Abstract
Under nitrogen-limiting conditions, legumes interact with symbiotic rhizobia to produce nitrogen-fixing root nodules. We have previously shown that glutathione and homoglutathione [(h)GSH] deficiencies impaired Medicago truncatula symbiosis efficiency, showing the importance of the low Mr thiols during the nodulation process in the model legume M. truncatula. In this study, the plant transcriptomic response to Sinorhizobium meliloti infection under (h)GSH depletion was investigated using cDNA-amplified fragment length polymorphism analysis. Among 6,149 expression tags monitored, 181 genes displayed significant differential expression between inoculated control and inoculated (h)GSH depleted roots. Quantitative reverse transcription polymerase chain reaction analysis confirmed the changes in mRNA levels. This transcriptomic analysis shows a down-regulation of genes involved in meristem formation and a modulation of the expression of stress-related genes in (h)GSH-depleted plants. Promoter-β-glucuronidase histochemical analysis showed that the putative MtPIP2 aquaporin might be up-regulated during nodule meristem formation and that this up-regulation is inhibited under (h)GSH depletion. (h)GSH depletion enhances the expression of salicylic acid (SA)-regulated genes after S. meliloti infection and the expression of SA-regulated genes after exogenous SA treatment. Modification of water transport and SA signaling pathway observed under (h)GSH deficiency contribute to explain how (h)GSH depletion alters the proper development of the symbiotic interaction.
Plants often establish symbiotic relationships with specific fungi and bacteria in order to acquire the nutrients to support growth and development. Leguminous plants are able to interact with soil bacteria, commonly named rhizobia, to produce a particular root organ, the nodule, in which the rhizobia reduce atmospheric N2 to ammonia that is subsequently exported and assimilated by the plant. In parallel, the host plant gives the symbiont the carbon source to support the energy-consuming nitrogen fixation.
Cross talk between the plant and bacteria is essential for the establishment of the symbiosis. Plant signal molecules attract the bacteria to the root surface and induce the biosynthesis of bacterial lipochitooligosaccharides, termed Nod factors. Nod factors recognized by the plant act as signaling molecules able to trigger the plant symbiotic program (Riely et al., 2004; Stacey et al., 2006a).
The first evident sign of symbiosis is the deformation of root hairs in the susceptible root infection zone (IZ). Within the curled root hairs, rhizobia promote the formation of plant-derived infection threads that contain the bacteria. Concomitantly, the cortical cells below the infection threads dedifferentiate and begin to divide to form a nodule primordium. The infection threads allow the transport of the bacteria from the root surface into the nodule primordium that give rise to the nitrogen-fixing organ (Patriarca et al., 2004).
The establishment of this symbiosis, including the regulation of bacterial infection of root tissues, is strictly controlled by the plant (Vasse et al., 1993; Penmetsa and Cook, 1997). Evidence suggests that the balance between the production of reactive oxygen species and their detoxification by antioxidant defense plays a role in the nodulation process. Reactive oxygen species have been shown to be produced during the early steps of the symbiotic interaction (Santos et al., 2001) and to modulate the nodulation-related processes (Ramu et al., 2002; D'Haeze et al., 2003; Shaw and Long, 2003; Lohar et al., 2007). Moreover, bacterial mutants deficient in antioxidant defense genes exhibit a reduced nodulation efficiency (Jamet et al., 2003; Harrison et al., 2005).
Glutathione (GSH) and its legume-specific homolog homoglutathione [(h)GSH], which are antioxidant molecules with important roles in plant defense against biotic and abiotic stresses, as well as in the maintenance of the redox state, play a major role in Medicago truncatula nodulation. Using both pharmacological and transgenic approaches, we showed that depletion in (h)GSH alters the formation of root nodules (Frendo et al., 2005). A strong reduction in the number of nascent nodules and in the expression of the early nodulin genes, Mtenod12 and Mtenod40, was observed in (h)GSH-depleted plants, suggesting that (h)GSH plays a critical role during the first steps of nodule organogenesis.
To understand the (h)GSH regulation of nodule formation, a transcript profiling analysis, using cDNA-amplified fragment length polymorphism (AFLP), was performed to study the effect of (h)GSH depletion on nodulation in M. truncatula. To identify (h)GSH-dependent genes, l-buthionine-[S-R]-sulfoximine (BSO), a specific inhibitor of (h)GSH synthesis, was added before infection with Sinorhizobium meliloti, and the expression profiles were compared to the profiles of untreated infected roots. A collection of 181 gene tags was obtained, classifiable in two clusters, with (h)GSH-dependent expression changes at different time points during the first 4 d of the nodulation process. The functional classification of the corresponding genes indicates that (h)GSH depletion results in down-regulation of genes involved in meristem formation and up-regulation of a wide variety of stress-related genes during the first hours following inoculation. Interestingly, this induction was associated with a modification of the SA perception in M. truncatula roots deficient in (h)GSH.
RESULTS AND DISCUSSION
In a previous study, we showed that (h)GSH plays a critical role in the proper development of the symbiotic interaction between M. truncatula and S. meliloti (Frendo et al., 2005). Indeed, a strong diminution in the number of nascent nodules and in the expression of the early nodulin genes, Mtenod12 and Mtenod40, was observed in GSH and (h)GSH-depleted plants. In contrast, modifications in the number of infection events or changes in the expression of the rhizobium-induced peroxidase rip1 were not observed. To characterize genes of which the expression is dependent upon (h)GSH levels during the first days of the nodulation process in M. truncatula, we compared the expression profiles from inoculated control and (h)GSH-deficient roots at different stages after infection using the cDNA-AFLP transcriptomics technology.
Effect of (h)GSH Depletion on Gene Expression
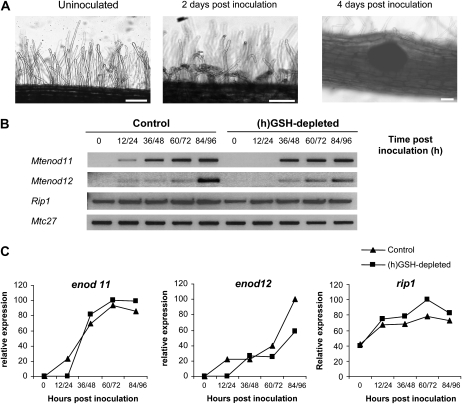
(h)GSH content was depleted by treatment with 0.1 mm BSO during 1 week (Frendo et al., 2005). HPLC analysis showed that this treatment led to an 85% reduction of total cellular (h)GSH in roots (data not shown). Control and (h)GSH-depleted roots were then inoculated with S. meliloti RCR2011 strain, and root samples corresponding to the zone susceptible for Rhizobium infection were collected for 96 h at 12-h intervals, resulting in nine sampling times for each treatment (a scheme of the experimental design and data analysis is presented in Supplemental Fig. S1). In parallel, samples, collected at the same time points as the transcript profiling, were checked by microscopy and reverse transcription (RT)-PCR analyses to verify the symbiotic progression. Typical morphological changes of the early nodulation events, root hair curling and nodule bumps, were visible in inoculated samples 48 and 96 h postinoculation (hpi), respectively (Fig. 1A), as previously reported (Timmers et al., 1999). No significant difference in the infection phenotype was observed between control and (h)GSH-depleted plants. RT-PCR analysis showed that expression of rip1, a marker of the early infection events (Cook et al., 1995), was induced similarly in the inoculated control and (h)GSH-depleted plants (Fig. 1, B and C). In contrast, expression of the early nodulin genes Mtenod11 (Journet et al., 2001) and Mtenod12 (Pichon et al., 1992) was delayed in (h)GSH-depleted plants when compared to control plants (Fig. 1, B and C).
Figure 1.
M. truncatula root responses to S. meliloti inoculation. A, Morphological analysis during nodulation. Pictures correspond to the control time point before inoculation (not inoculated), the central time point after inoculation (2 d postinoculation), and the last time point (4 d postinoculation) of the material harvested for cDNA-AFLP analysis. Root hair curling is clearly visible in the IZ 2 d postinoculation compared to the uninoculated root. Nodule primordia are visible 4 d postinoculation. Bars = 20 μm. B, The expression of enod11, enod12, and rip1 was analyzed by RT-PCR using the RNA samples collected to perform the cDNA-AFLP analysis. C, Expression levels of enod11, enod12, and rip1 analyzed by RT-PCR were standardized using Mtc27 expression as control and quantified as percentage of the maximum level of expression. Quantifications were performed using Image J software.
The expression pattern of the different samples was analyzed by cDNA-AFLP using 128 primer combinations with selectivity +2/+1. Gel images were analyzed quantitatively using AFLP-QUANTARPRO image analysis software, and 6,149 expression tags were scored, corresponding to around 23% of the M. truncatula genome. The raw intensity values were calculated and corrected for differences in total lane loading, and each individual gene expression profile was variance normalized. To select differentially expressed genes during the nodulation process, a coefficient of variance (CVnod) ≥0.4, corresponding to a 3.7-fold change between the maximum and the minimum tag expression value, was retained as a measure of differential expression. This threshold leads to the selection of 299 gene tags (group A) of which the expression is modulated during the nodulation process (see Supplemental Table S2, list A). To select genes affected by (h)GSH depletion, ANOVA (P ≤ 0.05), which allowed the comparison of gene expression values of the two sample groups corresponding to the inoculated control and (h)GSH-depleted plants, was performed. This resulted in group B, a collection of 181 gene tags (see Supplemental Table S2, list B). Hierarchical average linkage clustering analysis (Eisen et al., 1998) revealed two main clusters, one corresponding to genes down-regulated and the other to genes up-regulated as a consequence of (h)GSH depletion.
Tags with a significant differential expression level were isolated, sequenced, and analyzed. BLASTN against the database of MtGI (http://www.tigr.org) and MENS (http://medicago.toulouse.inra.fr) and BlastX algorithms against the nonredundant protein database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) were used to identify the gene tags, choosing as threshold similarity an e-value of 10−3 (Altschul et al., 1997). Out of the total, 61% were similar to genes with known function and 21% to genes with unknown function, and 18% did not exhibit any homology to known sequences in the databases. The differentially expressed genes were functionally classified according to the categories derived from the MENS database (Journet et al., 2002), showing that metabolic classes having a high relative abundance in comparison to the MENS libraries were “Defense and Cell Rescue,” “Chromatin and DNA Metabolism,” and “Secondary and Hormone Metabolism” categories (data not shown).
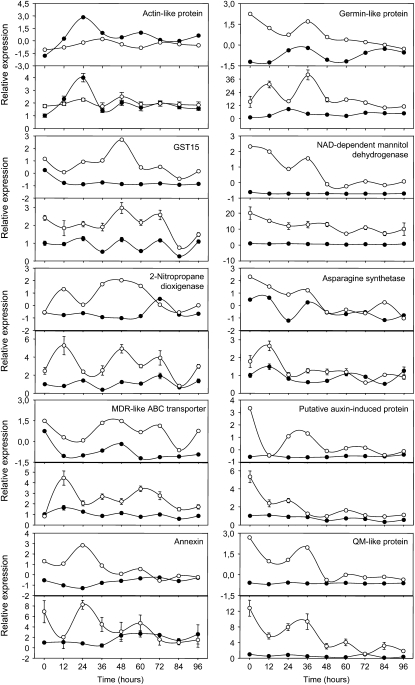
Quantitative RT-PCR (qRT-PCR) was performed on a subset of 10 genes using cDNA samples from the AFLP experiment and a biological repetition of templates to confirm the expression profiles obtained with the cDNA-AFLP analysis. The qRT-PCR analysis revealed relative transcript levels similar to those obtained with the cDNA-AFLP analysis (Fig. 2). Indeed, as seen in Figure 2, transcript levels corresponding to the GST15, the germin-like and the NAD-dependent mannitol dehydrogenase, are higher in (h)GSH-depleted root IZs than in control IZs during the full time-course analysis. Similarly, the transcript level corresponding to a putative multidrug resistance-like ATP binding cassette transporter is higher in (h)GSH-depleted IZs than in control IZs during the 4 d postinoculation. The level of transcript corresponding to the nitropropane dioxigenase is higher in (h)GSH-deficient IZs than in control IZs during the first 2 d of inoculation. A diminution of the transcript level corresponding to a QM-like protein and the putative auxin-induced protein is observed in (h)GSH-depleted IZ compared to the control IZ in which the transcript level of these proteins is not significantly modified. A higher level of transcript corresponding to an actin-like protein is higher in control IZ than in (h)GSH-deficient IZ 1 d postinoculation. Transcript levels of an Asn synthetase and an annexin are higher in the (h)GSH-depleted IZs than in the control IZs during the first 36 hpi. Taken together, these results confirm the expression profiles obtained by cDNA-AFLP analysis.
Figure 2.
qRT-PCR analysis confirming gene tag profiles obtained by cDNA-AFLP analysis. The cDNA-AFLP and the qRT-PCR profiles are presented on the top and bottom panels, respectively. Black circles correspond to inoculated control samples and white circles to inoculated (h)GSH-deficient plants. qRT-PCR levels represented the means ± se of six qRT-PCR analyses (technical triplicates of two biological experiments).
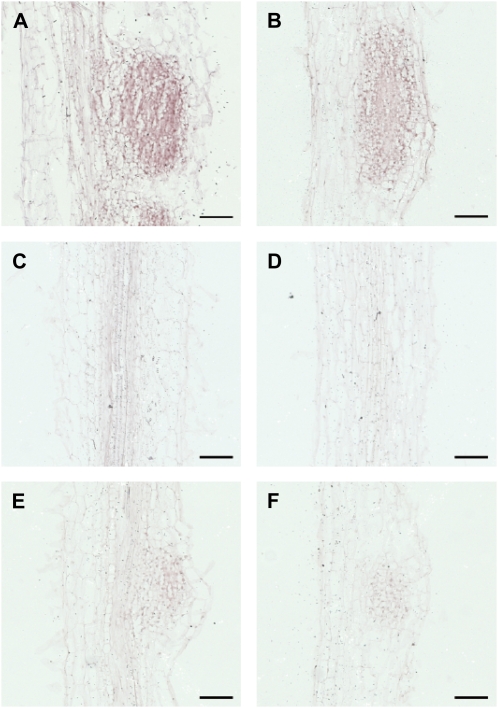
In this context, to test whether the gene tags for which the expression level was found to be up-regulated during the nodulation process (Supplemental Table S2, list A) were linked to the nodule development, the transcript localization of one gene tag belonging to group A was analyzed by in situ hybridization. The endo-1,4-β-glucanase gene (TC108882), found to be significantly induced during the nodulation process by the cDNA-AFLP analysis (CVnod = 1.05), was chosen to perform this experiment (Fig. 3). Control and (h)GSH-depleted plants were inoculated or not, and IZs were harvested 96 h later. Slides exposed to the sense probe showed a strong and unspecific staining in every tissue of the roots and the nodules (data not shown). Inoculated root slides exposed to an antisense probe showed coloration, both in the control and in (h)GSH-depleted plants (Fig. 3, A and B), whereas no staining was detected in uninoculated roots exposed to the antisense probe (Fig. 3, C and D). Staining was mainly localized in the cells of young developing nodules, indicating that the expression of the endo-1,4-β-glucanase is connected to nodule meristem formation. The control slides exposed to the digoxigenin antibody but to neither probe showed background staining (Fig. 3, E and F). These results are consistent with the transcriptomic analysis as this gene was more expressed in inoculated roots than in uninoculated ones. Moreover, they show that this endo-1,4-β-glucanase presents an expression pattern connected to nodule development.
Figure 3.
Localization of endo-1,4-β-glucanase transcripts in M. truncatula roots by in situ hybridization. A, Control plants 96 hpi showing a positive staining in developing nodule cells (purple coloration). B, (h)GSH-deficient plants (96 hpi) showing positive staining in developing nodule cells. C and D, Uninoculated root sections showing the absence of coloration in control plants and a (h)GSH-deficient plant, respectively. E and F, sections exposed to the antibody but to neither probe of control plants 96 hpi and (h)GSH-deficient plants 96 hpi, respectively. Bars = 100 μm. [See online article for color version of this figure.]
A low number of sequences corresponding to the nodule-specific or nodule-enhanced genes encoding nodulins was found in our analysis. This was due to the fact that only a few nodulin genes contain MseI and BstYI restriction sites, corresponding to the restriction enzymes used to perform the cDNA-AFLP analysis. Nevertheless, MtN6 was found to have a high expression at 84 and 96 hpi as described by Gamas et al. (1996). Moreover, the analysis of the RNA samples used for the cDNA-AFLP analysis clearly showed that Mtenod11 and Mtenod12 were expressed in our experiment (Fig. 1B).
Several studies have extensively described the expression profile of nodulated roots (Mitra et al., 2004; Lohar et al., 2006; Starker et al., 2006; Jones et al., 2008). In our study, among the 299 gene tags found to be differentially expressed during the first 4 d of the nodulation process (CVnod ≥0.4), >50% have been already described in the above reports, illustrating the reliability of the transcriptomic analysis. As previously described by Mitra et al. (2004) and Lohar et al. (2006), a transient accumulation of transcripts corresponding to putative defense and stress-related genes, such as a peroxidase precursor (TC100904) or a isoflavone reductase (TC96039), was observed around 12 to 24 hpi. Increased levels of transcripts corresponding to genes belonging to the cell wall (endo-1,4-β-glucanase already mentioned, TC108882), cytoskeleton (β-tubulin, TC106341), and chromatin/DNA metabolism (Histone H2B, TC94030) classes were also observed. The genes belonging to these metabolic classes are mainly involved in the formation of the infection thread and the development of nodule meristems (El Yahyaoui et al., 2004; Lohar et al., 2006).
The accumulation of transcripts of other genes involved in the cellular rearrangement and protein synthesis and processing, such as the putative microtubule organization protein (AF367245), putative kinesins (TC108829 and TC95595), and multiple putative ribosomal proteins (TC94277, TC92964, TC100590, TC107505, and TC94154), also confirm the modification of the cellular metabolism.
Analysis of Genes Regulated by (h)GSH Depletion during the Nodulation Process
Our goal was to identify genes of which the expression was directly or indirectly modified by (h)GSH in order to find the causes of the nodulation deficiency observed under these conditions. A total of 181 genes were identified by ANOVA (P < 0.05) to be differentially regulated under (h)GSH depletion (Supplemental Table S2, list B). Among them, 114 genes also belonged to group A (CVnod ≥0.4) showing that 38% (114/299) of group A are affected by (h)GSH depletion. The modification of the level of expression of numerous genes involved in the nodulation process reinforces the hypothesis for a key role of (h)GSH in the establishment of the symbiosis.
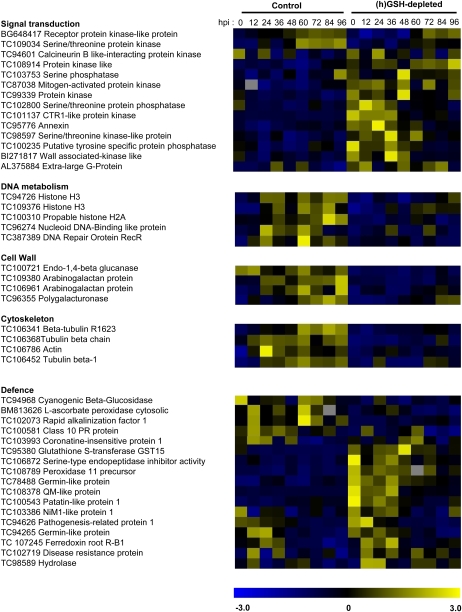
Modulation of GSH content has been suggested to affect different signaling mechanisms involved in perception and transduction of environmental cues (Gomez et al., 2004). Expression of genes corresponding to a putative Ser/Thr kinase (TC109034) and a receptor protein kinase-like protein (BG648417) was diminished in (h)GSH-depleted conditions (Fig. 4). In contrast, expression of multiple protein phosphatases (TC103753, TC102800, and TC100235), protein kinases (TC98597, TC87038, BI271817, TC101137, and TC108914), an extra large G-protein (AL375884), and an annexin (TC95776) was higher in (h)GSH-depleted plants than in controls. The modification of the level of expression of genes belonging to the signal transduction and posttranslational regulation class reflects the cellular adjustments occurring in (h)GSH-depleted plants.
Figure 4.
Functional gene categories modulated by hGSH depletion during the nodulation process. The normalized expression levels of genes were compared between inoculated control plants (C0 to C96) and inoculated (h)GSH-depleted plants (B0 to B96). The expression level ranged from −3 (blue) to +3 (yellow).
Moreover, the hormonal balance may also be disturbed by (h)GSH depletion; indeed, the expression of genes involved in auxin (multidrug resistance-like ATP binding cassette transporter, TC95110) and ethylene (Constitutive Triple Response1-like protein kinase, TC101137) signaling is modified under these conditions, and the role of both hormones in nodule development is well documented (Prayitno et al., 2006; Wasson et al., 2006; Zhang et al., 2009). In this way, gibberellin signaling may be also affected (see Gibberellin 2-oxidase, BI273162).
We have previously shown that the number of nodules and secondary meristems was reduced in (h)GSH-depleted plants (Frendo et al., 2005). GSH is known to be essential for the regulation of the cell cycle and for root meristem activity (Sanchez-Fernandez et al., 1997; Vernoux et al., 2000). With respect to this, the cDNA-AFLP analysis showed that transcripts of genes related to the chromatin and DNA metabolism class, such as a nucleoid DNA binding-like protein (TC96274), a DNA repair protein RecR (AL387389), two histones H3 (TC94726 and TC109376), and a histone H2A (TC100310), accumulate at higher levels in control plants compared to (h)GSH-depleted plants. Nodule organogenesis involves major cytoskeleton rearrangements and increased cell wall formation for infection thread formation and cell division. These biological responses involve the induction of genes associated with the cytoskeleton rearrangement and cell wall formation (El Yahyaoui et al., 2004; Lohar et al., 2006). Our results show that the expression of an actin (TC106786) and three β-tubulin (TC106452, TC106341, and TC106368) genes was affected by (h)GSH depletion, suggesting that the reorganization and proliferation of cytoskeletal elements occurring during early nodule development are directly or indirectly affected by (h)GSH depletion. Similarly, genes belonging to the cell wall metabolic class, such as arabinogalactan proteins (TC109380 and TC106961), polygalacturonase (TC96355), and a membrane-anchored endo-1,4-β-glucanase (TC100721), were less expressed following (h)GSH depletion.
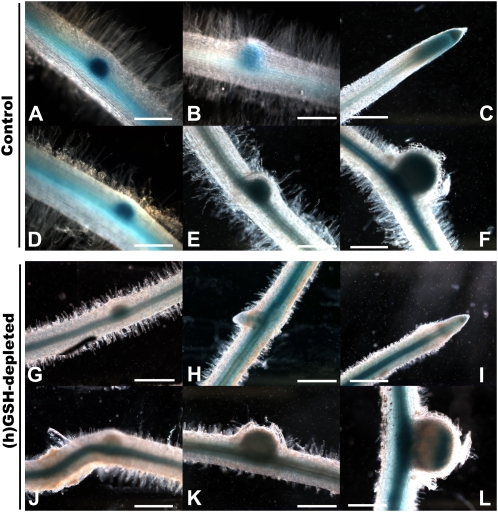
Some of these genes, encoding proteins involved in the connection between the extracellular matrix and the cytoplasm, such as a putative wall-associated kinase (BI271817), may be linked to infection thread growth. Recently, the plasma membrane aquaporin AtPIP2;1-GFP fusion was shown to label the apoplastic interface of the infection threads (Fournier et al., 2008). In this context, the expression of the putative aquaporin MtPIP2 (TC95581), which was up-regulated during nodulation (CVnod = 0.41) with a lower expression in (h)GSH-depleted plants (P < 0.05), was analyzed using a promoter-reporter fusion during 96 hpi. The putative promoter region of MtPIP2 was fused to a GUS reporter gene, and the histochemical analysis of GUS activity was followed after S. meliloti inoculation in control and (h)GSH-depleted roots of chimeric plants (Fig. 5). In control plants, staining was observed in the vascular tissue. Strong GUS expression was observed in the lateral root primordia (Fig. 5, A and B), becoming restricted to the root tip once the lateral roots grew out of the primary root (Fig. 5C). Strong GUS staining was also detected in the nodule primordia and in young nodules of control plants (Fig. 5, D–F). These results indicate that MtPIP2 expression might be expressed in the vascular tissue and up-regulated in mitotically activated cells. In (h)GSH-depleted roots, staining was also observed in vascular tissue, lateral root primordia (Fig. 5, G and H), root tip (Fig. 5I), nodule primordia (Fig. 5, J and K), and the young nodule (Fig. 5L). In contrast to control plants, GUS staining in (h)GSH-depleted roots was not higher in mitotically activated cells than in vascular tissue, showing that the up-regulation observed in mitotically activated cells in control roots is inhibited under (h)GSH depletion. These results, which strengthen the transcriptomic analysis, suggest that the putative MtPIP2 is involved in meristem formation. Moreover, the expression of MtPIP2 in the IZs of young nodules (Fig. 5L) suggest that MtPIP2 is implicated in the infection thread growth, as suggested by the localization of AtPIP2 (Fournier et al., 2008). The involvement of aquaporins in the nitrogen-fixing process has been demonstrated by the analysis of the NOD26 protein, a member of the aquaporin water channel family, which is a major protein component of the symbiosome membrane (Fortin et al., 1987; Sandal and Marcker, 1988). Here, we show that another aquaporin exhibits a modification of its expression during the earlier steps of the nodulation process. Taken together, these results highlight a possible regulatory role of (h)GSH on water exchange during the establishment of symbiosis.
Figure 5.
Transgenic M. truncatula roots for the PIP2 aquaporin promoter:GUS fusion. Blue staining indicates GUS expression. A to C, GUS expression at different stages of lateral root formation in control plants. D to F, GUS expression at different stages of nodule formation in control plants. G to I, GUS expression at different stages of lateral root formation in (h)GSH-depleted plants. J to L, GUS expression at different stages of nodule formation in (h)GSH-depleted plants. Bars = 100 μm.
The Expression of Defense and Cell Rescue Genes Is Modulated during Nodulation following (h)GSH Depletion
A significant modulation of the level of expression of genes belonging to the defense and cell rescue class was observed in (h)GSH-depleted plants. Moreover, several genes belonging to other metabolic classes but involved in multiple aspects of plant defense exhibited a similar behavior. Genes encoding a Ser-type endopeptidase inhibitor (PI; TC106872), a disease resistant protein (TC102719), a glutathione-S-transferase (GST; TC95380), a germin-like protein (TC95623), a peroxidase (TC108789), a QM-like protein (TC108378), a patatin-like protein (TC100354), and a germin-like protein (TC94265) were significantly more expressed in (h)GSH-depleted plants than in controls. In contrast, a higher expression of a cyanogenic β-glucosidase (TC94968), a cytosolic ascorbate peroxidase (BM813626), a class 10 Pathogenesis Related protein (TC100581), and Rapid Alkalinization Factor1 (MtRALF1; TC102073) was detected in inoculated control plants than in (h)GSH-depleted plants. Recently, expression analysis of MtRALFL1 confirmed that this gene is up-regulated during initial phases of nodulation (Combier et al., 2008). Moreover, the involvement of MtRALFL1 in symbiosis regulation was suggested as overexpression of this protein in M. truncatula roots resulted in a marked reduction in the number of nodules formed, as well as a strong increase in the number of aborted infection threads and in an abnormal nodule development (Combier et al., 2008).
Jasmonic acid (JA) is a key actor of the plant response to biotic stress (Kunkel and Brooks, 2002; Katagiri, 2004). It is worth noting that the significant up-regulation of a putative Coronatine Insensitive1 gene (TC103993), which is required for JA-dependent responses (Feys et al., 1994; Xie et al., 1998; Lorenzo and Solano, 2005), is observed 12 and 24 hpi in control plants and is delayed by (h)GSH depletion. The implication of JA during the nodulation process needs to be clarified, as multiple reports suggested its association with the symbiotic interaction. Interestingly, JA seems to regulate the infection process by inhibiting the plant's response to bacteria, with direct effects on Nod factor-induced calcium spiking (Sun et al., 2006). Moreover, mutation in the soybean (Glycine max) GmNARK encoding a CLAVATA1-like Ser/Thr receptor-like kinase, which causes excessive nodule development, induces about a 2-fold higher level of JA in the leaves of the mutant than in those of the wild-type plant (Seo et al., 2007).
On the other hand, a higher level of expression of a putative Nonexpressor of PR genes1 gene (NPR1; TC103386), encoding a central positive regulator of salicylic acid (SA)-induced gene expression (Dong, 2004), was observed in inoculated (h)GSH-depleted plants compared to inoculated control plants during the first 36 hpi. Likewise, the Pathogenesis-Related1 Protein gene (PR1; TC94626), a marker gene for the SA-dependent transduction pathway (Glazebrook et al., 2003), was more expressed in the (h)GSH-depleted plants than in the controls during the first 24 hpi. Thus, (h)GSH depletion appears to modify the SA-dependent response. SA has been extensively described as a plant hormone involved in the plant response to pathogens (Durrant and Dong, 2004; Loake and Grant, 2007) and abiotic stress (Borsani et al., 2001; Mateo et al., 2006). During symbiosis, the accumulation of SA by exogenous treatment inhibits the nodule primordia formation (Martinez-Abarca et al., 1998). Also, whereas an SA concentration of 0.1 mm completely blocks nodulation of vetch by Rhizobium leguminosarum bv viciae and inhibits the mitogenic effect of Nod factors, it does not affect Nod factor-induced root hair deformation (van Spronsen et al., 2003). Interestingly, this phenotype is similar to the one observed in (h)GSH-depleted plants, which show a strong reduction in the number of nascent nodules without modifying root hair curling (Frendo et al., 2005). Finally, the reduction of the endogenous SA levels in transgenic plants led to an enhanced nodulation and infection that pointed to an important role for SA-mediated plant defense pathways in controlling nodule formation (Stacey et al., 2006b).
To test whether the SA level was altered in (h)GSH-depleted roots, SA concentration was quantified by HPLC in these plants. HPLC analysis of SA and SA conjugate (SAG) content showed that SA concentration was not significantly higher in (h)GSH-deficient roots than in control roots with amounts of free SA ranging from 20 to 250 ng g−1 fresh roots.
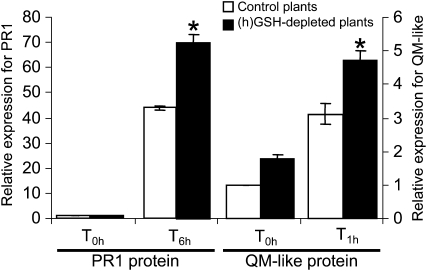
SA signaling is mediated by both NPR1-dependent mechanisms and NPR1-independent mechanisms (Shah, 2003), although the former pathway is better characterized. SA induces redox changes leading to the reduction of oligomeric NPR1 to active monomers. These monomers interact with the basic Leu zipper transcription factors, termed TGA factors, leading to the expression of SA-dependent genes (Mou et al., 2003). In this context, (h)GSH depletion in M. truncatula roots may alter the cellular redox state and modulate the expression of SA-dependent genes leading to an increased accumulation of their transcripts in (h)GSH-depleted plants. In a first step to test this hypothesis, the induction of a few marker genes found to be significantly induced in the (h)GSH-depleted background, namely, PI (TC106872), GST (TC95380), QM-like protein (TC108378), and PR1 (TC94626), by SA was tested by exogenous SA treatment of control roots. QM-like protein and the PR1 were found to be more expressed in SA-treated roots than in control roots, showing that these genes could be used as marker genes for the plant response to SA (data not shown). In a second step, the expression of genes encoding the PR1 protein and the QM-like protein was followed in M. truncatula control and (h)GSH-depleted roots after SA treatment (Fig. 6). The level of QM-like protein and PR1 transcripts was increased 3- and 40-fold, respectively, in roots of the control plants 1 and 6 h after SA treatment. In (h)GSH-depleted roots, the accumulation of QM-like protein and PR1 transcripts was increased 4.5- and 70-fold, respectively, after SA treatment. These results show that a higher gene expression is observed after SA treatment in the (h)GSH-deficient roots, suggesting that (h)GSH deficiency enhances the effect of SA in M. truncatula roots. As the SA transduction pathway is known to be redox regulated (Fobert and Despres, 2005; Rochon et al., 2006), (h)GSH deficiency may alter the cellular redox state, consequently modifying the SA-dependent response. These results are in agreement with the modulation of the SA signaling pathway observed in BSO-treated Arabidopsis (Arabidopsis thaliana) leaves (Koornneef et al., 2008).
Figure 6.
qRT-PCR analysis of the expression of QM-like protein and PR1 in (h)GSH-deficient roots treated with SA. Plants were grown on modified Farhaeus medium (control plants) and modified Farhaeus medium containing 0.1 mm BSO [(h)GSH-depleted plants] for 1 week and treated with 1 mm SA. Root tissue was collected 1 h (T1h) and 6 h (T6h) after treatment. The expression levels of QM-like protein and PR1 were standardized as described in “Materials and Methods.” Data are presented as the means ± se of nine qRT-PCR analysis (technical triplicates of three biological experiments). The asterisk indicates difference (P < 0.05).
In conclusion, the cDNA-AFLP analysis has shown that the (h)GSH depletion has a strong effect on gene expression during the establishment of the symbiosis. Genes involved in developmental processes and in plant stress response appeared to be regulated by (h)GSH levels. Modification of water transport and SA signaling pathway observed under (h)GSH deficiency may contribute to explain the critical role played by (h)GSH in the proper development of the symbiotic interaction.
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Growth Conditions
Medicago truncatula ecotype Jemalong was used for all experiments. Sterilized seedlings germinated for 48 h onto 0.5% agarose were plated onto modified Farhaeus medium (Boisson-Dernier et al., 2001) with 1.3% agar and grown for 7 d before inoculation. Plants were germinated and grown either in the presence or absence of 0.1 mmBSO, a specific inhibitor of (h)GSH synthesis. Plants were grown in a growth chamber with a day/night temperature of 22°C and with a photoperiod of 16 h. Sinorhizobium meliloti strain RCR2O11 expressing yellow fluorescent protein (Stuurman et al., 2000) was grown in Luria Broth medium in the presence of 10 μg/mL tetracycline as a selection marker. Bacteria were pelleted at 16,000g, washed two times with sterile water, and resuspended in sterile water to give a final OD600 of 0.1. Plants were inoculated with 300 μL/plant of the bacterial suspension per root. Inoculated root samples of control and (h)GSH-depleted roots (0.1 mm BSO treatment) were collected every 12 h during the first 96 hpi. For each time point, 15 root fragments corresponding to the IZs were harvested and frozen in liquid nitrogen for subsequent analysis. Root samples were harvested for microscopy observations of events following inoculation in control and (h)GSH-depleted plants.
For the SA treatment, control and (h)GSH-depleted plants were treated with modified Farhaeus liquid medium and modified Farhaeus liquid medium containing 1 mm SA. For each time point, roots were harvested and frozen in liquid nitrogen for subsequent analysis.
cDNA-AFLP Analysis
Total RNA was extracted with Trizol. cDNA synthesis and cDNA-AFLP transcript profiling were done as described in Breyne et al. (2003). The final amplification step was performed using MseI and BstYI primers with selectivity +2/+1.
Quantitative Measurements of the Expression Profiles and Data Analysis
Gel images were quantitatively analyzed with the AFLP-QUANTARPRO image analysis software (Keygene). The raw data were corrected for differences in total lane intensity and variance normalized. A coefficient of variance was calculated for control transcript profile. For statistical analysis of differentially expressed genes in control and (h)GSH-depleted samples, ANOVA (P < 0.05) using the (h)GSH deficiency as the variable factor was applied to the corrected-normalized values corresponding to the gene expression values. Hierarchical average linkage clustering (The Institute for Genomic Research Multiple Experiment Viewer) was performed on the gene tags showing a significant difference due to the treatment.
qRT-PCR Analysis
cDNA was synthesized with the Omniscript RT kit (Qiagen) following the manufacturer's protocol using 2 μg of total RNA. qRT-PCR was performed using a DNA Engine Opticon 2 continuous fluorescence detection system (MJ Research) and qPCR MasterMix Plus for SYBR green I (Eurogentec). In each reaction, 5 μL of 100-fold-diluted cDNA and 0.3 μm primer (sequences used are described in Supplemental Table S1) were used. The initial denaturing time was 10 min, followed by 40 PCR cycles of 95°C for 15 s and 60°C for 1 min. The specificity of the amplification was confirmed by a single peak in a dissociation curve at the end of the PCR reaction. Data were quantified using Opticon Monitor 2 (MJ Research) and normalized with the 2−ΔΔCT method (Livak and Schmittgen, 2001). Two constitutively expressed genes, Mtc27 (TC106535) and 40S Ribosomal Protein S8 (TC100533), were the endogenous controls (Van de Velde et al., 2006). PCR reactions for each of the two biological replicates were performed in triplicate. The absence of genomic DNA contaminations in the RNA samples was tested by PCR analysis of all samples using oligonucleotides bordering an intron in the glutathione synthetase gene of M. truncatula.
Promoter Fusion and Histochemical Localization of GUS Activity
A 1.3-kb DNA fragment upstream of the starting ATG of the putative aquaporin PIP2 (TC95581) was amplified by PCR and fused in the pGEM-T vector (Promega) according to the manufacturer's instructions. Using the native NcoI and NotI restriction sites, the fragment was subcloned into the entry vector pENTR4 (Gateway Cloning System; Invitrogen) and then fused to the GUS reporter gene in the destination vector pkGWFS7 (Karimi et al., 2002). The identity, integrity, and orientation of the promoter fragment were checked by sequencing. The destination vector carrying the GUS fusion was electroporated into the Agrobacterium rhizogenes strain A4S. M. truncatula roots were transformed with A. rhizogenes by the method described by Boisson-Dernier et al. (2001). Transformed roots were selected on kanamycin plates and transferred onto modified Fahraeus medium as previously described by Frendo et al. (2005). Transgenic roots were stained with GUS assay buffer as described by Hemerly et al. (1993). Roots from at least 30 plants from three biological experiments were observed.
Study of Spatiotemporal Gene Expression Using in Situ Hybridization
A fragment of 456 bp of an endo-β-1,4-glucanase (TC108882) was amplified from cDNA using specific oligonucleotides (left primer, 5′-TGGTTGGTTTTGGAGAGAAA-3′; right primer, 5′-TGACACTAGCAAAAGCATGTGA-3′). The resulting PCR product was subcloned in the pGEM-T vector (Promega). Sense and antisense probes were synthesized following the protocol of the digoxigenin labeling system (Roche) using T7 and SP6 promoters. IZs of M. truncatula roots 96 hpi were harvested under a binocular magnifier, fixed under vacuum in 2% (v/v) glutaraldehyde and 0.1 m sodium cacodylate buffer, pH 7.2, dehydrated in a graded ethanol series, embedded in paraffin, sectioned at 10 μm thickness, and finally fixed on slides. The slides containing the root sections were then hybridized with riboprobes following the protocol of de Almeida Engler et al. (2001). Controls included sections exposed to the sense probe and sections exposed to the antibody alone. Hybrid detection was performed by immunological detection with an antibody anti-digoxigenin linked to an alkaline phosphatase (Roche). Finally, the slides were incubated with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP), subtrates of alkaline phosphatase. They were subsequently mounted in distyrene, plasticiser, xylene reagent (Fluka) and observed under a light microscope. Roots from at least 20 plants were observed.
Quantification of SA
Control and BSO-treated roots (200–500 mg) were ground in liquid nitrogen. The powder was carefully dipped in methanol, approximately 500 μL/300 mg fresh material weight. After an overnight maceration, the suspension was filtered through a plug of glass wool jammed in a 1-mL pipette tip. After rinsing with 200 μL methanol, the filtered extract was adjusted to 20% (v/v) methanol with water and then to 20 mm Na acetate, pH 4. This fraction was passed through a prewetted reversed phase column (200 mg of Nucleodur C18 ec, 100 A, 30 μm, 4 mL cartridge; Macherey Nagel). The column was washed with 1 mL of 20% (v/v) aqueous methanol, 20 mm Na acetate, pH 4, unretained and washing fractions were pooled, evaporated under vacuum to dryness (water bath 35°C), and then were solubilized with 1 mL of water. The pH was shifted to approximately 2 with 5 μL of trifluoroacetic acid (TFA), and the fraction was passed through a second solid phase extraction column (100 mg of the same phase as above; 1-mL cartridge). After washing with 1 mL of water containing 0.1% (v/v) TFA, the column was eluted with 500 μL of methanol, and the fraction was directly analyzed by HPLC. HPLC was carried out on a Shimadzu Prominence system (high-pressure binary gradient, photodiode array, spectrofluorimeter). Samples were chromatographed on an Inertsil ODS 3 column (5 μm, 250 × 4.6 mm; Interchim) at a flow rate of 1 mL min−1 with a gradient of methanol in aqueous 0.1% (v/v) TFA (MeOH, %: 15, 1 min; 15–25, 3 min; 25–75, 27 min; 75–100, 3 min; and 100 for 3 min). SA was identified on the basis of its retention time and its typical UV spectrum and measured from a calibration curve obtained with commercial SA (Sigma-Aldrich).
Analysis of SAG was achieved by HPLC determination of SA after enzymatic hydrolysis of the previous analyzed fraction according to Lee and Raskin (1998). SA and SAG were estimated from at least triplicates of three independent experiments.
Statistical Analyses
All data presenting the quantification of SA and the relative gene expression values after SA treatment are given as means with se. The significance of the results was tested using the Student's t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Scheme of the experimental design and data analysis.
Supplemental Table S1. Sequences used for the qRT-PCR analysis.
Supplemental Table S2. Gene tags of which the expression is modulated.
Supplementary Material
Acknowledgments
We thank Janice de Almeida Engler and Eric Wajnberg for their help during the in situ hybridization experiments and the statistical analysis of the cDNA-AFLP results, respectively. We also thank David Barker and Terence Neil Ledger for critical reading of the manuscript and John Hancock for suggestions during the cDNA-AFLP gene screening. We gratefully acknowledge Wilson Ardiles Diaz for the sequencing during the cDNA-AFLP analysis and Stephanie Piardi for plant care. C.P. is recipient of a Marie Curie postdoctoral fellowship from the European Communities Training and Mobility of Researchers program (Contract MEIF–CT–2004–514982). This work was funded in part by the European Communities Improving Human Potential program (Contract HPRN–CT–2000–00094). We thank the Ministère des Affaires Etrangères for financial support through the Tournesol program (Contract 06744WF).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alain Puppo (puppo@unice.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogene-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14 695–700 [DOI] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Kuster H, Journet EP, Hohnjec N, Gamas P, Niebel A (2008) Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact 21 1118–1127 [DOI] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, Vandenbosh K (1995) Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J, De Groodt R, Van Montagu M, Engler G (2001) In situ hybridization to mRNA of Arabidopsis tissue sections. Methods 23 325–334 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, De Rycke R, Mathis R, Goormachtig S, Pagnotta S, Verplancke C, Capoen W, Holsters M (2003) Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc Natl Acad Sci USA 100 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7 547–552 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Despres C (2005) Redox control of systemic acquired resistance. Curr Opin Plant Biol 8 378–382 [DOI] [PubMed] [Google Scholar]
- Fortin MG, Morrison NA, Verma DP (1987) Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res 15 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier J, Timmers AC, Sieberer BJ, Jauneau A, Chabaud M, Barker DG (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo P, Harrison J, Norman C, Hernandez Jimenez MJ, Van de Sype G, Gilabert A, Puppo A (2005) Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol Plant Microbe Interact 18 254–259 [DOI] [PubMed] [Google Scholar]
- Gamas P, Niebel Fde C, Lescure N, Cullimore J (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9 233–242 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Metraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34 217–228 [DOI] [PubMed] [Google Scholar]
- Gomez LD, Vanacker H, Buchner P, Noctor G, Foyer CH (2004) Intercellular distribution of glutathione synthesis in maize leaves and its response to short-term chilling. Plant Physiol 134 1662–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, Puppo A, Frendo P (2005) Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 187 168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inze D (1993) cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A, Sigaud S, Van de Sype G, Puppo A, Herouart D (2003) Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol Plant Microbe Interact 16 217–225 [DOI] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci USA 105 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14 737–748 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Katagiri F (2004) A global view of defense gene expression regulation—a highly interconnected signaling network. Curr Opin Plant Biol 7 506–511 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CM (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol 147 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lee HI, Raskin I (1998) Glucosylation of salicylic acid in Nicotiana tabacum Cv. Xanthi-nc. Phytopathology 88 692–697 [DOI] [PubMed] [Google Scholar]
- Loake G, Grant M (2007) Salicylic acid in plant defence: the players and protagonists. Curr Opin Plant Biol 10 466–472 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Haridas S, Gantt JS, Vandenbosch KA (2007) A transient decrease in reactive oxygen species in roots leads to root hair deformation in the legume-rhizobia symbiosis. New Phytol 173 39–49 [DOI] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8 532–540 [DOI] [PubMed] [Google Scholar]
- Martinez-Abarca F, Herrera-Cervera JA, Bueno P, Sanjuan J, Bisseling T, Olivares J (1998) Involvement of salicylic acid in the establishment of the Rhizobium meliloti-alfalfa symbiosis. Mol Plant Microbe Interact 11 153–155 [Google Scholar]
- Mateo A, Funck D, Muhlenbock P, Kular B, Mullineaux PM, Karpinski S (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J Exp Bot 57 1795–1807 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Patriarca EJ, Tate R, Ferraioli S, Iaccarino M (2004) Organogenesis of legume root nodules. Int Rev Cytol 234 201–262 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiot. Science 275 527–530 [DOI] [PubMed] [Google Scholar]
- Pichon M, Journet EP, Dedieu A, de Billy F, Truchet G, Barker DG (1992) Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell 4 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prayitno J, Rolfe BG, Mathesius U (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu SK, Peng HM, Cook DR (2002) Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol Plant Microbe Interact 15 522–528 [DOI] [PubMed] [Google Scholar]
- Riely BK, Ane JM, Penmetsa RV, Cook DR (2004) Genetic and genomic analysis in model legumes bring Nod-factor signaling to center stage. Curr Opin Plant Biol 7 408–413 [DOI] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Despres C (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18 3670–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Fricker M, Corben LB, White NS, Sheard N, Leaver CJ, Van Montagu M, Inzé D, May MJ (1997) Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc Natl Acad Sci USA 94 2745–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal NN, Marcker KA (1988) Soybean nodulin 26 is homologous to the major intrinsic protein of the bovine lens fiber membrane. Nucleic Acids Res 16 9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Herouart D, Sigaud S, Touati D, Puppo A (2001) Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol Plant Microbe Interact 14 86–89 [DOI] [PubMed] [Google Scholar]
- Seo HS, Li J, Lee SY, Yu JW, Kim KH, Lee SH, Lee IJ, Paek NC (2007) The hypernodulating nts mutation induces jasmonate synthetic pathway in soybean leaves. Mol Cells 24 185–193 [PubMed] [Google Scholar]
- Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6 365–371 [DOI] [PubMed] [Google Scholar]
- Shaw SL, Long SR (2003) Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol 132 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, Libault M, Brechenmacher L, Wan J, May GD (2006. a) Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol 9 110–121 [DOI] [PubMed] [Google Scholar]
- Stacey G, McAlvin CB, Kim SY, Olivares J, Soto MJ (2006. b) Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiol 141 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Bras CP, Schlaman HR, Wijfjes AH, Bloemberg G, Spaink HP (2000) Use of green fluorescent protein color variants expressed on stable broad-host-range vectors to visualize rhizobia interacting with plants. Mol Plant Microbe Interact 13 1163–1169 [DOI] [PubMed] [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM (2006) Crosstalk between jasmonic acid, ethylene and Nod factor signalling allows integration of diverse inputs for regulation of nodulation. Plant J 46 961–970 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen PC, Tak T, Rood AM, van Brussel AA, Kijne JW, Boot KJ (2003) Salicylic acid inhibits indeterminate-type nodulation but not determinate-type nodulation. Mol Plant Microbe Interact 16 83–91 [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Truchet G (1993) Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompagnied by a hyprsensitive reaction. Plant J 4 555–566 [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inze D, May MJ, Sung ZR (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J 57 171–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.