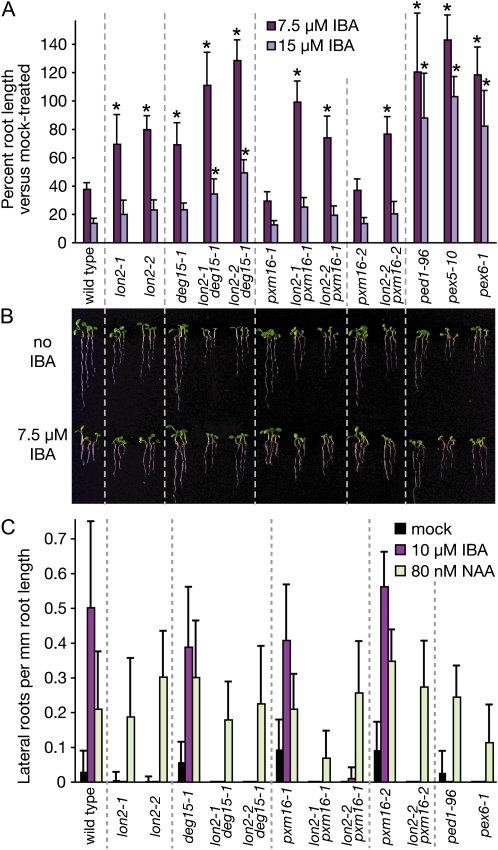
Many pex mutants and other β-oxidation mutants display apparent defects in converting the protoauxin IBA to the active auxin indole-3-acetic acid and therefore are resistant to both the inhibitory effects of IBA on root elongation and to the stimulatory effects of IBA on lateral root production (Zolman et al., 2000, 2005; Zolman and Bartel, 2004; Adham et al., 2005; Woodward and Bartel, 2005). We found that pxm16-1 and pxm16-2 resembled the wild type in the root elongation assay and that lon2-1, lon2-2, and deg15-1 were mildly resistant to low IBA concentrations compared to the strong IBA resistance of ped1-96, pex5-10, and pex6-1 (Fig. 3, A and B).
Figure 3.
The IBA resistance of lon2 mutants is enhanced when combined with deg15-1. A and B, IBA-resistant primary root elongation. After stratification for 1 d, seedlings were grown for 7 d under yellow light on Suc-containing media supplemented with the indicated concentration of IBA. Asterisks denote values significantly different than the wild type (Student's t test, P < 0.001). Three seedlings representing the range of observed phenotypes in A are shown in B. C, IBA promotion of lateral root formation. After stratification for 3 d, seedlings were grown for 4 d on hormone-free media and then transferred to media containing the indicated hormone. After four additional days, lateral roots were counted and primary root lengths were measured. Error bars represent sds of the means (n ≥ 12; A and C).
In contrast to the mild IBA resistance exhibited by lon2 mutants when assayed for root elongation inhibition (Fig. 3, A and B), lon2-1 and lon2-2 seedlings were both dramatically resistant to the stimulatory effects of IBA on lateral root formation, similar to ped1-96 and pex6-1 seedlings (Fig. 3C). In contrast, deg15 and pxm16 mutants resembled the wild type in the lateral root assay, efficiently producing lateral roots in response to IBA (Fig. 3C). All lines produced lateral roots when grown on the synthetic auxin 1-naphthaleneacetic acid, suggesting that the lateral root deficiency in the lon2 mutants stemmed from defective peroxisomal IBA metabolism, rather than a general inability to produce lateral roots or respond to auxin. The striking lon2 resistance to IBA-stimulated lateral root formation, accompanied by only mild resistance to the inhibitory effects of IBA on primary root elongation, suggested that LON2 may be more important for peroxisomal IBA metabolism as root cells mature and differentiate.
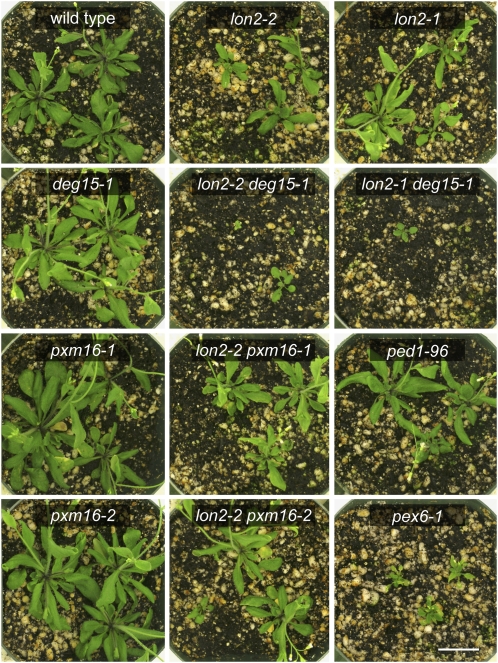
In addition to the defects displayed in specific assays of peroxisomal function described above, lon2-1 and lon2-2 mutants were smaller than the wild type both as seedlings (Fig. 2D) and as adult plants (Fig. 4), whereas deg15-1 and pxm16 seedlings and adult plants resembled the wild type.
Figure 4.
lon2 mutants display growth defects that are enhanced by deg15-1. Representative 28-d-old plants of the indicated genotypes are shown. Bar = 2 cm.
We examined mutants defective in two other putative peroxisomal proteases (At2g18080 and At2g35615; Table I) but did not find any differences from the wild type in IBA resistance or Suc dependence assays (data not shown). The only putative peroxisomal protease with a major peroxisome-targeting signal for which we did not identify a T-DNA insertion was the ovarian tumor-like Cys protease (At3g57810; Table I). At3g57810 is most highly expressed in flowers (Winter et al., 2007), which might explain the absence of this protein from peroxisomal proteome studies that have not sampled flowers (Fukao et al., 2002; Reumann et al., 2007; Arai et al., 2008; Eubel et al., 2008; Reumann et al., 2009).
lon2 Defects Are Enhanced by Disruption of DEG15
To assess genetic interactions among the peroxisomal proteases, we isolated lon2 deg15 and lon2 pxm16 double mutants. The deg15 mutant enhanced several lon2 defects. lon2-1 deg15-1 and lon2-2 deg15-1 double mutant seedlings (Figs. 2D and 3B) and adult plants (Fig. 4) were small and exhibited significantly enhanced Suc dependence both in the dark and in the light (Fig. 2). Similarly, the lon2-1 deg15-1 and lon2-2 deg15-1 double mutant seedlings were more resistant to the inhibitory effect of IBA on root elongation than the single mutants (Fig. 3A). The lon2 mutants were completely insensitive to the tested IBA concentration in the lateral root promotion assay; lon2 deg15 double mutants resembled lon2 single mutants in this assay (Fig. 3C). In contrast, the pxm16 mutants did not appear to enhance lon2 mutant defects; lon2 pxm16 double mutants resembled lon2 single mutants in Suc dependence (Fig. 2), IBA resistance (Fig. 3), and adult size (Fig. 4).
LON2, DEG15, and PXM16 Are Not Necessary for Degradation of Glyoxylate Cycle Enzymes
The physiological defects of the lon2 mutants suggested that LON2 was required for peroxisomal function. Peroxisomal proteases might function to degrade damaged or obsolete peroxisomal proteins, such as the glyoxylate cycle enzymes malate synthase (MLS) and isocitrate lyase (ICL), which are synthesized early in Arabidopsis postgerminative growth and then degraded after the onset of photoautotrophic growth (Cornah et al., 2004; Lingard et al., 2009). To determine whether LON2, DEG15, or PXM16 are necessary for the degradation of glyoxylate cycle enzymes during seedling development, we analyzed MLS and ICL stability in mutant seedlings. We found that MLS (Fig. 5) and ICL (Supplemental Fig. S1A) disappeared at similar rates in wild-type, lon2-1, lon2-2, and deg15-1 seedlings. Moreover, ICL disappeared at wild-type rates in pxm16-1 and pxm16-2 and in mutants defective in two other putative peroxisomal proteases (At2g18080 and At2g35615; Supplemental Fig. S1, B and C). As in the single mutants, MLS was degraded with approximately wild-type kinetics in the lon2 deg15 and lon2 pxm16 double mutants (Fig. 6). We concluded that LON2, DEG15, and PXM16 are not required for degradation of glyoxylate cycle enzymes during seedling development, consistent with recent data suggesting that ICL and MLS may be removed from the peroxisome for degradation in the cytosol (Lingard et al., 2009).
Figure 5.
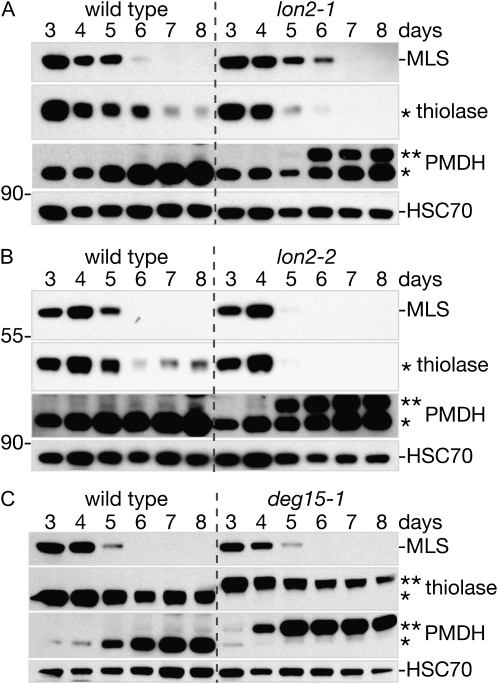
lon2 mutants degrade MLS normally but have defects in PTS2-containing protein accumulation and processing. Immunoblots of total protein extracted from wild-type and lon2-1 (A), lon2-2 (B), or deg15-1 (C) cotyledons (12 per lane) from 3- to 8-d-old seedlings. Blots were probed with antibodies raised against the PTS1-containing protein MLS, the PTS2-containing proteins PED1 (thiolase) and PMDH2, and HSC70 as a loading control. The positions of Mr markers (in kD) are indicated on the left; processed and unprocessed PTS2 proteins are marked by one or two asterisks, respectively.
Figure 6.
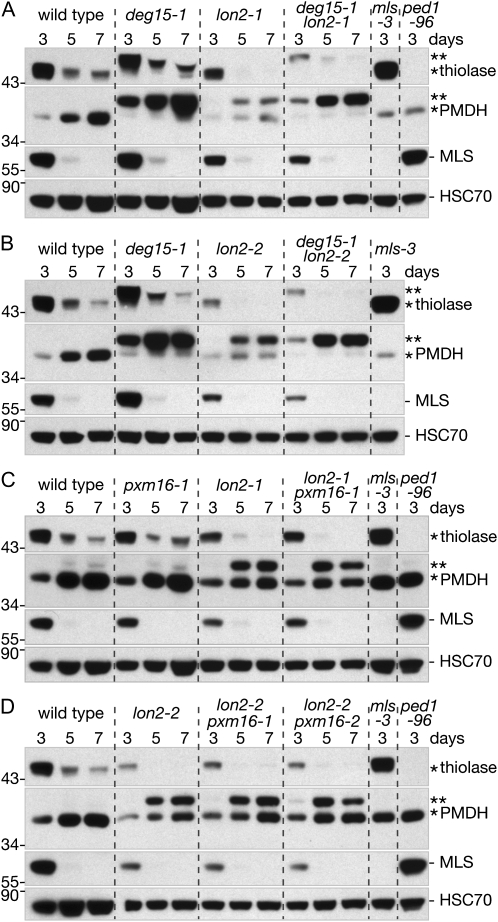
lon2 deg15 double mutants do not process PTS2-containing proteins and have thiolase accumulation defects. Immunoblots of protein extracted from 3-, 5-, and 7-d-old wild type and lon2 single and double mutants (seven seedlings per lane). Seeds were stratified for 6 d prior to plating. Blots were sequentially probed with anti-PED1 (thiolase), PMDH2, MLS, and HSC70 antibodies. The positions of Mr markers (in kD) are indicated on the left; processed and unprocessed PTS2 proteins are marked by one or two asterisks, respectively.
LON2 Is Necessary for Thiolase Accumulation and PTS2 Protein Processing
DEG15 is a PTS2-processing protease (Helm et al., 2007; Schuhmann et al., 2008), and we confirmed the previously reported deg15-1 defect in cleaving the PTS2-containing N-terminal region of thiolase (Fig. 5C; Schuhmann et al., 2008). Thiolase abundance peaked in 3- to 4-d-old wild-type seedlings before declining to a lower basal level in older seedlings (Fig. 5; Germain et al., 2001; Lingard et al., 2009). Unprocessed thiolase in deg15-1 accumulated similarly to processed thiolase in the wild type (Fig. 5C), suggesting that PTS2 removal does not appreciably affect peroxisomal thiolase stability. pxm16-1 and pxm16-2 displayed wild-type thiolase processing and accumulation (Supplemental Fig. S1B). In both lon2-1 and lon2-2, we observed processed thiolase in young seedlings, indicating that LON2 is not required for PTS2 processing. However, thiolase levels were reduced in 5- to 8-d-old lon2 mutants compared to the wild type (Fig. 5, A and B). We concluded that LON2 was necessary for normal thiolase accumulation during seedling development.
To determine whether the reduced thiolase accumulation in lon2 required PTS2 removal, we examined thiolase levels in lon2 deg15 double mutants. We found that, as in deg15-1, thiolase was wholly unprocessed in lon2-1 deg15-1 and lon2-2 deg15-1 and that the unprocessed thiolase appeared to be destabilized, as in lon2 (Fig. 6, A and B). Thus, the apparent thiolase instability in lon2 did not require PTS2 removal by DEG15. In the lon2 pxm16 double mutants, thiolase displayed similar accumulation and processing as in the lon2 single mutants (Fig. 6, C and D), consistent with the lack of enhancement of lon2 physiological phenotypes by the pxm16 mutants (Figs. 2–4).
We also examined processing and accumulation of a second PTS2-containing protein, peroxisomal malate dehydrogenase (PMDH), in the protease mutants. PMDH accumulated in a pattern reciprocal to thiolase during seedling development, with relatively low levels immediately following germination that increased as seedlings matured (Fig. 5). As previously reported (Helm et al., 2007; Schuhmann et al., 2008), PMDH was largely unprocessed in the deg15-1 mutant (Fig. 5C). In lon2-1 and lon2-2, the developmental increase in PMDH levels was accompanied by the appearance of unprocessed PMDH in 5- to 6-d-old seedlings (Fig. 5, A and B). Thus, whereas lon2 seedlings processed and accumulated the PTS2 proteins thiolase and PMDH normally for the first few days after germination, defects in accumulation (thiolase) or PTS2 removal (PMDH) became apparent with age in mutant seedlings.
PTS2-GFP Is Partially Mislocalized in lon2-2 Mutants
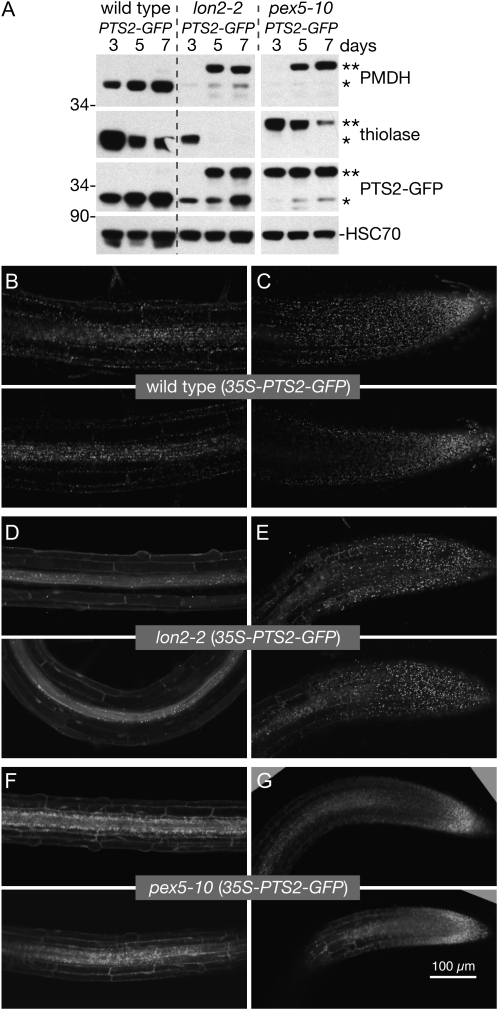
Because blocking PTS2 processing did not restore thiolase accumulation in the lon2 mutants, we tested whether the apparent thiolase instability and PMDH processing defects might reflect peroxisomal import defects. We examined a 35S-PTS2-GFP reporter (Woodward and Bartel, 2005) in the wild type, lon2-2, and pex5-10, a mutant with matrix protein import defects (Zolman et al., 2005; Lingard et al., 2009). 35S-PTS2-GFP includes the first 147 bp of PED1, encoding the PTS2 targeting signal and cleavage site, fused to the 5′ end of GFP (Woodward and Bartel, 2005); hence, PTS2-GFP has the same targeting signal and processing site as PED1, the predominant seedling thiolase isoform (Germain et al., 2001). In wild-type seedlings, PTS2-GFP was present and fully processed in 3- to 7-d-old seedlings, whereas in pex5-10, PTS2-GFP, like thiolase and PMDH, was almost completely unprocessed (Fig. 7A). In lon2-2, PTS2-GFP processing resembled PMDH: fully processed in 3-d-old seedlings but only partially processed in 5- and 7-d-old seedlings (Fig. 7A).
Figure 7.
lon2-2 roots have tissue-specific defects in PTS2-GFP import. A, Immunoblot of protein extracted from 3-, 5-, and 7-d-old wild-type (35S-PTS2-GFP), lon2-2 (35S-PTS2-GFP), and pex5-10 (35S-PTS2-GFP) seedlings (eight per lane). Seeds were stratified for 2 d prior to plating. Blots were probed with anti-PED1 (thiolase), PMDH2, GFP, and HSC70 antibodies. Positions of Mr markers (in kD) are indicated on the left; processed and unprocessed PTS2 proteins are marked by one or two asterisks, respectively. B to G, Root tip cells (C, E, and G) and maturing cells several millimeters above the tip (B, D, and F) of 6-d-old seedlings expressing PTS2-GFP were imaged via confocal microscopy. Micrographs are 11-μm-thick individual optical sections. Bar = 100 μm.
To determine the subcellular localization of PTS2-GFP and explore the apparent correlation between PTS2-GFP import defects and cell maturity, we analyzed PTS2-GFP via confocal fluorescence microscopy in wild-type, lon2-2, and pex5-10 root cells. In wild-type roots, PTS2-GFP displayed punctate fluorescence typical of peroxisomal localization in both root tips (Fig. 7C) and in maturing cells several millimeters above the tip (Fig. 7B), whereas in pex5-10, PTS2-GFP appeared to be partially cytosolic and peroxisomal in both root tip and maturation zone cells (Fig. 7, F and G). In lon2-2 root tip cells, PTS2-GFP appeared to be predominantly peroxisomal (Fig. 7E). However, PTS2-GFP displayed only partial peroxisomal localization in the lon2-2 vasculature, and PTS2-GFP appeared to be predominantly cytosolic in epidermis and cortex cells of the root maturation zone (Fig. 7D).
LON2 Facilitates Peroxisome Matrix Protein Import
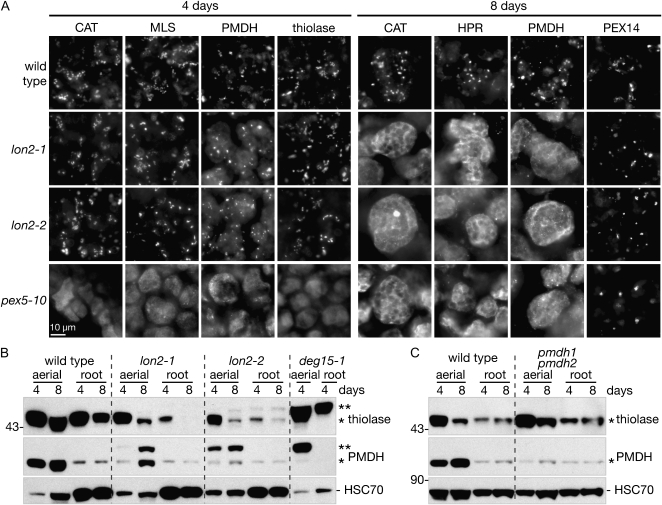
The defects in PMDH and PTS2-GFP processing and PTS2-GFP localization that we observed in older lon2 seedling extracts (Figs. 5 and 7A) and differentiated root cells (Fig. 7D) suggested that LON2 becomes necessary for peroxisomal import of matrix proteins as seedlings (or cells) mature. To determine whether LON2 also was necessary for import of native peroxisomal proteins, we immunolabeled cotyledon cells with antibodies to peroxisomal catalase (CAT), the PTS1 proteins MLS and hydroxypyruvate reductase (HPR), the PTS2 proteins thiolase and PMDH, and the membrane peroxin PEX14 (Fig. 8A). Cotyledons from 4- and 8-d-old seedlings were selected for labeling because Arabidopsis cotyledon cells do not divide during this period (Mansfield and Briarty, 1996), allowing observation of maturation-specific defects, and because MLS (Charlton et al., 2005) and HPR (Timm et al., 2008) are predominantly expressed in aerial tissues. Moreover, PMDH is not abundant in root tissue; the small amount of immunoreactivity observed in root tissue was also present in the pmdh1 pmdh2 double mutant and may result from a protein that cross-reacts with the PMDH antibody (Fig. 8B).
Figure 8.
lon2 mutant cotyledon cells import some matrix proteins normally at 4 d old, but matrix protein import is impaired at 8 d. A, Cotyledons from 4- and 8-d-old light-grown Arabidopsis seedlings were immunolabeled with antibodies raised against CAT, PTS2 proteins (PED1 and PMDH2), PTS1 proteins (MLS and HPR), and a membrane-bound peroxin (PEX14). Bar = 10 μm. B and C, Immunoblot of total protein extracted from wild-type, lon2-1, lon2-2, deg15-1, and pmdh1 pmdh2 aerial (eight seedlings, aerial tissue only) or root (16 roots per lane) tissue. Seeds were stratified for 1 d prior to plating. Blots were probed with anti-PED1 (thiolase), PMDH2, and HSC70 antibodies. Positions of Mr markers (in kD) are indicated on the left; processed and unprocessed PTS2 proteins are marked by one or two asterisks, respectively.
In cotyledons from both 4- and 8-d-old wild-type seedlings, we found the peroxisomal matrix and membrane proteins in punctate structures typical of peroxisomes, whereas all but the membrane protein PEX14 appeared to be largely cytosolic in the pex5-10 receptor mutant (Fig. 8A). In 4-d-old lon2-1 and lon2-2 seedlings, CAT, MLS, and thiolase appeared to be peroxisomal, whereas PMDH showed both peroxisomal and cytosolic labeling. The peroxisomal localization of thiolase and the dual localization of PMDH detected by immunolocalization at day 4 (Fig. 8A) were consistent with the presence of only processed thiolase but both processed and unprocessed PMDH detected by immunoblotting at day 4 (Fig. 8B). In 8-d-old seedlings, all of the monitored peroxisome matrix proteins (CAT2, HPR, and PMDH) appeared to be largely cytosolic in the lon2 mutants, although we observed occasional punctate labeling (Fig. 8A), consistent with the possibility that some peroxisomal import was still occurring and suggested by the detection of not only unprocessed but also processed PMDH in lon2 seedling extracts (Fig. 8B). Immunolabeling with anti-PEX14 antibodies revealed the presence of peroxisomes in the wild type, lon2-1, lon2-2, and pex5-10 (Fig. 8A), indicating that the mutant defects likely resulted from impaired import of peroxisomal matrix proteins, rather than impaired peroxisome formation. We concluded that the lon2 mutant import defect extends beyond PTS2 proteins, such as PMDH and PTS2-GFP, and includes a variety of matrix proteins, including at least one PTS1 protein (HPR) as well as catalase, which has an atypical PTS (Kamigaki et al., 2003).
DISCUSSION
Proteases function to degrade entire proteins and to process proproteins and can remove damaged or obsolete proteins and regulate metabolic and signaling pathways (Schaller, 2004). In this work, we demonstrated that the peroxisomal LON2 protease is needed for importing PTS1 and PTS2 proteins into the peroxisome matrix of maturing cotyledon and root cells of Arabidopsis seedlings.
One peroxisome-associated proteolytic process during Arabidopsis development is the degradation of the glyoxylate cycle enzymes ICL and MLS a few days after germination (Lingard et al., 2009). The five mutants that we identified in putative peroxisomal proteases (Table I) displayed wild-type degradation of these glyoxylate cycle enzymes (Fig. 5; Supplemental Fig. S1). Moreover, combining lon2 alleles with deg15-1 or pxm16 alleles did not result in MLS stabilization (Fig. 6), suggesting that these proteins do not function redundantly to degrade obsolete glyoxylate cycle enzymes. Because LON2, DEG15, and PXM16 are the only predicted Arabidopsis proteases that have been detected in peroxisomes (Reumann et al., 2004; Helm et al., 2007; Ostersetzer et al., 2007; Eubel et al., 2008; Schuhmann et al., 2008; Reumann et al., 2009), and none of the corresponding mutants had defects in glyoxylate cycle enzyme turnover, it may be that these proteases are not responsible for bulk degradation of damaged or obsolete proteins. Indeed, recent data suggest that peroxisome-associated degradation of damaged or obsolete matrix proteins may require the assistance of PEX4 and PEX6 and export to the cytosol (Lingard et al., 2009). Our failure to detect matrix protein degradation defects in lon2 or pxm16 mutants indicates that the substrates of these proteases remain to be identified and is consistent with the possibility that LON2 and PXM16 serve regulatory functions in peroxisomes.
Roles of DEG15 and PXM16 in Peroxisome Biogenesis
Our mutant analysis confirmed previous findings that DEG15 is required for PTS2 processing (Helm et al., 2007; Schuhmann et al., 2008). Although the deg15-1 mutant is severely compromised in PTS2 removal from thiolase and PMDH, deg15-1 displayed at most mild peroxisome-deficient phenotypes (Figs. 2 and 3). Because the deg15 seedling phenotypes are much less severe than those of mutants lacking functional PTS2 proteins, such as the PED1 thiolase (Figs. 2 and 3; Hayashi et al., 1998; Germain et al., 2001) or PMDH1 and PMDH2 (Pracharoenwattana et al., 2007), PTS2 removal does not appear to be necessary for stability or function of essential matrix proteins. Indeed, PTS2 signals are not removed from yeast peroxisomal proteins (for review, see Olsen, 1998). However, the deg15 mutant enhancement of lon2 growth defects (Fig. 4) and Suc dependence (Fig. 2) suggests that at least one of the PTS2 proteins needed for β-oxidation functions more effectively following PTS2 removal, at least in the context of lon2 mutations.
The pxm16 mutants showed no aberrant phenotypes and did not enhance lon2 defects under any of the examined conditions (Figs. 2–4). By contrast, other mutants with undetectable defects in peroxisome biogenesis can dramatically enhance moderate or severe defects in other mutants. For example, the pex22-1 mutant has no detectable peroxisome-defective phenotype but dramatically enhances pex4-1 mutant defects (Zolman et al., 2005), and the slight Suc dependence of lon2-1 and lon2-2 is markedly enhanced in lon2 deg15 double mutants (Fig. 2). The failure of pxm16 mutants to enhance lon2 mutant phenotypes suggests that PXM16 is not necessary for peroxisome biogenesis, IBA metabolism, or fatty acid β-oxidation. Given the role of PXM16 homologs in insulin degradation and wound response (Strassner et al., 2002; Schaller, 2004), it is possible that PXM16 functions in aspects of peroxisome metabolism not assayed here, such as pathogen or wound responses.
Roles of LON2 in Peroxisome Biogenesis
In contrast to deg15 and pxm16, the Arabidopsis lon2 mutants displayed severe defects in certain facets of peroxisome function and were smaller than the wild type following growth in soil (Fig. 4). Although the lon2 alleles displayed only minor defects in seedling growth without Suc supplementation (Fig. 2) and primary root elongation inhibition by IBA (Fig. 3, A and B), lon2 mutants were severely impaired in IBA-stimulated lateral root formation (Fig. 3C). In this regard, the lon2 mutant phenotype was unique compared to other peroxisome-defective mutants. Many mutants, like pex6-1 and pex5-10, show severe defects in both IBA-responsive root elongation inhibition and lateral root promotion (Zolman et al., 2000, 2005; Zolman and Bartel, 2004). However, previously characterized IBA response mutants with mild defects in primary root response to IBA generally show only partial defects in lateral root induction. For example, the pex7-1 peroxin mutant displays moderately IBA-resistant primary root elongation and partial defects in IBA-responsive lateral root promotion (Woodward and Bartel, 2005), and single mutants defective in three of the six Arabidopsis acyl-CoA oxidase genes display moderate IBA resistance in root elongation but no apparent defect in IBA-responsive lateral root promotion (Adham et al., 2005).
The lateral root assay, in which 4-d-old seedlings are transferred to IBA-containing medium for four additional days, may be particularly sensitive to developmentally delayed defects in peroxisome metabolism. The lon2 mutants displayed matrix protein import defects that appeared to correlate with cell age; differentiated root cells imported PTS2-GFP into peroxisomes much less efficiently than recently divided cells near the root tip (Fig. 7, D and E). Similarly, several PTS1 and PTS2 proteins were predominantly peroxisomal in 4-d-old lon2 cotyledon cells but largely cytosolic in 8-d-old cotyledon cells (Fig. 8A). These localization defects were accompanied by accumulation of both processed and unprocessed PMDH and PTS2-GFP in older lon2-2 seedlings (Figs. 5 and 7A). Although the T-DNA insertions that we identified in LON2 are near the C terminus and may not completely disrupt LON2 function, the lon2 protein products would lack the C-terminal PTS1 (Fig. 1A) and would be predicted to remain cytosolic. If present, it seems unlikely that cytosolic lon2 protein is responsible for the accelerated thiolase degradation we observed in lon2-1 and lon2-2, as thiolase stability in pex5-10 resembles the wild type (Fig. 7A) even though thiolase (and presumably LON2) is largely cytosolic in pex5-10 (Fig. 8A). Moreover, we cannot eliminate the possibility that some lon2 protein enters the peroxisome, either through a cryptic PTS1 provided by the T-DNA border or by association with an interacting peroxisome-bound protein. The future characterization of LON2 antibodies and demonstrated lon2 null alleles would aid in resolving these ambiguities.
The H. polymorpha Lon isoform Pln appears to degrade certain peroxisome-targeted, non-native proteins (Aksam et al., 2007). Although Arabidopsis lon2 mutants did not display notably reduced degradation rates of the matrix proteins that we examined (Figs. 5 and 6; Supplemental Fig. S1), it is possible that Arabidopsis LON2 degrades one or more peroxisomal proteins that we did not examine. In lon2, the absence of this hypothetical degradation might block further matrix protein import, either because LON2 targets a regulatory protein or because bulk undegraded protein in the peroxisome impairs further import.
Plant peroxisomal LON isoforms are more similar to chordate peroxisomal LON isoforms than either are to their yeast relatives (Fig. 1C). Moreover, phylogenetic analysis suggests that a subset of yeasts that includes H. polymorpha may have acquired a peroxisomal LON isoform in an evolutionary event distinct from the event in which the peroxisomal LON now found in plants and animals was obtained (Fig. 1C), consistent with the possibility that peroxisomal LON may function differently in these lineages. Arabidopsis lon2 mutants displayed striking matrix protein import defects (Figs. 7 and 8). Similarly, a mammalian cell line expressing a dominant negative pLon variant mis-sorts catalase to the cytosol (Omi et al., 2008). Rather than acting as a protease, Arabidopsis LON2 may assist in matrix protein import as a chaperone, perhaps aiding in dissociation of the PEX5 and PEX7 receptors following import or in dissociation of receptors and cargo, if such dissociations are necessary for receptor recycling. These models are consistent with the delayed onset of lon2 peroxisome-defective phenotypes. It is intriguing that certain eukaryotes, including Drosophila melanogaster, appear to lack peroxisomal LON isoforms (Fig. 1C). Further examination of peroxisomal Lon function in a variety of eukaryotes, as well as the identification of LON2 substrates, will assist in distinguishing among these possibilities.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The wild type and mutants were in the Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) accession. Seeds were surface sterilized, stratified in 0.1% [w/v] agar at 4°C for the indicated times, and germinated on nutrient media supplemented with Suc and hormones as previously described (Zolman and Bartel, 2004). In auxin response assays, seedlings were grown in continuous light filtered through yellow long-pass filters to slow photochemical breakdown of auxin (Stasinopoulos and Hangarter, 1990).
The wild type (35S-PTS2-GFP; Woodward and Bartel, 2005), pex5-10 (Zolman et al., 2005), pmdh1 pmdh2 (Pracharoenwattana et al., 2007), mls-3 (Lingard et al., 2009), and pex6-1 (Zolman and Bartel, 2004) were described previously. lon2-1 (SALK_128438), lon2-2 (SALK_043857), pxm16-1 (SALK_019128), pxm16-2 (SALK_023917), and deg15-1 (SALK_007184) were from the Salk Institute sequence-indexed insertion collection (Alonso et al., 2003). lon2-2 was backcrossed to Col-0 either once (Figs. 3C and 4–8) or twice (Figs. 2 and 3, A and B). The sites of T-DNA insertions were verified by sequencing junctional PCR amplicons. pex5-10 (35S-PTS2-GFP), lon2-2 (35S-PTS2-GFP), and the protease double mutants were isolated by crossing and following mutations and transgenes via PCR (Supplemental Table S1) in subsequent generations. We isolated ped1-96 from an ethyl methanesulfonate-mutagenized Col-0 population as a Suc-dependent and IBA-resistant seedling; the mutant harbors a C-to-T mutation in PED1/KAT2 (At2g33150) that destroys a HinP1I restriction enzyme site and replaces Gln 190 with a stop codon (Supplemental Table S1; Fig. 1B). deg15-1 and ped1-96 were backcrossed to Col-0 once prior to analysis.
Immunoblot Analysis
Cotyledons (Lingard et al., 2009) and whole seedlings (Zolman and Bartel, 2004) were prepared as previously described for western-blot analysis. Immunoblotting was conducted as previously described (Lingard et al., 2009). Primary antibodies were diluted as follows: 1:25,000 α-MLS (Olsen et al., 1993), 1:2,500 α-thiolase (Lingard et al., 2009), 1:2,000 α-PMDH2 (Pracharoenwattana et al., 2007), 1:500 α-GFP (632377; Clontech), and 1:8,000 α-HSC70 (SPA-817; StressGen Biotechnologies).
Microscopy
Samples for immunofluorescence microscopy were prepared as described previously (Lingard et al., 2009), except that 8-d-old cotyledon cell walls were permeabilized in 1.5% driselase (Sigma-Aldrich) for 1 h. Cotyledons were labeled with rabbit 1:2,000 α-CAT (Kunce et al., 1988), 1:24,000 α-MLS (Olsen et al., 1993), 1:2,000 α-ICL (Maeshima et al., 1988), 1:2,000 (4-d-old-seedlings), or 1:1,500 (8-d-old seedlings) α-PMDH2 (Pracharoenwattana et al., 2007), 1:2,500 α-PED1 (Lingard et al., 2009), 1:1,000 α-HPR (Kleczkowski and Randall, 1988), and 1:12,500 α-PEX14 (prepared from rabbits immunized with an Escherichia coli-produced fusion protein including the N-terminal 243 amino acids of Arabidopsis PEX14/At5g62810) for 4 h at 37°C. The secondary antibody was 1:1,500 goat α-rabbit Alexa 594 (Invitrogen).
A Zeiss Axioplan 2 fluorescence compound microscope equipped with narrow-band GFP filters (Chroma) and a 40× lens was used for imaging immunolabeled tissues. Images were acquired with a CoolSNAP HQ camera (Photometrics) using MetaMorph 7 imaging software (Molecular Devices). Confocal images of GFP-expressing tissues were acquired using a Zeiss LSM 510 confocal microscope (excitation, 488 nm; emission, 500–550 nm) equipped with a 20× lens. Images were cropped using NIH ImageJ and adjusted for brightness in Adobe Photoshop CS.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Table I and Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The glyoxylate enzyme isocitrate lyase is degraded with near wild-type kinetics in all identified putative peroxisomal protease mutants.
Supplemental Table S1. Primers used to determine mutant genotypes.
Supplemental Table S2. Accession numbers for LON homologs used in phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank John Harada, Masayoshi Maeshima, Douglas Randall, Steven Smith, and Richard Trelease for the MLS, ICL, HPR, PMDH, and CAT antibodies, respectively. We thank Randall Baldasarre for assistance in isolating double mutants, Andrew Woodward for assistance with figures, Steven Smith for pmdh1 pmdh2 seeds, the Arabidopsis Biological Resource Center at Ohio State University for seeds from Salk Institute insertion lines, and Naxhiely Martinez, Sarah Ratzel, Lucia Strader, Andrew Woodward, and Bethany Zolman for critical comments on the manuscript.
This research was supported by the National Science Foundation (MCB–0745122), the National Institutes of Health (R01GM079177), the Robert A. Welch Foundation (C–1309), and a postdoctoral fellowship to M.J.L. (USDA 2008–20659).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Bonnie Bartel (bartel@rice.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J 41 859–874 [DOI] [PubMed] [Google Scholar]
- Aksam EB, Koek A, Kiel JA, Jourdan S, Veenhuis M, van der Klei IJ (2007) A peroxisomal Lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3 96–105 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arai Y, Hayashi M, Nishimura M (2008) Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol 49 526–539 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton WL, Johnson B, Graham IA, Baker A (2005) Non-coordinate expression of peroxisome biogenesis, β-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep 23 647–653 [DOI] [PubMed] [Google Scholar]
- Cornah JE, Germain V, Ward JL, Beale MH, Smith SM (2004) Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J Biol Chem 279 42916–42923 [DOI] [PubMed] [Google Scholar]
- Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, et al (1996) Unified nomenclature for peroxisome biogenesis factors. J Cell Biol 135 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O'Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43 689–696 [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (2006) Origin and evolution of the peroxisomal proteome. Biol Direct 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Lorenzo M, Sjodin A, Jansson S, Funk C (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde J-P, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28 1–12 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Luck C, Prestele J, Hierl G, Huesgen PF, Frohlich T, Arnold GJ, Adamska I, Gorg A, Lottspeich F, et al (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki A, Man S, Terauchi K, Nishi Y, Tachibe-Kinoshita Y, Nito K, Kondo M, Hayashi M, Nishimura M, Esaka M (2003) Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J 33 161–175 [DOI] [PubMed] [Google Scholar]
- Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H (2004) Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J Biol Chem 279 421–428 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD (1988) Purification and characterization of a novel NADPH(NADH)-dependent hydroxypyruvate reductase from spinach leaves. Comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem J 250 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunce CM, Trelease RN, Turley RB (1988) Purification and biosynthesis of cottonseed (Gossypium hirsutum L) catalase. Biochem J 251 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurochkin IV, Mizuno Y, Konagaya A, Sakaki Y, Schonbach C, Okazaki Y (2007) Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in β-oxidation of fatty acids. EMBO J 26 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Suzuki CK (2008) Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates. Biochim Biophys Acta 1784 727–735 [DOI] [PMC free article] [PubMed]
- Lingard MJ, Monroe-Augustus M, Bartel B (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M, Yokoi H, Asahi T (1988) Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol 29 381–384 [Google Scholar]
- Mansfield SG, Briarty LG (1996) The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int J Plant Sci 157 280–295 [Google Scholar]
- Olsen LJ (1998) The surprising complexity of peroxisome biogenesis. Plant Mol Biol 38 163–189 [PubMed] [Google Scholar]
- Olsen LJ, Ettinger WF, Damsz B, Matsudaira K, Webb MA, Harada JJ (1993) Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi S, Nakata R, Okamura-Ikeda K, Konishi H, Taniguchi H (2008) Contribution of peroxisome-specific isoform of Lon protease in sorting PTS1 proteins to peroxisomes. J Biochem 143 649–660 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Kato Y, Adam Z, Sakamoto W (2007) Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol 48 881–885 [DOI] [PubMed] [Google Scholar]
- Platta HW, Erdmann R (2007) The peroxisomal protein import machinery. FEBS Lett 581 2811–2819 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK (2006) Peroxisomal β-oxidation--a metabolic pathway with multiple functions. Biochim Biophys Acta 1763 1413–1426 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50 381–390 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ (2008) MEROPS: the peptidase database. Nucleic Acids Res 36 D320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Ma CL, Lemke S, Babujee L (2004) AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol 136 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber AP, et al (2009) In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220 183–197 [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi H (2008) The peroxisome: still a mysterious organelle. Histochem Cell Biol 129 421–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I (2008) The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol 148 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki KI, Maeshima M, Yokota A, Tomizawa KI, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner J, Huet Y, Schaller A (2002) Cloning of tomato proteases by direct selection in yeast for enzymes that cleave the polypeptide wound signal systemin. In A Schmidt, B Mauch-Mani, eds, Induced Resistance in Plants against Insects and Diseases. IOBC/wprs Bulletin, Vol 25. IOBC/wprs, Dijon, France, pp 159–163
- Swofford DL (2001) PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods), Ed 4. Sinauer Associates, Sunderland, MA
- Timm S, Nunes-Nesi A, Parnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H (2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris V, Maenhaut-Michel G, Van Melderen L (2006) Biological roles of the Lon ATP-dependent protease. Res Microbiol 157 701–713 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.