Abstract
Major storage reserves of Arabidopsis (Arabidopsis thaliana) seeds are triacylglycerols (seed oils) and proteins. Seed oil content is severely reduced for the regulatory mutant wrinkled1 (wri1-1; At3g54320) and for a double mutant in two isoforms of plastidic pyruvate kinase (pkpβ1pkpα; At5g52920 and At3g22960). Both already biochemically well-characterized mutants were now studied by 13C metabolic flux analysis of cultured developing embryos based on comparison with their respective genetic wild-type backgrounds. For both mutations, in seeds as well as in cultured embryos, the oil fraction was strongly reduced while the fractions of proteins and free metabolites increased. Flux analysis in cultured embryos revealed changes in nutrient uptakes and fluxes into biomass as well as an increase in tricarboxylic acid cycle activity for both mutations. While in both wild types plastidic pyruvate kinase (PKp) provides most of the pyruvate for plastidic fatty acid synthesis, the flux through PKp is reduced in pkpβ1pkpα by 43% of the wild-type value. In wri1-1, PKp flux is even more reduced (by 82%), although the genes PKpβ1 and PKpα are still expressed. Along a common paradigm of metabolic control theory, it is hypothesized that a large reduction in PKp enzyme activity in pkpβ1pkpα has less effect on PKp flux than multiple smaller reductions in glycolytic enzymes in wri1-1. In addition, only in the wri1-1 mutant is the large reduction in PKp flux compensated in part by an increased import of cytosolic pyruvate and by plastidic malic enzyme. No such limited compensatory bypass could be observed in pkpβ1pkpα.
For many field crops, seeds are the most important part used for human and animal nutrition. Seed storage compounds such as triacylglycerol, proteins, and carbohydrates typically make up most of the mass of mature seeds, and the proportions of these components have large species-specific variations. Since seed composition and yield are important traits for breeding and agricultural research, partitioning of carbon and nitrogen into the major storage products within the developing seed is an important process. Based on knowledge of the biochemical pathways that convert maternal carbon and nitrogen supplies into storage products (Baud and Lepiniec, 2008), flux analysis with cultured developing seeds gave important insights into in vivo pathway usage and the integrated function and regulation of metabolic pathways in storage synthesis (Schwender and Ohlrogge, 2002; Schwender et al., 2003, 2004a, 2006; Sriram et al., 2004; Ettenhuber et al., 2005; Spielbauer et al., 2006; Alonso et al., 2007; Junker et al., 2007; Iyer et al., 2008; Allen et al., 2009). While studies that use network-scale flux analysis to analyze the effect of genetic perturbation on central metabolism are not uncommon for microorganisms (Fischer and Sauer, 2005), to our knowledge there are only a few studies of this kind in plants (Spielbauer et al., 2006). However, applying metabolic flux analysis to biochemically and genetically well-characterized mutants in plant central metabolism could help to better understand the plasticity of the central metabolism network.
Arabidopsis (Arabidopsis thaliana) is a well-established plant model organism that produces small seeds with oil and protein as major storage compounds and is closely related to the oleaginous species Brassica napus. Multiple mutations that affect seed storage metabolism in Arabidopsis have been isolated and studied (Meinke et al., 1994; Focks and Benning, 1998; Lin et al., 2004; Gomez et al., 2006). In this study, we characterize two Arabidopsis mutants with a severe reduction in seed oil content by metabolic flux analysis of cultured developing embryos. For this purpose, we adapted a methodology of embryo cultures established before for B. napus (Schwender and Ohlrogge, 2002; Schwender et al., 2003, 2004a, 2006; Junker et al., 2007).
For the flux studies, two well-studied mutants were chosen. The first is a mutation in the transcription factor WRINKLED1 (WRI1; At3g54320), which controls parts of seed storage metabolism (Focks and Benning, 1998; Ruuska et al., 2002; Cernac and Benning, 2004; Baud et al., 2007a). In mature seeds of the mutant allele wri1-1, seed oil content is reduced by about 80% (oil per seed) and sugar levels are increased, contributing to or causing the wrinkled appearance of the seeds (Focks and Benning, 1998). In developing seeds of wri1-1, the measurable enzyme activity of several glycolytic enzymes is strongly reduced (Focks and Benning, 1998; Baud and Graham, 2006). Expression profiling of wild-type and wri1-1 seeds during seed development indicated that WRI1 controls the expression of several genes encoding for glycolytic enzymes as well as of genes involved in fatty acid synthesis (Ruuska et al., 2002). Therefore, it appears that in wri1-1 developing seeds the capacity to convert Suc into triacylglycerol is strongly impaired by lack of induction of sufficient amounts of several metabolic enzymes of the lipid synthesis pathway.
One of the enzyme activities that is reduced in wri1-1 is that of pyruvate kinase (PK). PK is a glycolytic enzyme that converts phosphoenolpyruvate (PEP) into pyruvate, a precursor of fatty acid synthesis. Expression profiling of developing seeds of Arabidopsis clearly supported a major role of plastidic PK (PKp) in providing pyruvate for fatty acid synthesis (Ruuska et al., 2002). Flux studies on developing embryos of the related species B. napus had shown that PKp provides a large part of the pyruvate needed for fatty acid synthesis (Schwender and Ohlrogge, 2002; Schwender et al., 2006). Of the 14 putative isoforms of PK known in Arabidopsis, three genes encode plastidic subunits, two β-forms and one α-form, both of which combine to the functional enzyme (Andre et al., 2007; Baud et al., 2007b). One β-form, (PKp β1; At5g52920) appeared to be important in seed oil synthesis (Ruuska et al., 2002; Andre et al., 2007; Baud et al., 2007b). Accordingly, mutation in this gene has been shown to produce wrinkled seeds due to strong impairment of lipid synthesis with strong reduction of extractable PKp activity (Andre et al., 2007; Baud et al., 2007b). Since a double mutant in two isoforms of PKp (pkpβ1pkpα; At5g52920 and At3g22960) showed even more severe reduction in lipid content per seed (Baud et al., 2007b), we chose this double mutant for flux studies.
Based on a flux model established for developing embryos of B. napus (Schwender et al., 2006), we apply in this paper methods of steady-state metabolic flux analysis (for review, see Schwender et al., 2004b; Ratcliffe and Shachar-Hill, 2006; Rios-Estepa and Lange, 2007; Schwender, 2009) to cultured developing embryos of Arabidopsis. The two mutants are compared with their respective genetic wild-type backgrounds. The results shed light on (1) how the mutations affect the storage composition in embryos grown in culture as compared with mature seeds, (2) how strongly flux is reduced in the mutants due to loss of enzymatic capacity, and (3) how localized the flux perturbation might be and if compensatory bypassing of an impaired pathway can be observed.
RESULTS
Embryo Cultures
For growth of Arabidopsis embryos in liquid culture on 13C-labeled substrates, torpedo-stage embryos were dissected out of developing seeds. This developmental stage with a size of about 0.25 mm length is typically reached about 7 to 8 d after flowering, when seeds enter the phase of rapid storage accumulation (Focks and Benning, 1998; Baud et al., 2002). It was observed that embryos do not grow in darkness, only in the presence of light, which was kept continuously at 50 μmol m−2 s−1. The embryos were grown at 22°C with composition of the liquid culture medium (see “Materials and Methods”) similar to that used earlier for B. napus embryo cultures, where Glc, Suc, Gln, and Ala were the sole carbon sources (Schwender et al., 2006). In preliminary experiments with Arabidopsis embryos, both Suc and Glc were tested for their suitability as carbon sources. Both sugars were found to support the growth of Arabidopsis embryos in culture (data not shown). We could observe that with increasing Suc-to-Glc ratio, embryos grew to a slightly smaller final size with decreasing amounts of starch detectable (KI/I2 staining after 7 d of growth). If grown on Suc only, embryos were virtually starchless. This indicates that Suc as sole sugar carbon source in culture supports embryo maturation better than Glc, since mature wild-type seeds are almost free of starch and starch content in Arabidopsis embryos has been reported to decline during seed development (Baud and Graham, 2006). In addition, since during seed maturation Suc becomes more and more the dominant sugar in developing seeds (Baud et al., 2002), it was decided to use Suc as sole sugar carbon source in the labeling experiments.
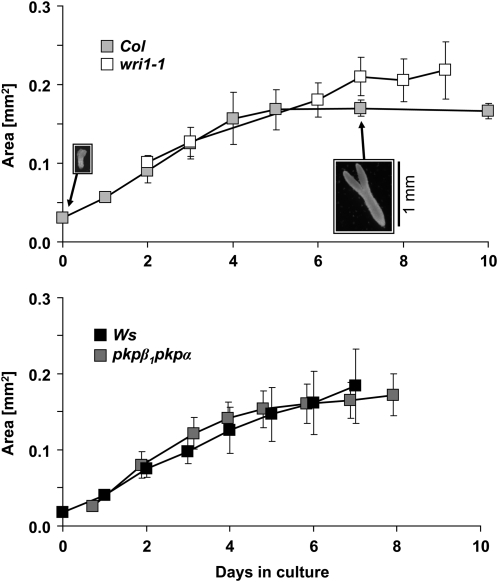
For comparison of the flux phenotypes of the two mutants pkpβ1pkpα and wri1-1 with their respective background ecotypes Wassilewskija (Ws) and Columbia (Col), developing embryos of each genotype were cultured in the presence of [U-13C12]Suc (12.5 mol % in unlabeled Suc), with Ala and Gln as additional carbon and nitrogen sources (see “Materials and Methods”). Figure 1 shows the growth of Arabidopsis embryos in culture under the conditions used for all labeling experiments. Embryos continue to grow for about 5 to 6 d (22°C, continuous light at 50 μmol m−2 s−1). After 7 d, the embryo tissue was harvested, the relative proportions of the major storage compounds were determined, and the labeling signatures in monomers of proteins, lipids, starch, and free Suc were analyzed by gas chromatography-mass spectrometry (GC-MS; see “Materials and Methods”). The fresh weight of Col embryos cultured for 7 d was 21 ± 0.6 μg embryo−1 (n = 3). Prolonged time in culture did not lead to continuation of growth, and the morphology of the embryos (Fig. 1) was not indicative of a transition into precocious germination, which would entail substantial changes of metabolism. In culture, precocious germination can be observed under reduced sugar concentrations or osmotic pressure, visible in B. napus or Arabidopsis embryos by gravitropic elongation of the root (data not shown).
Figure 1.
Growth of embryos of wild-type (Col and Ws) and respective mutant (wri1-1 and pkpβ1pkpα) genotypes in liquid culture under conditions used for labeling experiments. As a measure of embryo size, the surface area was determined from micrographs (area of embryo ± sd; n = 4). Two example images for Col embryos are shown. Embryos of wri1-1 at the beginning of culture (0 d of culture not shown) were of similar size to the wild-type embryos.
Biomass Proportions
Mutant embryos (pkpβ1pkpα and wri1-1) and embryos of their respective wild-type backgrounds (Ws and Col) were cultured in the presence of 13C-labeled Suc for 7 d (see “Materials and Methods”). Prior to analysis of the labeling signatures and estimation of fluxes, the proportions of the major storage compounds of cultivated embryos were determined. Lipids and polar metabolites were extracted from the embryos or seeds, and protein content was determined based on the nitrogen content in the residual insoluble material (elemental analysis; see “Materials and Methods”). Figure 2 shows the fractions of major biomass compounds in cultivated embryos and in mature seeds relative to total dry weight (Supplemental Tables S1 and S3). Starch was almost undetectable in the cultured embryos, while the maximal amount of starch found in mature seeds was 2.3% ± 0.1% (w/dry weight; wri1-1; Supplemental Table S2). The composition of the polar fractions (Fig. 2) was analyzed by GC-MS (see “Materials and Methods”), showing that Suc was the dominant component along with several amino acids and organic acids as additional major components (data not shown). Characteristic for Arabidopsis seed storage lipids is the occurrence of fatty acids of chain length longer than C18. In mature seeds of the Col ecotype, the C20 and C22 fatty acid species were reported to amount to 27 mol % of total fatty acids (Focks and Benning, 1998). In the lipid fraction of cultured embryos, between 16% and 18% (w/w) of fatty acids was C20 and C22 species, while in mature seeds, this fraction was somewhat higher, between 20% and 30% (Supplemental Tables S2 and S4).
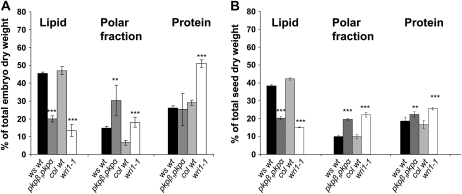
Figure 2.
Effects of pkpβ1pkpα and wri mutations on accumulation of total lipids, polar metabolites, and proteins in embryos cultured for 7 d as described in “Materials and Methods” (A) and in mature seeds (B). Values are means ± sd of three (Ws and wri1-1), four (Col), or six (pkpβ1pkpα) biological replicates. Significant differences between each wild type (wt) and corresponding mutant are indicated according to Student's t test (*** P < 0.001, ** P < 0.05).
Wild-type embryos cultivated for 7 d contained about 45% of lipid on a dry weight basis (Fig. 2A). This lipid level compares well with mature seeds (Fig. 2B). In cultivated embryos as well as in mature seeds, we observed for pkpβ1pkpα and wri1-1 a drastic reduction in oil content relative to their wild-type backgrounds Ws and Col, respectively (Fig. 2). For embryos, the lipid levels were reduced from about 45% (w/dry weight) in both wild types to 20% and 13% in pkpβ1pkpα and wri1-1, respectively (Fig. 2A). Similarly, for mature seeds the oil content was reduced from about 40% (w/dry weight) to about 20% and 15% in pkpβ1pkpα and wri1-1, respectively (Fig. 2B). If this reduction is expressed on a per seed basis, the amount of oil is reduced by 66% and 74% for pkpβ1pkpα and wri1-1, respectively (Supplemental Table S3). This confirms the reductions in oil by 70% and 80% reported before on a per seed basis for pkpβ1pkpα and wri1-1, respectively (Focks and Benning, 1998; Baud et al., 2007b). For mature seeds of both mutants, the reduction in the lipid fraction is balanced by an increase in the polar fraction and in lesser part by an increase in seed protein (Fig. 2B). A similar, but not identical, shift in biomass fractions can be observed in the cultured embryos. For the pkpβ1pkpα embryos, there is a large increase in the polar fraction, while protein does not change significantly. For wri1-1 embryos, protein increases more than the polar fraction (Fig. 2A).
Flux Analysis
After culture of wild-type and mutant embryos in the presence of 13C-labeled Suc and subsequent quantification of the biomass compounds and quantification of 130 mass isotopomer fractions in protein-derived amino acids, fatty acids, starch, and free Suc by GC-MS (Supplemental Table S9), the labeling data were interpreted by steady-state metabolic flux analysis. The metabolic network used here is based on former studies on developing embryos of B. napus (Schwender et al., 2006), where the quantification of cytosolic and plastidic PK fluxes was demonstrated. The derived metabolic network is extended by plastidic and cytosolic malic enzyme (Fig. 3). In addition, the subcellular compartmentation was modeled in more detail. In the B. napus model (Schwender et al., 2006), an a priori assumption had been made that, based on the labeling data available, glycolysis and the oxidative pentose phosphate pathway (OPPP) cannot be resolved as independent fluxes if duplicated in parallel in the plastidic and cytosolic compartments. Accordingly, a part of the metabolic network had been collapsed to a simpler uncompartmented view of glycolysis and OPPP (Schwender et al., 2006). The appropriateness of this model simplification in B. napus embryos had been justified in particular by experimental observations suggesting that isotopic label of several glycolytic intermediates was equilibrated across the plastid envelope, apparently due to highly active metabolite transport between cytosol and plastid (Schwender et al., 2003). In contrast to the former B. napus model, in this study we did not rely on an a priori assumption on whether cytosolic/plastidic compartmentalization can be resolved or not. The computational feasibility of the now extended compartmented model (Fig. 3) hinges on the calculation of statistical quality measures. While we used Monte Carlo stochastic simulation in this study, the statistics based on model linearization (Wiechert and de Graaf, 1997) in the former study do not tolerate statistically weakly determined fluxes (see “Materials and Methods”) and therefore encourages tailoring of isotopomer models toward a well-resolved topology.
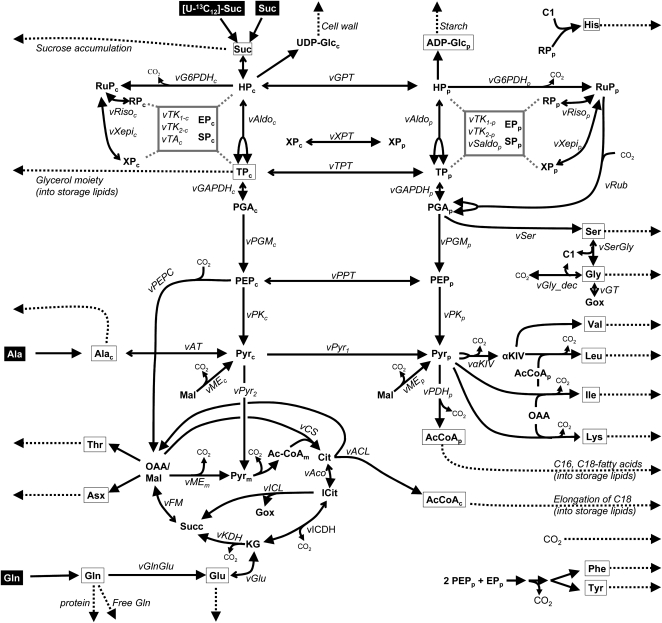
Figure 3.
Network model of central metabolism in developing Arabidopsis embryos, based on the network described earlier for flux studies with B. napus embryos (Schwender et al., 2006). Reactions and metabolite pools simulated by the network are shown. Cytosolic, plastidic, and mitochondrial reactions or metabolites are specified by subscripts c, p, and m, respectively, if the model distinguishes such subcellular species. For boxed metabolites (or direct products thereof), labeling information was experimentally determined and included into the model. Suc, Ala, and Gln are inputs, while dashed arrows indicate reactions where metabolites leave the system (stored in biomass). In the model, Suc is represented as hexose units, and the interconversion of Suc and hexose phosphates (HPc) is simplified as one bidirectional reaction. Double-headed arrows indicate reactions that are modeled reversibly. The pentose phosphate subnetwork is not shown in detail (shaded in gray). Reaction abbreviations are as follows: vACL, ATP:citrate lyase; vAco, aconitase; vAldo, Fru-1,6-bisphosphate aldolase, phosphofructokinase, Fru-1,6-bisphosphatase; vAT, Ala transaminase; vCS, citrate synthase, pyruvate dehydrogenase; vFM, succinate dehydrogenase, fumarase, malate dehydrogenase; vGAPDH, glyceraldehyde 3-phosphate dehydrogenase, phosphoglycerate kinase; vG6PDH, Glc-6-P dehydrogenase, 6-phosphogluconate dehydrogenase; vGlnGlu, Gln into Glu by glutaminase or Glu synthase; vGlu, interconversion of Glu and ketoglutarate by transaminase or Glu synthase; vGly_dec, Gly decarboxylase; vGPT, Glc-6-P phosphate antiporter; vGT, Gly transaminase; vICDH, isocitrate dehydrogenase; vICL, isocitrate lyase; vαKIV, α-ketoisovalerate synthesis; vKDH, ketoglutarate dehydrogenase, succinate kinase; vME, malic enzyme; vPDH, pyruvate dehydrogenase; vPEPC, phosphoenolpyruvate carboxylase; vPGM, phosphoglycerate mutase, enolase; vPK, pyruvate kinase; vPPT, phosphoenolpyruvate phosphate antiporter; vPyr1, pyruvate transport cytosol into plastid; vPyr2, pyruvate transport cytosol into mitochondrion; vRiso, Rib-5-P isomerase; vRub, ribulose 1,5-bisphosphate carboxylase, phosphoribulokinase; vSaldo, sedoheptulose 1,7-bisphosphate aldolase, sedoheptulose bisphosphatase; vSer, Ser synthesis; vSerGly, Ser hydroxymethyltransferase; vTA, transaldolase; vTK1 and vTK2, transketolase reactions; vTPT, triose phosphate/phosphate antiporter; vXepi, xylulose 5-phosphate epimerase; vXPT, xylulose 5-phosphate/phosphate antiporter. Metabolite abbreviations are as follows: AcCoA, acetyl-CoA; Asx, carbon chain of Asp, Asn, and Met; αKIV, α-ketoisovalerate; C1, 5,10-methylene- or 5-formyl-tetrahydrofolate; Cit, citrate; EP, erythrose 4-phosphate; Gox, glyoxylate; HP, hexose phosphates; ICit, isocitrate; KG, ketoglutarate; Mal, malate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PGA, 3-phosphoglycerate; Pyr, pyruvate; RP, Rib-5-P; RuP, ribulose 5-phosphate; SP, sedoheptulose 7-phosphate; Succ, succinate; TP, triose phosphate; XP, xylulose 5-phosphate.
For each of the four genotypes (pkpβ1pkpα, wri1-1, Ws, and Col), a respective model variant was derived and simulated to estimate intracellular fluxes during storage synthesis (see “Materials and Methods”; for labeling data, see Supplemental Table S9). For comparison of flux results between genotypes, all flux values are given in relative units (Table I; Fig. 4), with the total flow of carbon into biomass kept constant across the genotypes. In particular, flux values were normalized according to the carbon equivalent of 100 mol of Glc (600 mol of carbon) being accumulated in biomass. By normalization relative to the total biomass accumulation rate, we base the flux comparison across genotypes on equal growth speed of the four genotypes. It should be noted that the conclusions drawn from comparison of normalized flux values across genotypes might be dependent on the choice of normalization. Another normalization often used is to scale all fluxes relative to the uptake flux of the main carbon source Suc (Iyer et al., 2008). When this different normalization was applied to the flux values in this study, the major conclusions drawn from the comparisons stayed the same (data not shown). In addition, the interpretation of the flux results was also based on unit less flux ratios, which were compared across genotypes (Table II).
Table I.
Values of net fluxes for the four Arabidopsis genotypes analyzed in this study
Fluxes are given in relative units, normalized to 600 mol of carbon (100 mol of hexose) accumulated in biomass. For abbreviations of flux names, see Figure 3. Values show best fit of flux estimation ± sd as determined based on Monte Carlo stochastic simulation (see “Materials and Methods”). Boldface values indicate that the difference of flux from the wild type is significant (P = 95%; t test with n = 3 for Ws, n = 6 for pkpβ1pkpα, n = 4 for Col, and n = 3 for wri1-1).
| Reaction | Genotype |
|||
|---|---|---|---|---|
| Ws | pkpβ1pkpα | Col | wri1-1 | |
| Uptake reactions with Ala and Glu catabolism | ||||
| Uptake Suca | 100.76 ± 9.06 | 95.03 ± 8.44 | 105.78 ± 5.20 | 82.78 ± 11.48 |
| Uptake Ala | 24.33 ± 6.07 | 12.06 ± 2.44 | 11.35 ± 2.89 | 27.11 ± 4.40 |
| Uptake Gln | 13.67 ± 3.19 | 21.36 ± 2.48 | 15.83 ± 2.66 | 52.19 ± 15.86 |
| vGlnGlu | 8.08 ± 3.18 | 11.20 ± 2.30 | 12.08 ± 2.64 | 42.47 ± 15.95 |
| vGlu | 3.37 ± 3.20 | 5.45 ± 2.68 | 6.93 ± 2.57 | 31.63 ± 15.93 |
| vAT | 21.80 ± 6.08 | 8.97 ± 2.69 | 8.58 ± 2.92 | 21.28 ± 4.54 |
| Lower glycolysis- and pyruvate-related reactions | ||||
| vRub | 73.30 ± 19.48 | 74.37 ± 13.11 | 89.24 ± 7.49 | 89.07 ± 33.48 |
| vPGMc+p | 175.70 ± 7.08 | 121.17 ± 8.90 | 198.25 ± 5.18 | 119.80 ± 12.75 |
| vPKc | 32.29 ± 7.94 | 28.73 ± 6.27 | 22.50 ± 5.95 | 62.45 ± 10.26 |
| vPKp | 128.20 ± 12.93 | 73.12 ± 9.92 | 159.69 ± 8.73 | 28.67 ± 9.36 |
| vPEPC | 12.27 ± 5.33 | 15.75 ± 3.52 | 6.64 ± 1.97 | 21.93 ± 8.11 |
| vPyr1 | 43.49 ± 14.46 | 20.62 ± 9.63 | 17.33 ± 10.22 | 46.65 ± 14.09 |
| vPyr2 | 10.59 ± 2.48 | 17.07 ± 4.72 | 13.76 ± 3.82 | 44.36 ± 12.54 |
| vMEp | 3.80 ± 6.49 | 6.71 ± 4.87 | 3.29 ± 3.99 | 12.12 ± 6.84 |
| vMEc | 0.00 ± 2.31 | 0.00 ± 2.22 | 0.00 ± 1.01 | 7.29 ± 7.74 |
| vPDHp | 164.02 ± 1.84 | 86.45 ± 9.55 | 167.76 ± 3.98 | 61.04 ± 8.00 |
| TCA cycle reactions | ||||
| vCS | 15.19 ± 3.29 | 22.73 ± 4.78 | 16.13 ± 4.53 | 61.85 ± 24.82 |
| vAco | 12.20 ± 3.29 | 21.12 ± 4.75 | 13.38 ± 4.51 | 60.73 ± 24.88 |
| vICDH | 10.73 ± 3.42 | 20.15 ± 4.71 | 12.05 ± 4.52 | 57.51 ± 25.27 |
| vKDH | 14.09 ± 6.04 | 25.61 ± 5.84 | 18.98 ± 6.90 | 89.13 ± 40.73 |
| vFM | 15.56 ± 6.00 | 26.57 ± 5.89 | 20.31 ± 6.89 | 92.36 ± 40.37 |
| vMEm | 4.59 ± 2.29 | 5.66 ± 2.94 | 2.37 ± 1.59 | 17.49 ± 16.35 |
| vICL | 1.47 ± 0.71 | 0.96 ± 0.77 | 1.34 ± 0.36 | 3.22 ± 1.48 |
| vACL | 2.99 ± 0.13 | 1.61 ± 0.23 | 2.74 ± 0.43 | 1.12 ± 0.27 |
| Ser/Gly metabolism | ||||
| vGT | 1.47 ± 0.71 | 0.96 ± 0.77 | 1.34 ± 0.36 | 3.22 ± 1.48 |
| vSer | 4.04 ± 0.79 | 5.34 ± 1.51 | 4.55 ± 0.79 | 9.37 ± 1.67 |
| vSerGly | 1.37 ± 0.38 | 2.08 ± 0.65 | 1.63 ± 0.26 | 3.23 ± 0.76 |
| vGly_dec | −0.95 ± 0.38 | −1.57 ± 0.62 | −1.17 ± 0.21 | −2.26 ± 0.81 |
| Fluxes into biomass and CO2 efflux | ||||
| His into protein | 0.42 ± 0.15 | 0.51 ± 0.22 | 0.46 ± 0.20 | 0.96 ± 0.40 |
| Val into protein | 2.08 ± 0.13 | 2.54 ± 0.60 | 2.28 ± 0.20 | 4.79 ± 0.43 |
| Leu into protein | 2.25 ± 0.14 | 2.75 ± 0.72 | 2.46 ± 0.17 | 5.18 ± 0.38 |
| Ile into protein | 1.49 ± 0.08 | 1.82 ± 0.55 | 1.63 ± 0.14 | 3.43 ± 0.26 |
| Lys into protein | 1.32 ± 0.07 | 1.60 ± 0.42 | 1.44 ± 0.10 | 3.03 ± 0.23 |
| Phe into protein | 0.97 ± 0.05 | 1.19 ± 0.27 | 1.06 ± 0.09 | 2.24 ± 0.15 |
| Tyr into protein | 0.49 ± 0.05 | 0.60 ± 0.13 | 0.54 ± 0.05 | 1.14 ± 0.10 |
| Ala into protein | 2.53 ± 0.13 | 3.09 ± 0.67 | 2.77 ± 0.19 | 5.83 ± 0.52 |
| Thr into protein | 1.11 ± 0.09 | 1.35 ± 0.36 | 1.21 ± 0.10 | 2.55 ± 0.24 |
| Asx into protein | 3.32 ± 0.24 | 4.05 ± 0.79 | 3.63 ± 0.40 | 7.65 ± 0.70 |
| Gln into protein/free Gln | 5.59 ± 0.22 | 10.17 ± 1.12 | 3.75 ± 0.35 | 9.73 ± 0.84 |
| Glu into protein | 4.71 ± 0.31 | 5.75 ± 1.40 | 5.15 ± 0.40 | 10.84 ± 0.84 |
| Suc accumulationa | 7.19 ± 0.32 | 15.46 ± 2.21 | 3.13 ± 0.58 | 10.27 ± 1.24 |
| HPp into starch | 0.12 ± 0.04 | 1.29 ± 0.45 | 1.12 ± 0.20 | 0.67 ± 0.15 |
| Glycerol in lipids | 6.04 ± 0.15 | 3.12 ± 0.42 | 6.16 ± 0.30 | 2.08 ± 0.43 |
| AcCoAc into elongation of C18 | 2.99 ± 0.13 | 1.61 ± 0.23 | 2.74 ± 0.43 | 1.12 ± 0.27 |
| AcCoAp into C16 and C18 fatty acids | 161.77 ± 1.84 | 83.71 ± 9.78 | 165.29 ± 4.00 | 55.86 ± 8.19 |
| CO2 efflux |
145.92 ± 59.14 |
113.16 ± 38.70 |
147.90 ± 34.44 |
238.99 ± 118.84 |
Suc expressed in mol hexose units.
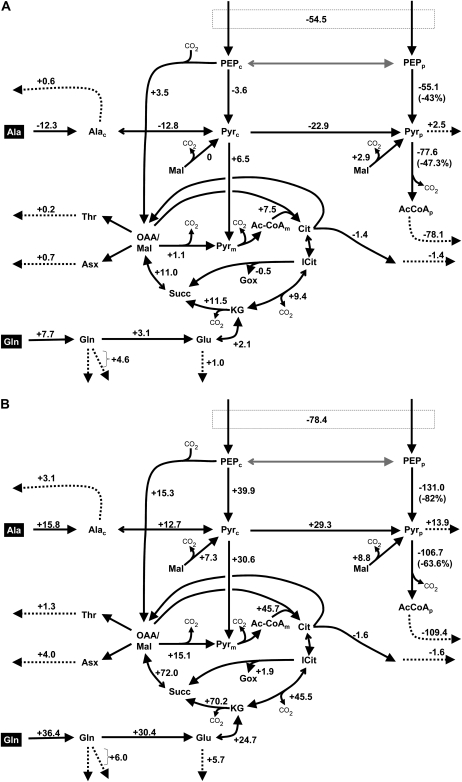
Figure 4.
Effects of pkpβ1pkpα and wri1-1 mutations on fluxes visualized for the lower part of the metabolic network shown in Figure 3. Positive or negative changes in flux values as derived from the flux values in Table I are shown. A, Flux changes in pkpβ1pkpα versus Ws. B, Flux changes in wri1-1 versus Col. For a few fluxes, the change is also given as percentage reduction of the wild-type value. The fluxes of Pyrp into Val, Leu, Ile, and Lys (Fig. 3) are condensed into one efflux. For abbreviations of metabolite and flux names, see Figure 3.
Table II.
Selected flux ratios as determined from values in Table I
For abbreviations of flux names, see Figure 3. sds are based on Monte Carlo stochastic simulation (see “Materials and Methods”). Boldface values indicate that the difference of flux from the wild type is significant (P = 95%; t test with n = 3 for Ws and wri1-1, n = 4 for Col, and n = 6 for pkpβ1pkpα).
| Parameter | Genotype |
|||
|---|---|---|---|---|
| Ws | pkpβ1pkpα | Col | wri1-1 | |
| Percentage of total carbon uptake by amino acidsa | 19 ± 2 | 20 ± 2 | 15 ± 1 | 41 ± 4 |
| Percentage of total PGA formed by Rubiscob | 82 ± 22 | 118 ± 20 | 91 ± 8 | 138 ± 54 |
| Percentage of catabolized hexose phosphate released as CO2 by TCA cyclec | 8 ± 2 | 16 ± 3 | 8 ± 2 | 52 ± 17 |
| Percentage of PEPc into OAAd | 28 ± 12 | 35 ± 8 | 23 ± 8 | 26 ± 7 |
| Percentage of Pyrp contributed by PKpe | 73 ± 7 | 73 ± 8 | 89 ± 4 | 33 ± 11 |
| vPEPC/vFM | 0.79 ± 0.09 | 0.59 ± 0.06 | 0.33 ± 0.03 | 0.24 ± 0.03 |
| vPyr1/vPyr2 |
4.11 ± 1.73 |
1.21 ± 0.79 |
1.26 ± 0.82 |
1.05 ± 0.51 |
100 × (3 × uptake Ala + 5 × uptake Gln)/(3 × uptake Ala + 5 × uptake Gln + 6 × uptake Suc).
100 × 2 × vRub/(2 × vRub + vGAPDHc+p). vRub, Stoichiometric coefficient for PGAp is 2.
100 × (CO2 production by mitochondrial pyruvate dehydrogenase, isocitrate dehydrogenase, 2-ketoglutarate dehydrogenase, malic enzyme)/(6 × Suc uptake – Suc accumulation – hexose into cell walls – hexose into starch).
vPEPC/(vPEPC + vPKc).
100 × vPKp/(vPKp + vPyr1 + vMEp).
Metabolic and Isotopic Steady State during Storage Synthesis
Embryos were taken into culture at 7 to 8 d after flowering, at the stage when seeds enter the phase of rapid storage accumulation (Baud et al., 2002). The embryos then continue to grow in culture for about 5 d (Fig. 1). We assume that during this growth in culture, a stationary metabolic state is maintained that parallels the continuous deposition of storage oil and proteins in seeds between about 8 and 17 d after flowering (Baud et al., 2002). In planta, the rapid storage deposition leads into a phase of seed maturation (Baud et al., 2002). Since embryo growth in culture comes to an end by about day 5 of culture (Fig. 1), storage synthesis is likely to cease as well by this time. Based on this growth characteristic, we consider steady state as follows. Steady-state flux analysis requires that the metabolic fluxes are constant over a time period that allows the labeling signatures in the metabolites to become stationary, which can be expected after multiple turnovers of the metabolite pools. Troufflard et al. (2007) determined for developing linseed embryos isotopic steady state of central metabolites to be realized after 18 h. The 7-d duration of embryo culture in our experiments exceeds this time by about 9-fold. As a precondition for continuous steady state during the 7-d culture, the environmental conditions (light and temperature) were kept constant and embryos were cultured in excess of culture medium in order to limit changes in nutrient concentration (see “Materials and Methods”). In addition, it is important to note that the experiments of this study are based on the analysis of 13C label in storage products. These are end products that are biosynthetically derived from central metabolites deposited in vacuoles and are not be expected to have metabolic turnover. In consequence, the labeling in storage products reflects the labeling signatures of central metabolites during storage synthesis. If embryos are harvested by day 7, storage metabolism may have ceased, causing central metabolism to come to a different state. However, the labeling signatures reflecting this change will not contribute the label in storage compounds.
To further support the notion of steady state during storage synthesis, Col embryos were cultured for 3 d under conditions identical to the 7-d labeling experiments. The labeling signatures of protein-bound amino acids were measured (see “Materials and Methods”) and compared with those obtained for Col after 7 d of culture (Supplemental Table S9). Initial comparison revealed substantial differences in labeling signatures, but most of it could be explained by the influence of unlabeled inoculum biomass. During growth, newly synthesized labeled biomass is continuously added to the initial unlabeled embryo inoculum. In consequence, we observed that the average 13C enrichments in the amino acids considerably increased between 3 and 7 d. To account for the effect of isotopic dilution by unlabeled biomass, mass isotopomer fractions from the 7-d culture were numerically adjusted by considering 7-d culture to be mixed with unlabeled amino acids (1.07% natural 13C enrichment) in a ratio of 84:16. Based on this dilution adjustment, only 23 out of 152 compared mass isotopomer abundances (P = 95%) were significantly different between 3 and 7 d, with all differences being less than 3% on the fractional 13C enrichment scale. Also, for Suc, only one out of six mass isotopomers was significantly different. We therefore consider the labeling signatures largely the same between day 3 and day 7 and consider metabolic and isotopic steady state reasonably well approximated.
Statistical Quality of the Flux Values
The statistical quality measures for the fluxes were derived by Monte Carlo stochastic simulation, based on addition of normally distributed random noise to the labeling data and biomass fluxes, according to the experimentally derived sd values (see “Materials and Methods”). The validity of this method was verified by an independent approach as described in “Materials and Methods.” The stochastically dispersed flux values obtained from the stochastic simulation allowed calculating sd values of all fluxes as well as of sums or ratios of fluxes of interest. Not all flux values are reproduced in Table I; in particular, those with very large statistical deviation are only reported in Supplemental Table S8. We rejected fluxes as not trustworthy if, across all genotypes, at least one sd value is larger than 100% of the flux value or if it is more than 100 relative flux units.
Substrate Uptake and Biomass Effluxes
If the uptake fluxes are compared between the mutants and their respective wild types, wri1-1 has a noticeably reduced Suc uptake while, at the same time, it takes up more amino acids (Table I; Fig. 4). Expressed by ratios, in Col 15% of total carbon uptake is in the form of amino acids, while for wri1-1 this number rises to 41% (Table II). This large increase in amino acid uptake is expected, since with the large increase in the protein fraction for wri1-1 (Fig. 2), more nitrogen has to be imported. The increased amino acid uptake flux resulting from isotopomer balancing is validated by the increased protein fraction. In the flux model, the amino acid uptake fluxes are free variable and nitrogen is not balanced. Although these fluxes are only constrained by the 13C-labeling signatures, the model does predict increased uptake of nitrogen sources, as expected with an increased protein fraction.
The changes in lipid, protein, and free metabolite fractions between wild-type and mutant embryos are translated by the modeling process into changes in biomass fluxes. The largest decrease in biomass flux values (mutant versus wild type) is found in fatty acid biosynthesis (Table I). Fluxes into free Suc as well as the many effluxes of amino acids into protein correspondingly increase, but by much smaller values.
Upper Glycolysis and the Pentose Phosphate Pathway
Twenty fluxes of the cytosolic and plastidic pentose phosphate pathways, the glycolytic cleavage of hexose phosphates, and the triose, pentose, and hexose phosphate transport have low statistical confidence and are not reproduced in Table I. Fluxes through the OPPP were not resolved, which can be explained since this reaction is typically not well resolved using uniformly 13C-labeled substrates, like [U-13C12]Suc in this study. Across the genotypes, the two fluxes through cytosolic and plastidic phosphoglycerate mutase (vPGMc and vPGMp; Supplemental Table S8) had large sd values between 20% and 800% of the mean value, indicating very low statistical confidence in the flux values. However, the sum vPGMc + vPGMp can be given with good statistical precision (Table I), with the sd being maximum at 11% of the mean value. This indicates that the labeling data do not allow distinguishing the fluxes for the two compartmentalized parallel reactions separately (Fig. 3).
The flux through Rubisco (vRub; Table I) appears to be of considerable size for all four genotypes. In all cases, more than 80% of the total phosphoglycerate (PGAc+p) is formed by Rubisco (Table II), which indicates substantial contribution of the Rubisco bypass of glycolysis (Schwender et al., 2004a) to hexose breakdown. In contrast to the upper part of the network, the flow of carbon that enters lower glycolysis through the cytosolic and plastidic PEP pools is better resolved. The combined flux through vPGMc and vPGMp is reduced in the mutants relative to the wild types (Table I). The fluxes connecting from PEPc and PEPp to the lower parts of the metabolic network (Fig. 3), namely vPEPc, vPKp, and vPKp, as well as tricarboxylic acid (TCA) cycle-related fluxes are better resolved. In addition to the flux values shown in Table I and selected flux ratios shown in Table II, Figure 4 shows the changes in flux values in this lower part of the metabolic network.
TCA Cycle
In Arabidopsis wild-type developing embryos, 8% of the carbon entering catabolism as hexose phosphate was released as CO2 by mitochondrial pyruvate dehydrogenase and the TCA cycle reactions, and this number increased substantially for the two mutants (Table II). Also, the values for particular TCA cycle fluxes like vICDH or vKDH considerably increased in the mutants (Table I; Fig. 4). This indicates that there is substantial cyclic catabolic activity of the TCA cycle in the wild type that increases in the mutants. As a reaction leading into the TCA cycle, the flux though PEP carboxylase (vPEPC) increased about 3-fold between Col and wri1-1 (Table I). However, the flux ratio at the PEPc branch point in wri1-1 (i.e. the relative amount of the metabolite PEPc converted by PEPC) was rather unchanged (Table II).
Formation of Plastidic Pyruvate
Since both mutants are impaired in fatty acid synthesis, detailed analysis of the mass balance around plastidic pyruvate reveals the most important features of the flux phenotypes. Among the flux values determined for the Arabidopsis ecotypes Ws and Col (Table I), the generation of plastidic pyruvate (vPKp) and its conversion into fatty acids (vPDHp; flux of AcCoAp into C18 fatty acids) is of dominating magnitude. With the strong reduction in oil content in both mutants (Fig. 2), vPDHp is reduced by 47.3% and 63.6% in pkpβ1pkpα and wri1-1, respectively (Fig. 4). The generation of plastidic pyruvate, in turn, is shared by three reactions, namely plastidic pyruvate kinase (vPKp), pyruvate import into the plastids (vPyr1), and plastidic malic enzyme (vMEp; Fig. 3). PKp provides 73% and 89% of the plastidial pyruvate in Ws and Col, respectively (Table II). Realizing that in the wild type most of the flux directed toward fatty acid synthesis is carried by PKp, it is not surprising that the strong reduction in oil content in the mutants mainly affects this step: for pkpβ1pkpα and wri1-1, vPKp is reduced by 43% and 82% of the wild-type value, respectively (Fig. 4). The relative contribution of PKp to plastidic pyruvate stays constant at 73% for pkpβ1pkpα and Ws (Table II). In contrast, while in Col 89% of the plastidic pyruvate is provided by PKp, this is only 33% in wri1-1 (Table II). As visualized in Figure 4, in the case of wri1-1 the fluxes vPKc, vPyr1, and vMEp increase and partially compensate for the loss in PKp flux.
Due to the unexpectedly moderate reduction of vPKp in the PK mutant, we tested for pkpβ1pkpα a model variant where the PKp reaction was deleted from the network by constraining vPKp to zero. After repeated optimization of the model from 100 random start points, a minimal sum of squared residuals of 104.9 was repeatedly found. This value is high, since according to the χ2 test for goodness of fit the sum of squared residuals should be less than 75 (see “Materials and Methods”). Closer inspection of the model-predicted labeling signatures showed that, in particular, the labeling in plastidic fatty acids, Ala and Val, cannot be matched by the model if vPKp is removed. The error sum for these three metabolites was 49.6 (i.e. about half of the total residual error sum). Therefore, we conclude that the flux value found for vPKp in pkpβ1pkpα (Table I) is realistic and determined mainly by the labeling signatures in the plastidic fatty acids, Ala and Val.
We also tested if some of the differences in flux phenotype observed between pkpβ1pkpα and wri1-1 could be related to the differences in the embryo biomass composition (Fig. 2A). Considering the relatively large statistical error in the free metabolite and protein fractions for pkpβ1pkpα (Fig. 2A), one might suspect a gross error in the respective weight measurements to have major impact on the pkpβ1pkpα flux phenotype. Accordingly, we redetermined all flux estimates in all four genotypes by substituting the biomass composition of the embryos (Fig. 2A) by the biomass composition of mature seeds (Fig. 2B). This means that all biomass fluxes were redetermined based on the biomass ratios in mature seeds. Then, for all four model variants, best-fit flux values were estimated as before (see “Materials and Methods”). According to the resulting fluxes (data not shown), for both mutants the observations on TCA cycle fluxes, on the relative reductions in PKp flux, as well as on the flux ratios related to plastidic pyruvate still hold.
DISCUSSION
Limitations of the Experimental Approach
In this study, we establish that mutant Arabidopsis embryos grown in liquid culture replicate the main storage composition in the wild type as well as the low-oil phenotype in the mutants, as observed for mature seeds developed in planta. Therefore, the observations on flux distribution in culture should be relevant for the in planta seed development. Also, the mutant low-oil phenotype in culture clearly confirms the underlying assumption that the reduction in seed oil content in pkpβ1pkpα and in wri1-1 mutants is caused mainly by the disruption of the embryo metabolism rather than by effects in the maternal plant. However, the relevance of the results from the cultured embryos to plant-grown seeds should be carefully considered, keeping in mind that the cultures do not perfectly mimic in planta growth. For example, a day/night light regime as usually applied to plants was not used in culture; therefore, its potential influence on seed development is neglected. Also, while the supply of Suc and of two amino acids to the embryo in culture is kept constant, a more diverse nutrient mix might be available in planta. The endosperm nutrient supply might change during seed development and have specific effects on seed maturation. Furthermore, the apparently retarded growth and reduced final size of pkpβ1pkpα and wri1-1 mutant embryos as observed in planta (Baud et al., 2007a, 2007b) could not be detected in culture (Fig. 1). In this study, we base our comparison of flux values across genotypes on relative flux units (see “Results”), which does not account for differences in growth speed in culture, if they occur. If the apparent in planta different growth rates and final seed size of the mutants are to be considered, particular alternative conclusions might be possible. For example, based on this study, the flux into amino acid and protein biosynthesis is substantially increased in wri1-1 (Table I), which implies a redirection of carbon resources from lipid synthesis to protein synthesis. However, wri1-1 seeds are smaller than Col seeds, and on a per seed basis, Focks and Benning (1998) found that in wri1-1 seeds the amount of protein is unaltered. In contrast to our conclusion, this suggests that protein synthesis is rather unaffected by the wri mutation. Alongside the following discussion of the flux results based on relative flux units, such possible alternative interpretations should be kept in mind.
Flux analysis was carefully carried out with three to six biological replicates per genotype, with about 1,000 embryos to be manually dissected per culture replicate. The time-consuming and difficult experimental procedure ruled out the use of different substrate labeling in addition to [U-13C12]Suc. Since no additional tracers could be used, the number of fluxes in the flux model that could be determined with adequate accuracy might be lower than before for B. napus (Schwender et al., 2006). Nevertheless, several key fluxes and flux ratios related to the mutations studied here could be well resolved.
The Role of the PEPc Branch Point and the TCA Cycle
For cultured wri1-1 embryos, a distinct shift from oil to protein synthesis can be observed (Table I). This coincides with increased uptake of the nitrogen sources Ala and Gln. At the same time, an increased flux through PEPC is found (Table I; Fig. 4), which, at first sight, is in accordance with the expected role of PEPC in carbon partitioning (Smith et al., 1989; Golombek et al., 2001; Tajuddin et al., 2003; Rolletschek et al., 2004). PEPC produces the TCA cycle intermediate oxaloacetate (OAA), linking carbohydrate catabolism to nitrogen fixation and amino acid synthesis from TCA cycle intermediates. However, carbon partitioning at the PEPc branch point is unchanged if the ratio vPEPC/(vPKc + vPEPC) is considered (Table II). The increase in vPEPC might also be seen as being part of a metabolic bypass of the inhibited PKp reaction. For each OAA produced by PEPC, malate can be removed from the TCA cycle and converted to pyruvate by malic enzyme. This kind of flux rerouting appears to be the case for wri1-1 (Fig. 4) and has been observed before for a double knockout of the two PK genes in Escherichia coli (Emmerling et al., 2002).
With an increase in vPKc and vPyr2 in wri1-1, more carbon is entering the TCA cycle as pyruvate, destined for oxidative degradation (Fig. 4). Substantial cyclic flux and catabolic TCA cycle activity was found, which increased in both mutants. For flux analysis of B. napus developing embryos grown under culture conditions similar to this study, a smaller catabolic activity of the TCA cycle was observed (Schwender et al., 2006). There was no substantial cyclic flux, and only 3.7% of hexose carbon entering catabolism was released as CO2 by TCA cycle reactions, which is a smaller catabolic activity than found in other plant flux studies (Schwender et al., 2006). The strongly increased catabolic TCA cycle activity together with markedly increased protein accumulation in wri1-1 might be explained by substantially higher ATP demands for protein synthesis in comparison with lipid synthesis (see Fig. 1 in Schwender, 2008). Also, since the PK reaction produces ATP, the reduction in vPKp might substantially reduce the availability of ATP, which in turn would explain an increase in TCA cycle activity and mitochondrial ATP production.
Impairment of Plastidic Pyruvate Kinase and Compensatory Adjustments by Metabolic Bypasses
In Arabidopsis wild-type embryos, most of the pyruvate destined for plastidic fatty acid synthesis is produced by PKp (Table II). This agrees with the results from expression profiling during seed development in Arabidopsis (Ruuska et al., 2002), which suggested the importance of PKp in seed lipid synthesis. Similarly, in B. napus developing embryos, PKp was found to provide most of the pyruvate to lipid synthesis (Schwender et al., 2006). In both mutants of this study, the levels of PKp enzyme are clearly reduced. In developing pkpβ1pkpα seeds, no full-length transcripts of the two mutated PKp genes (At3g22960 and At5g52920) were detectable (Baud et al. 2007b), causing the plastid-specific PK enzyme activity to be reduced by 75% of the wild-type value (Baud et al. 2007b). In wri1-1, the transcripts for PKpα (At3g22960) and PKpβ1 (At5g52920) were reduced less than 2-fold and 2- to 3-fold, respectively (Supplemental Table S2 in Ruuska et al., 2002). This means that in wri1-1 there should be more PKp activity left than in pkpβ1pkpα. In this study, assuming for pkpβ1pkpα a close to 75% reduction of PKp enzyme, the observed reduction of flux by 43% of the wild-type value (Fig. 4) appears to be unexpectedly low. In comparison, in wri1-1, flux through PKp was found to be reduced by 82% of the wild-type value, although the reduction in PKp enzyme activity must be less severe than in pkpβ1pkpα. As an explanation for this apparent inconsistent relation between enzyme levels and pathway flux in the two mutants, one might consider the realization of one of the major paradigms that emerged from metabolic control analysis: that one enzyme step in a metabolic pathway tends to have only limited control over pathway flux (Fell, 1997; Morandini and Salamini, 2003). A reduction of enzyme capacity (Vmax) in a single step of a pathway is likely to cause a less than proportional reduction in pathway flux and sometimes has almost no effect. For example, at moderate light levels, the reduction of Rubisco levels by more than 50% in transgenic tobacco (Nicotiana tabacum) plants had only a marginal effect on the photosynthesis rate (Stitt et al., 1991). Similarly, for several enzymes related to starch synthesis, there was little effect on starch production in potato (Solanum tuberosum) tubers if their level was largely reduced by antisense inhibition (Geigenberger et al., 2004). In essence, this consideration can be applied to the results of this study. In the case of the mutation in a single enzyme step (pkpβ1pkpα), large reduction in PKp catalytic capacity appears to be the case (Baud et al., 2007b) and there is limited response of flux through the enzyme. For the global regulator (wri1-1) in our study, it was reported that multiple glycolytic enzyme activities are reduced. Hexokinase and pyrophosphate-dependent phosphofructokinase were reduced the most (by 83% and 62% of the wild-type activity, respectively), while total PK activity was reduced by 34% of the wild-type activity (Focks and Benning, 1998). Therefore, given reductions in several glycolytic enzymes in wri1-1, a moderate reduction in PKp could have a more severe impact on the flux through PKp than in the case of pkpβ1pkpα.
Unlike the wild type and pkpβ1pkpα, where most of the plastidic pyruvate is produced by PKp, this is not the case for wri1-1 (Table II). Based on the network structure (Fig. 3), the reduction in PKp catalytic capacity might be compensated by various reactions: flux through either PKc and Pyr1 or through a bypass via PEPC, malate dehydrogenase, and MEp could increase (Fig. 3). Even increased uptake of Ala and conversion to pyruvate might be possible (Fig. 3). For pkpβ1pkpα, the relative contribution of PKp to plastidic pyruvate is unchanged (Table II). This means that no increased bypass flux is observed. For wri1-1, however, the relative contribution of PKp to plastidic pyruvate is decreased (Table II), and apparently bypass fluxes via both vPKc and vPEPC are increased (Fig. 4). The reason for the difference in bypass activation between pkpβ1pkpα and wri1-1 is unclear. The observed flux adjustments could be related to an increase in levels of cellular PEPc, the common substrate for PKc and PEPC. Also, the complex allosteric regulation of both PKc and PEPC as described for B. napus (Smith et al., 2000) should be relevant.
CONCLUSION
In this study, two different Arabidopsis mutants with low seed oil phenotype were compared with their respective wild-type genetic backgrounds by flux analysis of cultured developing embryos. The mutants markedly differed in fluxes from the wild types and from each other. These differences relate to uptake fluxes, biomass effluxes, as well as central metabolism, including the TCA cycle and lower glycolysis. The study of a mutation in a single enzyme step and a mutation in a transcriptional regulator in developing embryos has demonstrated the power of metabolic flux analysis to better understand the potential of reorganization of central metabolism in response to genetic perturbations and to generate valuable hypotheses about the regulation of carbon partitioning in developing seeds. For example, the relative constant ratio of the fluxes vPEPC and vPKc across the genotypes (Table II) indicates branch point rigidity for the PEPc branch point. This rigid flux control might be based on the complex biochemical regulation of this branch point, as described for B. napus (Smith et al., 2000). To further dissect the control of this branch point and its role in carbon partitioning, similar gene mutations could be generated in an Arabidopsis relative with larger seeds. This should allow us to measure enzyme and metabolite levels in parallel to the flux studies (Junker et al., 2007).
MATERIALS AND METHODS
Chemicals
[U-13C12]Suc (99 mol % 13C), [1-13C]palmitic acid, and [1,2-13C2]palmitic acid were purchased from Sigma. [2-13C]Palmitic acid was purchased from C/D/N Isotopes.
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) ecotypes Ws and Col were obtained from the Arabidopsis Biological Resource Center. Homozygous seeds of the wri1-1 mutant (Focks and Benning, 1998) were obtained from Christoph Benning. Homozygous seeds of the pkpβ1pkpα double mutant (Baud et al., 2007b) were obtained from Christine Rochat. Surface-sterilized seeds were plated on half-strength Murashige and Skoog (1962) medium, solidified with phytagel (2 g L−1), with addition of Suc (10 g L−1) in the case of wri1-1 and pkpβ1pkpα. Also, kanamycin (50 μg mL−1) was added for germination of pkpβ1pkpα. After 2 d of cold treatment (4°C), the seeds were germinated during 10 d at 22°C under continuous light (50 μmol m−2 s−1). The seedlings were then grown in Premier Pro-Mix Bx soil (Premier Horticulture) under controlled conditions (16-h light photoperiod, 150 μmol m−2 s−1, 20°C night/20°C day) in a CMP 4030 growth chamber (Conviron).
Dissection and Culture of Embryos in Early Maturation Phase
Embryos in torpedo stage at early maturation phase, at about 0.25 mm length, were dissected from siliques about 7 d after flowering. Selected siliques were harvested and immediately treated for 2 min with diluted commercial bleach (about 1% [w/v] NaOCl as active ingredient), then rinsed several times in sterile water, and developing seeds were removed. Using fine forceps and tungsten needles, embryos were dissected aseptically and placed immediately into growth medium. About 200 freshly dissected embryos were grown in one vessel (Falcon no. 353107 Tissue Culture Flasks with vented cap; Becton-Dickinson) in 3.0 mL volume of growth medium for 7 d at 22°C and continuous light (50 μmol m−2 s−1). Finally, about 5 mg of embryo tissue was obtained. As a prerequisite for steady state, the amount of embryo biomass formed during culture was close to 0.5 mg dry weight mL−1 culture medium, which for Brassica napus embryos is estimated to result in no more than 10% depletion of any of the major organic substrates (Schwender et al., 2006). About 25 mg of embryo tissue was pooled to one biological replicate. Three to six biological replicates were made for each genotype.
The liquid growth medium contained 20% (w/v) polyethylene glycol 4000 and the carbon and nitrogen sources Suc (80 mm), Gln (35 mm), and Ala (10 mm). Labeling experiments contained unlabeled Suc (70 mm) and [U-13C12]Suc (10 mm). Inorganic nutrients were used similarly to Schwender et al. (2003): CaCl2 (5.99 mm), MgSO4 (1.5 mm), KCl (4.69 mm), KH2PO4 (1.25 mm), Na2EDTA (14.9 mg L−1), FeSO4·7H2O (11.1 mg L−1), H3BO3 (12.4 mg L−1), MnSO4H2O (33.6 mg L−1), ZnSO4·7H2O (21 mg L−1), KI (1.66 mg L−1), Na2MoO4·2H2O (0.5 mg L−1), CuSO4·5H2O (0.05 mg L−1), CoCl2·6H2O (0.05 mg L−1), nicotinic acid (5 mg L−1), pyridoxine HCl (0.5 mg L−1), thiamine HCl (0.5 mg L−1), folic acid (0.5 mg L−1). pH was adjusted to 5.8 using KOH, and growth medium was sterilized by 0.22-μm sterile vacuum filter units (Stericup; Millipore).
Growth Kinetics of Embryos in Culture
In order to follow the growth of Arabidopsis embryos in culture, fresh weight determination was impracticable. As an approximation for biomass, the visible area of an image of an embryo was taken. Photographs were taken daily with a digital camera (Spot Diagnostics Instruments) attached to a microscope at 25× magnification. All images were analyzed using Adobe Photoshop CS2 and the image-processing software Image J (version 1.41o; http://rsb.info.nih.gov) by calculating the area of each embryo image, referenced to millimeter scales photographed under identical conditions.
Embryo Harvest and Biomass Fractionation
Embryos were harvested in 1.5-mL Eppendorf tubes and repeatedly washed in NaCl solution (0.33 m) approximately isotonic to growth medium. Embryos were stored at −20°C prior to extraction. The following extraction steps were performed while keeping the quantitative recovery of all fractions in mind. Twenty-five to 50 mg of embryo tissue was homogenized in 3 mL of prechilled methanol:water (4:3, v/v) at 0°C to 4°C using an Omni TH Tissue Grinder (Omni International). The homogenate was heated to 70°C for 15 min to quench enzymatic activities. For extraction of lipids and nonlipid metabolites, 3.4 mL of CHCl3 was added, resulting in a biphasic solvent system (CHCl3:methanol:water, 8:4:3 [v/v]; Folch et al., 1957). After heating (70°C, 5 min) and cooling to room temperature, the solvent phases were separated by centrifugation at 3,270g for 60 min (Allegra X-12R; Beckman Coulter) and the upper methanol:water phase was removed. Methanol:water (4:3, v/v) was added, the phase separation was repeated, and a combined methanol:water phase was obtained. After the lipid phase was removed, the remaining cell pellet was extracted with methanol and a combined lipid phase was obtained. Finally, after evaporation of solvents in a stream of nitrogen (50°C; Reacti-Therm III; Pierce) and complete drying of samples under vacuum in a Centrivap concentrator (Labconco), the weights of lipid, methanol:water, and cell pellet fractions were determined.
Elemental Analysis of Cell Pellet Fraction
Protein determination was based on elemental analysis of the pellet fraction, using a vario MICRO cube CHNS Analyzer in a CHN configuration (Elementar Americas). The measured weight fraction of nitrogen in the pellet was converted into protein by a conversion factor of 5.64 (g protein g−1 nitrogen). This factor was calculated from the molar fractions of amino acids in seed protein used in the model and the elemental composition of protein-bound amino acids (Supplemental Table S3).
Amino Acid and Fatty Acid Composition
Analysis of amino acids in the cell pellet fraction was performed as reported earlier (Schwender et al., 2006). The cell pellet fraction was hydrolyzed in 6 n HCl for 24 h at 100°C. HCl was evaporated at 50°C under a stream of nitrogen. Part of the amino acid fraction from hydrolysis of the cell pellet fraction was lyophilized and then derivatized to their N,O-tert-butyldimethylsilyl derivatives by adding 100 μL of N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide:acetonitrile (1:1, v/v) to 100 μg of amino acids and heating at 120°C for 1 h (Schwender et al., 2003). The amino acid derivatives were then analyzed by GC-MS for measurement of labeling signatures in selected fragments (see below).
For flux analysis, the amino acid composition of embryo biomass has to be known. For estimation of the amino acid composition in storage proteins, samples were derivatized and analyzed in parallel with an amino acid standard of known composition. Due to destruction of some amino acids during hydrolysis, this analysis allowed us to determine the relative amounts of 14 amino acids (Supplemental Table S5). The abundance of additional proteinogenic amino acids was estimated from gene sequence statistics over 10 Arabidopsis genes of 12S globulins and 2S albumins, being predominant types of seed storage proteins in Arabidopsis seeds (Heath et al., 1986), as well as three oleosin genes (Supplemental Table S6).
Starch Analysis
Starch in the pellet fraction was degraded enzymatically, and free Glc was quantified using a starch assay kit (Sigma; SA-20) according to the manufacturer's protocol but with sample volumes decreased to one-fifth of the original and using a multifunctional Ultra 384 Microplate Reader (Tecan Group). The Glc fraction obtained from starch degradation was also derivatized to Glc methoxime penta-acetate. One milliliter of methoxyamine hydrochloride in pyridine (20 mg mL−1) was added to 50 to 100 μg of Glc and heated to 50°C for 1 h. After cooling to room temperature, 1 mL of acetic acid anhydride was added and the sample was again heated to 50°C for 1 h. Finally the derivative was extracted with toluene after adding 1 volume of water to the reaction, dried, dissolved in toluene, and analyzed by GC-MS (Schwender et al., 2003).
Analysis of Biomass Composition in Seeds
Fifty milligrams of wild-type or mutant mature seeds was extracted, and biomass fractions were determined as outlined above.
Lipid Analysis
Lipids were transesterified by heating to 90°C in 5% (w/v) HCl in methanol for 1 h. After cooling to room temperature, 1 volume of water was added and fatty acid methyl esters were extracted with hexane (Browse et al., 1986) and analyzed by GC-MS for determination of fatty acid composition. The fatty acid methyl ester fraction was then reduced under hydrogen [Pt(IV) oxide catalyst; Bao et al., 2000], and the labeling signatures in saturated fatty acids were analyzed by GC-MS (Schwender et al., 2003). The glycerol fraction remaining from transesterification of the lipids was freeze dried, and the residue containing glycerol was derivatized with trifluoroacetic acid anhydride for 1 h at room temperature to obtain glycerol trifluoroacetate, residual derivatization reagent was removed at room temperature with a stream of nitrogen, and the derivatives were dissolved in toluene and analyzed by GC-MS (Schwender et al., 2003).
GC-MS Analysis of Free Metabolites
The free metabolite fraction was analyzed as methoxyamine-trimethylsilyl derivatives by GC-MS based on the protocol by Roessner et al. (2000). The dry samples were incubated for 90 min at 30°C in 100 μL of methoxyamine HCl (20 mg mL−1 pyridine). Then, 160 μL of N-methyl-N-trifluoroacetamide was added and incubated at 37°C for 30 min, then at room temperature for 120 min. The derivatized samples were analyzed by injection of 1 μL into the GC-MS device in splitless mode (see below).
GC-MS Analysis of Labeling Profiles
In the following, the carbon atoms of a metabolite or a fragment are referred to as demonstrated by the following example: Glc(1–3) refers to carbons 1 to 3 of Glc or a molecule fragment that contains these atoms.
The different metabolites were analyzed by GC-MS (6890N GC/5975 quadrupole mass spectrometer; Agilent Technologies). All analyses were in splitless mode, with 1-μL injection volume, and the carrier gas was helium at a flow rate of 1 mL min−1 with a DB1 column (30 m, 0.25 mm; J&W Scientific). Trimethylsilyl derivatives of free metabolites were analyzed as follows: injector temperature of 250°C; initial, 70°C, 4 min; to 310°C at 5°C min−1; final time, 10 min. For amino acid analysis, the injector temperature was set to 275°C and the column temperature was programmed as follows: initial, 100°C, 4 min; to 200°C at 5°C min−1; to 300°C at 10°C min−1; final time, 5 min. For fatty acid methyl ester analysis, the injector temperature was 250°C and the column temperature was programmed as follows: initial, 90°C, 4 min; to 240°C at 10°C min−1; final time, 15 min. For analysis of Glc methoxime acetate, the injector temperature was 250°C and the column temperature was programmed as follows: initial, 100°C, 4 min; to 300°C at 20°C min−1; final time, 2 min. For analysis of glycerol trifluoroacetate, the injector temperature was 250°C and the column temperature was programmed as follows: initial, 60°C, 4 min; to 250°C at 10°C min−1. By chromatographic separation using the scan mode of the mass analyzer, peak purity and retention time were inspected. During additional chromatographic separation, for each derivative mass isotopomers of selected fragments were monitored using the selected ion monitoring mode of the mass spectrometer, with mass-to-charge ratio (m/z) values set as close as possible to the exact mass of the fragment. All molecule fragments analyzed and used for flux modeling in this study were validated by unlabeled and 13C-labeled standards on the instrument used for all analyses. For this purpose, for each fragment the isotope peak abundance was compared with the theoretically expected abundance according to the elemental composition and the natural enrichment of heavy isotopes in carbon, hydrogen, nitrogen, oxygen, silicon, and sulfur. For the t-butyldimethylsilyl derivatives of amino acid, 32 fragments were monitored in 15 amino acids as described earlier (Schwender et al., 2006; Junker et al., 2007). For the saturated fatty acid methyl esters, the fragment m/z 74 was monitored (see below). For glycerol trifluoroacetate, the fragments m/z 267 [C7H5O4F6, glycerol(1–3)] and m/z 253 [C6H3O4F6, glycerol(1–2)] were monitored (Schwender et al., 2006). For Glc methoxime penta-acetate, the ion m/z 360 [C15H22O9N, Glc(1–6)] was monitored (Schwender et al., 2003). In the analysis of free metabolites, the fragment m/z 361 of the Suc trimethylsilyl derivative was monitored. This abundant ion results from fragmentation at the glycosidic linkage, leading to m/z 361 (C15H33O4Si3) representing all six carbons of the glucosyl moiety of the Suc molecule (Fuzfai et al., 2008). Mass spectral data were extracted using the ChemStation Program (MSD ChemStation; Agilent Technologies). Chromatographic peaks were integrated, and background subtraction was used to correct for the baseline signal of the mass spectrometer. The contribution of naturally occurring isotopes of hydrogen, nitrogen, oxygen, and silicon in measured fragments and of carbon in derivative parts of the fragments was corrected by isotope distribution matrices that were computed according to Wahl et al. (2004). Then for each fragment, the measured mass isotopomer values were expressed as fractional abundances. In order to derive statistical errors in the MS measurements, the entire labeling experiment and analysis were repeated at least three times for embryo cultures initiated from separately grown plants.
Label in Fatty Acids
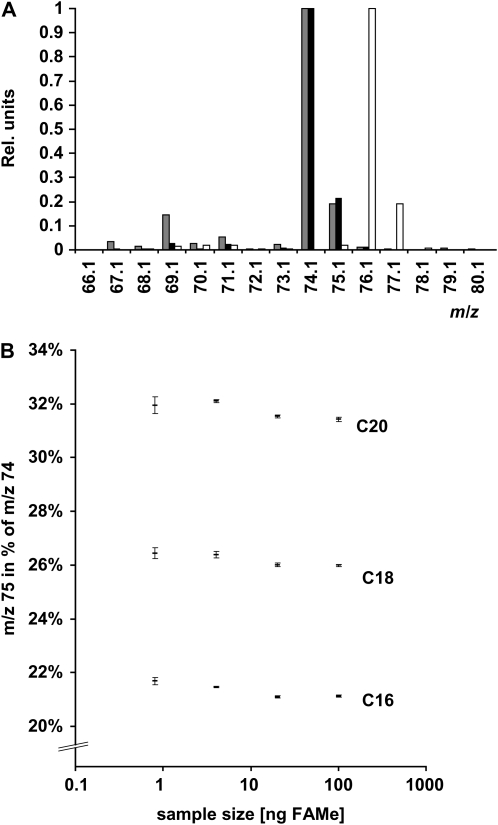
13C label in fragment m/z 74, comprising carbons 1 and 2 of the fatty acid chain and the methyl ester group, was determined as described earlier (Schwender and Ohlrogge, 2002; Schwender et al., 2003, 2006) with the modification that the ionization energy for the quadrupole mass analyzer was reduced from the standard 70 eV to 15 eV in order to suppress fragmentations that lead to ions close to m/z 74. In mass spectra of fatty acid methyl esters under electron-impact ionization, the base peak at m/z 74 has been identified as a fragment (C3H6O2+) due to a McLafferty rearrangement with cleavage of the bond between carbons 2 and 3 of the fatty acid (Murphy, 1993). In addition, small-intensity ions m/z 69, 70, and 71 have been identified in oleic acid methyl ester as C5H9+, C5H10+, and C5H11+, respectively (Schwender and Ohlrogge, 2002). If multiple 13C nuclei are present in these C5 fragments, isotopic peaks might significantly overlap with the m/z 74 ion cluster. As shown for unlabeled palmitic acid methyl ester in Figure 5A, the 15-eV ionization energy reduces the intensity of the ions m/z 69, 70, and 71 to less than 3% of the abundance of m/z 74. Therefore, the possibility of contamination of the ion cluster m/z 74 to 77 by mass isotopomers of fragments C5H9+, C5H10+, and C5H11+ is reduced by this configuration. In addition, fragment m/z 73 was found to be 0.7% of m/z 74 and therefore is considered an insignificant source of fragment impurity.
Figure 5.
Overlapping fragments m/z 74 (C3H6O2+) and m/z 75 (C3H7O2+) in the low-mass region of electron-impact mass spectra of saturated fatty acid methyl ester (FAMe). A, Mass spectra of palmitic acid methyl ester. Intensities are normalized to the base peak of the spectrum. An unlabeled standard was analyzed with 70 eV (gray bars) and 15 eV (black bars) ionization energy. For 15 eV, the ions m/z 69, 70, and 71 are reduced to less than 3% of the abundance of m/z 74. Fragment m/z 73 is 0.7% of m/z 74 and therefore most likely an insignificant source of fragment impurity. For [1,2-13C2]palmitic acid methyl ester (white bars), the cluster of fragments shifts by 2 mass units in accordance with the identity of fragments 74 and 75. Rel. units, Relative units. B, Relative abundance of m/z 75 as a function of sample size and fatty acid chain length. The ion cluster m/z 74 to 77 was corrected for natural isotopes in C3H6O2. As a result, the total intensity of the cluster was found in m/z 74 and 75.
The quantification of carbon isotope label in the ion cluster m/z 74 to 77 is complicated due to the overlap of two fragments, m/z 74 (C3H6O2+) and m/z 75 (C3H7O2+), the latter of which derives by a hydrogen transfer to C3H6O2+ (Murphy, 1993). Hence, m/z 75 is much more intense than expected by natural isotope content in C3H6O2+. However, the overlapping fragments differ by only one hydrogen, and the fragment overlap can be corrected for provided that the relative intensity of the two fragments is reproducible between measurements. This was verified for unlabeled saturated fatty acid methyl ester of four different chain lengths for different sample sizes injected (Fig. 5B). The abundance of C3H7O2+ relative to C3H6O2+ was highly reproducible over 2 orders of magnitude sample size. Therefore, for correction of the ion cluster m/z 74 to 77 in the methyl esters of saturated C18, C20, and C22 fatty acids, labeled samples are analyzed in parallel with unlabeled standards with injection of similar sample size. The ion correction takes place in three steps. (1) The relative abundance of m/z 75 (C3H7O2+) is derived from the unlabeled standards by correction of ion cluster m/z 74 to 77 for natural isotopes in C3H6O2. (2) The ion cluster m/z 74 to 77 in labeled samples is corrected for the ion m/z 75 according to the ratio in the unlabeled standard. (3) The resulting ion cluster is corrected for natural isotope levels in C1H6O2 (correction for the methyl group, hydrogen, and oxygen). The validity of this correction method is demonstrated in Table III, where standards of [1-13C]-, [2-13C]-, and [1,2-13C2]palmitic acid methyl ester were analyzed.
Table III.
Correction of the m/z 74 ion cluster in unlabeled and labeled standards of palmitic acid methyl ester for ion overlap and for natural isotopes in hydrogen, oxygen, and the methyl group
Corrected relative intensity ± sd (n = 3). Note that the standards are not expected to correct to exact 100% intensities in m/z 75 ([1-13C]- and [2-13C]palmitate) or in m/z 76 ([1,2-13C2]palmitate), since the labeled positions are enriched only to about 99% 13C abundance. Also, natural 13C in the unlabeled position is not corrected for.
| m/z | Label in Palmitic Acid |
|||
|---|---|---|---|---|
| Unlabeled | 1-13C | 2-13C | 1,2-13C2 | |
| 74.1 | 97.74 ± 0.01 | 0.75 ± 0.01 | 0.78 ± 0.01 | 0.12 ± 0.00 |
| 75.1 | 2.12 ± 0.01 | 97.94 ± 0.02 | 98.48 ± 0.04 | 1.76 ± 0.02 |
| 76.1 | 0.13 ± 0.01 | 1.19 ± 0.03 | 0.68 ± 0.05 | 98.18 ± 0.03 |
| 77.1 |
0.02 ± 0.01 |
0.12 ± 0.00 |
0.06 ± 0.02 |
−0.06 ± 0.04 |
Flux Modeling
The simulation and analysis of the metabolic model were performed using the software package 13CFLUX obtained from Dr. W. Wiechert (Institut für Biotechnologie 2, Forschungszentrum Jülich; Wiechert et al., 2001). In applying the modeling software, the values of 40 free flux variables were determined by the following constraints: (1) the topology and stoichiometry of the metabolic network at steady state; (2) irreversibility and unidirectionality of 16 internal reactions as well as of four uptake reactions and 20 effluxes into biomass; (3) the fixed values for 20 effluxes into biomass, as derived from the biomass composition of the embryos; and (4) the measured 13C labeling patterns in metabolites after labeling with [U-13C12]Suc. Flux parameter estimation was performed using the evolutionary algorithm of 13CFLUX as well as the Sequential Quadratic Programming algorithm by Peter Spellucci (Technical University Darmstadt). Computation was performed on a Linux-based computer with dual Intel xeon processors as well as on the Brookhaven Linux Cluster with 150 processors.
Model Structure
Central metabolism of developing Arabidopsis embryos was modeled, including reactions of glycolysis, OPPP, Rubisco, and TCA cycle. A total of 88 metabolic reactions were formulated by carbon transitions, and the model simulates fractional abundance of 13C-labeled isotopomers in 67 metabolic pools (Supplemental Tables S9 and S10). The structure of the flux model in this study (Fig. 3) is based on a flux model of developing embryos of B. napus (Schwender et al., 2006), a close relative to Arabidopsis. While the B. napus flux model is only in part compartmentalized (Schwender et al., 2006), in the current version the plastidic and cytosolic parts of upper glycolysis and the pentose phosphate pathway were modeled in both cytosol and plastid and respective metabolite transporters for hexose, pentose, triose phosphates, as well as PEP were added. The increased complexity is also motivated by the use of labeling measurements in Suc in the model (Fig. 3) in addition to the measurements in starch, which had been considered before in the B. napus flux model. Since Suc is derived from cytosolic hexose phosphate and starch is derived from plastidial hexose phosphate pools (Fig. 3), a full compartmentation of glycolysis and the pentose phosphate pathway was warranted. In the former B. napus flux model (Schwender et al., 2006), this complexity was assumed but the model was tailored to an uncompartmented model in anticipation that the reduced complexity can be resolved.
The presence of all major pathways and intracellular metabolite transporters shown in Figure 3 is supported by the gene expression pattern in developing Arabidopsis seeds (Ruuska et al., 2002; Schmid et al., 2005). In addition, the expression and presence of enzyme activities of both the cytosolic and plastidic Glc-6-P dehydrogenase isoforms in developing seeds of Arabidopsis has been well documented (Wakao et al., 2008) and suggests that the OPPP is operational in both compartments. However, the presence of nonoxidative parts of the pentose phosphate pathway in the plant cytosol is not clearly established and still debated (Kruger and von Schaewen, 2003). In Arabidopsis, the genes encoding for cytosolic transketolase and transaldolase might be missing (Kruger and von Schaewen, 2003). Without these enzymes in the cytosol, the presence of a xylulose 5-phosphate/phosphate translocator (XPT) in the plastidic envelope (Eicks et al., 2002) would allow pentose phosphates generated in the cytosol by the cytosolic OPPP to be further metabolized in the plastids (Eicks et al., 2002; Kruger and von Schaewen, 2003). Expression of XPT (At5g17630) in developing seeds was demonstrated in the expression data of Schmid et al. (2005). To account for the uncertainty about cytosolic tansketolase and transaldolase, we implemented both enzymes in the flux model as well as the XPT. This allows the model to simulate the complete cytosolic pentose phosphate pathway but also to assume states where cytosolic tansketolase and transaldolase are not used.
At present, simulation of lumped metabolic pools (e.g. triose phosphates, hexose phosphates) is widely used in 13C-tracer-based flux modeling in order to limit the model complexity. This model simplification can be justified by isotopic equilibration between metabolic pools due to fast interconversion. Accordingly, such assumptions in the B. napus model (Schwender et al., 2006) are made for the current model as well. Also, fast interconversion by transaminase activity of pyruvate and Ala was evident in B. napus embryos by labeling with [15N]Ala and [15N]Gln (Schwender et al., 2006) and also considered in this model. Another characteristic from the B. napus model that is maintained is the in vivo reversibility of isocitrate dehydrogenase established for B. napus embryos (Schwender et al., 2006). Being important for the determination of cytosolic and plastidic fluxes in lower glycolysis, the import of pyruvate into the plastids and mitochondria was modeled as unidirectional (nonreversible), because of a supposedly electro-neutral pH-driven uptake. For several plants, a pH-driven pyruvate transport into mitochondria has been described with biochemical properties similar to the mammalian mitochondrial pyruvate carrier (Laloi, 1999). In addition, uptake kinetics with isolated plastids of B. napus embryos suggest the presence of a plastidic pyruvate carrier (Eastmond and Rawsthorne, 2000) and give justification to the directional restriction of the plastidial pyruvate carrier.
Since the disruption of PK by mutation was studied here, it is important to consider the possibility of a bypass of PK via PEPC, malate dehydrogenase, and malic enzyme. In Arabidopsis, four different genes coding for NADP-malic enzyme have been identified, biochemically characterized, and their subcellular localization determined (Wheeler et al., 2005). Both plastid- and cytosol-localized isoforms of NADP-malic enzyme are expressed in developing embryos, and in particular a plastidic isoform (At1g79750) is strongly expressed in all embryo stages (Wheeler et al., 2005). Also, NADP-malic enzyme activity has been found to peak 12 d after flowering in developing Arabidopsis embryos (Baud and Graham, 2006). Therefore, both plastidic and cytosolic malic enzyme reactions were added to the model in addition to the already existing mitochondrial isoform.
Additional Model Adjustments
Initial modeling of the Arabidopsis labeling experiments did not result in a good fit for the labeling signature in Gly. Therefore in our network model, the biosynthesis of Gly was extended. During seed development, up-regulation of isocitrate lyase (At3g21720) and glyoxylate transaminase activity (GGT2; At1g70580) is indicated by developmental expression profiling data (Schmid et al., 2005) and might contribute to Gly formation. Furthermore, for Gly decarboxylase, two reaction directions were allowed, since reversibility of this enzyme complex has been found at least for in vitro assays (Sarojini and Oliver, 1983; Walker and Oliver, 1986). Adding the two reactions (vICL and vGT; Fig. 3) to the network and allowing Gly decarboxylase reversibility resulted in an acceptable fit of the Ser and Gly labeling signatures by the model. Also, Gly could be formed directly from Thr by Thr aldolase, which is known to be present in developing Arabidopsis seeds (Joshi et al., 2006). This reaction was not further tested. More studies are needed for full clarification of Gly biosynthesis in Arabidopsis seeds.
Flux Estimation
For each of the four genotypes (pkpβ1pkpα, wri1-1, Ws, and Col), a respective model variant was derived and simulated to estimate intracellular fluxes during storage synthesis. For the pkpβ1pkpα model variant, no a priori assumption about the missing PK activity was made. The model variants differ according to the label measurements for each genotype. As a consequence of differences in the biomass composition (Fig. 2A), the model variants differ in the values of the 20 biomass fluxes (Fig. 3). Most outstanding is the change in lipid biosynthetic flux between the wild types and mutants. For each model variant, a total of 81 reaction rates in the model are estimated based on nonlinear optimization with iterative adjustment of the values for 15 free net fluxes and 25 exchange fluxes (reaction reversibilities). The process minimizes the difference between 130 model-predicted mass spectral isotopomer signals and the respective experimental values obtained from GC-MS analysis (Supplemental Table S9). The goodness of fit is judged by a sum of squares function:
 |
where n represents the number of label measurements; mjpredicted and mjexperimental are predicted and experimental values for the labeling measurement j, and σj is the sd of measurement j.
In order to ensure that a global optimum has been found, the flux parameter estimation was started 200 times, each time with random start values for the free fluxes. Typically, the optimization algorithm reached an optimum after 200 to 500 improvements of the initial flux values. Assuming that the studied biological process is adequately described by the metabolic model and that there are no gross measurement errors or other artifacts, the optimal χ2 value should represent a sum of random noise. Based on the general model dimensions (130 modeled MS signals − 29 molecular fragments − 40 free fluxes = 61 degrees of freedom), the χ2 value should be less than 75 (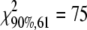 ; 90% confidence level). Initial optima first indicated that two measurements consistently had large error residuals: the m5 isotopomer fraction derived from ion m/z 360 [Glc(1–6)] of Glc methoxime penta-acetate and m5 of m/z 361 (C15H33O4Si3) of the Suc-trimethylsilyl derivative. These could be removed from the data set without the optimum solution being different. As a result, each model consistently converged to a minimum value for χ2 (30.15, 52.48, 37.82, and 33.75 for Ws, pkpβ1pkpα, Col, and wri1-1, respectively) for which the values for multiple fluxes were reproduced with good precision. Accordingly, the statistical test of goodness of fit passes (Supplemental Table S9).
; 90% confidence level). Initial optima first indicated that two measurements consistently had large error residuals: the m5 isotopomer fraction derived from ion m/z 360 [Glc(1–6)] of Glc methoxime penta-acetate and m5 of m/z 361 (C15H33O4Si3) of the Suc-trimethylsilyl derivative. These could be removed from the data set without the optimum solution being different. As a result, each model consistently converged to a minimum value for χ2 (30.15, 52.48, 37.82, and 33.75 for Ws, pkpβ1pkpα, Col, and wri1-1, respectively) for which the values for multiple fluxes were reproduced with good precision. Accordingly, the statistical test of goodness of fit passes (Supplemental Table S9).
Statistical Quality Measures
For the comparison of flux phenotypes, it is important to have statistical confidence limits for the flux values. In the statistical approach based on model linearization realized in 13CFLUX (Wiechert and deGraaf, 1997), the computation of the model statistics by the software tool EstimateStat is only possible if the information contained in the labeling allows us to determine all free fluxes in the model (more specifically, if the free flux/measurement sensitivity matrix has full numerical rank). EstimateStat identifies fluxes for which there is no information in the labeling data and suggests declaring those fluxes as exactly known in the model in order to allow the computation to be completed. This essentially excludes the variability and statistical uncertainty of those fluxes from the model. Multiple net and exchange fluxes were not or only poorly determined by the labeling data. Neglecting the statistical uncertainty in these fluxes might lead to significant underestimation of the size of confidence limits for the remaining fluxes of interest. Therefore, the statistical quality measures were computed by Monte Carlo stochastic simulation, which is computationally much more expensive than EstimateStat but can be performed with consideration of the variability of all free fluxes in the network. For each genotype model variant, the model was reoptimized 50 times after random resampling of the labeling data and the flux measurements (fluxes into biomass). To the original mean measurement values, random noise was added with Gaussian distribution according to the estimated sd values (Box and Muller, 1958). Repeated reoptimizing resulted in 50 flux vectors. For each flux, mean and sd were calculated. For the fluxes reproduced in Table I, the mean values from the stochastic simulation were very close to the best-fit flux values.
In order to validate the Monte Carlo approach, the results were compared with another alternative method to derive statistical quality measures. Here, the confidence limits of a selected flux are determined by repeated reoptimization of the model (Antoniewicz et al., 2006): The flux parameter-fitting procedure is repeated multiple times, with each time the selected flux step by step being removed from its optimum value. This means that all other fluxes are redetermined by minimization of the χ2 sum. As the value of the selected flux is removed from its optimum value, the χ2 sum increases relative to the minimum value at the optimum (Fig. 6). The selected flux is within a 1 − α confidence interval, as long as the increase of the sum of squared residuals is less than the value of 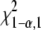 (χ2 distribution with 1 degree of freedom; Antoniewicz et al., 2006). This method has been shown to produce confidence limits that coincide with exact analytically derived confidence intervals (Antoniewicz et al., 2006). It was used to derive 90% confidence limits for a number of selected fluxes (Table IV). sd values from Monte Carlo stochastic simulation were transformed into 90% confidence intervals according to the relation (Wiechert et al., 1997):
(χ2 distribution with 1 degree of freedom; Antoniewicz et al., 2006). This method has been shown to produce confidence limits that coincide with exact analytically derived confidence intervals (Antoniewicz et al., 2006). It was used to derive 90% confidence limits for a number of selected fluxes (Table IV). sd values from Monte Carlo stochastic simulation were transformed into 90% confidence intervals according to the relation (Wiechert et al., 1997):
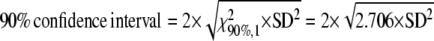 |
Figure 6.
Determination of 90% flux confidence interval for metabolic fluxes according to Antoniewicz et al. (2006). For the Col wild-type model, the flux vPKp was removed from its optimum value by small increments, and each time all other fluxes were redetermined by flux optimization (i.e. by minimization of the sum of squared residuals). On the abscissa, the flux confidence interval (gray arrow) is given by intersection with a line defined by the χ2 critical value for 1 degree of freedom. For vPKp > 180.3, no points are shown, since due to the directionality constraints in the flux model those do not exist.
Table IV.
Comparison of two methods for the determination of statistical confidence limits of fluxes
The size of the 90% confidence intervals was calculated for selected fluxes by stepwise removal from the flux optimum value and reoptimizing of the model (“Step”; see text). These values are compared with 90% confidence intervals derived by Monte Carlo stochastic simulation (“MC”; see text).
| Genotype | Flux | Step | MCa | MC/Step |
|---|---|---|---|---|
| Ws | vPKc | 24.00 | 26.13 | 109% |
| vPKp | 32.00 | 42.52 | 133% | |
| vPEPC | 20.00 | 17.55 | 88% | |
| vPyr1 | 39.00 | 47.58 | 122% | |
| pkpβ1pkpα | vPKc | 32.00 | 20.62 | 64% |
| vPKp | 28.00 | 32.65 | 117% | |
| vPEPC | 20.00 | 11.59 | 58% | |
| vPyr1 | 40.00 | 31.70 | 79% | |
| Col | vPKc | 29.50 | 19.57 | 66% |
| vPKp | 37.49 | 28.73 | 77% | |
| vPEPC | 10.00 | 6.49 | 65% | |
| vPyr1 | 40.00 | 33.63 | 84% | |
| wri1-1 | vPKc | 34.00 | 33.75 | 99% |
| vPKp | 34.00 | 30.79 | 91% | |
| vPEPC | 18.00 | 26.68 | 148% | |
| vPyr1 |
57.00 |
46.36 |
81% |
Size of the 90% confidence interval is two times the 90% deviation.
As shown in Table IV, the 90% confidence intervals generated by both methods are in good agreement for most fluxes.
The significance of differences between fluxes or flux ratios was judged with Student's t test. Here, sample size (n) refers to the number of biological replicates averaged in the MS labeling signatures and is different for the four genotypes (Table I). Therefore, the t test for unequal sample size and unequal variance was used.
Definition of Biomass Fluxes
The model contains 20 effluxes into biomass that are constant in the simulations and separately defined for the each of the four genotypes. They were derived considering (1) the weight fractions in dry weight of protein, storage oils, free metabolites, and starch (Supplemental Table S1), (2) breaking down each biomass fraction into precursor fractions according to the composition of molecule constituents or biosynthetic monomers (Supplemental Tables S2 and S7), and (3) considering the molar weight of each precursor. The amino acid composition in protein was estimated based on the amino acid composition in cell pellet hydrolysate of Ws ecotype (Supplemental Tables S5–S7). Lipid composition was estimated according to the fatty acid profile for each genotype (Supplemental Table S3). The free metabolite fraction was further analyzed as methoxyamine-trimethylsilyl derivatives by GC-MS based on the protocol by Roessner et al. (2000). Accordingly, Suc, Glu, Gln, and other amino acids were found as dominant compounds. By integration of the chromatograms, the Suc peak had around 50% of the total peak area. Since this GC-MS analysis did not allow accurate quantification of the components, the free metabolite fraction was defined to consist approximately of 50% (w/w) Suc and 50% (w/w) Gln. In addition, after accounting for protein, lipid, free metabolites, and starch in all four genotypes, between 15% and 25% of the embryo dry weight remained as not further characterized insoluble cell pellet fraction. Its weight was accounted for in the model by assuming it to be hexose polymer in cell wall (Supplemental Table S1).
Flux Units of the Simulation Results
The best-fit flux values resulting from model simulation (Supplemental Table S8) might be presented in units that reflect the rate of biomass accumulation if respective growth measurements are available. Since the growth kinetics of cultured Ws, Col, wri1-1, and pkpβ1pkpα embryos did not indicate drastic differences in growth speed (Fig. 1), it was decided to normalize the flux values relative to biomass units. The flux data shown in Table I and Figure 4 are scaled in such a way that for each genotype, 600 mol of carbon (the equivalent of 100 mol of Glc) flows into biomass.
Bias of Labeling Signatures by Unlabeled Biomass
The growth curves of cultured Arabidopsis embryos shown in Figure 1 suggest that up to 20% of the embryo mass harvested after 7 d might be from the initial embryo inoculum. If the final labeled embryo tissue is analyzed, the unlabeled inoculum storage compounds still present will dilute the newly formed labeled material. This constitutes a bias in the labeling signatures that is particularly relevant for mass spectrometric measurements, since these account for all 13C/12C isotopomers. In contrast to former studies with B. napus embryos (Schwender et al., 2003, 2006), where this effect was much less pronounced, our flux model was extended to explicitly model unlabeled biomass. For each network metabolite Mi for which label measurements are considered in the model, the network is extended by two auxiliary reactions that mix Mi with an unlabeled metabolite Mui according to:
 |
and
 |
with vx being the actual rate by which Mi is accumulated in biomass and α being a dilution coefficient applied to all Mi in the model. In the section LABEL_MEASUREMENTS of the 13CFLUX model file, M′i is declared as label measurement. Each Mui has to be defined by natural 13C abundance in the LABEL_INPUT section. The exact implementation of α in the model is described in Supplemental Model S1. Note that this model configuration is only meaningful if each reaction Mi → M′i is an efflux out of the metabolic network. The value for α can be optimized like other free flux parameters in the model. Due to problems with the optimizing algorithms under this model configuration, the four genotype model variants were simulated by fixing α to a value of 0.1, which corresponds to assuming 9.1% unlabeled biomass to be present. For all four model variants, this configuration in the model improved the best fit.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Biomass fractions in cultivated embryos.
Supplemental Table S2. Fatty acid composition of embryo lipid fractions, as determined by GC-MS analysis of fatty acid methyl ester.
Supplemental Table S3. Biomass fractions in mature seeds.
Supplemental Table S4. Fatty acid composition of lipid fraction in mature seeds, as determined by GC-MS analysis of fatty acid methyl ester.
Supplemental Table S5. Amino acid composition in hydroylsates of extracted embryo tissue of ecotype Ws.
Supplemental Table S6. Amino acid composition averaged over 13 gene sequences of Arabidopsis seed storage proteins.
Supplemental Table S7. Amino acid composition (mol %) in biomass protein used for flux analysis.
Supplemental Table S8. Scaled flux values for best fit: net fluxes, exchange (XCH) fluxes, and sd.
Supplemental Table S9. Mass spectrometric measurements of fractional 13C enrichment and model-predicted values for best fit.
Supplemental Table S10. Metabolite names.
Supplemental Model S1. Arabidopsis developing seed flux model in .ftbl format (13CFLUX model file).
Supplementary Material
Acknowledgments
We thank Christoph Benning (Michigan State University) for wri1-1 seeds as well as Christine Rochat and Sebastien Baud (INRA/AgroParisTech, Versailles, France) for pkpβ1pkpα seeds. We also thank Jordan Hay (Brookhaven National Laboratory) for manuscript reading and critical discussion.
This work was supported by the U.S. Department of Energy, Office of Basic Energy Sciences.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jörg Schwender (schwend@bnl.gov).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen DK, Ohlrogge JB, Shachar-Hill Y (2009) The role of light in soybean seed filling metabolism. Plant J 58 220–234 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y (2007) Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J 52 296–308 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2006) Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8 324–337 [DOI] [PubMed] [Google Scholar]
- Bao X, Focke M, Pollard M, Ohlrogge J (2000) Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J 22 39–50 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype Ws. Plant Physiol Biochem 40 151–160 [Google Scholar]
- Baud S, Graham IA (2006) A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild-type and mutant embryos of Arabidopsis using in situ histochemistry. Plant J 46 155–169 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L (2008) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47 448–455 [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007. a) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007. b) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52 405–419 [DOI] [PubMed] [Google Scholar]
- Box GEP, Muller ME (1958) A note on the generation of random normal deviates. Ann Math Stat 29 610–611 [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152 141–145 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40 575–585 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryo. Plant Physiol 122 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M, Maurino V, Knappe S, Flügge UI, Fischer K (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol 128 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wuthrich K, Bailey JE, Sauer U (2002) Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol 184 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettenhuber C, Spielbauer G, Margl L, Hannah LC, Gierl A, Bacher A, Genschel U, Eisenreich W (2005) Changes in flux pattern of the central carbohydrate metabolism during kernel development in maize. Phytochemistry 66 2632–2642 [DOI] [PubMed] [Google Scholar]
- Fell DA (1997) Understanding the Control of Metabolism. Portland Press, London
- Fischer E, Sauer U (2005) Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nat Genet 37 636–640 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226 497–509 [PubMed] [Google Scholar]
- Fuzfai Z, Boldizsar I, Molnar-Perl I (2008) Characteristic fragmentation patterns of the trimethylsilyl and trimethylsilyl-oxime derivatives of various saccharides as obtained by gas chromatography coupled to ion-trap mass spectrometry. J Chromatogr A 1177 183–189 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27 655–673 [Google Scholar]
- Golombek S, Rolletschek H, Wobus U, Weber H (2001) Control of storage protein accumulation during legume seed development. J Plant Physiol 158 457–464 [Google Scholar]
- Gomez LD, Baud S, Gilday A, Li Y, Graham IA (2006) Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46 69–84 [DOI] [PubMed] [Google Scholar]
- Heath JD, Weldon R, Monnot C, Meinke DW (1986) Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169 304–312 [DOI] [PubMed] [Google Scholar]
- Iyer VV, Sriram G, Fulton DB, Zhou R, Westgate ME, Shanks JV (2008) Metabolic flux maps comparing the effect of temperature on protein and oil biosynthesis in developing soybean cotyledons. Plant Cell Environ 31 506–517 [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Lonien J, Heady LE, Rogers A, Schwender J (2007) Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68 2232–2242 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, von Schaewen A (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6 236–246 [DOI] [PubMed] [Google Scholar]
- Laloi M (1999) Plant mitochondrial carriers: an overview. Cell Mol Life Sci 56 918–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cluette-Brown JE, Goodman HM (2004) The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol 135 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini P, Salamini F (2003) Plant biotechnology and breeding: allied for years to come. Trends Plant Sci 8 70–75 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays in tobacco tissue culture. Physiol Plant 15 473–497 [Google Scholar]
- Murphy CM (1993) Mass spectrometry of lipids. In F Snyder, ed, Handbook of Lipid Research, Vol 7. Plenum Press, New York, pp 71–130
- Ratcliffe RG, Shachar-Hill Y (2006) Measuring multiple fluxes through plant metabolic networks. Plant J 45 490–511 [DOI] [PubMed] [Google Scholar]
- Rios-Estepa R, Lange BM (2007) Experimental and mathematical approaches to modeling plant metabolic networks. Phytochemistry 68 2351–2374 [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23 131–142 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Radchuk R, Miranda M, Heim U, Wobus U, Weber H (2004) Seed-specific expression of a bacterial phosphoenolpyruvate carboxylase in Vicia narbonensis increases protein content and improves carbon economy. Plant Biotechnol J 2 211–219 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojini G, Oliver DJ (1983) Extraction and partial characterization of the glycine decarboxylase multienzyme complex from pea leaf mitochondria. Plant Physiol 72 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schwender J (2008) Metabolic flux analysis as a tool in metabolic engineering of plants. Curr Opin Biotechnol 19 131–137 [DOI] [PubMed] [Google Scholar]
- Schwender J (2009) Isotopic steady-state flux analysis. In J Schwender, ed, Plant Metabolic Networks. Springer, New York, pp 245–284
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004. a) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge J, Shachar-Hill Y (2004. b) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7 309–317 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB (2002) Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol 130 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278 29442–29453 [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB (2006) Mitochondrial metabolism in developing embryos of Brassica napus. J Biol Chem 281 34040–34047 [DOI] [PubMed] [Google Scholar]
- Smith AJ, Rinne RW, Seif RD (1989) Phosphoenolpyruvate carboxylase and pyruvate-kinase involvement in protein and oil biosynthesis during soybean seed development. Crop Sci 29 349–353 [Google Scholar]
- Smith CR, Knowles VL, Plaxton WC (2000) Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures: implications for the integration of glycolysis with nitrogen assimilation. Eur J Biochem 267 4477–4485 [DOI] [PubMed] [Google Scholar]
- Spielbauer G, Margl L, Hannah LC, Romisch W, Ettenhuber C, Bacher A, Gierl A, Eisenreich W, Genschel U (2006) Robustness of central carbohydrate metabolism in developing maize kernels. Phytochemistry 67 1460–1475 [DOI] [PubMed] [Google Scholar]
- Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou R, Westgate ME, Spalding MH, Shanks JV (2004) Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional 13C labeling, two-dimensional [13C,1H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiol 136 3043–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Quick WP, Schurr U, Schulze ED, Rodermel SR, Bogorad L (1991) Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with antisense rbcs. 2. Flux control coefficients for photosynthesis in varying light, CO2, and air humidity. Planta 183 555–556 [DOI] [PubMed] [Google Scholar]
- Tajuddin T, Watanabe S, Yamanaka N, Harada K (2003) Analysis of quantitative trait loci for protein and lipid contents in soybean seeds using recombinant inbred lines. Breed Sci 53 133–140 [Google Scholar]
- Troufflard S, Roscher A, Thomasset B, Barbotin JN, Rawsthorne S, Portais JC (2007) In vivo 13C NMR determines metabolic fluxes and steady state in linseed embryos. Phytochemistry 68 2341–2350 [DOI] [PubMed] [Google Scholar]
- Wahl SA, Dauner M, Wiechert W (2004) New tools for mass isotopomer data evaluation in (13)C flux analysis: mass isotope correction, data consistency checking, and precursor relationships. Biotechnol Bioeng 85 259–268 [DOI] [PubMed] [Google Scholar]
- Wakao S, Andre C, Benning C (2008) Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiol 146 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JL, Oliver DJ (1986) Glycine decarboxylase multienzyme complex: purification and partial characterization from pea leaf mitochondria. J Biol Chem 261 2214–2221 [PubMed] [Google Scholar]
- Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flugge UI, Maurino VG (2005) A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol 139 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechert W, deGraaf AA (1997) Bidirectional reaction steps in metabolic networks. 1. Modeling and simulation of carbon isotope labeling experiments. Biotechnol Bioeng 55 101–117 [DOI] [PubMed] [Google Scholar]
- Wiechert W, Mollney M, Petersen S, de Graaf AA (2001) A universal framework for 13C metabolic flux analysis. Metab Eng 3 265–283 [DOI] [PubMed] [Google Scholar]
- Wiechert W, Siefke C, deGraaf AA, Marx A (1997) Bidirectional reaction steps in metabolic networks. 2. Flux estimation and statistical analysis. Biotechnol Bioeng 55 118–135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.