Abstract
Nodulation is the result of a symbiosis between legumes and rhizobial bacteria in soil. This symbiosis is mutually beneficial, with the bacteria providing a source of nitrogen to the host while the plant supplies carbon to the symbiont. Nodule development is a complex process that is tightly regulated in the host plant cell through networks of gene expression. In order to examine this regulation in detail, a library of quantitative reverse transcription-polymerase chain reaction primer sets was developed for a large number of soybean (Glycine max) putative regulatory genes available in the current expressed sequence tag collection. This library contained primers specific to soybean transcription factor genes as well as genes involved in chromatin modification and translational regulation. Using this library, we analyzed the expression of this gene set during nodule development. A large number of genes were found to be differentially expressed, especially at the later stages of nodule development when active nitrogen fixation was occurring. Expression of these putative regulatory genes was also analyzed in response to the addition of nitrate as a nitrogen source. This comparative analysis identified genes that may be specifically involved in nitrogen assimilation, metabolism, and the maintenance of active nodules. To address this possibility, the expression of one such candidate was studied in more detail by expressing in soybean roots promoter β-glucuronidase and green fluorescent protein fusions. This gene, named Control of Nodule Development (CND), encoded a Myb transcription factor gene. When the CND gene was silenced, nodulation was reduced. These results, associated with a strong expression of the CND gene in the vascular tissues, suggest a role for CND in controlling soybean nodulation.
Nodulation involves the intimate relationship between soil bacteria (rhizobia) and legume plants, which results in the formation of a novel organ, the nodule, in which the bacteria reside and provide a steady source of nitrogen to the plant. Control of the initial host-symbiont interaction and subsequent nodule development is complex and appears to require a continuous exchange of chemical signals between the partners. For example, the compatible symbiont initially recognizes flavonoid molecules released by the host. This, in turn, leads to the production of a specific lipochitin nodulation signal (Nod factor) that is required for rhizobial infection and also triggers the development of the nodule primordium (Oldroyd and Downie, 2008).
Primarily through an examination of plant mutants defective in nodulation, the basic steps in the Nod factor signaling pathway have been elucidated (Oldroyd and Downie, 2008). Initial Nod factor recognition is mediated through two LysM receptors that likely directly interact with the compatible Nod factor (Amor et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003). The resulting signaling cascade ultimately leads to activation of specific transcription factors (TFs), a few of which have been identified. For example, the Medicago truncatula MtNSP1 and MtNSP2 genes encode two GRAS family TFs (Catoira et al., 2000; Oldroyd and Long, 2003; Kalo et al., 2005; Smit et al., 2005) that are essential to root hair infection. MtERN, a member of the AP2-EREBP family (Middleton et al., 2007), was shown to play a key role in the initiation and maintenance of rhizobial infection. In Lotus japonicus, the NIN gene encodes a putative TF gene (Schauser et al., 1999). Mutants in the L. japonicus NIN gene, or the Pisum sativum ortholog Sym35, fail to support rhizobial infection and do not show cortical cell division upon inoculation (Schauser et al., 1999; Borisov et al., 2003). In contrast, the L. japonicus astray mutant exhibits hypernodulation. The ASTRAY gene encodes a bZIP TF (Nishimura et al., 2002).
The expression of some of these key TF genes is affected by rhizobial inoculation. For example, NSP2, ERN, and NIN expression was induced in response to root inoculation by the compatible symbiont (Kalo et al., 2005; Marsh et al., 2007; Middleton et al., 2007). Such results suggest that TFs involved in nodulation may be identified by directly analyzing their transcriptional response to inoculation. Based on such facts, large-scale DNA microarray studies identified a variety of putative TF genes that were differentially expressed during nodule initiation and development (El Yahyaoui et al., 2004; Kouchi et al., 2004; Benedito et al., 2008; Brechenmacher et al., 2008). For example, in the study of El-Yahyaoui et al. (2004), 24 M. truncatula TF genes were found to respond to rhizobial inoculation. Among these genes was the M. truncatula HAP2-1 gene, a member of the CCAAT-binding TF family. Combier et al. (2006) showed that MtHAP2-1 expression was regulated through the action of a specific microRNA (miR169). Boualem et al. (2008) identified a second microRNA in M. truncatula, miR166, targeting at least three class III HD-ZIP TF genes (MtCNA1, MtCNA2, and MtHB8). The down-regulation of these three genes affected the number of infection initiation events in the symbiosis between M. truncatula and Sinorhizobium meliloti. Collectively, these studies reflect the important roles of TF genes in nodulation.
Due to the low global expression level of many TF genes, DNA microarrays lack the sensitivity to accurately measure their expression (Czechowski et al., 2004; Libault et al., 2007). Czechowski et al. (2004) created a resource that utilized highly sensitive quantitative reverse transcription (qRT)-PCR to accurately analyze the expression of more than 2,000 Arabidopsis (Arabidopsis thaliana) TF genes. This study clearly demonstrated that the use of such qRT-PCR primer libraries provides 100- to 1,000-fold greater sensitivity in accurately measuring TF gene expression compared with DNA microarray technologies. Since this initial paper, other primer libraries have been developed for the various putative regulatory genes in Oryza sativa (Caldana et al., 2007) and Medicago truncatula (Kakar et al., 2008). These libraries, as well as the original Arabidopsis library, were used in large-scale analyses to identify putative regulatory genes that are differentially expressed in different plant tissues, in response to pathogens and microbial elicitors, and after treatment with nitrogen and phosphorous (Czechowski et al., 2004; Scheible et al., 2004; McGrath et al., 2005; Caldana et al., 2007; Libault et al., 2007; Morcuende et al., 2007; Kakar et al., 2008; Verdier et al., 2008; Gruber et al., 2009).
In this study, by mining public EST databases, 2,586 putative soybean (Glycine max) regulatory genes (i.e. TF genes as well as those involved in chromatin modification and translational regulation) were identified. PCR primers were designed and validated to create 1,149 specific primer sets, which can be used to specifically quantify gene expression by qRT-PCR. To demonstrate the usefulness of this resource, the expression of these genes was profiled during soybean nodule development by comparing nodulated and nodule-free roots. The results showed that 126 putative regulatory genes were differentially regulated during nodulation, including putative orthologs of MtHAP2.1 and MtERN. Most of these genes were regulated at the later stages when active nitrogen fixation is occurring. Further analysis distinguished those putative regulatory genes strictly regulated during nodulation, after nitrogen starvation, and/or those responsive to the addition of nitrate as a nitrogen source. Control of Nodule Development (CND), a Myb TF gene, was studied in more detail through the construction of GUS and GFP promoter fusions. Silencing of CND resulted in a decrease in nodule numbers. Our data clearly indicate that a specific, temporal pattern of putative regulatory gene expression correlates with nodule development.
RESULTS AND DISCUSSION
Identification of 2,586 Putative Soybean Regulatory Genes in Public EST Databases
To begin compiling a library of putative regulatory genes in soybean, the sequences of predicted TF genes in Arabidopsis (2,290 and 2,250 predicted TF transcripts from the Database of Arabidopsis Transcription Factors [http://datf.cbi.pku.edu.cn/] and the Plant Transcription Factor Database [http://plntfdb.bio.uni-potsdam.de/v2.0/index.php?sp_id=ATH], respectively) were used for similarity searches to identify homologous soybean genes. Because the soybean genome sequences were not available at the time of the design of the primer sets, the identified sequences were compared against the National Center for Biotechnology Information G. max Unigene database (with BLAST e-value ≤ 1e-12) to identify homologous soybean sequences. In addition, The Institute for Genome Research soybean Gene Indices (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi? species=soybean) and soybean EST databases (http://www.ncbi.nlm.nih.gov/dbEST) were mined to identify 1,321 tentative consensus sequences and 1,043 ESTs, respectively, that are annotated as regulatory genes. Finally, the annotation of the soybean Affymetrix arrays (Robert Goldberg, personal communication) was used to identify additional putative soybean regulatory genes. Altogether, after identifying and removing redundant sequences between the different databases, these different approaches identified 2,586 transcripts (Supplemental Table S1). In cases of ambiguity, the predicted protein sequence was analyzed using Interproscan (http://www.ebi.ac.uk/Tools/InterProScan/) for final domain prediction and validation of TF family.
Creation and Validation of the qRT-PCR Primer Set Library
To accurately quantify the expression levels of the identified putative regulatory genes, specific qRT-PCR primers were designed from each of the putative regulatory gene sequences using PRIMEGENS (Xu et al., 2002). The specificity of each primer set was validated based on the number of amplicons produced after RT-PCR and qRT-PCR (i.e. only one amplicon was expected per RT/qRT-PCR at the average size of 100 bp; Supplemental Fig. S1, A and B). Moreover, 96 different amplicons were randomly selected for sequencing. This analysis returned 90 correct sequences out of the 96 analyzed (data not shown). Despite resequencing the last six amplicons, the sequences were of insufficient quality and, therefore, were discarded without a conclusion as to their identity. These data suggested that at least 94% of the primer sets are specific for the targeted putative regulatory genes. Since the 96 amplicons were randomly selected for sequencing, similar results should be expected for the entire primer library. Altogether, 1,280 primer sets were validated by these approaches.
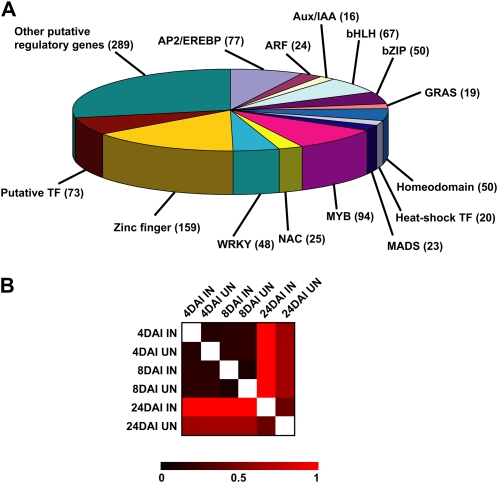
The lack of specificity of the other primer sets is likely the result of the large number of soybean paralogs that arose from the whole genome duplications detected from analysis of the soybean genome (Schlueter et al., 2004, 2007). Although PRIMEGENS attempts to compensate for these effects, genome duplication is an additional difficulty in the design of specific primer sets against soybean transcripts. Based on the low divergence of homeologous genes, we assumed that amplicon size and dissociation curve profiles might not be sufficiently stringent to validate the primer set specificity. Accordingly, to confirm primer set specificity, we took advantage of the recent release of the soybean genome sequence (http://www.phytozome.net/soybean) to BLAST the 1,280 qRT-PCR primer sets. Such analysis also allowed us to attribute a soybean gene annotation number to each of the specific qRT-PCR primer sets. A total of 131 primer sets matched against two or three different soybean transcripts. Such primer sets were discarded, finally leading to the selection of 1,149 primer sets in the present soybean qRT-PCR library (Supplemental Table S2). These 1,149 primer sets allowed the quantification of 1,034 soybean genes (Fig. 1A).
Figure 1.
The creation of a soybean qRT-PCR primer set library allowed the accurate quantification of the expression of more than 1,043 soybean putative regulatory genes in eight different conditions. A, Distribution on the soybean genes quantifiable by qRT-PCR according to their family membership. Only the main TF families are indicated. B, Comparison of the transcriptomes of the 1,043 putative soybean regulatory genes during nodulation. Using a pair-wise Pearson correlation coefficient, a heat map was generated using MeV software (http://www.tm4.org/mev.html). Black color shows strong correlation, while red color shows low correlation between the eight conditions tested (inoculated [IN] and uninoculated [UN] at 4, 8, and 24 DAI).
In addition to their specificity, primer efficiency is another important characteristic to validate suitable qRT-PCR primer sets. Theoretically, under ideal conditions, the number of amplicons should increase by two after each PCR cycle. Based on an earlier study (Libault et al., 2007), only a few primer sets are highly efficient across a set of tested conditions. In this study, the efficiency of each putative soybean regulatory gene primer set was measured using the LinRegPCR software (Ramakers et al., 2003). The average primer set efficiency was calculated when gene expression was detected in at least 20 out of the 24 different cDNA samples tested. Out of the 1,149 primer sets used, 1,043 were retained and exhibited an average primer efficiency of 1.97 ± 0.05 (Supplemental Table S3). None of the primer sets displayed an efficiency of less than 60%, and 953 primer sets (91.4% of the library) had an efficiency of greater than 90% (Supplemental Fig. 1C). These data provide confidence that the primer set library is of high specificity and can be used to accurately quantify putative regulatory gene mRNA expression levels.
Expression Pattern of Putative Soybean Regulatory Genes during Root and Nodule Development
The putative regulatory gene primer set library was first used to profile gene expression during nodule development. To identify regulated genes during nodulation, we compared gene expression in soybean roots inoculated with Bradyrhizobium japonicum and mock-inoculated roots at different stages of nodule development (4, 8, and 24 d after inoculation [DAI]). In our conditions, at 4 DAI, nodules were not visible but extensive root hair cell deformation could be seen, indicative of a strong plant response to B. japonicum. At 8 DAI, nodule primordia emerged, while nodules were fully developed at 24 DAI (i.e. leghemoglobin was present in the nodule [pink-red color], and nitrogen fixation occurred based on the absence of leaf chlorosis). Overall analysis of the expression pattern of soybean regulatory genes in mock-inoculated roots highlighted strong correlations between 4 and 8 DAI (Fig. 1B). A lower correlation between 4- to 8-DAI and 24-DAI mock-inoculated roots suggested that gene expression was modified during root development. This analysis suggests that the regulatory gene expression profiles were well established and stably maintained during the first stages of root development.
During nodulation, the putative soybean regulatory gene expression profile was not affected during the first days following root inoculation by B. japonicum (i.e. the Euclidian distance between 4- and 8-DAI inoculated and mock-inoculated roots was low) but was strongly disrupted at 24 DAI (Fig. 1B). Such results suggested that a significant number of soybean regulatory genes were specifically expressed in mature nodules but not in developing nodules. On the other hand, based on our experience, mature nodules are plant organs extremely rich in mRNA. Consequently, comparisons between inoculated and mock-inoculated roots between 4 to 8 DAI and 24 DAI might also be explained by a strong dilution of the 4- to 8-DAI nodule mRNA by the overall root system mRNA, a dilution that was limited at 24 DAI when individual nodules can be harvested for analysis. These observations support the notion that a large number of putative regulatory genes were differentially expressed at 24 DAI.
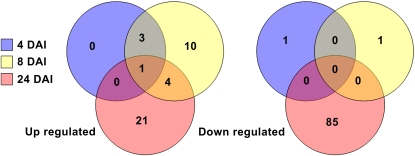
We identified for each time point the differentially expressed putative soybean regulatory genes. For example, 126 soybean regulatory genes were differentially expressed during soybean nodule development (P < 0.05 and fold change < 0.5 or > 2; Supplemental Table S4). Among them, 31% (39 genes) were up-regulated, while 69% (87 genes) were down-regulated (Fig. 2). As expected based on the calculation of the correlation between the different conditions tested, most of the putative regulatory genes were differentially expressed at 24 DAI (five, 19, and 111 genes were regulated at 4, 8, and 24 DAI, respectively; Fig. 2). Among the 126 genes, only eight genes were found to be differentially regulated at multiple time points of the time course: three CCAAT-Box, one AP2/EREBP, one Myb, one C2H2 Zinc Finger domain, one bHLH, and one RNA-dependent RNA polymerase gene. All eight of these genes were exclusively up-regulated. Such genes might have an important role in multiple stages of nodule development (e.g. infection of plant cells by bacteroids or control of nodule development).
Figure 2.
Identification of putative soybean regulatory genes expressed differentially during nodulation. Venn diagrams showing the number of soybean genes significantly up-regulated (left) and down-regulated (right) during nodulation at 4 (yellow), 8 (red), and 24 (blue) d after B. japonicum inoculation. This analysis was performed on 1,043 different putative soybean regulatory genes. Genes regulated during nodulation were validated according to their fold change between inoculated (nodulated) and mock inoculated (nodule-free) roots (fold change > 2 or < 0.5). Student's t test (P < 0.05) was performed between the three independent biological materials to ensure the significance of the differences.
Characterization of the Roles of Putative Regulatory Genes in Nitrogen Assimilation/Metabolism and Nodule Development
A difficulty in profiling gene expression is choosing which tissues to compare. For example, during our analysis, it appears that 24-d mock-inoculated roots, in addition to lacking nodules, also exhibited symptoms of nitrogen starvation, such as chlorosis and leaf senescence. This stress condition might affect the expression pattern of the putative soybean regulatory genes, especially with respect to the comparisons between mock-inoculated and inoculated plants. Consequently, at 24 DAI, we made the assumption that some of the regulatory genes were regulated in response to nitrogen starvation in the mock-inoculated plants and did not have a specific role in nodule development. In order to obtain a clearer view of those genes that may be responding to the nitrogen provided by actively fixing bacteroids versus those that are strictly involved in the nodulation response, 24-DAI gene expression (inoculated versus mock inoculated) was compared in roots grown with and without nitrogen (KNO3 versus KCl; Supplemental Table S4).
Of the 111 genes regulated in 24-DAI nodulated roots, only 12 were also responsive to root nitrogen status (1.2% of the 1,034 genes analyzed; Supplemental Table S4). Among these 12 genes, 10 were repressed during both nodulation and KNO3 treatments. The remaining two genes were strongly up-regulated at 24 DAI but repressed in roots treated with KNO3. The remaining 99 genes (9.6%; Supplemental Table S4) were regulated specifically in nodulated roots, thereby supporting a role of these putative regulatory genes in the maintenance and activity of the nodule. The comparison of KNO3- and KCl-treated roots identified 38 genes responsive to nitrogen status but not involved in the nodulation process (3.7%; Supplemental Table S4). Recently, Ruffel et al. (2008) also studied the response of M. truncatula roots to different nitrogen treatments. In that study, plants were grown with or without 1 mm KNO3 as well as under conditions where atmospheric nitrogen was present or absent in the environment of nodulated roots. Among the 752 TF genes analyzed, Ruffel et al. (2008) identified 101 TF genes regulated specifically in response to KNO3 treatment (13.4%), 37 differentially expressed TF genes in response to the presence of atmospheric nitrogen (4.9%), and only 11 TF genes differentially regulated in response to both treatments (1.5%). This distribution was very different from our results with soybean. Such differences might be explained by the fact that in both studies the qRT-PCR primer libraries used only sampled a subset of the total TF genes that may be responding to nitrogen status. Of course, the data may also suggest fundamental differences by which these two plant species respond to nitrogen status. Finally, the experimental designs used by Ruffel et al. (2008) and in our experiments are also quite different, which could also explain the differences in the magnitude of the responses seen. While we compared B. japonicum inoculated with mock-inoculated roots, Ruffel et al. (2008) compared the expression patterns of TF genes on nodulated roots with or without atmospheric nitrogen.
Putative Soybean Orthologs of M. truncatula and L. japonicus Nodulation-Related Genes Are Regulated during Soybean Nodulation
We next compared the sequences of the 126 genes that we identified as differentially regulated in soybean nodule development with sequences of TFs from L. japonicus and M. truncatula with known roles in nodule development. Among them, Glyma02g35190 and Glyma10g10240 are homologous to M. truncatula HAP2.1 and its L. japonicus homologs CBF-A01 and CBF-A22 (Asamizu et al., 2008). To enlarge our investigation to other soybean genes sharing sequence similarity to MtHAP2.1, we BLAST searched the MtHAP2.1 protein sequence against soybean genomic sequences. Based on our search criteria (e-value < 10−12 and score > 100), we identified four soybean homologous genes (Glyma03g36140, Glyma02g35190, Glyma19g38800, and Glyma10g10240). These same four genes were also identified by BLAST searching LjCBF-A01 and LjCBF-A22 protein sequences. To support the putative orthology between MtHAP2.1 and these four soybean genes, we examined the syntenic relationship based on gene content, order, and orientation in the corresponding genomic regions, as previously performed by Zhang et al. (2007). Despite chromosome rearrangements (e.g., inversion of gene orientation), gene content was similar between the different chromosome regions analyzed, supporting that these genes are likely orthologous (Supplemental Fig. S2). While LjCBF-A22 was slightly induced several hours after bacterial inoculation, MtHAP2.1 and LjCBF-A01 expression was strongly induced in nodules (Combier et al., 2006; Asamizu et al., 2008). Among the four soybean genes identified, Glyma10g10240 was strongly and consistently up-regulated during soybean nodulation, while Glyma02g35190 expression was significantly induced during the earlier steps of nodule development (Supplemental Table S4). Glyma03g36140 and Glyma19g38800 were not represented in the current qRT-PCR primer library. However, based on Illumina-Solexa sequencing of soybean cDNA, we confirmed that both genes were preferentially expressed in soybean root tissues including nodules (Supplemental Table S5).
In addition to the identification of the four putative soybean orthologs of MtHAP2.1, we also identified several soybean putative orthologs of MtERN (Glyma16g04410, Glyma16g27040, Glyma19g29000, Glyma02g08020, and Glyma08g12130). Based on this study, the expression of Glyma16g04410 was induced during early nodulation (Supplemental Table S4). An examination of the genomic regions of MtERN and Glyma16g04410 supports the notion that these genes are likely orthologous (Supplemental Fig. S3). A similar approach also identified Glyma19g29000 and Glyma08g12130 as putative MtERN orthologs. However, the genomic regions of Glyma16g27040 and Glyma02g08020 genes showed no microsynteny to the MtERN genome locus (Supplemental Fig. S3). Glyma16g27040, Glyma19g29000, Glyma02g08020, and Glyma08g12130 were not represented in the current qRT-PCR primer library. However, Glyma19g29000 and Glyma02g08020 were strongly expressed in soybean nodules, while Glyma16g27040 and Glyma08g12130 were not (Supplemental Table S5). Such observations reinforced the putative orthologous relationship between MtERN, Glyma16g04410, and Glyma19g29000. Note that even though the genome regions of Glyma08g12130 and MtERN showed significant microsynteny, Glyma08g12130 was not induced during nodulation. This result suggests that the regulation of Glyma08g12130 likely diverged from its progenitor following the duplication that gave rise to this paralog.
The Expression of Soybean Homeologous Regulatory Genes during Nodulation
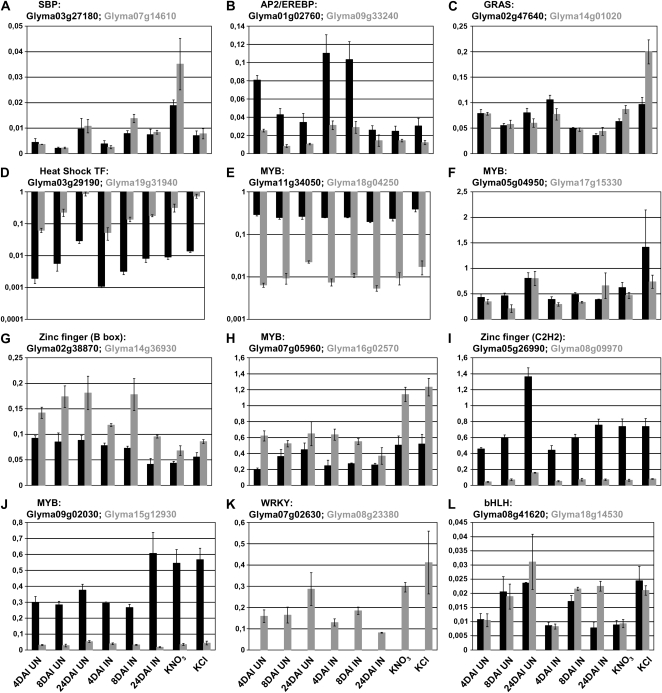
The identification of several putative soybean orthologs of MtHAP2.1 and MtERN is likely the result of the whole genome duplication events that occurred in soybean (Schlueter et al., 2004, 2007). The paralogs that arose from these duplications may show functional redundancy, which, among other issues, could present challenges to efforts to silence specific genes using RNA interference (RNAi) approaches in order to study nodulation phenotypes. A key question is whether these various paralogs show similar expression patterns suggestive of functional redundancy. To address this in a more general way, we identified 46 pairs of soybean homeologous genes (92 genes) represented in the qRT-PCR primer library (M. Libault, A. Farmer, G.D. May, and G. Stacey, unpublished data). Among them, we identified four pairs with both putative regulatory genes regulated during nodulation, eight pairs with one of the two homeologs regulated during nodulation, and finally 34 pairs of putative regulatory genes without any of the two homeologous genes regulated during nodulation or in response to KNO3 treatment. Looking at their expression patterns, the four pairs responding to inoculation shared very similar expression patterns and also similar expression levels, with the exception of Glyma03g29190 and Glyma19g31940 (Fig. 3). With regard to the eight pairs where only one of the two genes was regulated during nodulation (Fig. 3), the data show a significant divergence in the expression profiles of the various paralogs (e.g. little or no expression of Glyma07g02630, Glyma18g04250, Glyma08g09970, and Glyma15g12930, while Glyma08g23380, Glyma11g34050, Glyma05g26990, and Glyma09g02030 were expressed at stronger levels; Fig. 3). In rare cases, the two homeologous genes shared very similar expression patterns, with the exception of one condition leading to the nodulation-specific expression of one of these genes (e.g. Glyma08g41620 and Glyma18g14530; Fig. 3). The remaining 34 pairs, in addition to showing no significant response to inoculation, also exhibited significant divergence of expression under the other conditions tested (Supplemental Fig. S4). These results support the notion that the expression of paralogs can diverge significantly after duplication and suggest that RNAi silencing of single genes in soybean will likely not be hampered to a large degree by the presence of paralogs showing functional redundancy (at least at the level of similar expression).
Figure 3.
Expression of soybean homeologous genes during nodulation and in response to KNO3 and KCl treatments. The expression of each soybean gene (black bars) and its respective homeolog (gray bars) was quantified by qRT-PCR after normalization with the soybean reference gene Cons6 (y axis). Eight different conditions were used to quantify expression (4, 8, and 24 DAI inoculated [IN] and mock inoculated [UN] and in response to KNO3 and KCl treatments; x axis). These 12 pairs of homeologous genes were divided in two subgroups (A–D, both homeologs were significantly regulated during nodulation; E–L, only one of the two homeologs was significantly regulated during nodulation; for details, see Supplemental Table S2).
Putative Soybean Regulatory Genes Have Temporally Defined Expression Patterns
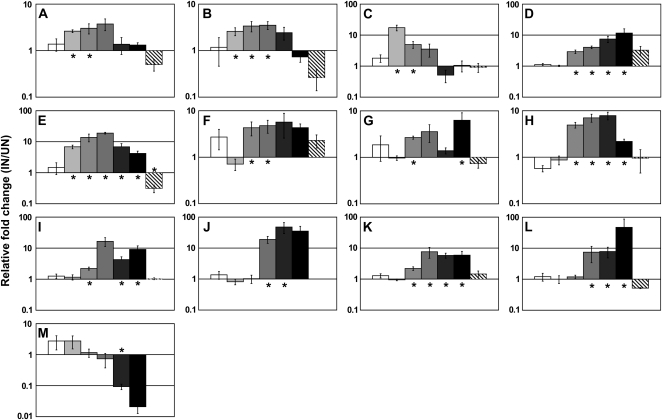
In order to examine in detail the response of a select number of putative regulatory genes during nodulation, the expression patterns of the eight genes found to be differentially expressed at multiple time points were enlarged by including additional time points (0, 16, and 32 DAI; for details, see Supplemental Table S6). In addition, the expression patterns of five additional genes (Glyma18g49360, Glyma19g34380, Glyma03g27250, Glyma03g31980, and Glyma06g08610) were also further analyzed. The detailed time-course analysis grouped these 13 putative regulatory genes into three classes based on their expression patterns (Fig. 4). Three genes were up-regulated only during the early stages of nodulation (Glyma16g04410, Glyma02g35190, and Glyma12g34510; Fig. 4, A–C), while six were up-regulated throughout all stages of nodulation (Glyma16g26290, Glyma10g10240, Glyma03g31980, Glyma06g08610, Glyma13g40240, and Glyma01g01210; Fig. 4, D–I). A third category of genes were regulated specifically during the later stages of nodulation (Glyma18g49360, Glyma17g07330, Glyma19g34380, and Glyma03g27250; Fig. 4, J–M). These three distinctive groups of genes clearly showed a temporally defined expression pattern that may reflect a specific developmental role for each gene. Based on these expression patterns, we hypothesized that the early induced putative regulatory genes might be involved in bacterial infection or in controlling the first steps of nodule development (e.g. cortical cell division). The late induced putative regulatory genes or those induced throughout nodule development might be involved in controlling the later steps of nodule development (e.g. cell elongation, bacteroid development, nitrogen metabolism, or control of nodule growth).
Figure 4.
Regulation of the expression of putative soybean regulatory genes during nodule development. The fold changes between the expression levels of inoculated and mock roots through different stages of nodule development (0 [white bars], 4 [light gray bars], 8 [gray bars], 16 [dark gray bars], 24 [darkest gray bars], and 32 [black bars] d after B. japonicum inoculation) and in response to KNO3 treatment (striped bars) were investigated for 13 different putative soybean regulatory genes. A, Glyma16g04410 (AP2/EREBP). B, Glyma02g35190 (CCAAT box). C, Glyma12g34510 (CCAAT box). D, Glyma16g26290 (bHLH). E, Glyma10g10240 (putative TF). F, Glyma03g31980 (CND; Myb). G, Glyma06g08610 (DNA methyltransferase). H, Glyma13g40240 (zinc finger). I, Glyma01g01210 (RNA-dependent RNA polymerase). J, Glyma18g49360 (Myb). K, Glyma17g07330 (Myb). L, Glyma19g34380 (Aux/IAA). M, Glyma03g27250 (zinc finger; GATA). Error bars show se. Asterisks indicate Student's t test results (P < 0.05) when comparing inoculated (IN) versus uninoculated (UN) or KNO3 versus KCl roots.
Investigation of Myb Gene Expression Patterns Using Promoter-Reporter Gene Fusions
Our basic strategy is to identify TF genes important to soybean nodulation by examining their differential expression in response to inoculation. The identification of putative MtHAP2.1 and MtERN soybean orthologous genes (Glyma02g35190, Glyma16g04410, and Glyma10g10240) induced during nodulation provides some validation to this approach. Among the different genes regulated, Glyma03g31980 and Glyma17g07330, two Myb TF genes, were found to be regulated at several time points of the time course (Fig. 4, F and K; Supplemental Table S6).
Based on further characterization (see below), Glyma03g31980 was named CND. The expression profiles of CND and Glyma17g07330 during nodulation were similar. In a previous study, Koltai et al. (2001) reported the specific expression of the M. truncatula PHANTASTICA gene encoding a Myb TF in nodulated roots. Comparison of the M. truncatula PHANTASTICA protein sequence with that of the soybean CND and Glyma17g07330 MYB proteins using EMBOSS Pairwise Alignment Algorithms (http://www.ebi.ac.uk/Tools/emboss/align/) showed low degrees of identity and similarity (23.3% and 33.7% with CND and 20.3% and 34% with Glyma17g07330, respectively). In addition, no microsynteny exists between these three genes (data not shown). Therefore, to better characterize the role of these two TF genes, we investigated their expression patterns in a variety of soybean tissues. Interestingly, despite similar expression during nodule development, CND and Glyma17g07330 expression showed tissue-specific differences. Whereas CND was mainly expressed in root hair cells and the shoot apical meristem, the expression of Glyma17g07330 was very low in most of the tissues tested, with the exception of stripped root (i.e. root devoid of root hair cells) and root tissues (Supplemental Fig. S5).
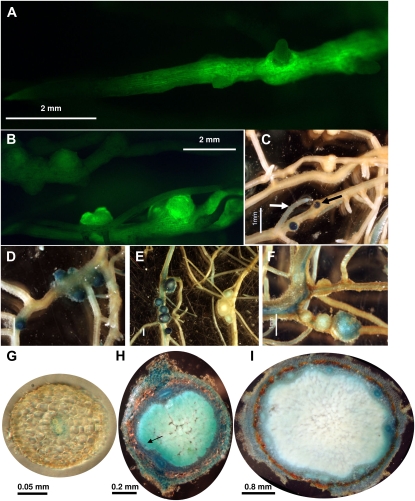
To further examine the expression of CND during nodule development, promoter-GUS and -GFP fusions were constructed and introduced into soybean roots by Agrobacterium rhizogenes hairy root transformation. A 2-kb region 5′ of the CND gene was identified from the Phytozome Web site and cloned upstream of the GFP and GUS reporter genes to construct transcriptional fusions. Early during nodule development (13 DAI), transformed roots showed CND promoter-GFP expression in emerging nodules and, at a lower level, in root cells (Fig. 5, A–D). Although CND expression was apparent throughout the root, GFP signal was noticeably absent in emergent secondary root tips (Fig. 5, A and B). This result was also supported by the analysis of roots expressing the CND promoter-GUS construct, although in rare occasions secondary root tips did show some GUS staining (Fig. 5C, white arrow). CND expression was induced very early during nodule primordium formation, at the location of the infected zone (Fig. 5, C and H, black arrows). Later during nodule development (28 DAI and later), CND expression decreased in the infection zone of the nodule (Fig. 5, E, F, and I). Looking at transverse sections of GUS-stained transgenic roots and developing and mature nodules, CND was strongly expressed in the vascular tissues of all organs (Fig. 5, G–I). At a finer level, GUS staining was also observed in epidermal, endodermal, and cortical cells. These observations support a change in the expression pattern of CND during nodule development, especially in the infected zone of the nodule.
Figure 5.
Localization of GmCND promoter-GFP and promoter-GUS expression in inoculated soybean roots by B. japonicum. A and B, Expression pattern of the GmCND promoter-GFP 13 d after B. japonicum inoculation. C to F, Expression pattern of the GmCND promoter-GUS 13 (C and D), 28 (E), and 39 (F) d following B. japonicum inoculation. G to I, Expression pattern of the GmCND promoter-GUS in root sections (G) and 18 and 40 d after B. japonicum inoculation in nodules (H and I, respectively). The white arrow in C highlights a GUS-stained secondary root tip. Black arrows in C and H highlight the expression of CND in the infected zone of the nodule. Bars = 2 mm (A and B), 1 mm (C–F), 0.05 mm (G), 0.2 mm (H), and 0.8 mm (I).
Characterization of the Role of CND during Nodule Development
RNAi silencing is a common strategy to characterize gene function when mutants are not available. Several studies focusing on legume nodulation have successfully used this strategy to investigate gene function. For example, silencing of MtDMI2 led to 75% of roots with no nodules compared with 0% to 20% of control roots with no nodulation (Limpens et al., 2003). Using a similar RNAi strategy, Vernié et al. (2008) showed an increase in nodule formation and infection threads after rhizobial inoculation coincident with the silencing of MtEFD, an MtERF TF gene regulated during M. truncatula nodulation (El Yahyaoui et al., 2004). However, the authors concluded that silencing of MtEFD created a weak allele relative to the efd null mutant. In addition, the silencing of MtCDPK1 led to smaller roots and decreases in nodule formation and mycorrhizal infection (Ivashuta et al., 2005).
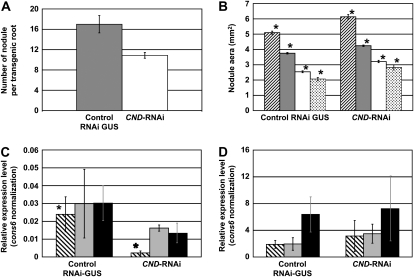
Because the transcriptional response of a gene to a specific treatment does not necessarily imply an essential role in this process, we further investigated the role of CND in nodule formation. To specifically silence CND, a DNA fragment corresponding to the predicted 3′ untranslated region (UTR) was cloned to create a hairpin structure (see “Materials and Methods”). As a control, pCGT5200 carrying an RNAi construct specific to the GUS gene was used. Three independent experiments were performed allowing the counting of nodule numbers on 44 CND-RNAi and 36 control RNAi-GUS roots. The silencing of CND consistently affected nodule development by significantly reducing nodule numbers by approximately 40% compared with the RNAi-GUS control roots (Student's t test, P ≤ 0.03; Fig. 6A). There was also a slight increase in nodule size that coincided with the drop in nodule numbers in the CND-silenced roots (i.e. nodule size increased by around 20% in CND-RNAi roots when compared with the control (Student's t test, P < 0.001; Fig. 6B). A relationship between nodule size and nodule number was previously investigated in the supernodulating soybean mutant nts382 and its wild-type parent (soybean cv Bragg; Day et al., 1987). In this case, more nodules in the mutant roots resulted in a smaller average nodule size. Similarly, the hypernodulation mutant Ljtml also exhibited smaller nodules (Magori et al., 2009). In keeping with this relationship, the increase in nodule size in the CND-RNAi-silenced roots is likely an indirect effect of the total decrease in nodule numbers.
Figure 6.
Silencing of the CND gene affects soybean nodulation. A, Comparison of nodule number between RNAi-GUS (gray bar) and CND-RNAi (white bar) soybean roots. Error bars show se. P < 0.04. B, Comparison of nodule size between RNAi-GUS (left bars) and CND-RNAi (right bars) roots. According to their size, nodules were divided in four categories: large (striped bars), medium (gray bars) and small nodules with leghemoglobin (white bars) and immature nodules (i.e. lacking leghemoglobin; dotted bars). Asterisks indicate Student's t test results (P < 0.05) when comparing control and CND-RNAi for each nodule category. C, Gene expression analysis of CND in RNAi-GUS (left bars) and CND-RNAi (right bars) nodules. Transcriptomic analysis was performed on small, medium, and large nodules (striped, gray, and black bars, respectively). Gene expression was normalized against the expression of the constitutive Cons6 gene. Asterisks indicate Student's t test results (P < 0.05) when comparing control and CND-RNAi. D, Confirmation of the specificity of RNAi silencing of CND. Gene expression levels of Glyma19g34740, which shares strong nucleotide sequence homology with CND, were quantified by qRT-PCR on RNAi-GUS (left bars) and CND-RNAi (right bars) nodules: small, medium, and large nodules (striped, gray, and black bars, respectively).
Based on the expression of CND in the infected zone of young nodules, we investigated more closely the infection of CND-RNAi nodules by B. japonicum. CND-RNAi and RNAi-GUS nodules were stained with SYTO-13, a nucleic acid-binding dye (Veereshlingam et al., 2004). By laser-scanning confocal microscopy, we did not observe a significant modification in the pattern of infected cells coincident to the silencing of the CND gene (Supplemental Fig. S6). Both the CND-RNAi nodules and the control nodules exhibited a clearly distinguishable infected zone.
To confirm the silencing of CND, we quantified gene expression in RNAi-GUS control and CND-RNAi isolated nodules using qRT-PCR (i.e. for each of the three biological replicates of CND-RNAi and RNAi-GUS roots, small, medium, and large nodules were pooled and RNA was extracted). These experiments confirmed the silencing of CND at all stages of nodule development (Fig. 6C). We assume that the fact that nodule numbers were only reduced by 40% is likely due to the fact that 100% silencing of CND gene expression was not achieved. We also cannot rule out the possibility of functionally redundant TF genes that were not silenced by the CND-RNAi construct.
Although primers were designed to specifically silence CND expression, the similarity of sequences between the members of the Myb TF family may have resulted in silencing of more than one gene. To test this hypothesis, the CND nucleotide sequence was BLAST searched against the available soybean genomic sequence (http://www.phytozome.net/soybean.php). The closest homolog was Glyma19g34740 (score = 379, e-value = 6.2e−103). Based on the strong identity between CND and Glyma19g34740 (83% and 87% identity of their nucleotide and amino acid sequences, respectively), we hypothesized that Glyma19g34740 might be homeologous to CND and that the function of these two Myb genes might be redundant. To better establish the relationship between these two genes, we mined soybean genome sequences to identify genes surrounding both Myb genes. Not surprisingly, the environment of these two genes in terms of gene content, order, and orientation was very similar, supporting their homeology (Supplemental Fig. S7). The strong nucleotide sequence identity between CND and Glyma19g34740 suggested that both genes might be silenced during our experiments. Accordingly, we also quantified the expression levels of Glyma19g34740 in the CND-RNAi and RNAi-GUS control nodules. The data showed that our construct specifically silenced the expression of the CND Myb gene (Fig. 6D). In addition, these analyses also demonstrated the strong expression of Glyma19g34740 in nodules, supporting a possible partial redundancy in the function of these two Myb genes. This result might be a second factor explaining the mild phenotype of the CND-RNAi plants.
CONCLUSION
The development and use of our soybean qRT-PCR library allowed quantification of the expression of 1,034 soybean putative regulatory genes. By focusing on nodule development, we validated the use of this approach by identifying putative soybean orthologs of MtHAP2.1 and MtERN as regulated during nodulation. In addition, using RNAi silencing, we identified a Myb TF gene, CND, involved in nodule formation. In addition to PHANTASTICA, our data suggest that probably several Myb genes including CND might be involved in nodule development.
MATERIALS AND METHODS
Bacterial Culture
DH5α Escherichia coli and K599 Agrobacterium rhizogenes were grown in Luria-Bertani medium supplemented with appropriate antibiotics at 37°C and 30°C, respectively. Bradyrhizobium japonicum USDA110 was grown at 30°C for 3 d in HM medium (Cole and Elkan, 1973) supplemented with yeast extract (0.025%), d-Ara (0.1%), and chloramphenicol (0.004%). Before plant inoculation, B. japonicum cells were pelleted (4,000 rpm for 10 min), washed, and diluted with sterile water to the appropriate concentration according to the experiments (i.e. optical density at 600 nm [OD600] = 0.1 or 0.3 when inoculated with wild-type soybean [Glycine max] seedlings and hairy roots, respectively).
Cloning
To silence the CND Myb gene, a 120-bp fragment specific to the 3′ UTR of GmCND was amplified from soybean cDNA using the following primers: GmMyb-RNAifor (5′-TCGAGTAACAGTCGTAATGGACA-3′) and GmMyb-RNAirev (5′-GACCAAGTCCTTCATTCAACG-3′). The amplified PCR fragment was cloned into the entry vector CGT11050 by TA cloning using two AhdI sites (engineered to produce 3′ T overhangs). The RNAi entry vector was recombined with the destination vector CGT11017A (Supplemental Fig. S8) using the LR Clonase reaction (Gateway LR Clonase II enzyme mix; Invitrogen). The LR Clonase reaction combining the gene fragment in the entry vector and destination vector CGT11017A was used to produce GmCND-RNAi vector. The RNAi control vector used in these studies (CGT5200) was previously described by Govindarajulu et al. (2009). The fidelity of the clones was verified by sequencing, and the clones were electroporated into A. rhizogenes strain K599 and used for composite plant production (see below).
The Gateway system (Invitrogen) was used to clone GmMyb promoter upstream to the GFP and GUS cDNAs. By mining genomic sequences available on the Phytozome Web site (http://www.phytozome.net/soybean.php), we identified a 1,967-bp DNA fragment 5′ to the first codon of the CND gene. By two independent PCRs, we created the AttB sites at the extremities of the promoter sequences. We used soybean cv Williams 82 genomic DNA as template and the following primers: GmMybpromoAttB-for (5′-AAAAAGCAGGCTCCTACATGAATATGTGTTCAAAATA-3′) and GmMybpromoAttB-rev (5′-AGAAAGCTGGGTTTTGATGACTTAGACTACTCCTTC) first and the universal AttB primers attB1adaptor (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and attB2adaptor (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) second.
Using the Gateway BP Clonase II enzyme mix, the GmMyb promoter fragment was introduced first into the pDONR-Zeo vector (Invitrogen) and then into pYXT1 or pYXT2 destination vector using the Gateway LR Clonase II enzyme mix (Invitrogen). The pYXT1 and pYXT2 destination vectors carry the GUS and GFP reporter genes, respectively (Xiao et al., 2005).
A. rhizogenes (strain K599) was transformed by electroporation with pCGT11017-3′UTR-GmMyb, GmMybpromoter-pYXT1, and GmMybpromoter-pYXT2 vectors.
Plant Culture
Soybean seeds were surface sterilized according to Wan et al. (2005) and then placed on moist filter paper for germination in dark conditions, 80% humidity, and 27°C. Three-day-old seedlings were grown on moist filter paper and transferred into vermiculite-perlite (3:1) supplied with B&D liquid medium (Broughton and Dilworth, 1971), while nodulated or nodule-free roots produced to quantify gene expression during nodule development were watered with B&D medium not supplemented. Seedlings were inoculated with 1 mL of B. japonicum suspension in water (OD600 = 0.1) or only water (mock). B&D medium was supplemented with KNO3 or KCl (10 mm final concentration) when analyzing putative regulatory gene repression in response to nitrogen starvation. Nitrogen (KNO3) and nitrogen-free (KCl) roots, nodulated or nodule free, were harvested 24 d after treatment and 0, 4, 8, 16, 24, and 32 DAI. In all cases, root apices (around 1 cm) were removed prior to freezing in liquid nitrogen. Two to six plants were harvested by biological replicates. These experiments were repeated at least three times on different set of plants to ensure reproducibility in the plant tissues analyzed.
Plant Transformation and Nodule Observation
Soybean hairy root transformation was done essentially as described by Taylor et al. (2006). Two-week-old soybean shoots were cut between the first true leaves and the first trifoliate leaf and placed into rock-wall cubes (Fibrgro). Each shoot was inoculated with 4 mL of A. rhizogenes (OD600 = 0.3) and then allowed to dry for approximately 3 d (23°C, 50% humidity, long-day conditions) before watering with deionized water. After 1 week, the plants were then transferred to pots with vermiculite:perlite mix (3:1) wetted with nitrogen-free plant nutrient solution (Lullien et al., 1987). One week later, the shoots were transferred to the greenhouse (27°C, 20% humidity, long-day conditions). Two weeks after vermiculite-perlite transfer, the shoots were inoculated with B. japonicum (10 mL, OD600 = 0.08).
CND-RNAi and control nodulated transgenic roots were isolated (these roots expressed the GFP reporter gene), and the nodules were isolated and then categorized based on the presence of leghemoglobin (i.e. a pink-red color in the infected zone of the nodule was used as a marker of nodule maturity; leghemoglobin gene expression increases proportionally with the age of the nodule [Marcker et al., 1984], and its induction occurs before the detection of nitrogenase activity [Verma et al., 1979]) and nodule size (i.e. leghemoglobin-free nodules and small [<3.5 mm2], medium [≥3.5 mm2 and <4.5 mm2], and large [≥4.5 mm2] mature nodules with leghemoglobin).
For each category, nodule numbers were counted with a Leica MZFLIII stereomicroscope equipped with epifluorescence excitation and a GFP long-pass filter (500 nm long pass; no. 41018; Chroma Technology). Molecular Devices MetaMorph 7.5 was used to measure the area of each nodule (the areas of 73 large, 182 medium, 109 small mature, and 33 immature nodules from RNAi-GUS and 54 large, 178 medium, 88 small mature, and 32 immature nodules from CND-RNAi were measured).
To observe B. japonicum infection in transgenic nodules, CND-RNAi and RNAi-GUS nodules were embedded in paraffin before sectioning. Five-micrometer sections were stained during 20 min with 10 μm SYTO13 in 1 m sodium phosphate buffer (pH 7.0). Stained sections were observed with a Zeiss LSM 510 META confocal microscope (488 nm excitation wavelength and 500- to 550-nm emission filter).
GmMyb promoter-GUS and -GFP transgenic roots were observed 13, 18, 29, and 40 DAI. GmMyb promoter-GUS roots were fixed and stained as described by Govindarajulu et al. (2008). To section roots and nodules stained with GUS, 80-μm-thick sections were obtained using a fully automated Vibratome 3000 Plus after embedding the tissues in 6% low-melting agarose. Roots with GUS-stained nodules were observed using a SMZ1500 Nikon microscope equipped with a DXM1200 Nikon digital camera, while GmFWL1promoter-GFP roots were observed using a Leica MZFLIII stereomicroscope equipped with epifluorescence excitation and a GFP long-pass filter (500 nm long pass; no. 41018; Chroma Technology).
Illumina-Solexa Sample Preparation, Read Alignment, and Statistical Analysis
mRNAs isolated from different soybean tissues were reverse transcribed using standard protocols. After first- and second-strand cDNA synthesis, the cDNAs were end repaired prior to ligation of Solexa adaptors. The products were sequenced on an Illumina-Solexa platform.
Illumina Genome Analyzer II image data were base called and quality filtered using the default filtering parameters of the Illumina GA Pipeline GERALD stage. Alignments of passing 36-mer reads to all contigs of the Glyma1 8× Soybean Genome assembly (Soybean Genome Project, Department of Energy Joint Genome Institute) were performed using GSNAP, an alignment program derived from GMAP (Wu and Watanabe, 2005) with optimizations for aligning short transcript reads from next-generation sequencers to genomic reference sequences. Alignments were processed using the Alpheus pipeline (Miller et al., 2008), keeping only alignments that had at least 34 out of 36 identities and had no more than five equivalent best hits. Read counts used in expression analyses were based on the subset of uniquely aligned reads that also overlapped the genomic spans of the Glyma1 gene predictions.
qRT-PCR Primer Design
To ensure maximum specificity of the designed primers, we used PRIMEGENS (http://digbio.missouri.edu/primegens/; Xu et al., 2002). This software compares the sequence of each primer designed against a database of the 2,258 putative soybean regulatory genes identified in this study as well as the G. max Unigene database to ensure that the designed primers are unique to the sequence and are likely to give specific amplification. The following criteria were used to design the primers: melting temperature of 60°C, PCR amplicon lengths of 80 to 120 bp, and yielding primer sequences with lengths of 19 to 23 nucleotides with an optimum at 21 nucleotides and GC contents of 40% to 60%. The list of the validated primers described in this study is available in Supplemental Table S2.
To quantify specifically the expression of the Glyma19g34740 Myb gene, Glyma19g34740-for (5′-CCAAGACGCCAAACTAGAACA-3′) and Glyma19g34740-rev (5′-TCGCTAGAACAATGAGGAGGA-3′) primers were designed. Primer specificity was confirmed by dissociation curve profile after qRT-PCR amplification.
RNA Extraction, DNase Treatments, and RT
Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instructions followed by chloroform extraction to improve their purity. Purified RNA was treated with TURBO DNase (Ambion) to remove any contaminating genomic DNA according to the manufacturer's instructions. In to the experiments, 100 μg of DNA-free total RNA was used for first-strand cDNA synthesis using Moloney murine leukemia virus (Promega) according to the manufacturer's instructions. The lack of genomic DNA contamination was verified by qRT-PCR using primers designed against soybean genomic DNA. Moreover, cDNA synthesis efficiency was analyzed by amplifying by qRT-PCR Cons6 cDNA with two different primer sets separated by 1 kb (Cons6-3′ forward, 5′-AGATAGGGAAATGGTGCAGGT-3′, and reverse, 5′-CTAATGGCAATTGCAGCTCTC-3′; Cons6-5′ forward, 5′-AAAGGTGAAATTGCCTCTTCC-3′, and reverse, 5′-CCCAAAGATCTGCCAAATGTA-3′). For each condition and replicate tested, less than one PCR cycle difference was measured between both primer sets, supporting the high quality of the cDNA synthesis.
qRT-PCR Conditions and Data Analysis
qRT-PCR was performed as described by Libault et al. (2008). Primer set specificity was confirmed by analyzing the dissociation curve profile of each qRT-PCR amplicon, and the efficiency of primers (Peff) was quantified using LinRegPCR (Ramakers et al., 2003). Cons6, encoding an F-box protein (Libault et al., 2008), was used to normalize the expression levels of putative soybean regulatory genes. The cycle threshold (Ct) value of the reference gene was subtracted from the Ct values of the test gene analyzed (ΔCt). The expression level (E) of each gene was calculated according to the equation E = Peff(−ΔCt). The fold change of the expression level between inoculated versus uninoculated and KNO3 versus KCl was calculated at different time points for the three different biological replicates. The average of the fold changes between three different replicates was calculated at each time point and statistically validated using Student's t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of the specificity and efficiency of the qRT-PCR primers designed to quantify putative soybean regulatory genes.
Supplemental Figure S2. Syntenic relationship between putative soybean orthologous genes to MtHAP2-1 and MtHAP2-1.
Supplemental Figure S3. Syntenic relationship between putative soybean orthologous genes to MtERN and MtERN.
Supplemental Figure S4. Expression of soybean homeologous genes during nodulation and in response to KNO3 and KCl treatments.
Supplemental Figure S5. Expression patterns of Glyma03g31980 (CND) and Glyma17g07330 in 10 soybean tissues.
Supplemental Figure S6. Confocal light micrographs of CND-RNAi nodule (left) and RNAi-GUS (right) sections.
Supplemental Figure S7. Syntenic relationship between Glyma03g31980 and Glyma19g34740.
Supplemental Figure S8. Construction of the RNAi vector CGT11017A.
Supplemental Table S1. Identification of putative soybean regulatory genes from public cDNA databases.
Supplemental Table S2. Gene expression analysis for the 1,149 putative regulatory genes during nodulation and in response to KNO3.
Supplemental Table S3. Quantification of primer set efficiency in a large number of independent qRT-PCRs.
Supplemental Table S4. Expression of 162 soybean putative regulatory genes during nodulation and in response to KNO3 and KCl treatments.
Supplemental Table S5. Expression of putative soybean orthologs of MtHAP2.1 and MtERN genes in different soybean tissues.
Supplemental Table S6. Time course of expression of the putative soybean regulatory genes during nodulation (i.e. 0–32 d after B. japonicum inoculation) or after the KNO3 treatment.
Supplementary Material
Acknowledgments
We thank Sandra Thibivilliers, Jeff Anderson, and Aaron J. Witte for critical reading of the manuscript. We also thank Yong-Li Xiao and Christopher Town for providing the pYXT1 and pYXT2 binary vectors, Christopher Taylor for providing CGT11050 and CGT11017 vectors, and Andrew Farmer and Gregory May for their help in the Illumina-Solexa sequencing analysis.
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. DBI–0421620).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gary Stacey (staceyg@missouri.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34 495–506 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Shimoda Y, Kouchi H, Tabata S, Sato S (2008) A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol 147 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55 504–513 [DOI] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54 876–887 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, et al (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact 21 631–645 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MA, Elkan GH (1973) Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother 4 248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Day DA, Price GD, Schuller KA, Gresshoff PM (1987) Nodule physiology of a supernodulating soybean (Glycine max) mutant. Aust J Plant Physiol 14 527–538 [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Elmore JM, Fester T, Taylor CG (2008) Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol Plant Microbe Interact 21 1027–1035 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Kim SY, Libault M, Berg RH, Tanaka K, Stacey G, Taylor CG (2009) GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol 149 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber V, Blanchet S, Diet A, Zahaf O, Boualem A, Kakar K, Alunni B, Udvardi M, Frugier F, Crespi M (2009) Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol Genet Genomics 281 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, VandenBosch KA, Vance CP, Harrison MJ, Gantt JS (2005) RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17 2911–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, et al (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Koltai H, Dhandaydham M, Opperman C, Thomas J, Bird D (2001) Overlapping plant signal transduction pathways induced by a parasitic nematode and a rhizobial endosymbiont. Mol Plant Microbe Interact 14 1168–1177 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1 44–54 [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20 900–911 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9 469–478 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M (2009) Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact 22 259–268 [DOI] [PubMed] [Google Scholar]
- Marcker A, Lund M, Jensen EO, Marcker KA (1984) Transcription of the soybean leghemoglobin genes during nodule development. EMBO J 3 1691–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, Kingsmore SF, Farmer AD, Langley RJ, Mudge J, Crow JA, Gonzalez AJ, Schilkey FD, Kim RJ, van Velkinburgh J, et al (2008) Management of high-throughput DNA sequencing projects: Alpheus. J Comp Sci Sys Biol 1 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30 85–112 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Kawaguchi M (2002) The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43 853–859 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Freixes S, Balzergue S, Tillard P, Jeudy C, Martin-Magniette ML, van der Merwe MJ, Kakar K, Gouzy J, Fernie AR, et al (2008) Systemic signaling of the plant nitrogen status triggers specific transcriptome responses depending on the nitrogen source in Medicago truncatula. Plant Physiol 146 2020–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47 868–876 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Lin JY, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O'Bleness M, Roe BA, Nelson RT, Scheffler BE, et al (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Taylor CG, Fuchs B, Collier R, Lutke WK (2006) Generation of composite plants using Agrobacterium rhizogenes. Methods Mol Biol 343 155–167 [DOI] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J, Kakar K, Gallardo K, Le Signor C, Aubert G, Schlereth A, Town CD, Udvardi MK, Thompson RD (2008) Gene expression profiling of M. truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol Biol 67 567–580 [DOI] [PubMed] [Google Scholar]
- Verma DPS, Ball S, Guerin C, Wanamaker L (1979) Leghemoglobin biosynthesis in soybean root nodules: characterization of the nascent and released peptides and the relative rate of synthesis of the major leghemoglobins. Biochemistry 18 476–483 [DOI] [PubMed] [Google Scholar]
- Vernié T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P (2008) EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Torres M, Ganapathy A, Thelen J, DaGue BB, Mooney B, Xu D, Stacey G (2005) Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol Plant Microbe Interact 18 458–467 [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21 1859–1875 [DOI] [PubMed] [Google Scholar]
- Xiao YL, Smith SR, Ishmael N, Redman JC, Kumar N, Monaghan EL, Ayele M, Haas BJ, Wu HC, Town CD (2005) Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol 139 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li G, Wu L, Zhou J, Xu Y (2002) PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18 1432–1437 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.