Abstract
Vitamin A deficiency, a global health burden, can be alleviated through provitamin A carotenoid biofortification of major crop staples such as maize (Zea mays) and other grasses in the Poaceae. If regulation of carotenoid biosynthesis was better understood, enhancement could be controlled by limiting β-carotene hydroxylation to compounds with lower or no nonprovitamin A activity. Natural maize genetic diversity enabled identification of hydroxylation genes associated with reduced endosperm provitamin A content. A novel approach was used to capture the genetic and biochemical diversity of a large germplasm collection, representing 80% of maize genetic diversity, without having to sample the entire collection. Metabolite data sorting was applied to select a 10-line genetically diverse subset representing biochemical extremes for maize kernel carotenoids. Transcript profiling led to discovery of the Hydroxylase3 locus that coincidently mapped to a carotene quantitative trait locus, thereby prompting investigation of allelic variation in a broader collection. Three natural alleles in 51 maize lines explained 78% of variation and approximately 11-fold difference in β-carotene relative to β-cryptoxanthin and 36% of the variation and 4-fold difference in absolute levels of β-carotene. A simple PCR assay to track and identify Hydroxylase3 alleles will be valuable for predicting nutritional content in genetically diverse cultivars found worldwide.
Vitamin A deficiency is a global health problem affecting 140 to 250 million children and accounts for increased childhood mortality and disease (World Health Organization, 1995; Underwood, 2004; Black et al., 2008). Humans and animals are unable to synthesize their own vitamin A and rely on dietary provitamin A carotenoid pigments; plant-derived carotenes are metabolized to produce vitamin A, two molecules from β-carotene and only one from α-carotene or β-cryptoxanthin. Seed endosperm tissue of maize (Zea mays) and other grasses (Poaceae) represents 70% of worldwide food production (Chandler and Brendel, 2002), but has limited provitamin A value. Improvement of provitamin A content in staple crops is therefore a critical step toward alleviating vitamin A deficiency worldwide. Maize, a major food staple in vitamin A-deficient Sub-Saharan Africa and Latin America, exhibits a wide range of carotenoid content and composition. The underlying basis for this variation has been the subject of quantitative trait locus (QTL) and association studies (Palaisa et al., 2003; Wong et al., 2004; Chander et al., 2007; Harjes et al., 2008). Given the genetic and biochemical diversity of maize, opportunity exists for developing breeding markers, based on candidate genes of the carotenoid biosynthetic pathway, to select for predicted nutritional content.
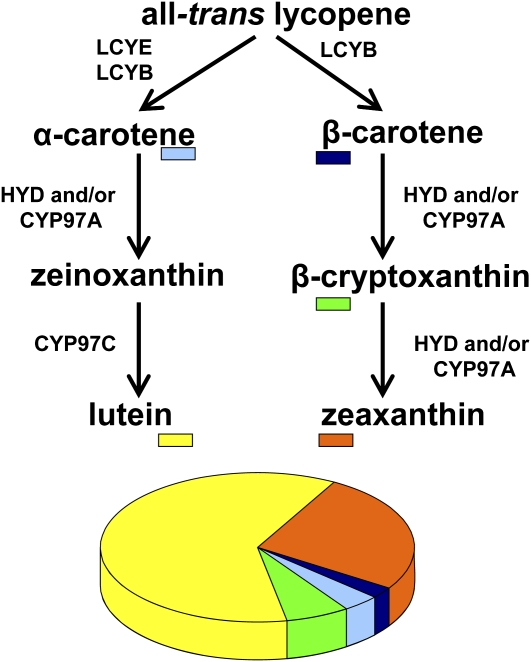
The carotenes are intermediates in the (recently revised) carotenoid biosynthetic pathway (Li et al., 2007; Matthews and Wurtzel, 2007), leading to the predominant nonprovitamin A xanthophylls in maize kernels (Fig. 1), which is why maize kernels generally have limited provitamin A content despite being rich in carotenoids (unlike rice [Oryza sativa], which does not naturally accumulate endosperm carotenoids; Islam, 2004). Lycopene, the immediate precursor of provitamin A carotenes, represents a branch point in the carotenoid biosynthetic pathway. The linear lycopene molecule is further modified by lycopene β-cyclase and lycopene ε-cyclase (LCYE) enzymes that catalyze formation of terminal β-ionone rings and ε-rings, respectively. Depending on relative activity levels of the cyclases, the products are β-carotene with two β-ionone rings, the most potent dietary source of vitamin A, and α-carotene, with only one β-ionone ring and therefore half the provitamin A potential compared to β-carotene; the hydroxylation intermediate β-cryptoxanthin, with only one β-ionone ring, has half the provitamin A activity compared to β-carotene. The pathway continues with hydroxylation of the carotenes that depletes the provitamin A pool by converting provitamin A compounds to nonprovitamin A xanthophylls (Matthews and Wurtzel, 2007). Therefore, pathway branching and hydroxylation are key determinants in controlling provitamin A levels. Both of these aspects have been targets for metabolic engineering of endosperm carotenoids in other species (Diretto et al., 2006, 2007). However, transgenic solutions to manipulate the carotenoid biosynthetic pathway, as achieved in Golden Rice (Ye et al., 2000) and other plants (Giuliano et al., 2008), are not always acceptable as observed by the overwhelming public resistance to genetically modified organism food crops. Moreover, transgene incorporation into one genetic variety used for laboratory transformation produces variable carotenoid phenotypes (Aluru et al., 2008), and the desired transgene-produced trait is not predictably transferred to other genetic backgrounds of cultivars worldwide. This lack of predictability could be overcome given a better understanding of the rate-controlling steps and tools to genotype and/or phenotype varieties for predicting the outcome of transgene introduction as we report here. Given the extensive natural diversity inherent in maize, it is conceivable to predictably breed high provitamin A maize in a wide range of genotypes given a thorough understanding of pathway bottlenecks and development of corresponding breeding alleles.
Figure 1.
Conversion of provitamin A carotenes to nonprovitamin xanthophylls. Relative abundance of mature kernel carotenoids in maize inbred B73. LCYB, Lycopene β-cyclase; HYD, β-carotene hydroxylase (diiron); CYP97A, β-carotene hydroxylase (P450); CYP97C, ε-carotene hydroxylase (P450). [See online article for color version of this figure.]
Our collaborative effort in the recent development of LCYE-based breeding markers for maize demonstrated feasibility of a nontransgenic, traditional breeding approach to control the pathway branching step and force pathway flux toward β-carotene and its nonprovitamin A derivatives (the β-branch; Harjes et al., 2008). However, unless hydroxylation is also controlled, nonprovitamin A xanthophyll compounds will predominate. Given that lycopene cyclization can be forced toward the β-branch, the next challenge in breeding high β-carotene in maize endosperm is to block β-carotene hydroxylation to increase levels of β-carotene relative to β-cryptoxanthin and downstream xanthophylls. Therefore, we embarked on a study capitalizing on maize germplasm resources to characterize the maize β-carotene hydroxylase genes and to discover breeding markers for enhancing the relative levels of seed β-carotene.
RESULTS AND DISCUSSION
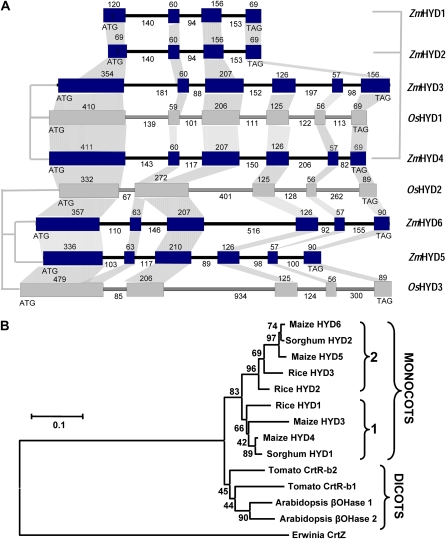
Two structurally distinct classes of carotene hydroxylases are known: the P450 heme-thiolate CYP97A and CYP97C enzymes and the nonheme diiron monoxygenases (for review, see Matthews and Wurtzel, 2007; Giuliano et al., 2008). As found in rice (Quinlan et al., 2007), maize contained one gene each for CYP97A and CYP97C, respectively. A total of six unlinked maize paralogs encoding nonheme diiron β-carotene hydroxylases (HYD) were identified in contrast to three rice genes (Fig. 2; Supplemental Tables S1 and S2); phylogenetic analysis suggested that the gene duplications found in the grasses occurred after the monocot and dicot evolutionary split. Maize HYD1 and HYD2 are pseudogenes, while HYD3, HYD4, HYD5, and HYD6 encode enzymes with characteristic hydroxylase domains and plastid-targeting signals (Sun et al., 1996; Supplemental Fig. S1). HYD3 and HYD4 are syntenous with rice HYD1 (Fig. 3) and are predicted to encode proteins with markedly different pIs, but functional testing confirmed that both encode carotene β-ring hydroxylases (Supplemental Fig. S2). The CYP97A and CYP97C genes have been functionally tested in rice (Quinlan et al., 2007); phylogenetic analysis of this ancient evolutionary clade would suggest that the maize encoded enzymes behave similarly.
Figure 2.
HYD gene family structure and phylogeny. A, Gene organizational structure showing paralogs and orthologs of the HYD gene family in maize (Zm) and rice (Os) species representing two subfamilies of the Poaceae (grasses). Boxes and lines indicate exons and introns, respectively, sizes for which are in bp. Vertical lines connecting paralogs represent a subfamily. B, Phenetic analysis showing the evolution and duplication of carotenoid hydroxylase amino acid sequences. Monocot sequences fall into two major groups labeled 1 and 2. GenBank accessions are in parentheses: Erwinia (crtZ AAA64983), Arabidopsis (βOHase1, AT4G25700; βOHase2, AT5G52570), tomato (Solanum lycopersicum; crtR-b1, Y14809; crtR-b2, Y14810); “Materials and Methods” and Supplemental Table S1 list monocot accession numbers. [See online article for color version of this figure.]
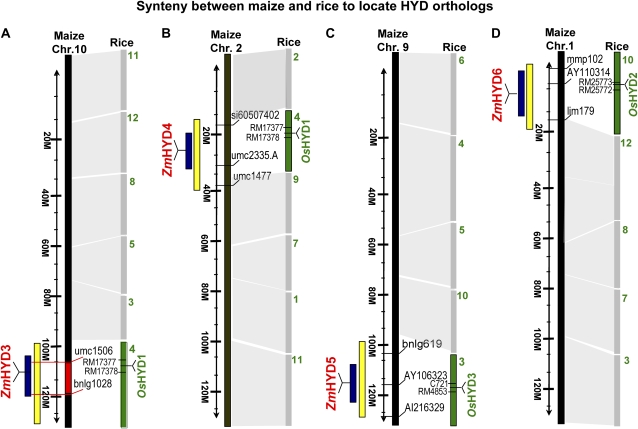
Figure 3.
Synteny between maize and rice to map maize HYD orthologs. Orthologous genes encoding maize and rice nonheme diiron β-carotene hydroxylases were compared by synteny, where each chromosome of maize is syntenous to multiple rice chromosome regions numbered in green. Each maize chromosome is marked by bin (yellow box) and contig (dark-blue box), with its respective maize gene and linked genetic markers. Maize HYD3 (ZmHYD3) maps to chromosome 10 between DNA markers umc1506 and bnlg1028 that flank a β-carotene QTL (Chander et al., 2007) colored in red. A, ZmHYD3 (BAC clone no. c0156B23; contig no. 415; chromosome 10.05). B, ZmHYD4 (BAC clone no. c0112K16; contig no. 73; chromosome 2.03). C, ZmHYD5 (BAC clone no. b0589L12; contig no. 391; chromosome 9.07). D, ZmHYD6 (BAC clone no. b0072L05; contig no. 03; chromosome 1.01). [See online article for color version of this figure.]
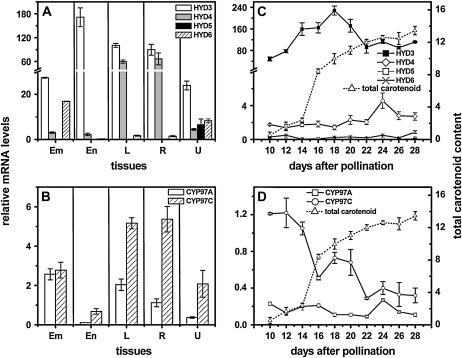
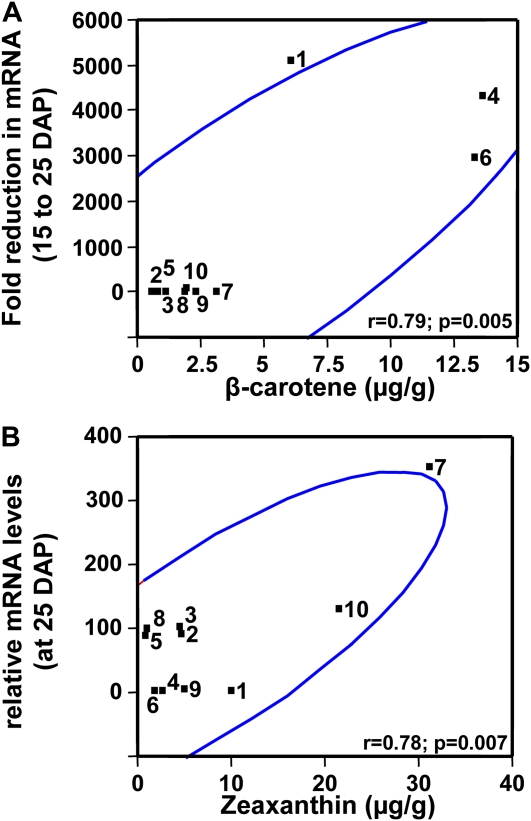
In the maize inbred B73 that has been used extensively for investigating regulation of endosperm carotenogenesis (Li et al., 1996, 2008a, 2008b, 2009; Matthews et al., 2003; Vallabhaneni and Wurtzel, 2009), mRNA abundance for the six functional carotene hydroxylases genes varied among tissues and during endosperm development, where comparison was also made with carotenoid accumulation (Fig. 4). To test which, if any, carotene hydroxylase gene showed a statistical correlation between mRNA levels and endosperm provitamin A β-carotene content, transcript abundance was quantified in a genetically diverse maize germplasm collection (Liu et al., 2003; Islam, 2004; Harjes et al., 2008). However, it was impractical to screen endosperm developmental samples in staged, hand-pollinated plants for the entire collection, representing 80% of maize genetic diversity. Therefore, a uniquely selected subset of lines for testing was chosen from 148 lines with known carotenoid content and composition (Islam, 2004). From existing metabolite data, we sorted lines for highest total carotenoid content, highest ratio of β-carotene to β-cryptoxanthin, highest ratio of β-cryptoxanthin to zeaxanthin, highest ratio of β-carotene to zeaxanthin, highest ratio of α-carotene to lutein, and highest ratio of lutein to zeaxanthin. The resulting 10-line subset (Supplemental Tables S3 and S4) exhibited the most extreme carotenoid biochemical phenotypes and likely contained the most favorable and informative alleles for breeding carotenoid content and composition. For example, a block in carotene hydroxylase activity would be predicted to manifest as a high ratio of β-carotene to β-cryptoxanthin, while factors that controlled flux would be predicted to influence levels of total carotenoids. The lines are genetically diverse, spanning four major genetic diversity groups and eight subgroups (Supplemental Table S4) among the 260 Goodman diversity lines (Liu et al., 2003). The 10 lines were then used for quantitative transcript profiling of endosperm samples at different developmental stages (days after pollination [DAP]). To demonstrate adequacy of sample size and to validate use of this biochemically diverse subset to infer pathway regulation, we applied Pearson correlation analysis to show that PSY1 but not PSY2 or PSY3 transcript levels showed statistically significant correlation with seed carotenoid content (Li et al., 2008b; Vallabhaneni and Wurtzel, 2009). The findings were consistent with molecular and genetic studies associating PSY1 and control of pathway flux in endosperm (Randolph and Hand, 1940; Palaisa et al., 2003; Wong et al., 2004; Pozniak et al., 2007; Li et al., 2008b). The subset has also been recently tested to examine additional genes that encode enzymes for steps that represent carotenoid biosynthetic pathway bottlenecks and those steps that do not (Vallabhaneni and Wurtzel, 2009). For example, the finding of carotenoid bottlenecks in the upstream isoprenoid biosynthetic pathway is consistent with other studies in the literature. The validated germplasm subset was then used to assess the possible role of carotene hydroxylase gene expression in controlling seed β-carotene composition. Transcript abundance was quantitatively measured, over a range of endosperm developmental stages, for the five carotene β-ring hydroxylase genes, HYD3, HYD4, HYD5, HYD6, and CYP97A, and the one ε-ring hydroxylase gene, CYP97C. Time points chosen were based on earlier studies that investigated carotenoid accumulation during endosperm development (Li et al., 2008b). HYD3 was the only gene for which transcripts were found to correlate with carotenoids (Supplemental Table S3). Interestingly, some of the lines showed a steep reduction in HYD3 transcripts between 15 and 25 DAP, ranging from 0.7- to >5,000-fold. A Pearson correlation analysis comparing β-carotene content and transcript levels for all carotene hydroxylase genes was conducted and HYD3 was the only gene for which there was a statistically significant (P ≤ 0.05) correlation between transcript levels and provitamin A β-carotene content (Fig. 5). In the correlation plots shown, where the colored lines denote a 95% confidence interval, the fold reduction of HYD3 transcripts over endosperm development (15–25 DAP) positively correlated with seed β-carotene (μg/g) levels (r = 0.79, P = 0.005; Fig. 5A). As would be predicted for a critical endosperm hydroxylase gene, HYD3 transcripts at 25 DAP also positively correlated with zeaxanthin (μg/g) levels (r = 0.78, P = 0.007; Fig. 5B). That is, the greater the reduction in transcripts over development, the higher the β-carotene content, as would be predicted from a correspondingly lower level of hydroxylase activity; conversely, the higher the transcripts at 25 DAP, the greater the level of zeaxanthin, as would be predicted from increased hydroxylase activity.
Figure 4.
Transcript profiling of hydroxylase gene family members in maize standard, B73 inbred. Transcript levels of HYD (A) and P450-type (B) hydroxylase genes in different maize tissues: embryo (Em) and endosperm (En) collected at 20 DAP from field-grown plants; leaf (L) and root (R) samples collected from seedlings at the six-leaf stage; unfertilized ovule (U). Transcript levels were expressed relative to endosperm levels at 20 DAP. Transcript abundance in developing endosperm is compared to accumulation of total carotenoids (C and D). Mean of three replicates ± sd.
Figure 5.
Correlation analysis showing HYD3 mRNA levels correlate with endosperm provitamin A content. A, Provitamin A β-carotene levels correlate with the fold reduction in HYD3 transcripts between 15 and 25 DAP. B, Nonprovitamin A zeaxanthin levels correlate with HYD3 transcripts at 25 DAP. Pearson correlation (r) and statistical significance (P) for comparison of HYD3 transcript levels and kernel carotenoid composition (taken from Supplemental Table S3) in maize inbred lines (Harjes et al., 2008) performed using JMP v. 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship at a 95% confidence interval (blue line). Inbred lines are: 1, A619; 2, B73; 3, B37; 4, CI.7; 5, C131A; 6, DE3; 7, KUI2007; 8, NC300; 9, SD44; 10, TZI18. [See online article for color version of this figure.]
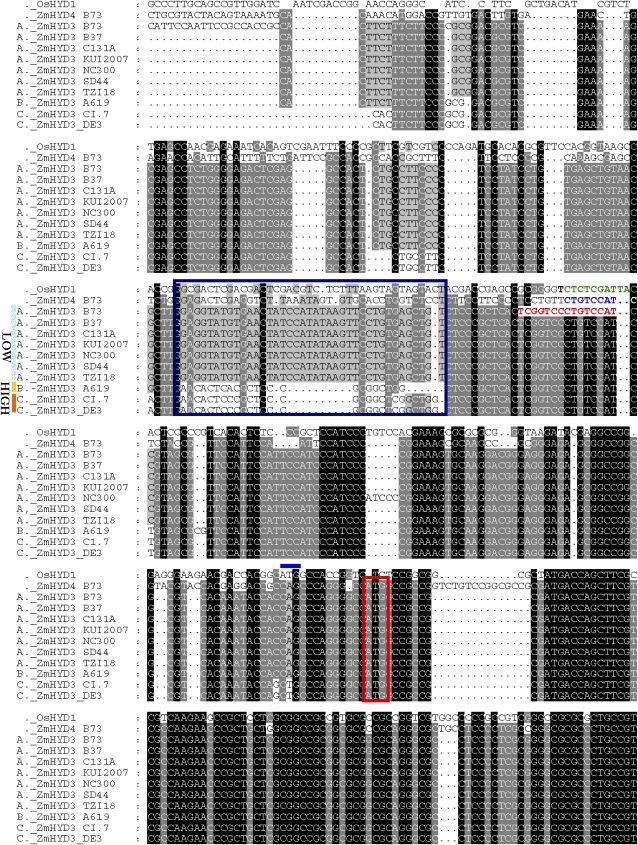
We noted the presence of a QTL for endosperm β-carotene content (Chander et al., 2007) that maps together with HYD3, between markers umc1506 and bnlg1028 on maize chromosome 10.05 (Fig. 3). Therefore, we sequenced all HYD3 alleles in the 10-line subset. Variation consistently seen in the high β-carotene lines was found in an approximately 40-bp region adjacent to the transcript start site (Fig. 6, blue box). A conserved transcript start site and first ATG were mapped by aligning available paralog-specific ESTs and genomic DNA for maize B73 HYD3 and sister paralog HYD4, and rice synteny partner HYD1 (Fig. 6). B73 and the other six low β-carotene lines had identical sequence in this region (variant A); A619 had sequence variant B, while CI.7 and DE3 shared variation C. B and C in the high β-carotene lines appear to contain a duplicated sequence from a downstream region that replaced the progenitor sequence seen in A, although allele C shows a more complete match (14/17 nucleotides) as compared to allele B (10/17 nucleotides). Diversity analysis (Liu et al., 2003) of the 260 Goodman lines placed A619 and CI.7, which carried different HYD3 variations, in two genetically distinct subgroups of the maize non-Stiff stalk lines; such diversity grouping is consistent with finding that they do not share the same polymorphism. The identical C polymorphism seen in DE3 and CI.7 may be a result of a possible overlapping pedigree since DE3 was placed in a mixed group as an indication of genetic structure shared with more than one maize diversity group. However, further sequencing of the entire HYD3 gene of DE3 and CI.7 revealed some minor sequence differences between HYD3 alleles in these two inbreds (Supplemental Fig. S3).
Figure 6.
Alignment of maize sister paralogs HYD3 and HYD4 plus their rice synteny partner HYD1. DNA sequences were aligned for HYD3 and HYD4, their rice synteny partner HYD1, and the HYD3 sequences of all of the maize inbreds used in this study. Transcription start sites (colored nucleotides) for B73 HYD3, B73 HYD4, and rice HYD1 were estimated by aligning ESTs. The first ATG for HYD1 is denoted by a blue overline and contained within a red box for the remaining HYD3 and HYD4 sequences. A blue box encompasses the variant region that distinguishes the high and low β-carotene lines. Black, dark-gray, and light-gray shading indicate degree of conserved nucleotides where black is the highest (100%) match. [See online article for color version of this figure.]
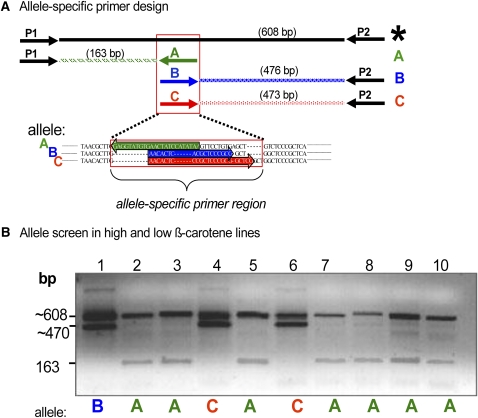
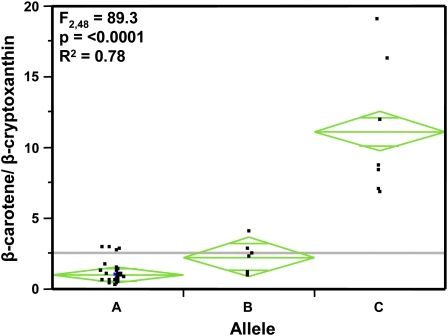
To test whether HYD3 allelic variation could explain β-carotene variation, a PCR assay was developed to rapidly genotype a broader maize collection. The assay distinguished between A and C or between A and B alleles and generated a common HYD3 paralog-specific product that could be sequenced to reveal new alleles; the assay correctly genotyped all 10 lines for their HYD3 allele (Fig. 7 We then selected an additional 41 lines spanning maize genetic diversity and range of carotenoid content and composition (Supplemental Table S4). The combined group of 51 lines included nine new lines identified by metabolite sorting to exhibit high ratios of β-carotene to β-cryptoxanthin, as possible indicator of a block in β-carotene hydroxylation. The common HYD3 paralog-specific product was amplified and sequenced to characterize HYD3 alleles in this broader maize collection (Fig. 7). In the combined 51 lines, we identified 14 high-carotene alleles: seven C alleles, including one new C-like allele, and seven B alleles. The 14 B and C alleles were widely distributed among genetic diversity groups, as were the 37 A alleles (Supplemental Table S4). One-way ANOVA showed that the mean ratios of β-carotene to β-cryptoxanthin exhibited a statistically significant difference among the three HYD3 alleles (F2,48 = 89.3, P < 0.0001) and that the HYD3 allele explained 78% of the variation (R2) in the β-carotene-to-β-cryptoxanthin ratio (Fig. 8). The absolute amount of β-carotene (μg/gm) also showed a statistically significant difference among the three alleles (A–C) in one-way ANOVA (F2,48 = 13.7, P < 0.0001) and HYD3 allele explained 36% of total variation (R2) in level of β-carotene (μg/gm). It is reasonable that HYD3 allele will explain less of the variation in absolute levels of β-carotene compared to ratio of β-carotene to β-cryptoxanthin. Absolute leaves of β-carotene will be influenced not only by LCYE and HYD3 activities, but also by multiple factors controlling pathway flux (Vallabhaneni and Wurtzel, 2009). In comparison, β-carotene-to-β-cryptoxanthin ratio will be influenced most directly by the HYD3 enzyme activity.
Figure 7.
Multiplex PCR diagnosis of HYD3 alleles in high and low β-carotene lines. The sequence variation in HYD3 (Fig. 6, blue box) was used to develop a multiplex PCR assay to track HYD3 alleles (B and C) in the high β-carotene lines and to distinguish these from the A allele in the low β-carotene line. With this simple PCR assay, natural alleles can be tracked using traditional, nontransgenic breeding. The assay contained four primers to (1) amplify a control, HYD3-specific product (*), which was conserved in all alleles and encompassed the allele-specific region (Figs. 7A and 6 [blue box]), and (2) to amplify and distinguish specific alleles A from C or A from B. The multiplex PCR assay was next used to genotype all of the 10 inbred lines used in the correlation study (Fig. 7); as predicted, all lines having B or C alleles also had a high β-carotene content (21%–55% of total carotenoids) as compared to maize B73 (A allele) where kernels have only 3% β-carotene. A, Allele-specific primer design. Top, HYD3 primers (P1 and P2) amplify all alleles, as product (*), the size of which is shown based on the A allele (B and C alleles generate a * product that differs by a few bp); allele-specific primers (A, B, or C) amplify maize allele A from low β-carotene line B73, allele B from high β-carotene line A619, or allele C from high β-carotene lines CI.7 and DE3, respectively. Bottom, The boxed region used to design the allele-specific primers is shown in more detail. Sequences and amplification direction are shown in corresponding colors for the three alleles. PCR product sizes are based on the A allele; products from the other alleles differ slightly. B, Screening of alleles in the maize diversity lines. Primers used were P1/P2/A plus either B (lane 1) or C (lanes 2–10). Inbred lines are: 1, A619; 2, B73; 3, B37; 4, CI.7; 5, C131A; 6, DE3; 7, KUI2007; 8, NC300; 9, SD44; 10, TZI18. [See online article for color version of this figure.]
Figure 8.
One-way ANOVA between HYD3 alleles (A, B, C) and the ratio of β-carotene to β-cryptoxanthin in 51 genetically diverse maize lines. The mean ratios of β-carotene to β-cryptoxanthin showed a statistically significant difference among the three HYD3 alleles in one-way ANOVA (F2,48 = 89.3, P < 0.0001), and the HYD3 allele explained 78% of the variation (R2) in the β-carotene-to-β-cryptoxanthin ratio. Three different alleles denoted as A, B, and C are plotted on the x axis and ratios of β-carotene (μg) to β-cryptoxanthin (μg) are plotted on the y axis. Gray line indicates the grand mean of the entire sample. The green diamonds indicate 95% confidence interval of the mean, and the dots represent each inbred line in study. [See online article for color version of this figure.]
We next compared the means of the three alleles using the Tukey-Kramer honestly significant difference test at a 0.05 level. We observed a significant difference (P < 0.0001) between alleles C and B, and C and A for the ratio of β-carotene and β-cryptoxanthin and β-carotene alone. No significant difference was observed between alleles B and A for both the ratio of β-carotene and β-cryptoxanthin and β-carotene alone.
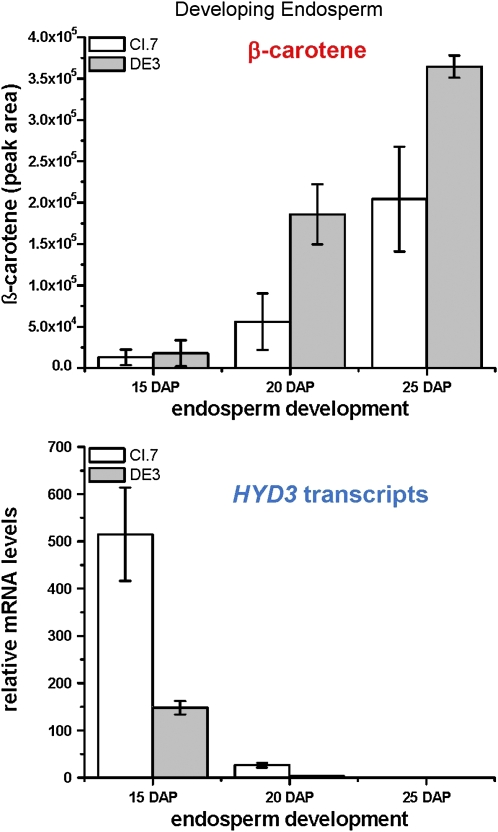
The most favorable HYD3 C allele was associated with approximately 11-fold higher levels of β-carotene relative to β-cryptoxanthin and 4-fold higher levels of β-carotene (μg/gm). The most likely explanation is that during endosperm development a reduction in HYD3 transcripts leads to reduced conversion of β-carotene to downstream xanthophylls, causing β-carotene to accumulate. To test this possibility, we selected CI.7 and DE3, lines carrying the C allele, and measured β-carotene at each of three developmental stages and compared to HYD3 transcript level for triplicate replicates of endosperms collected at 15, 20, and 25 DAP. As shown in Figure 9, levels of β-carotene increased during endosperm development (for equal amounts of extracted carotenoids), as HYD3 mRNA levels decreased in the same endosperm developmental samples. Therefore, there is a direct link between HYD3 transcript level and accumulated β-carotene.
Figure 9.
Relative amount of β-carotene increases and HYD3 transcript levels decrease during development of maize endosperm in lines carrying a C allele of HYD3. Total carotenoids were extracted and equal amounts of total carotenoids subjected to UPLC separation and quantification of β-carotene in triplicate. Samples from the same endosperm samples were subjected to quantitative RT-PCR (see Supplemental Table S3). Means of three replicates are shown with sd. [See online article for color version of this figure.]
Enhancement of provitamin A levels necessitates control of both pathway branching and carotene hydroxylation in addition to elevating pathway flux and minimizing product degradation. Previous development of LCYE markers (Harjes et al., 2008) provided the first step in tracking alleles to control pathway branching through traditional nontransgenic breeding selection. However, the LCYE markers were insufficient. Even if pathway branching could be controlled through selection of optimal LCYE alleles, there remained the problem that the provitamin A carotenes produced in the pathway would automatically be hydroxylated to nonprovitamin A compounds, as typically seen for maize endosperm composition of zeaxanthin or lutein (Kurilich and Juvik, 1999). Other upstream steps may influence carotenoid content (Vallabhaneni and Wurtzel, 2009), but enhancement of provitamin A composition will specifically require the combined use of tools that target both LCYE and HYD3.
The C allele can be monitored by the simple PCR assay and its presence in genetically diverse lines will facilitate use in different geographical regions. This PCR assay may now be used to test predictability of the HYD3 allele in controlling β-carotene content in diverse cultivars. With 51 lines analyzed for HYD3 allele, LCYE allele data was available for 48 of those lines (Harjes et al., 2008). However, none carried optimal alleles for both HYD3 and LCYE. Therefore, the HYD3 assay may be used in combination with the previously described LCYE assays (Harjes et al., 2008) to select parental lines containing optimal HYD3 or LCYE alleles and to screen progeny at the seedling stage and identify those that are homozygous for optimal alleles of both genes. Thus it is predicted that the combination of HYD3 and LCYE alleles will lead to higher β-carotene levels in maize endosperm than having optimal alleles of either gene alone.
Many ongoing studies have investigated pathway regulation through transgene expression in one cultivar (Giuliano et al., 2008); the resulting phenotype is dependent on the genotype used and the resulting data are not predictably transferred to other genotypes, perhaps because of limited understanding of pathway regulation. Here we took advantage of natural variation inherent in a large germplasm collection; by judicious sampling of the collection, we were able to pinpoint a specific gene family member for which favorable alleles were discovered for predicting enhanced β-carotene composition. Metabolite-based sorting of a germplasm collection provided accessibility to a valuable resource that yielded critical knowledge and tools that will be needed for breeding high provitamin A in maize, an important food staple in vitamin A-deficient Sub-Saharan Africa and Latin America, where vitamin A deficiency is a serious health problem. This approach can be easily adapted to other metabolite targets in other species to rapidly discover pathway regulation and to develop tools for predictive breeding.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays) inbred lines (A619, B73, B37, CI.7, C131A, DE3, KUI2007, NC300, SD44, and TZI18; Harjes et al., 2008) were field grown in Bronx, NY, and sibling pollinated. Unfertilized ear, embryo (20 DAP), and endosperm (10–30 DAP) were collected from field-grown plants, whereas leaf and root samples were collected from six-leaf-stage seedlings grown in a greenhouse (16-h day at 25°C). Samples were stored at −80°C prior to use.
Sequence Analysis and Chromosome Mapping
Cloning and DNA Sequence Analysis
A maize B73 genomic bacterial artificial chromosome (BAC) library (Gallagher et al., 2004) was probed with the maize HYD4 cDNA, GenBank AY844956. Eight BAC clones representing three groups were identified and a representative of each group was sequenced by primer walking (DNA Sequencing Facility, University of Chicago Research Center) and the data deposited in GenBank (HYD1, EU638325; HYD2, EU638326; HYD3, AY844957). The HYD3 cDNA sequence (GenBank no. AY844958) was used as a query to identify additional maize paralogs and orthologs from rice (Oryza sativa; www.gramene.org) and Sorghum bicolor (www.phytozome.org). Maize methyl-filtered contig sequences and corresponding BAC clones harboring carotene hydroxylase genes (diiron type) were identified (Supplemental Table S1). Gene models were drawn using Genscan (www.genscan.com), Softberry (www.softberry.com), and Vector NTI Suite 9.0 (Invitrogen). Transcription start sites were estimated by EST and promoter analysis (www.softberry.com). Maize ESTs corresponding to CYP97A (DV169913) and CYP97C (BE552887) were found by searching plantGDB maize database using orthologous gene sequences of CYP97A (LOC_Os02g57290) and CYP97C (LOC_Os10g39930) from rice (Quinlan et al., 2007). As found for rice and sorghum, only one maize ortholog was found for each gene encoding these two CYP97 enzymes (Supplemental Table S1).
Mapping and Synteny
Chromosomal positions of maize genes in the maize B73 inbred were mapped either by utilizing tools available at WebAGCoL package (Pampanwar et al., 2005) or MaizeGDB. Orthologous genes from rice Japonica were identified through synteny comparisons with maize (www.tigr.org/tdb/synteny/).
Phenetic and Protein Analysis
HYD amino acid sequences were aligned using ClustalW and a neighbor-joining tree was constructed with bacterial crtZ from Erwinia herbicola (GenBank no. AAA64983) as an out group with 500 bootstrap replication support using MEGA3 software (Kumar et al., 2001). Chloroplast transit peptide signal and transmembrane analysis were predicted using ChloroP 1.1 (Emanuelsson et al., 1999) and TMHMM 2.0 (Krogh et al., 2001), respectively.
Transcript and Total Carotenoid Analysis
RNA extraction and quantitative reverse transcription (RT)-PCR were performed using gene-specific primers (Supplemental Table S1) and normalized to actin, as described (Li et al., 2008a). Values are expressed as the mean of three RT-PCR replicates ±sd. Total carotenoids were extracted as previously described (Li et al., 2008b) and the concentration (shown in Fig. 4) was calculated using the Lambert-Beer equation (Schiedt and Liaaen-Jensen, 1995).
Plasmids and Functional Complementation
Escherichia coli BL21 (DE3) cells (Novagen) containing pAC-BETA-04 accumulate β-carotene and were used to test hydroxylase function (Sun et al., 1996). BM382572 (HYD3) and BG320875 (HYD4) cDNAs were subcloned for functional analysis into the EcoRI/NotI and EcoRI/XhoI sites of the pET23c expression vector (Novagen), and renamed as pTHYD3 and pTHYD4, respectively. Transformants carrying the test plasmids or empty vector were grown on selective medium and pigments extracted and analyzed by HPLC as described (Gallagher et al., 2004).
Total Carotenoid Content Measurement
The carotenoid extraction procedure was based on Kurilich and Juvik (1999). Five hundred milligrams of maize endosperm was ground in ethanol and incubated for 6 min (6 mL ethanol, 0.1% butylated hydroxytoluene) at 85°C, followed by 10 min saponification with 120 μL (1 g/mL) KOH. Samples were vortexed, placed on ice, and 4 mL cold distilled water added. Three milliliters of 2:1 petroleum ether:diethyl ether (PE:DE) (v/v) were added to each sample, vortexed, and centrifuged for 10 min at 3,500 rpm. The upper layer was retrieved and the separation was repeated twice with 3 mL of 2:1 PE:DE (v/v). The combined fractions were made up to 10 mL, and carotenoids were measured spectrophotometrically at OD 450 nm using a Lambda UV/VIS spectrophotometer (Perkin Elmer Life Sciences), and the concentration of carotenoids were calculated using the Lambert-Beer equation (Schiedt and Liaaen-Jensen, 1995).
Measurement of β-Carotene in Developing Endosperm of High β-Carotene Lines
The carotenoid extraction procedure was based on Kurilich and Juvik (1999). Five hundred micrograms of maize endosperm was ground in ethanol and incubated for 6 min (6 mL ethanol, 0.1% butylated hydroxytoluene) at 85°C, followed by 10 min saponification with 120 μL (1 g/mL) KOH. Samples were vortexed, placed on ice, and 4 mL cold distilled water added. Three milliliters of 2:1 PE:DE (v/v) were added to each sample, vortexed, and centrifuged for 10 min at 3,500 rpm. The upper layer was retrieved and transferred to a tube with a known weight. The separation was repeated twice with 3 mL of 2:1 PE:DE (v/v) and upper layers combined and dried under nitrogen gas. Total weight of the tube was measured to obtain the net weight of dried extract in each tube. All extractions and measurements were performed in triplicates, and finally dried extract was redissolved appropriately in HPLC grade isopropanol to maintain equal concentration (100 μg/μL) in all samples. Four microliters of each sample was injected onto an Acquity ultra performance LC (UPLC) BEH C18 1.7 μm 2.1 × 100 mm (part no. 186002352) column kept at 65°C and attached to a Waters Acquity UPLC with a photodiode array detector. Separation was carried out using a linear gradient from 100% acetonitrile to 80% acetonitrile/20% isopropanol over 6 min, followed by 100% acetonitrile at 6.5 min. The solvent flow rate was 0.3 mL/min. Data were collected for 7 min total and analyzed with Empower Pro software to determine β-carotene peak area for each sample. All samples were run in triplicates and sd calculated.
Statistical Analyses
Pearson correlation analysis of transcript and carotenoid composition from 10-line subset of maize inbreds was performed using JMP v. 5.1.2 (SAS Institute) to test the statistical significance (P ≤ 0.05) of the relationship. One-way ANOVA and Tukey-Kramer honestly significant difference test for comparisons between the alleles (A–C) and ratio of β-carotene to β-cryptoxanthin or absolute levels of β-carotene in 51 lines was performed using JMP 5.1 (SAS institute Inc.).
Multiplex PCR Assay for Tracking HYD3 Alleles
A multiplex PCR assay based on HYD3 promoter variation (A, B, or C) was developed to distinguish between HYD3 allele A (in the low-carotene B73 inbred) and the HYD3 alleles B and C found in the high-carotene inbreds A619 (allele B), CI.7 (allele C), and DE3 (allele C). The assay involved amplification of one HYD3 paralog-specific control PCR product produced by all alleles and a second product that distinguished allele A from C or allele A from B, in both homozygous and heterozygous samples. The PCR assay utilized four primers: (1) two external primers P1 (no. 1,595, GACTTGTGAGCAAGGGGAAG) and P2 (no. 1,592, GACGTGACTCCGAGGCTAGA) for amplification of a control product that was conserved in all alleles; and (2) two internal primers added for amplification of the HYD3 specific alleles. To track alleles C and A, the specific allele forward primer was C (no. 2,111, AACACTCCCGCTCCCGCGGCTCG, allele C), and the reverse primer was A (no. 2,116, TTATATGGATAGTTCACATACCTC, allele A). Alternatively, to distinguish allele B from A, the forward primer used was primer B (no. 2,109, AACACTCACGCTCCCGCG, allele B) together with reverse primer A. The 50-μL PCR reaction contained: 1× PCRx amplification buffer (Invitrogen), 0.2 mm deoxyribonucleotide triphosphates (Invitrogen), 1.5 mm MgSO4 (Invitrogen), 2.5 units iTaq DNA polymerase (BIO-RAD), 0.6 μm each of two external primers; 0.4 μm each of two internal primers, 0.5 μg genomic DNA template, 3× final concentration of PCRx enhancer solution (Invitrogen). Thermal cycling conditions were: one cycle (94°C for 3 min) followed by 40 cycles (94°C for 45 s, 54°C for 45 s, and 72°C for 1 min) and one cycle (72°C for 10 min). Amplified products were separated on 1% agarose gels and validated by sequencing. Maize inbred sequences used for developing the multiplex primers were: FJ228406, FJ228407, FJ228408, FJ228409, FJ228410, FJ228411, FJ228412, FJ228413, FJ228414, and FJ228415.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY844956, AY844957, AY844958, EU638325, EU638326, FJ228406, FJ228407, FJ228408, FJ228409, FJ228410, FJ228411, FJ228412, FJ228413, FJ228414, and FJ228415.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Conserved motifs in HYD proteins of maize and rice.
Supplemental Figure S2. Functional complementation of HYD genes.
Supplemental Figure S3. Alignment of maize HYD3 in 51 lines.
Supplemental Table S1. Primer and gene accession numbers.
Supplemental Table S2. Predicted structural characteristics of HYD proteins.
Supplemental Table S3. Transcript profiling of HYD genes in maize diversity lines and Pearson correlation values.
Supplemental Table S4. Genotyping of HYD3 alleles and comparison with carotenoid composition of diverse maize germplasm accessions.
Supplementary Material
Acknowledgments
We thank Dr. Dwight Kincaid (Lehman College, City University of New York, New York) for advice on statistical analysis and Wurtzel lab members (Dr. Louis Bradbury, Dr. Faqiang Li, Dr. Maria Shumskaya, and Oren Tzfadia) for helpful advice. Maize lines were obtained from the National Germplasm Collection, except for the initial 10-line subset (Li et al., 2008b).
This work was supported by the National Institutes of Health (grant nos. GM08225 and GM081160 to E.T.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eleanore T. Wurtzel (wurtzel@lehman.cuny.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aluru M, Xu Y, Guo R, Wang Z, Li S, White W, Wang K, Rodermel S (2008) Generation of transgenic maize with enhanced provitamin A content. J Exp Bot 59 3551–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RE, Allen LH, Bhutta ZA, Caulfield LE, de Onis M, Ezzati M, Mathers C, Rivera J, Maternal Child Undernutrition Study Group (2008) Maternal and child undernutrition: global and regional exposures and health consequences. Lancet 371 243–260 [DOI] [PubMed] [Google Scholar]
- Chander S, Guo YQ, Yang XH, Zhang J, Lu XQ, Yan JB, Song TM, Rocheford TR, Li JS (2007) Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor Appl Genet 116 223–233 [DOI] [PubMed] [Google Scholar]
- Chandler VL, Brendel V (2002) The Maize Genome Sequencing Project. Plant Physiol 130 1594–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, Papacchioli V, Beyer P, Giuliano G (2006) Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol 6 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto G, Welsch R, Tavazza R, Mourgues F, Pizzichini D, Beyer P, Giuliano G (2007) Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol 7 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CE, Matthews PD, Li F, Wurtzel ET (2004) Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses (Poaceae). Plant Physiol 135 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26 139–145 [DOI] [PubMed] [Google Scholar]
- Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, et al (2008) Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam SN (2004) Survey of carotenoid variation and quantitative trait loci mapping for carotenoid and tocopherol variation in maize. Master's thesis. University of Illinois at Urbana-Champaign, Urbana-Champaign
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305 567–580 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17 1244–1245 [DOI] [PubMed] [Google Scholar]
- Kurilich A, Juvik J (1999) Quantification of carotenoid and tocopherol antioxidants in Zea mays. J Agric Food Chem 47 1948–1955 [DOI] [PubMed] [Google Scholar]
- Li F, Murillo C, Wurtzel ET (2007) Maize Y9 encodes a product essential for 15-cis zetacarotene isomerization. Plant Physiol 144 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Tzfadia O, Wurtzel ET (2009) The Phytoene Synthase gene family in the grasses: subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal Behav 4 208–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Wurtzel ET (2008. a) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic-stress-induced root carotenogenesis. Plant Physiol 146 1333–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008. b) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress-tolerance. Plant Physiol 147 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Matthews PD, Burr B, Wurtzel ET (1996) Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol 30 269–279 [DOI] [PubMed] [Google Scholar]
- Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165 2117–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PD, Luo R, Wurtzel ET (2003) Maize phytoene desaturase and zetacarotene desaturase catalyze a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot 54 2215–2230 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Wurtzel ET (2007) Biotechnology of food colorant production. In C Socaciu, ed, Food Colorants: Chemical and Functional Properties. CRC Press, Boca Raton, FL, pp 347–398
- Palaisa KA, Morgante M, Williams M, Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampanwar V, Engler F, Hatfield J, Blundy S, Gupta G, Soderlund C (2005) FPC web tools for rice, maize, and distribution. Plant Physiol 138 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak CJ, Knox RE, Clarke FR, Clarke JM (2007) Identification of QTL and association of a phytoene synthase gene with endosperm colour in durum wheat. Theor Appl Genet 114 525–537 [DOI] [PubMed] [Google Scholar]
- Quinlan R, Jaradat T, Wurtzel ET (2007) Escherichia coli as a platform for functional expression of plant P450 carotene hydroxylases. Arch Biochem Biophys 458 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph LF, Hand DB (1940) Relation between carotenoid content and number of genes per cell in diploid and tetraploid corn. J Agric Res 60 51–64 [Google Scholar]
- Schiedt K, Liaaen-Jensen S (1995) Chapter 5: isolation and analysis. In G Britton, S Liaaen-Jensen, H Pfander, eds, Carotenoids—Isolation and Analysis, Vol 1A. Birkhäuser, Basel, pp 81–108
- Sun Z, Gantt E, Cunningham JFX (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271 24349–24352 [DOI] [PubMed] [Google Scholar]
- Underwood BA (2004) Vitamin A deficiency disorders: international efforts to control a preventable “pox”. J Nutr 134 231S–236S [DOI] [PubMed] [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2009) Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol 150 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Lambert RJ, Wurtzel ET, Rocheford TR (2004) QTL and candidate genes phytoene synthase and zetacarotene desaturase associated with the accumulation of carotenoids in maize. Theor Appl Genet 108 349–359 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1995) Global Prevalence of Vitamin A Deficiency. Micronutrient Deficiency Information System. Working Paper No. 2. WHO/NUT/95.3. World Health Organization, Geneva
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287 303–305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.