Abstract
A novel lachrymatory factor synthase (LFS) was isolated and purified from the roots of the Amazonian medicinal plant Petiveria alliacea. The enzyme is a heterotetrameric glycoprotein comprised of two α-subunits (68.8 kD each), one γ-subunit (22.5 kD), and one δ-subunit (11.9 kD). The two α-subunits are glycosylated and connected by a disulfide bridge. The LFS has an isoelectric point of 5.2. It catalyzes the formation of a sulfine lachrymator, (Z)-phenylmethanethial S-oxide, only in the presence of P. alliacea alliinase and its natural substrate, S-benzyl-l-cysteine sulfoxide (petiveriin). Depending on its concentration relative to that of P. alliacea alliinase, the LFS sequesters, to varying degrees, the sulfenic acid intermediate formed by alliinase-mediated breakdown of petiveriin. At LFS:alliinase of 5:1, LFS sequesters all of the sulfenic acid formed by alliinase action on petiveriin, and converts it entirely to (Z)-phenylmethanethial S-oxide. However, starting at LFS:alliinase of 5:2, the LFS is unable to sequester all of the sulfenic acid produced by the alliinase, with the result that sulfenic acid that escapes the action of the LFS condenses with loss of water to form S-benzyl phenylmethanethiosulfinate (petivericin). The results show that the LFS and alliinase function in tandem, with the alliinase furnishing the sulfenic acid substrate on which the LFS acts. The results also show that the LFS modulates the formation of biologically active thiosulfinates that are downstream of the alliinase in a manner dependent upon the relative concentrations of the LFS and the alliinase. These observations suggest that manipulation of LFS-to-alliinase ratios in plants displaying this system may provide a means by which to rationally modify organosulfur small molecule profiles to obtain desired flavor and/or odor signatures, or increase the presence of desirable biologically active small molecules.
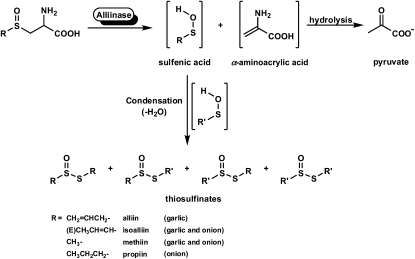
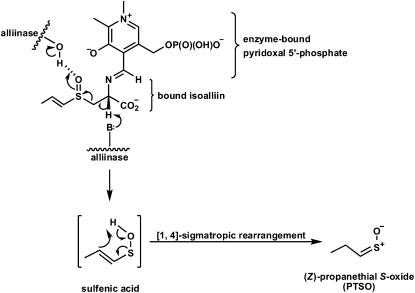
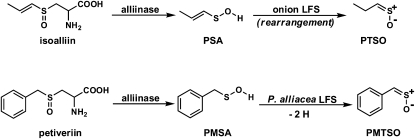
Lachrymatory factor synthase (LFS) is the term coined to refer to the recently discovered enzyme shown to catalyze the formation of the sulfine responsible for the lachrymatory effect of onion (Allium cepa), (Z)-propanethial S-oxide (PTSO; Imai et al., 2002). Until the discovery of the onion LFS, the formation of the onion lachrymatory factor (LF) was thought to be mediated by only a single enzyme, onion alliinase. Alliinases, which are pyridoxal 5′-P (PLP)-dependent Cys sulfoxide lyases most often found in members of the Allium genus, catalyze the breakdown of Cys sulfoxide derivatives to yield fleeting sulfenic acid intermediates and α-aminoacrylic acid (Scheme 1; Block, 1992; Shimon et al., 2007). Once formed, the sulfenic acids are most often observed to spontaneously condense with loss of water to form thiosulfinates, whereas the α-aminoacrylic acid is further hydrolyzed with loss of ammonia to form pyruvate. The S-substituted Cys sulfoxides that are acted upon by alliinases differ from one another by the identity of the sulfur-bound R group. In Allium plants, the R groups are alk(en)yl, with R = methyl and 2-propenyl appearing in large quantities in garlic (Allium sativum) and R = methyl and (E)-1-propenyl preponderating in onion (Scheme 1). The Cys sulfoxide that serves as the precursor of the onion lachrymator is (E)-S-(1-propenyl)-l-Cys sulfoxide (isoalliin). It is structurally distinct from other naturally occurring S-substituted Cys sulfoxides so far reported in that it is α,β-unsaturated. This structural feature affords its corresponding 1-propenylsulfenic acid (PSA) the possibility of undergoing a [1,4]-sigmatropic rearrangement that, in principle, would furnish the onion lachrymator, PTSO. Indeed, the formation of the onion lachrymator was proposed to occur by such a mechanism (Scheme 2; Block, 1992). Thus, it was surmised that were the α,β-unsaturation to be absent in the precursor S-substituted Cys sulfoxide, the [1,4]-sigmatropic rearrangement that would lead to sulfine formation could not occur. Consequently, it was not surprising that other S-substituted Cys sulfoxides constitutively present in garlic, onion, and other alliinase-containing plants, but devoid of this α,β-unsaturation in the sulfur-bound R group, did not themselves yield lachrymators on plant tissue wounding. It has since been discovered, however, that formation of the onion lachrymator is not catalyzed by onion alliinase, but instead by a novel class of enzyme—LFS. Imai et al. (2002) observed that although a crude preparation of onion alliinase yielded both the LF and the corresponding thiosulfinate, the protein fraction with lachrymator-forming ability could be completely separated from that with alliinase activity by passing the crude onion protein preparation through a hydroxyapatite column. The LFS was subsequently purified and shown to be highly substrate specific, producing the LF from only (E)-S-(1-propenyl)-l-Cys sulfoxide (isoalliin), which occurs constitutively in onion. Interestingly, the LF was detected only when three components, namely, the purified onion alliinase, isoalliin, and the onion LFS, were present in the reaction mixture simultaneously (Imai et al., 2002). Omission of the LFS from the reaction mixture resulted in an increased yield of thiosulfinates, but no LF. Although the complete cDNA sequence of the onion LFS has been determined (Imai et al., 2002), to our knowledge, full biochemical characterization of the enzyme has yet to be reported.
Scheme 1.
Alliinase-mediated formation of thiosulfinates from Cys sulfoxide precursors (Block, 1992; Shimon et al., 2007). Alliin is S-allyl-l-Cys sulfoxide, isoalliin is (E)-S-(1-propenyl)-l-Cys sulfoxide, methiin is S-methyl-l-Cys sulfoxide, and propiin is S-propyl-l-Cys sulfoxide.
Scheme 2.
Mechanism advanced by Block (1992) to account for formation of the onion lachrymator, PTSO. Alliinase-bound PLP forms a Schiff base with bound isoalliin. General base catalysis at the active site yields an α,β-unsaturated sulfenic acid that can undergo a [1,4]-sigmatropic rearrangement to furnish the sulfine.
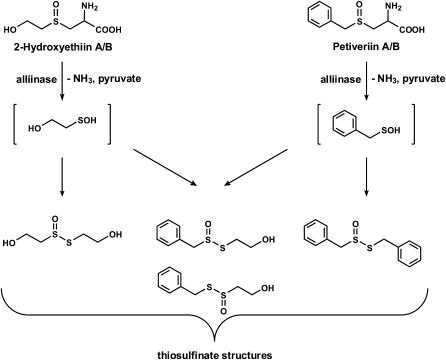
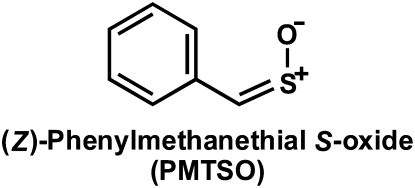
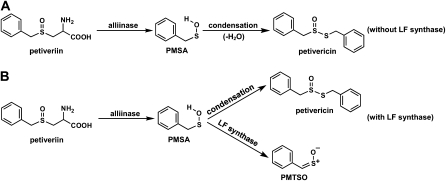
In the course of our studies on the organosulfur chemistry of non-Allium plants, we isolated and characterized the S-benzyl-l-Cys sulfoxides (petiveriins) and S-(2-hydroxyethyl)-l-Cys sulfoxides (2-hydroxyethiins) from the Amazonian medicinal plant Petiveria alliacea (Fig. 1; Kubec and Musah, 2001; Kubec et al., 2002). These compounds are S-substituted Cys sulfoxide derivatives with R = benzyl and 2-hydroxyethyl, respectively, that, to our knowledge, had never before been isolated from plants. We showed that, as has been observed in garlic and onion, symmetrical and mixed thiosulfinate derivatives of the corresponding petiveriin and 2-hydroxyethiin precursors could be extracted with ether solvent (Fig. 1; Kubec et al., 2002) upon root tissue disruption. We have also shown that an alliinase that mediates the transformation of the petiveriins and 2-hydroxyethiins to their corresponding thiosulfinates is present in P. alliacea (Musah et al., 2009). Interestingly, while working with P. alliacea root extracts, we noted the presence of a potent lachrymator that we subsequently determined to be a sulfine—(Z)-phenylmethanethial S-oxide (PMTSO; Fig. 2; Kubec et al., 2003). However, the biochemical precursor of PMTSO and the pathway(s) leading to its formation upon disruption of P. alliacea tissue remain to be determined. Given that the onion LF (PTSO), whose formation is mediated by an LFS, is also a sulfine, we were prompted to investigate the possibility of the presence of a LFS in P. alliacea. In this report, we describe our confirmation of the existence of a LFS in P. alliacea, and detail biochemical characterization of this novel class of enzymes.
Figure 1.
Cys sulfoxides and their corresponding thiosulfinate derivatives isolated from the Amazonian medicinal plant P. alliacea. The breakdown of the Cys sulfoxides is mediated by P. alliacea alliinase.
Figure 2.
Lachrymatory sulfine isolated from P. alliacea.
RESULTS
Confirmation of the Existence of a LFS in P. alliacea
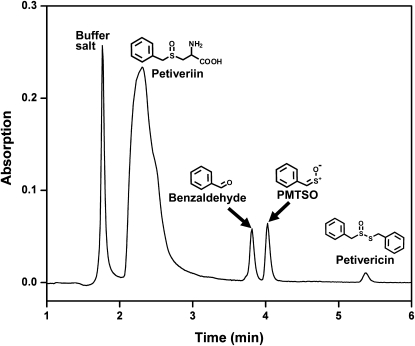
Since PMTSO production was only observed when LFS was exposed to both petiveriin and P. alliacea alliinase, the presence of the LFS in protein fractions was determined by tracking which protein fractions, when combined with a solution of petiveriin and P. alliacea alliinase in buffer, reliably produced PMTSO, as monitored by reversed-phase (RP) C-18 HPLC (Fig. 3). Over the course of these experiments, a chromatographic protocol was developed that resulted in the isolation of purified LFS whose activity in the production of PMTSO could be completely separated from the activity of the P. alliacea alliinase.
Figure 3.
Confirmation of the production of PMTSO and benzaldehyde by P. alliacea LFS action on the PMSA generated through P. alliacea alliinase-catalyzed breakdown of petiveriin using RP C-18 HPLC. A 10 μL aliquot of a reaction mixture comprised of 1.0 mL of 10 mm phosphate buffer at pH 8.0, 25 μm PLP, 1.5 mm petiveriin, and P. alliacea-derived protein was, after incubation for 10 min at room temperature, analyzed by HPLC (flow rate: 1.0 mL min−1; mobile phase: water:acetonitrile [30:70, v/v]; detection wavelength: 210 nm).
Isolation and Purification of P. alliacea LFS
The LFS was purified from homogenized P. alliacea roots through the sequential use of ammonium sulfate precipitation and anion-exchange, hydroxyapatite (twice), and gel-filtration chromatographies. The results for a typical purification of the P. alliacea LFS are summarized in Table I. A crude protein sample derived from 60% ammonium sulfate precipitation of a 150 g P. alliacea root macerate in phosphate buffer was subjected to anion-exchange chromatography. The protein eluted between 65 and 120 mm NaCl. The eluent was subjected to hydroxyapatite chromatography (second column) where it eluted between 160 and 220 mm phosphate. Further purification of the eluent obtained from the first hydroxyapatite column by a second hydroxyapatite column yielded the LFS, which eluted between 88 and 134 mm phosphate. The LFS eluent from the second hydroxyapatite chromatographic separation was subjected to further purification by gel-filtration chromatography (fourth column), which furnished purified LFS.
Table I.
Purification of the LFS from 150 g of P. alliacea roots
| Fraction | Volume | Total Protein | Total Activitya | Specific Activity | Recovery | Purification |
|---|---|---|---|---|---|---|
| mL | mg | nkat | nkat mg−1 | % | fold | |
| Homogenate | 230 | 289 | – | – | – | – |
| 60% (NH4)2SO4 | 20 | 152 | – | – | – | – |
| Anion exchange | 4.0 | 26 | 140 | 5.5 | 100 | 1.0 |
| Hydroxyapatite (1st) | 4.0 | 9.2 | 58 | 6.3 | 40 | 1.1 |
| Hydroxyapatite (2nd) | 2.6 | 2.9 | 23 | 7.9 | 16 | 1.4 |
| Gel filtration |
2.6 |
1.0 |
12 |
12 |
8 |
2.2 |
1 nkat = 1 nmol PMTSO s−1.
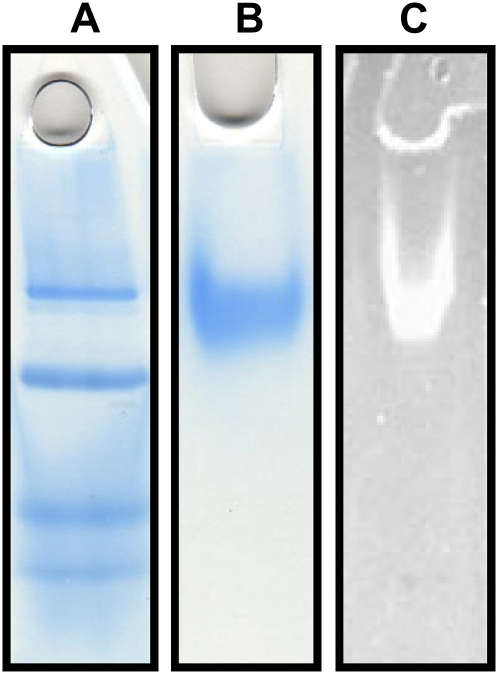
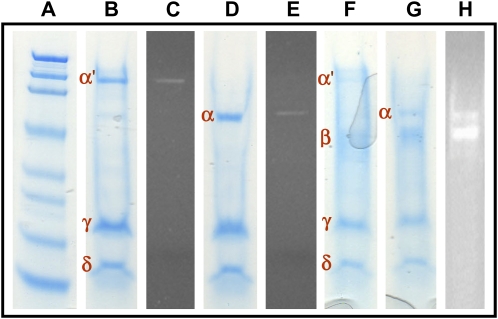
Analysis of the protein by native PAGE using Coomassie Brilliant Blue G-250 stain showed a single band (Fig. 4B), indicating the success of the purification. The diffuse appearance of the band even at low protein concentrations suggested that the protein might be glycosylated, which was confirmed through the in-gel detection of carbohydrate by oxidation of the protein-bound sugars within the gel to aldehydes, followed by reaction of the aldehyde with a hydrazide, which produced an easily detectable fluorescent conjugate (Fig. 4C). The molecular mass of the protein was determined by Ferguson plot analysis to be 217.7 kD (Supplemental Fig. S1), and its pI was observed by chromatofocusing to be 5.2 (Supplemental Fig. S2). SDS-PAGE analysis in the absence of β-mercaptoethanol (BME; Fig. 5B) showed three bands termed α′ (136.0 kD), γ (22.5 kD), and δ (11.9 kD). In the presence of BME, SDS-PAGE analysis revealed that although the γ and δ bands were retained, the α′ band had collapsed to a new band, termed α, of molecular mass 68.8 kD (Fig. 5D). Thus, in the native protein, two α-subunits are linked together by a disulfide bond to form the subunit α′. In-gel fluorescent glycoprotein detection indicated that the source of the glycosylation observed by native PAGE was the α-subunits (Fig. 5, C and E).
Figure 4.
Native PAGE characterization of P. alliacea LFS. A, Native PAGE molecular mass marker. B, Native PAGE with Coomassie Brilliant Blue staining showing the LFS enzyme purified to homogeneity. C, Native PAGE of the LFS after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate (i.e. a positive test for the presence of sugars).
Figure 5.
SDS-PAGE analysis of P. alliacea LFS and P. alliacea alliinase. A, Molecular mass marker. B, SDS-PAGE of P. alliacea LFS in the absence of BME, with Coomassie Brilliant Blue staining showing the three bands representing the subunits of which P. alliacea LFS is comprised. C, SDS-PAGE of P. alliacea LFS in the absence of BME after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate. The results show that the α′-subunit is glycosylated. D, SDS-PAGE of P. alliacea LFS in the presence of BME showing that the α′ band in B collapses into the α band seen in D. E, SDS-PAGE of P. alliacea LFS in the presence of BME after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate. The results show that the α-subunits are glycosylated. F, SDS-PAGE of P. alliacea alliinase in the absence of BME showing the bands representing the subunits of which P. alliacea alliinase is comprised. G, SDS-PAGE of P. alliacea alliinase in the presence of BME showing that the α′ band in F collapses into the α band. Comparison of D and G reveals that the P. alliacea LFS shares with the P. alliacea alliinase subunits α, γ, and δ, but not its β-subunit. H, SDS-PAGE of P. alliacea alliinase in the presence of BME after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate. The results show that its α- and β-subunits are glycosylated.
Characterization of P. alliacea LFS Activity
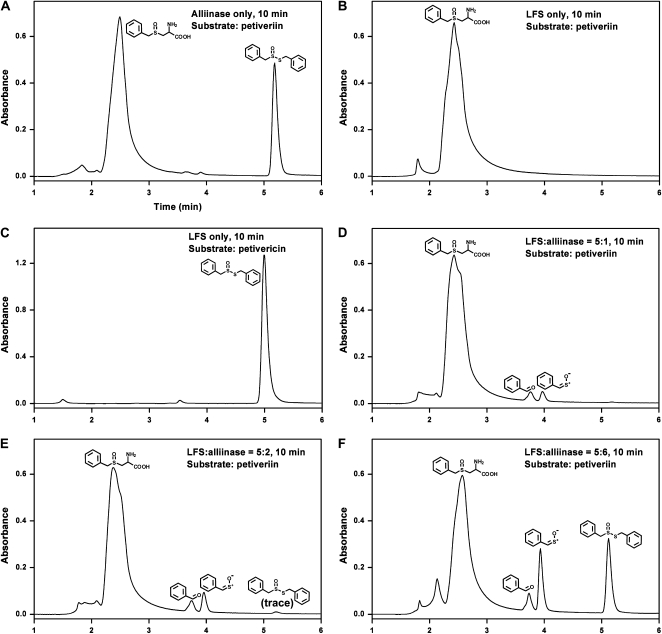
The conditions under which LFS mediates the formation of PMTSO were determined through monitoring the formation of PMTSO under a variety of conditions by HPLC. Specifically, PMTSO formation as a function of (1) ratio of LFS to alliinase; and (2) LFS in the presence of petiveriin, thiosulfinate (i.e. petivericin), and/or P. alliacea alliinase, was determined. The results of these experiments are shown in Figure 6. In the absence of LFS, alliinase, when exposed to petiveriin, produced only S-benzyl phenylmethanethiosulfinate (petivericin; Fig. 6A). LFS in the presence of petiveriin or petivericin, but in the absence of alliinase, produced no product (Fig. 6, B and C). When petiveriin was exposed to LFS:alliinase of 5:1, only PMTSO along with its hydrolysis product, benzaldehyde, were formed (Fig. 6D). When LFS:alliinase was changed to 5:2, and in the presence of petiveriin, the amount of PMTSO and benzaldehyde increased when compared with the case when LFS:alliinase was 5:1, and trace amounts of petivericin were observed (Fig. 6E). With a change in LFS:alliinase to 5:6, substantial amounts of both petivericin and PMTSO were formed (Fig. 6F).
Figure 6.
Substrate determination for P. alliacea LFS using RP C-18 HPLC. A 20 μL reaction mixture comprised of 1.0 mL of 10 mm phosphate buffer, pH 8.0, 25 μm PLP, 1.5 mm petiveriin (or 0.25 mm petivericin), and 5.7 μg of purified LFS (approximately 100 nm) at several LFS-to-alliinase molar ratios was incubated for 10 min at room temperature and then analyzed by HPLC to reveal what products, if any, were formed. The substrate used in the reaction for A, B, D, E, and F was petiveriin, and the substrate for C was petivericin. A, Only alliinase was present in the reaction; B and C, only LFS was present in the reaction; D, both LFS and alliinase were present in the reaction, and LFS:alliinase molar ratio was 5:1; E, LFS:alliinase molar ratio was 5:2; F, LFS:alliinase molar ratio was 5:3.
DISCUSSION
In this work, we report the successful isolation and purification of an enzyme exhibiting activity similar to that observed for the LFS in onion (Fig. 3). The protein, purified from pulverized root extracts by a sequence of steps comprised of ion-exchange, hydroxyapatite, and gel-filtration chromatographies (Table I), exhibits a single band by SDS-PAGE (Fig. 4B).
SDS-PAGE analysis of the protein showed it to consist of a total of four subunits: two α-subunits connected via a disulfide bond, and two additional subunits termed γ and δ (Fig. 4). The molecular mass estimates based on SDS-PAGE in the absence of BME, from largest to smallest, are approximately 138 kD (from two α-subunits of approximately 69 kD each), 22.5 kD, and 11.9 kD for the α′-, γ-, and δ-subunits, respectively. This gives a molecular mass estimate for the whole protein of approximately 172 kD. However, the molecular mass of the protein observed by Ferguson plot analysis was 217.7 kD (Supplemental Fig. S1), approximately 46 kD more than was estimated by SDS-PAGE. This discrepancy was not unexpected. It is well documented that glycoproteins as well as proteins with rigid disulfide linkages often exhibit anomalous migration profiles in SDS-PAGE (Segrest et al., 1971; Marciani and Papamatheakis, 1978). Additionally, we observed similar discrepancies with Mr determination for P. alliacea alliinase, which shares with the P. alliacea LFS four of its five protein subunits (Musah et al., 2009). In-gel carbohydrate detection experiments showed that the α′-subunit is glycosylated (Fig. 4C). The pI of the LFS was determined by chromatofocusing to be 5.2 (Supplemental Fig. S2).
After comparison of the SDS-PAGE results obtained for the LFS with those determined for the P. alliacea alliinase reported in the companion article (Musah et al., 2009), we were surprised to observe that the LFS appears to be identical to the alliinase, with the exception of the absence in the LFS of the β-subunit that is present in the alliinase (Fig. 5F). The native PAGE of both proteins shows an α′-subunit (approximately 130 to 140 kD) which, in the presence of BME, collapses into a single band of molecular mass 68 to 69 kD, and both proteins exhibit two additional subunits of similar molecular mass (approximately 24 and 13 kD for the γ- and δ-subunits, respectively). It is possible that these subunits have very slight differences that are a consequence of alternative gene splicing. This would result in small differences in protein sequences that would yield unique protein isoforms. However, the low level of resolution of the SDS-PAGE method used here does not permit us to state this with certainty. Additionally, the limited availability of protein did not allow us to explore this hypothesis further. Our results may imply though that each of these units is encoded by a single gene. Indeed, a single gene product can be a member of multiple complexes, such that the same protein or peptide can be utilized in different ways, depending upon the other peptides or proteins with which it is complexed. Surprisingly, even though the P. alliacea alliinase contains all the subunits of which the LFS is comprised, it is totally devoid of LFS activity (Musah et al., 2009). Likewise, the presence in the LFS of four of the five subunits contained in the P. alliacea alliinase does not render the LFS able to catalyze the breakdown of S-substituted Cys sulfoxides. These results are in sharp contrast to what has been shown in onion. Unlike the P. alliacea LFS, that of onion is a monomeric protein of approximately 18.6 kD whose primary structure is totally unlike that of the onion alliinase. Additionally, the onion alliinase functions as a monomer or homomultimer (Clark et al., 1998). Since detailed biochemical characterization of the onion LFS has heretofore not been reported, we are unable to further compare and contrast the two LFS enzymes.
The pI of the P. alliacea LFS was determined to be 5.2 by chromatofocusing (Supplemental Fig. S2), higher than the 4.78 value observed for the P. alliacea alliinase. This fact facilitated separation of the two proteins during the purification process. When the protein precipitate obtained by treatment of macerated P. alliacea roots with ammonium sulfate was subjected to anion-exchange chromatography, the pH used was 7.6. Thus, the LFS with its higher pI would be expected to have less affinity for the ion-exchange column than the alliinase, with the consequence that it should elute at a lower salt concentration than the alliinase. This was observed to be true, with the LFS eluting at 65 to 120 mm NaCl, and the alliinase eluting at 220 to 265 mm NaCl.
When petiveriin was exposed to P. alliacea alliinase with no LFS present, no LF (i.e. PMTSO) was formed, and only the expected thiosulfinate petivericin was observed (Fig. 6A). This is analogous to what has been reported for onion, in which exposure of isoalliin to purified alliinase yields only the corresponding thiosulfinate (Imai et al., 2002). When the P. alliacea LFS was incubated with petiveriin, as expected, no products were formed (Fig. 6B). Exposure of petivericin to the LFS in the absence of alliinase also did not result in the formation of any product (Fig. 6C). Although we expected that exposure of petiveriin to a combination of LFS and alliinase would yield the LF, we were surprised by the observation that the relative amounts of petivericin and PMTSO shifted as a function of LFS-to-alliinase ratios. Thus, when LFS:alliinase was 5:1, only PMTSO and its decomposition product, benzaldehyde, were observed (Fig. 6D). When LFS:alliinase was reduced to 5:2, increased amounts of PMTSO and benzaldehyde were observed, along with a trace amount of petivericin (Fig. 6E). When LFS:alliinase was further reduced to 5:6, substantial amounts of both PMTSO and petivericin were formed (Fig. 6F).
Given that, consistent with what has been observed in onion, the LFS does not act directly on a Cys sulfoxide precursor and only forms PMTSO when in the presence of alliinase, we conclude that the substrate of the LFS is phenylmethanesulfenic acid (PMSA) generated by alliinase-mediated breakdown of petiveriin (Fig. 7). In the absence of LFS, the sulfenic acid produced by alliinase action on petiveriin spontaneously condenses with loss of water to form petivericin (Fig. 7A). However, the presence of LFS in the milieu provides the opportunity for a second fate for the sulfenic acid (Fig. 7B). When the amount of LFS far exceeds that of the alliinase, all of the sulfenic acid produced by the alliinase is sequestered by the LFS, which catalyzes its conversion to PMTSO. Such a scenario is represented in Figure 6D, in which the only products produced are PMTSO and its decomposition product, benzaldehyde. As the amount of LFS is decreased relative to that of the alliinase (Fig. 6, E and F), the LFS is unable to sequester all of the sulfenic acid, and that which escapes condenses to form substantial amounts of petivericin. The effective entrapment of the sulfenic acid by the LFS to form PMTSO exclusively implies that the LFS and alliinase function not only in tandem, but are in close proximity, and may in fact be conjoined in the intact plant tissue.
Figure 7.
Fate of the sulfenic acid intermediate produced by alliinase-catalyzed breakdown of petiveriin. A, When no LFS is present, no PMTSO is formed from PMSA. B, When LFS is present, and depending on its concentration relative to that of the alliinase, the PMSA that is formed can be intercepted by the LFS and converted to PMTSO.
It is worth noting that our analyses of P. alliacea plants from both Florida and the Caribbean have consistently shown the amount of LFS to exceed that of the alliinase, with the actual ratios of the two enzymes varying depending upon the location and time of the year when the plants are harvested. Indeed, prior to our appreciation of the extent to which PMTSO and petivericin formation were influenced by the ratio of LFS to alliinase, we assumed, in every case in which root tissue disruption did not result in formation of significant quantities of petivericin, that the alliinase that mediates its formation had been destroyed or inactivated by some unknown mechanism. Our studies have now revealed that this notion was erroneous, and that the presence of a large excess of LFS relative to the alliinase can effectively eclipse thiosulfinate formation. In such cases, it is the formation of the sulfine lachrymator (PMTSO) rather than that of thiosulfinates that serves as evidence of the presence of the alliinase, since the alliinase is what furnishes the sulfenic acid substrate that is acted upon by the LFS to produce the sulfine.
It is also worth mentioning that although both onion and P. alliacea LFSs mediate formation of sulfines (PTSO and PMTSO, respectively), there is a significant difference in their action. The onion LFS catalyzes only a rearrangement of PSA into the isomeric sulfine (PTSO). On the other hand, the P. alliacea LFS mediates the dehydrogenation of PMSA to yield the sulfine (PMTSO; Fig. 8).
Figure 8.
The difference in the mode of action of the onion LFS and P. alliacea LFS.
Our observations may have important implications for onion, the plant in which the presence of a LFS was first reported. The ability to rationally modify the organosulfur small molecule profile of agriculturally and/or medicinally important plants is a desired goal. It would permit the capability of directing the cascade of enzyme-mediated reactions to produce a range of predicted secondary compounds with unique and desirable sensory, odor, and flavor notes, as well as health-promoting attributes. Although, to our knowledge, biochemical characterization of the onion LFS has not been reported, the connection between the discovery of the LFS and the possibility of creating an onion whose pungency can be reduced and thiosulfinate profile modified through down-regulation of the onion LFS gene has not been lost on researchers. For example, Eady et al. (2008) have shown that RNAi silencing of the onion LFS gene yields an onion with substantially reduced pungency, this attribute being replaced by a sweeter aroma reminiscent of cooked onions. Because of the dramatically reduced level of the LFS in the genetically modified onion, much of the isoalliin-derived sulfenic acid (PSA) remained unsequestered by the LFS. As predicted, this resulted in the formation of much higher concentrations of isoalliin-derived thiosulfinates. These thiosulfinates reacted further to form zwiebelanes, which were present in higher concentrations relative to what is observed in regular onion. Zwiebelanes have been shown to possess antiplatelet aggregation activity (Block, 1992; Block et al., 1996; Keusgen, 2002). In fact, the bulk of the plethora of biological activities reported for Allium crops such as garlic and onion have been ascribed to the organosulfur small molecule derivatives of precursor S-substituted Cys sulfoxides, most notably the thiosulfinates and complex compounds that are formed from them that appear further downstream, such as di-, tri-, and polysulfides, cepaenes, and the aforementioned zwiebelanes. These molecules are reported to exhibit cardiovascular effects, antimicrobial activity, and anticancer activity (Bayer et al., 1989; Dorsch et al., 1990; Block, 1992; Iranshahi et al., 2008; Block, 2010). We have shown that petivericin and other thiosulfinates produced from P. alliacea S-substituted Cys sulfoxide precursors also exhibit significant antibacterial and antifungal activity (Kim et al., 2006). As more information becomes available on the unique structural features that render many of these organosulfur derivatives biologically active, it will become increasingly desirable to have ways in which molecules with the desired features can be acquired. Our results suggest that in addition to RNAi suppression of LFS gene expression, another way in which to modify the pungency and thiosulfinate profile (and by extension the formation and identity of other organosulfur molecules further downstream) in plants containing LFS/alliinase-type systems, may be through manipulation of LFS-to-alliinase ratios. Studies on the possibility of influencing the thiosulfinate/LF profile in P. alliacea and onion, and the effects that such manipulations may have on organosulfur small molecule signatures, are currently under way.
MATERIALS AND METHODS
Plant and Materials
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich chemical company. Whole fresh plants of Petiveria alliacea were obtained from Native Habitat Landscaping and Barbados. They were collected in Vero Beach, Indian River County, FL, and stored at −30°C until analysis. A voucher specimen is deposited at the herbarium PIHG at the Florida Department of Agriculture and Consumer Services, Division of Plant Industry, Gainesville, Florida, under accession number 7801.
Reference Compounds
S-Substituted-l-Cys derivatives and the corresponding S-substituted-l-Cys sulfoxides, as well as petivericin, were synthesized according to the method of Kubec and Musah (2001) and Kubec et al. (2002).
Analytical Methods
HPLC separations were performed on a Dynamax SD-200 binary pump system, employing a Varian PDA 330 detector.
Native PAGE Analysis
Native PAGE was performed according to the method of Davis (1964) with 10% Tris-HCl Ready gels (Bio-Rad Laboratories). Prestained SDS-PAGE broad range standards (Bio-Rad Laboratories) were used as molecular mass markers. One part protein sample was mixed with two parts sample buffer. The sample buffer was comprised of 62.5 mm Tris, 40% (v/v) glycerol, and 0.01% (w/v) bromphenol blue at pH 6.8. The running buffer was comprised of 0.1 m Tris and 0.1 m Tricine at pH 8.3. The gel was run at a constant voltage of 120 V, with a starting current of 63 mA, to a final current of 32 mA. The total run time was 45 min.
SDS-PAGE Analysis
SDS-PAGE was carried out by the method of Laemmli (1970) using 10% Tris-HCl Ready gels (Bio-Rad Laboratories). Prestained SDS-PAGE broad range standards (Bio-Rad Laboratories) were used as molecular mass markers. One part protein sample was mixed with two parts sample buffer, and the resulting solution was heated at 100°C for 15 min. The sample buffer was made from 980 μL Tricine sample buffer (Bio-Rad Laboratories) and 20 μL BME. The running buffer was comprised of 0.1 m Tris, 0.1 m Tricine, and 0.1% (w/v) SDS at pH 8.3. The gel was run at a constant voltage of 120 V, with a starting current of 63 mA, to a final current of 32 mA. The total run time was 45 min.
In-Gel Staining of Proteins
To detect the proteins, Bio-Safe Coomassie G-250 stain (Bio-Rad Laboratories) was used after native PAGE and SDS-PAGE.
Molecular Mass Determination
Ferguson plot analysis (Ferguson, 1964) was used to determine the molecular mass of P. alliacea LFS: Bio-Rad's Precision Plus Protein All Blue standards (50 to 150 kD range) were used as the molecular mass markers. Native PAGE, according to the method described above, was run at three different gel concentrations (5%, 10%, 15%). The molecular mass of the protein was estimated from plotting the logarithm of the protein's mobility as a function of the gel concentration. The slope of the resulting line, i.e. the retardation coefficient (Kr), is proportional to the protein's molecular mass. The subunit molecular masses were estimated by SDS-PAGE as already described.
Isoelectric Point Measurement
The pI was determined using a chromatofocusing column (5.0 mm i.d. × 200 mm Mono P 5/200 GL, Amersham Biosciences) according to the manufacturer's specifications. The pH interval was 5.7 to 3.5. The flow rate was 0.5 mL min−1. The UV monitor wavelength was set at 280 nm.
Carbohydrate Detection
Protein glycosylation was detected using a high-sensitivity fluorescent glycoprotein detection kit (Sigma-Aldrich) according to the manufacturer's specifications. Following PAGE, the proteins were fixed in the gel with an acetic acid:methanol:water (3:50:47, v/v/v) solution. The carbohydrates on the proteins are oxidized to aldehydes with periodic acid. A hydrazide dye was reacted with the aldehydes, forming a stable fluorescent conjugate that was viewed using a standard fluorescent UV transilluminator with emission at 312 nm.
Purification of LFS from P. alliacea
P. alliacea LFS was purified following a procedure modeled after published protocols for the isolation of alliinase from shiitake mushrooms (Lentinus edodes; Kumagai et al., 2002) and LFS from onion (Imai et al., 2002). All steps were carried out at 4°C. Fresh P. alliacea roots (150 g) were carefully cleaned in water and homogenized with a blender in 400 mL of 20 mm phosphate buffer at pH 7.0, containing 20 μm PLP, 10% (v/v) glycerol, 5% (w/v) NaCl, 5% (w/v) polyvinylpolypyrrolidone, 5 mm EDTA, and 0.05% (v/v) BME. The homogenate was filtered through four layers of gauze, and the filtrate was centrifuged at 15,000g for 30 min. The 250 mL of supernatant obtained was brought to 60% (w/v) saturation with ammonium sulfate. The precipitate obtained after centrifuging at 15,000g for 30 min was resuspended in 15 mL of 20 mm Tris-HCl at pH 7.6, containing 20 μm PLP and 10% (v/v) glycerol, and dialyzed overnight against the same buffer. The dialysate obtained (20 mL) was centrifuged at 10,000g for 10 min, and then applied by HPLC in 5.0 mL aliquots to a HiPrep 16/10 DEAE fast-flow column (16 mm i.d. × 100 mm, Amersham Biosciences) that had been preequilibrated with 20 mm Tris-HCl buffer at pH 7.6 containing 10% (v/v) glycerol. The protein sample was washed first with 65 mL 20 mm Tris-HCl buffer at pH 7.6, containing 10% (v/v) glycerol. Then the column was washed again using a linear gradient, over 100 min as follows: solvent A: 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP, 10% (v/v) glycerol; solvent B: 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP, 10% (v/v) glycerol, and 280 mm NaCl. Finally, the column was washed with 30 mL of 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP, 10% (v/v) glycerol, and 280 mm NaCl. The flow rate for this column was 2.0 mL min−1. Active fractions (see “Enzyme Activity Detection”) were pooled and concentrated to 4.0 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD), and then dialyzed overnight against 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The dialysate obtained was applied in 2 mL aliquots to a hydroxyapatite column (15 mm i.d. × 113 mm, Bio-scale CHT20-I, Bio-Rad Laboratories) that had been preequilibrated with 20 mm phosphate buffer at pH 7.0, containing 20 μm PLP and 10% (v/v) glycerol. The column was washed first with 60 mL 20 mm phosphate buffer at pH 7.0, containing 20 μm PLP and 10% (v/v) glycerol, then with the following solvent combination over a linear gradient lasting 100 min: solvent A: 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol; solvent B: 400 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol, and finally with 30 mL 400 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The flow rate for this column was 1.0 mL min−1. Active fractions were pooled and concentrated to 4.0 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD), then dialyzed overnight against a 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The dialysate obtained was applied to the same hydroxyapatite column that had been equilibrated with 20 mm phosphate buffer pH 7.0, containing 20 μm PLP and 10% (v/v) glycerol. The column was washed first with 60 mL of 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol, then with the following solvent combination over a linear gradient lasting 100 min: solvent A: 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP, 10% (v/v) glycerol; solvent B: 200 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol, and finally with 30 mL of 200 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The flow rate for this column was 1.0 mL min−1. Active fractions were pooled and concentrated to 2.6 mL. The resulting sample was applied in 200 μL aliquots to a gel filtration column (7.8 mm i.d. × 300 mm, Bio-Sil SEC-250, Bio-Rad Laboratories) that had been preequilibrated with 20 mm phosphate buffer at pH 6.8 containing 0.15 m NaCl and 10% (v/v) glycerol. The loaded protein was washed with the same phosphate buffer at a flow rate of 1.0 mL min−1. Active eluents were pooled and concentrated to 2.6 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD). Protein concentrations were estimated based on the sample's A260 and A280 at a 1.0 cm path length, using the following equation: protein concentration (mg mL−1) = 1.55 × A280 − 0.75 × A260 (Simpson, 2004).
Enzyme Activity Detection
LFS activity detection was carried out in vitro in solution with the substrate petiveriin. The reaction mixture in 10 mm phosphate buffer, pH 8.0 (in a total volume of 1.0 mL), was comprised of 1.5 mm petiveriin, 25 μm PLP, 1.0 μg purified alliinase (approximately 6.9 nm), and 20 to 60 μL of LFS extract in the protein concentration range of approximately 0.4 to 7.6 mg mL−1, depending on the stage of purification and activity of the sample (see “Purification of LFS from P. alliacea”). The mixtures were incubated for 10 min at room temperature, and then 10 to 20 μL of the reaction solution was analyzed by HPLC using an analytical RP C-18 column (Microsorb-MV 100Å, 250 × 4.6 mm, 5 μm, Varian) under the following conditions: flow rate: 1.0 mL min−1; mobile phase: water:acetonitrile (30:70, v/v); detection wavelength: 210 nm. Under these conditions, benzaldehyde, the PMTSO hydrolysis product, elutes at 3.75 min, whereas PMTSO elutes at 3.94 min. Both are observed if the fraction being analyzed exhibits LFS activity. The concentration of PMTSO and benzaldehyde were calculated from their molar extinction coefficients (4,387 L mol−1 cm−1 for PMTSO and 11,040 L mol−1 cm−1 for benzaldehyde) at 210 nm. The identity of eluted PMTSO and benzaldehyde peaks were verified by ESI-HRMS and by comparison with authentic compounds.
Enzyme Substrate Determination
The enzyme substrate was determined by a method similar to that described above (see “Enzyme Activity Detection”) except that: (1) the amount of purified LFS used was 5.7 μg (approximately 34 nm), and several different LFS:P. alliacea alliinase molar ratios (5:0, 5:1, 5:2, 5:6, and 0:6) were used; and (2) the reaction mixture was incubated for 10 min with and without petiveriin and petivericin (0.25 mm).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular mass determination of P. alliacea LFS by Ferguson plot analysis.
Supplemental Figure S2. P. alliacea LFS pI determination by chromatofocusing.
Supplemental Materials and Methods S1. Supplemental Materials and Methods corresponding to Supplemental Figures S1 and S2.
Supplementary Material
Acknowledgments
The technical assistance provided by the Scripps Research Institute Center for Mass Spectrometry is appreciated. The authors thank Distinguished Professor Dr. Eric Block for helpful discussions and critical reading of the manuscript.
This work was supported by the National Science Foundation (grant no. 0239755) and the Research Foundation of the State University of New York.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rabi A. Musah (musah@albany.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bayer T, Wagner H, Block E, Grisoni S, Zhao SH, Neszmelyi A (1989) Zwiebelanes: novel biologically active 2,3-dimethyl-5,6-dithiabicyclo[2.1.1]hexane 5-oxides from onion. J Am Chem Soc 111 3085–3086 [Google Scholar]
- Block E (1992) The organosulfur chemistry of the genus Allium—implications for the organic chemistry of sulfur. Angew Chem Int Ed Engl 31 1135–1178 [Google Scholar]
- Block E (2010) Garlic and Other Alliums: The Lore and the Science. Royal Society of Chemistry, Cambridge, UK
- Block E, Bayer T, Naganathan S, Zhao SH (1996) Allium chemistry: synthesis and sigmatropic rearrangements of alk(en)yl 1-propenyl disulfide S-oxides from cut onion and garlic. J Am Chem Soc 118 2799–2810 [Google Scholar]
- Clark SA, Shaw ML, Every D, Lancaster JE (1998) Physical characterization of alliinase, the flavor generating enzyme in onions. J Food Biochem 22 91–103 [Google Scholar]
- Davis B (1964) Disk electrophoresis-II method and application to human serum proteins. Ann N Y Acad Sci 121 404–427 [DOI] [PubMed] [Google Scholar]
- Dorsch W, Schneider E, Bayer T, Breu W, Wagner H (1990) Anti-inflammatory effects of onions: inhibition of chemotaxis of human polymorphonuclear leukocytes by thiosulfinates and cepaenes. Int Arch Allergy Appl Immunol 92 39–42 [DOI] [PubMed] [Google Scholar]
- Eady CC, Kamoi T, Kato M, Porter NG, Davis S, Shaw M, Kamoi A, Imai S (2008) Silencing onion lachrymatory factor synthase causes a significant change in the sulfur secondary metabolite profile. Plant Physiol 147 2096–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KA (1964) Starch-gel electrophoresis: application to the classification of pituitary proteins and polypeptides. Metabolism 13 985–1002 [DOI] [PubMed] [Google Scholar]
- Musah RA, He Q, Kubec R, Jadhav A (2009) Studies of a novel cysteine sulfoxide lyase from Petiveria alliacea: the first heteromeric alliinase. Plant Physiol 151 1304–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Tsuge N, Tomotake M, Nagatome Y, Sawada H, Nagata T, Kumagai H (2002) An onion enzyme that makes the eyes water. Nature 419 685. [DOI] [PubMed] [Google Scholar]
- Iranshahi M, Fata A, Emami B, Shahri BMJ, Bazzaz BSF (2008) In vitro antifungal activity of polysulfides-rich essential oil of Ferula latisecta fruits against human pathogenic dermatophytes. Nat Prod Commun 3 1543–1546 [Google Scholar]
- Keusgen M (2002) Health and Alliums. In HD Rabinowitch, L Currah, eds, Allium Crop Science: Recent Advances. CAB International, Wallingford, UK, pp 357–378
- Kim S, Kubec R, Musah RA (2006) Antibacterial and antifungal activity of sulfur-containing compounds from Petiveria alliacea L. J Ethnopharm 104 188–192 [DOI] [PubMed] [Google Scholar]
- Kubec R, Kim S, Musah RA (2002) S-substituted cysteine derivatives and thiosulfinate formation in Petiveria alliacea—part II. Phytochemistry 61 675–680 [DOI] [PubMed] [Google Scholar]
- Kubec R, Kim S, Musah RA (2003) The lachrymatory principle of Petiveria alliacea. Phytochemistry 63 37–40 [DOI] [PubMed] [Google Scholar]
- Kubec R, Musah RA (2001) Cysteine sulfoxide derivatives in Petiveria alliacea. Phytochemistry 58 981–985 [DOI] [PubMed] [Google Scholar]
- Kumagai H, Kono H, Sakurai H, Tokimoto K (2002) Comparison of C-S lyase in Lentinus edodes and Allium sativum. Biosci Biotechnol Biochem 66 2560–2566 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Marciani DJ, Papamatheakis JD (1978) Anomalous behavior of the major avian myeloblastosis virus glycoprotein in the presence of sodium dodecyl sulfate. J Virol 26 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest JP, Jackson RL, Andrews EP, Marchesi VT (1971) Human erythrocyte membrane glycoprotein: a re-evaluation of the molecular weight as determined by SDS polyacrylamide gel electrophoresis. Biochem Biophys Res Commun 44 390–395 [DOI] [PubMed] [Google Scholar]
- Shimon LJW, Rabinkov A, Shin I, Miron T, Mirelman D, Wilchek M, Frolow F (2007) Two structures of alliinase from Allium sativum L.: apo form and ternary complex with aminoacrylate reaction intermediate covalently bound to the PLP cofactor. J Mol Biol 366 611–625 [DOI] [PubMed] [Google Scholar]
- Simpson RJ (2004) Purifying Proteins for Proteomics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 676
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.