Legume seeds are an important source of food and feed, and have been principally studied with a view to optimizing the composition of reserve substances starch, oligosaccharides, oil, and protein. The availability of genome and transcribed genome sequences for the legume model species Medicago truncatula and Lotus japonicus has stimulated the application of the -omics technologies to understanding legume seed development in these and closely related crop species. The -omics techniques that we shall consider here are transcriptomics, proteomics, metabolomics, and ionomics (Fig. 1). Transcriptomics and proteomics methodologies attempt to describe the global complement of transcripts and proteins, respectively, of an organism, and utilize directly primary genomic sequence or expressed sequence tag information as reference. In contrast, metabolomics analyses determine organic metabolite constitution, and ionomics the inorganic ion contents of the organism. They are thus distinct but complementary sources of information about different levels of the phenome. The full exploitation of -omics depends on the establishment of interactive databases that facilitate connecting these different sources of information obtained without any a priori knowledge (Dondrup et al., 2009; Truncatulix: http://lily.cebitec.uni-bielefeld.de/truncatulix/app; Henckel et al., 2009; Legoo: http://www.legoo.org/). This backbone of technology is being applied to legume seeds in a number of ways: to dissect gene expression at different levels of detail (whole seed, component tissues, organelles, etc.), to study different physiological phenomena such as the different phases of seed development, maturation and germination, or biotic and abiotic stresses, and to evaluate the effects of specific mutations on phenotypes.
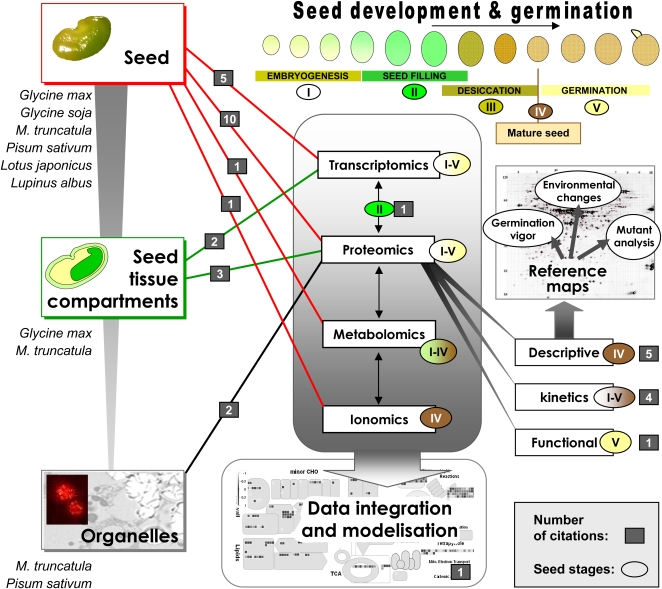
Figure 1.
Post-genomics strategies applied to legume seeds for dissecting their composition and/or identifying regulatory and metabolic networks during developmental processes. Gray squares represent the number of citations for each strategy. For the entire seed, the citations refer to post-genomics experiments carried out in various legume species at the level of the transcriptome (Firnhaber et al., 2005; Buitink et al., 2006; Gallardo et al., 2007; Benedito et al., 2008; Verdier et al., 2008), metabolome (Vigeolas et al., 2008), ionome (Sankaran et al., 2009), and proteome. At the level of the proteome, some citations refer to the mature seed (Watson et al., 2003; Magni et al., 2007; Bourgeois et al., 2009; Krishnan et al., 2009; Natarajan et al., 2009), to the kinetics of seed development (Hajduch et al., 2005; Gallardo et al., 2007; Agrawal et al., 2008; Dam et al., 2009), and to a functional study for identifying targets of thioredoxin (Alkhalfioui et al., 2007). For the separated seed tissues, the citations refer to transcriptome and proteome analyses in soybean (Le et al., 2007) and M. truncatula (Boudet et al., 2006; Zhang et al., 2006; Gallardo et al., 2007). For the seed organelles, the citations refer to proteome analyses of pea mitochondria (Bardel et al., 2002) and M. truncatula nuclei (Repetto et al., 2008). The citation referring to the comparison of transcriptome and proteome datasets is Gallardo et al. (2007) and that referring to data integration is Goffard and Weiller (2006).
THE -OMICS TECHNOLOGIES APPLIED TO LEGUME SEEDS
These studies began historically with proteome analyses on mature, and subsequently, developing seeds. The results typically identified a few hundred abundant proteins including the major storage proteins, structural proteins, chaperonins, and certain enzymes. The techniques were limited by the necessity of isolating individual polypeptides for sequencing, usually from two-dimensional gel electrophoretic separations. More recently, techniques of shotgun protein sequencing have developed which, with appropriate informatics support, can provide a much more comprehensive picture of the proteome (Malmstrom et al., 2007). These analyses have established reference maps of the protein complement of developing or mature seed in model and crop species, allowing for the identification of species-specific features (Agrawal et al., 2008; Gallardo et al., 2008). The methods have been applied to total protein extracts, or in some cases to subcellular fractions, as for example in the analysis of nuclear proteins (Repetto et al., 2008). By sampling the nuclear proteome just before seed filling, the most numerous class of nuclear proteins detected was found to be that involved in ribosome biogenesis. Besides numerous ribosomal proteins, this class includes several nucleolus-specific proteins, such as a PESCADILLO-like protein, which may play a role in ribosomal precursor processing and subunit assembly. The accumulation of these proteins precedes massive protein synthesis activity later during seed filling. Other approaches taken to reduce the complexity of the data analysis have been to focus on specific size fractions, e.g. in studies of the peptidome (Zhang et al., 2006), or to focus on specific functional properties of the proteins, e.g. their redox state or their interaction with other proteins such as thioredoxin (Alkhalfioui et al., 2007) or heat-stable proteins for identifying desiccation tolerance-associated proteins (Boudet et al., 2006). Furthermore, several methods of quantifying proteomics analyses have been devised that can greatly reinforce interpretation of the data produced (Oeljeklaus et al., 2009). More comprehensive sequence information also permits the analysis of various posttranslational modifications that occur and increase the number of polypeptides arising from a single sequence. A reconstruction of posttranslational modifications leading to the deposition of mature globulins was elegantly demonstrated by Dam et al. (2009) in L. japonicus and by Bourgeois et al. (2009) in pea (Pisum sativum). Dam et al. (2009) also compared the protein compositions of green and mature seeds using gel filtration liquid chromatography/tandem mass spectrometry (MS/MS) and identified 920 proteins from green and 264 proteins from mature seeds. The difference in complexity may be due to the dominance of globulins in the mature seed spectra. Their results are accessible at the Web interface: http://www.cbs.dtu.dk/cgi-bin/lotus/db.cgi. Among interesting findings were the identification of a plastid-localized Gln synthetase in green seeds, indicating a possible seed plastid source of Gln, and identification of enzymes of the irreversible hydrolytic starch breakdown pathway, e.g. β-amylase. Thus a different starch breakdown route is used in developing seeds to that in germinating seeds, where the phosphorolytic pathway is employed (Smith et al., 2005). Weigelt et al. (2009) have used transcriptomic and proteomic profiling to monitor the effects of reducing carbon flux into starch in ADP-Glc pyrophosphorylase-repressed pea plants. Reduced starch accumulation was accompanied by diverse metabolic responses including increased carbohydrate oxidation and reserve protein accumulation, cell proliferation, and production of reactive oxygen species.
These proteomics analyses have established reference maps of the protein complement of developing or mature seed in several model and crop species (see Fig. 1), allowing for the identification of species-specific features (Gallardo et al., 2008). They have further revealed a series of orchestrated stages of storage protein biosynthesis during seed development and specific pathways activated at these stages.
The most extensive post-genomic analyses have been carried out at the transcriptomic level, either in isolation or in combination with proteomic analyses on the same sample batches. These began with the construction of microarray chips of a few thousand cDNA-derived sequences, now replaced by the Affymetrix chips for L. japonicus and M. truncatula (Lotus1a520343 and Medicago Genome Genechips) bearing probes derived from all of the expressed sequences, or those predicted on the basis of the genome sequence. Usually the data is obtained as an expression profile for a given gene, e.g. during seed development. The recently published Medicago Gene Atlas of transcriptome data, accessible at http://bioinfo.noble.org/gene-atlas/, includes analyses of RNA from immature seeds harvested at 10, 12, 16, 20, 24, and 36 d after pollination (dap), and from a pod sample, screened with 50,900 probe sets representing a large portion of the expressed M. truncatula genome (Benedito et al., 2008). Of these, nearly half were expressed in seeds and at least one other tissue, and only 584 were completely seed specific, i.e. not expressed in any other tissue. Sixty-two percent were expressed differentially during seed development. The most highly expressed genes encoded storage proteins and late embryogenesis abundant proteins. A hierarchical cluster analysis of expression profiles yielded four clusters, corresponding to genes expressed, respectively, in the late embryogenesis, start of seed filling, mid seed filling, and desiccation phases. A functional classification of these sequences using the Kyoto Encyclopedia of Genes and Genomes pathway database (http://www.genome.jp/kegg/pathway.html) indicated that certain cellular activities were predominant at particular developmental stages. For example, transcripts of cell division-related genes were restricted to the 10 and 12 dap RNA population, those involved in metabolic pathways being more highly represented from 12 dap onwards, and those involved in desiccation in the 36 dap sample. A comparison of seed transcriptome profiles was used by Buitink et al. (2006) to identify gene expression changes associated with the acquisition of desiccation tolerance in M. truncatula. A cluster analysis identified groups of genes corresponding to discrete stages of adaptation: stress responses, acquisition of desiccation tolerance, and entry into quiescence marked by down-regulation of the cell cycle and metabolic activity.
While chip hybridizations give an indication of transcript abundance, very low abundance transcripts cannot be accurately quantified with this technique. To overcome this difficulty, panels of low-abundance sequences (e.g. transcription factors [TFs]) have been analyzed by quantitative real-time PCR assays (Kakar et al., 2008; Verdier et al., 2008). A recent alternative for transcript quantification, also suitable for low-abundance sequences, is the approach of massively parallel sequencing of cDNA (Brautigam et al., 2008).
To date there have been relatively few published analyses at the metabolomic and ionomic levels for legume seeds. Vigeolas et al. (2008) compared the seed metabolome of pea lines possessing and lacking a major reserve protein, pea albumin 2 (PA2), produced by introgression of a naturally occurring PA2 locus deletion into a standard genetic background. They showed that the absence of this protein was associated with differences in amino acid and polyamine contents in the seed. This was attributed in part to decreases in activities of two enzymes of polyamine synthesis, spermidine synthase and Arg decarboxylase. How PA2 controls the activity of these enzymes is not yet clear. PA2 contains four copies of a hemopexin-like repeat, implicated in mammalian systems in regulation of extracellular protease activity, possibly via protein-protein interaction. The other metabolomic studies in legumes have been largely restricted to the model species and to non-seed tissues. To date, the most comprehensive metabolomics analyses of seeds have been performed in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). In rice, a metabolite profiling study of seed germination based on gas chromatography-MS detected 615 constituents, of which 174 were identified (Shu et al., 2008). In Arabidopsis, metabolomics analyses of seed-expressed chalcone synthase and chalcone isomerase mutants using liquid chromatography electrospray ionization quadrupole time-of-flight MS, which detects more than 1,000 compounds, have revealed an unexpected plasticity in metabolic adaptation, with the synthesis of about 40 seed compounds not previously known for Arabidopsis (Böttcher et al., 2008). Non-targeted profiling of metabolites has been made easier by the development of MS/MS-based spectral tag annotation to identify peaks in Arabidopsis extracts (Matsuda et al., 2009). Given the technical developments in metabolomics methodology and informatics analysis, the paucity of metabolite profiling studies in legume seeds will undoubtedly soon change.
Ionomics has received a boost through the establishment of spectroscopic techniques (inductively coupled plasma MS, inductively coupled optical emission spectroscopy), permitting the simultaneous analysis of tens of different elements (Baxter, 2009). The composition of the ionome varies according to environmental conditions, plant developmental status, and genetic constitution. Such changes often affect many elements simultaneously, emphasizing the importance of considering the ionome rather than changes in any one element alone. Baxter et al. (2008) identified multivariable signatures corresponding to the ionome's response to variation in phosphorus or iron status. The establishment of ionome signatures for legume seed tissues is in its infancy. It has been performed for mineral concentrations and content in mature seeds of a recombinant inbred population in M. truncatula (Sankaran et al., 2009) and for screening mutants for shoot mineral concentrations in L. japonicus (Chen et al., 2009). Several quantitative trait loci (QTL) affecting seed mineral concentrations and content were identified, which provide a basis for identifying the genes responsible with a view to improving the mineral content of various pulses. The rapid acquisition of genomics data for crop species using recently developed techniques (e.g. Timko et al., 2008) and the expansion of genetic resources should facilitate ionomic gene discovery in crop legume species.
SEED PROTEOME AND TRANSCRIPTOME EXHIBIT TISSUE-SPECIFIC FEATURES
The seed consists of three principal components, embryo, endosperm, and integument, and within each entity functionally distinct domains may also exist. These domains can be individually studied using laser capture microdissection (LCM). RNA obtained from eight LCM-dissected soybean (Glycine max) seed regions by Le et al. (2007) was hybridized to Genechip arrays to generate global gene activity profiles, accessible at http://estdb.biology.ucla.edu/seed. Gallardo et al. (2007) separated by manual dissection the embryos, endosperm, and integument from immature M. truncatula seed tissues and analyzed them separately. The protein profiles were highly distinct, with few proteins highly expressed in two or more tissues. The results have underlined the metabolic interdependence of the component tissues of the seed, certain enzymes/metabolic steps, or processes being restricted to one or other tissue compartment. This asymmetric distribution of enzyme activity may increase the rate of solute transfer into the embryo by shifting reaction equilibria. The most energy-intensive processes, such as lipid biosynthesis, tend to be concentrated at the exterior of the embryo or in embryo-surrounding tissues to profit from light-dependent ATP generation.
The embryo-surrounding tissues fulfil transient roles during seed development in supplying the embryo with nutrients and protecting it from pathogen ingress. The nutrient contribution of maternal tissues to embryo development was illustrated by in vitro culture of seeds (Gallardo et al., 2006). Whereas intact seeds were able to accumulate reserve proteins at similar rates to those operating in vivo, this process depends on a supply of nitrogen remobilized from embryo-surrounding tissues, as the isolated embryos, lacking surrounding endosperm and integument, did not accumulate reserve proteins unless nitrogen was added to the medium. Consistent with these observations, one subtilisin gene was expressed within the endosperm during seed development, which may play a role in mobilization of nitrogen for the embryo (Gallardo et al., 2007). A second subtilisin expressed later in the seed coat may mobilize nitrogen for the late desiccation phase of seed maturation. Temporary carbon stores, in the form of lipids or starch, are deposited in the seed envelopes during seed development and mobilized to furnish the embryo with carbon or energy. Starch synthesis in the embryo is fueled in part by embryonic photosynthesis, and key enzymes have been identified by proteomics (e.g. Suc synthase, starch synthase, chlorophyll a/b-binding protein, oxygen-evolving enhancer protein; Gallardo et al., 2003). A β-amylase involved in the irreversible pathway of starch breakdown was identified by Dam et al. (2009) in developing L. japonicus seeds that may play a role in starch remobilization with the seed.
A partitioning of enzymes involved in sulfur metabolism between seed tissues also exists. An AdoHcy hydrolase was specifically detected in the embryo during mid seed filling. This enzyme ensures transmethylations in embryo cells in the course of the activated methyl cycle. Its disappearance in late seed filling, and thus the loss of the active methyl cycle, is consistent with the establishment of a metabolically quiescent state that persists in the mature seed until germination. This cycle may regenerate Met from Hcy produced by the activated methyl cycle. Consistently, a Met synthase was expressed in the embryo in mid-term seed filling. Met is also partly synthesized from S-methylmethionine, by Hcy S-methyltransferase located in the seed coat. The abundance of Hcy S-methyltransferase over Met synthases in the seed coat suggests that most Met is synthesized from an S-methylmethionine pool delivered by the phloem.
The tissue-type specificity of TF gene expression was evaluated by quantitative reverse transcription-PCR on mRNA from hand-separated integument, endosperm, and embryo (Verdier et al., 2008). This allowed the classification of 41 TFs that were specifically or preferentially expressed in one of the component tissues of the seed. These include likely orthologs of previously identified master regulators of seed development, such as LEC1 and B3-domain factors, as well as representatives of other major TF families. Le et al. (2007) took another approach to identify sites of TF expression, in using LCM to isolate probes from embryo proper and suspensor of scarlet runner bean and hybridizing to the heterologous Soybean Affymetrix Genechip. Among the 37,593 soybean probe sets, 157 TF sequences could be detected in embryo, suspensor, or both tissues.
INTEGRATING THE -OMICS DATA
While comprehensive -omics information is gradually becoming available for legume seeds, the challenge remains to organize and integrate the data. Moreover, when used in combination, the -omics technologies generate new levels of information, which require new bioinformatic tools to integrate them. The strategies used for the organization of -omics data and the first attempts made to integrate the data are described hereafter.
The transcriptomics and proteomics data can be organized in functional classes using the MapMan ontology initially developed for Arabidopsis (Thimm et al., 2004). This ontology has been extended to soybean and M. truncatula (Goffard and Weiller, 2006; http://bioinfoserver.rsbs.anu.edu.au/), and combined with the MapMan software it provides a biologically structured overview of changes in transcripts and proteins in the main metabolic pathways. Such -omics datasets can also be analyzed by a cluster analysis to identify the most related groups of values obtained for one or several samples. The results, presented as networks of related values, can give valuable clues about regulation events in the samples analyzed. By using this strategy, a cluster analysis of the entire seed-expressed transcriptome with the corresponding proteome data revealed different modes of regulation (Gallardo et al., 2007). In about 50% of the cases, the RNA and protein accumulation were coordinately expressed throughout seed development. This class includes examples of genes whose products accumulate during seed filling, those which are being degraded during this period, and a few whose protein and mRNA levels did not vary greatly. Other profile types included a class showing rapid RNA turnover but stable protein accumulation. This class included nuclear-encoded mitochondrial and chloroplast proteins. Another class was represented by genes whose protein products turned over while the transcript accumulates. This class includes the stored mRNAs deposited in the mature seed and utilized upon germination; it may also include proteins that are posttranslationally processed, if the products are not detected or associated with the same accession number. This study was complemented by a large-scale, quantitative real-time PCR profiling of TF gene expression throughout M. truncatula seed development using a panel of 712 putative TF sequences (Verdier et al., 2008). By clustering these results with those obtained for the entire seed-expressed transcriptome, candidate TFs for the regulation of different developmental stages could be pinpointed. This study identified good candidates for the functional orthologs of the master regulators LEC1, FUS3, and ABI3 of Arabidopsis. Unlike the situation in Arabidopsis, M. truncatula legumin and vicilin-type storage protein classes accumulate sequentially and a cluster analysis permitted to associate specific TFs with each of the differentially accumulating classes.
A further level of exploitation of -omics data can be obtained by combining -omics results with quantitative traits data. Thus the expression properties of a gene can be compared with traits that map at the same genetic locus, mapped by QTL or by association genetics.
CONCLUSION AND PERSPECTIVES
Genomics techniques are in the process of revolutionizing legume biology, but they need to be complemented by other studies, and their limits should not be overlooked. Frequently -omics results are correlative: gene function is inferred from its expression at a crucial stage of development, in a given cell type, etc. There is still the need for functional confirmation from analyses of mutants obtained from resources such as TILLING or TnT1 or generated by transgenesis. Additional supporting evidence for the role of a gene can also come from colocalization with a corresponding QTL. Various approaches toward modeling seed filling (e.g. Larmure and Munier-Jolain, 2004) and seed metabolism (e.g. Sulpice et al., 2009) will be increasingly used as frameworks integrating -omics with other data sources, and for deducing relationships between -omics values and determinism of seed traits. It may also be necessary to complete the picture of gene function provided by snapshots at the -omics levels by kinetic studies such as physiological analyses of metabolic flux, or ontogenetic studies of organ development.
Omics data are often relative and do not yield absolute quantities. Estimations of transcript abundance using microarrays typically show high variability and are limited in application to genes transcribed at .1 copy per cell. The quantitative reverse transcription-PCR method is up to 100-fold more sensitive but not on the same scale of probe throughput, as it demands individual assays for each gene. Data is often a snapshot in isolation of real kinetic/flux changes occurring, so that insufficient data may be available to conclude about temporal modulation of metabolic or regulatory pathways. Despite some efforts to identify metabolic/expression profiles in cells of differing cell types, most data is the sum of values for different cell types in one organ or tissue. The definition and application of a series of -omics standards (Field and Sansone, 2006; Taylor et al., 2008; Jorrín-Novo et al., 2009) should overcome the problem of assembling/comparing datasets produced under diverse experimental conditions.
While globally the process of seed development in legumes adheres closely to that exhibited by the Arabidopsis seed, an interesting perspective are certain legume-specific features that have emerged from -omics studies: families of proteins with putative defense roles and therefore evolving rapidly (Silverstein et al., 2006), staging of expression of different storage protein fractions (Gallardo et al., 2007), proliferation of the suspensor (Le et al., 2007), and different metabolite compositions notably of secondary metabolites such as isoflavones (Veitch, 2009). Finally, an area meriting more intensive study is the effect of rhizobial or mycorrhizal symbioses on seed filling.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard Thompson (thompson@dijon.inra.fr).
References
- Agrawal GK, Hajduch M, Graham K, Thelen JJ (2008) In-depth investigation of the soybean seed-filling proteome and comparison with a parallel study of rapeseed. Plant Physiol 148 504–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Vensel WH, Wong J, Tanaka CK, Hurkman WJ, Buchanan BB, Montrichard F (2007) Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol 144 1559–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2 880–898 [DOI] [PubMed] [Google Scholar]
- Baxter I (2009) Ionomics: studying the social network of mineral nutrients. Curr Opin Plant Biol 12 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, Guerinot ML, Salt DE (2008) The leaf ionome as a multivariable system to detect a plant's physiological status. Proc Natl Acad Sci USA 105 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55 504–513 [DOI] [PubMed] [Google Scholar]
- Böttcher C, von Roepenack-Lahaye E, Schmidt J, Schmotz C, Neumann S, Scheel D, Clemens S (2008) Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol 147 2107–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larré C, Satour P, Leprince O (2006) Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol 140 1418–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois M, Jacquin F, Savois V, Sommerer N, Labas V, Henry C, Burstin J (2009) Dissecting the proteome of pea mature seeds reveals the phenotypic plasticity of seed protein composition. Proteomics 9 254–271 [DOI] [PubMed] [Google Scholar]
- Brautigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM (2008) Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol 136 44–53 [DOI] [PubMed] [Google Scholar]
- Buitink J, Leger JJ, Guisle I, Vu BL, Wuillème S, Lamirault G, Le Bars A, Le Meur N, Becker A, Küster H, et al (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J 47 735–750 [DOI] [PubMed] [Google Scholar]
- Chen Z, Watanabe T, Shinano T, Okazaki K, Osaki M (2009) Rapid characterization of plant mutants with an altered ion-profile: a case study using Lotus japonicus. New Phytol 181 795–801 [DOI] [PubMed] [Google Scholar]
- Dam S, Laursen BS, Ornfelt JH, Jochimsen B, Staerfeldt HH, Friis C, Nielsen K, Goffard N, Besenbacher S, Krusell L, et al (2009) The proteome of seed development in the model legume Lotus japonicus. Plant Physiol 149 1325–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondrup M, Albaum SP, Griebel T, Henckel K, Jünemann S, Kahlke T, Kleindt CK, Küster H, Linke B, Mertens D, et al (2009) EMMA 2-A MAGE-compliant system for the collaborative analysis and integration of microarray data. BMC Bioinformatics 10 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Sansone SA (2006) A special issue on data standards. OMICS 10 84–93 [Google Scholar]
- Firnhaber C, Puhler A, Küster H (2005) EST sequencing and time course microarray hybridizations identify more than 700 Medicago truncatula genes with developmental expression regulation in flowers and pods. Planta 222 269–283 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Firnhaber C, Zuber H, Hericher D, Belghazi M, Henry C, Küster H, Thompson RD (2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds. Mol Cell Proteomics 6 2165–2179 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Kurt C, Thompson RD, Ochatt S (2006) In vitro culture of immature M. truncatula grains under conditions permitting embryo development comparable to that observed in vivo. Plant Sci 170 1052–1058 [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Thompson RD, Burstin J (2008) Reserve accumulation in legume seeds. C R Biol 331 755–762 [DOI] [PubMed] [Google Scholar]
- Goffard N, Weiller G (2006) Extending MapMan: application to legume genome arrays. Bioinformatics 1 2958–2959 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henckel K, Runte KJ, Bekel T, Dondrup M, Jakobi T, Kster H, Goesmann A (2009) TRUNCATULIX—a data warehouse for the legume community. BMC Plant Biol 9 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrín-Novo JV, Maldonado AM, Echevarría-Zomeño S, Valledor L, Castillejo MA, Curto M, Valero J, Sghaier B, Donoso G, Redondo I (2009) Plant proteomics update (2007-2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics 72 285–314 [DOI] [PubMed] [Google Scholar]
- Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, et al (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Oehrle NW, Natarajan SS (2009) A rapid and simple procedure for the depletion of abundant storage proteins from legume seeds to advance proteome analysis: a case study using Glycine max. Proteomics 9 3174–3188 [DOI] [PubMed] [Google Scholar]
- Larmure A, Munier-Jolain NG (2004) A crop model component simulating N partitioning during seed filling in pea. Field Crops Res 85 135–148 [Google Scholar]
- Le BH, Wagmaister JA, Kawashima T, Bui AQ, Harada JJ, Goldberg RB (2007) Using genomics to study legume seed development. Plant Physiol 144 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni C, Scarafoni A, Herndl A, Sessa F, Prinsi B, Espen L, Duranti M (2007) Combined 2D electrophoretic approaches for the study of white lupin mature seed storage proteome. Phytochemistry 68 997–1007 [DOI] [PubMed] [Google Scholar]
- Malmstrom J, Lee H, Aebersold R (2007) Advances in proteomic workflows for systems biology. Curr Opin Biotechnol 18 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K (2009) MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J 57 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan SS, Krishnan HB, Lakshman S, Garrett WM (2009) An efficient extraction method to enhance analysis of low abundant proteins from soybean seed. Anal Biochem 394 259–268 [DOI] [PubMed] [Google Scholar]
- Oeljeklaus S, Meyer HE, Warscheid B (2009) Advancements in plant proteomics using quantitative mass spectrometry. J Proteomics 72 545–554 [DOI] [PubMed] [Google Scholar]
- Repetto O, Rogniaux H, Firnhaber C, Zuber H, Küster H, Larré C, Thompson R, Gallardo K (2008) Exploring the nuclear proteome of Medicago truncatula at the switch towards seed filling. Plant J 56 398–410 [DOI] [PubMed] [Google Scholar]
- Sankaran RP, Huguet T, Grusak MA (2009) Identification of QTL affecting seed mineral concentrations and content in the model legume Medicago truncatula. Theor Appl Genet 119 241–253 [DOI] [PubMed] [Google Scholar]
- Shu XL, Frank T, Shu QY, Engel KH (2008) Metabolite profiling of germinating rice seeds. J Agric Food Chem 56 11612–11620 [DOI] [PubMed] [Google Scholar]
- Silverstein KAT, Graham MA, VandenBosch KA (2006) Novel paralogous gene families with potential function in legume nodules and seeds. Curr Opin Plant Biol 9 142–146 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56 73–98 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al (2009) Starch as a main integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, et al (2008) Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotechnol 26 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Timko MP, Rushton PJ, Laudeman TW, Bokowiec MT, Chipumuro E, Cheung F, Town CD, Chen X (2008) Sequencing and analysis of the gene-rich space of cowpea. BMC Genomics 9 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch NC (2009) Isoflavonoids of the Leguminosae. Nat Prod Rep 26 776–802 [DOI] [PubMed] [Google Scholar]
- Verdier J, Kakar K, Gallardo K, Le Signor C, Aubert G, Schlereth A, Town CD, Udvardi MK, Thompson RD (2008) Gene expression profiling of M-truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol Biol 67 567–580 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Chinoy C, Zuther E, Blessington B, Geigenberger P, Domoney C (2008) Combined metabolomic and genetic approaches reveal a link between the polyamine pathway and albumin 2 in developing pea seeds. Plant Physiol 146 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Küster H, Rutten T, Fait A, Fernie AR, Miersch O, Wasternack C, Emery RJN, Desel C, Hosein F, et al (2009) ADP-glucose pyrophosphorylase-deficient pea embryos reveal specific transcriptional and metabolic changes of carbon-nitrogen metabolism and stress responses. Plant Physiol 149 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KR, McKinlay C, Hocart CH, Djordjevic MA (2006) The Medicago truncatula small protein proteome and peptidome. J Proteome Res 5 3355–3367 [DOI] [PubMed] [Google Scholar]