Abstract
Cleavage and polyadenylation of precursor mRNA is an essential process for mRNA maturation. Among the 15 to 20 protein factors required for this process, a subgroup of proteins is needed for both cleavage and polyadenylation in plants and animals. This subgroup of proteins is known as the cleavage and polyadenylation specificity factor (CPSF). To explore the in vivo structural features of plant CPSF, we used tandem affinity purification methods to isolate the interacting protein complexes for each component of the CPSF subunits using Arabidopsis (Arabidopsis thaliana ecotype Landsberg erecta) suspension culture cells. The proteins in these complexes were identified by mass spectrometry and western immunoblots. By compiling the in vivo interaction data from tandem affinity purification tagging as well as other available yeast two-hybrid data, we propose an in vivo plant CPSF model in which the Arabidopsis CPSF possesses AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF100, AtCPSF160, AtFY, and AtFIPS5. Among them, AtCPSF100 serves as a core with which all other factors, except AtFIPS5, are associated. These results show that plant CPSF possesses distinct features, such as AtCPSF73-II and AtFY, while sharing other ortholog components with its yeast and mammalian counterparts. Interestingly, these two unique plant CPSF components have been associated with embryo development and flowering time controls, both of which involve plant-specific biological processes.
Cleavage and polyadenylation of precursor mRNA (pre-mRNA) is a critical process during mRNA biogenesis. It consists of cleavage at the cleavage site and polyadenylation that adds a tract of adenosines [poly(A)] to the newly generated 3′ end. Correct formation of mRNA 3′ end with a poly(A) tail is essential for mRNA functions, such as mRNA stability (Holec et al., 2006), exportation (Hammell et al., 2002), and translatability (Buratowski, 2005). Cleavage and polyadenylation is a seemingly simple process, but it is associated with many aspects of mRNA biogenesis and functions, including transcription initiation, elongation (Proudfoot, 2004) and termination (Hammell et al., 2002), pre-mRNA splicing (Rigo and Martinson, 2008), and translation initiation (Hammell et al., 2002; Buratowski, 2005). Based on the variety of processes involved in mRNA 3′ end formation, it has been proposed that polyadenylation may work as a hub for an integrated network of cotranscriptional mRNA processing events. It is through this integrated network that multiple regulatory mechanisms, each exerting its own level of mRNA biogenesis, work together to fine-tune gene expression (Danckwardt et al., 2008).
The 15 to 20 protein factors that are required for cleavage and polyadenylation are organized into complexes, as they were originally isolated based on chromatography from yeast and mammals (Zhao et al., 1999; for a list of these factors, see Supplemental Table S1). In the case of mammals, the 3′ end-processing complex contains several subcomplexes, including the cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I, and cleavage factor II. Cleavage factor II contains two subunits, hPcf11 and hClp1, which are yeast orthologs of Pcf11p and Clp1p (both belong to yeast cleavage factor IA [CF-IA]), respectively. Pcf11p contains a conserved RNA polymerase II C-terminal domain-interacting domain, mutations of which do not affect 3′ end processing but instead cause incorrect transcriptional termination. Immunodepletion of hClp1 abolishes cleavage activity, but it does not affect polyadenylation (Mandel et al., 2008). In addition, poly(A) polymerase (PAP), poly(A)-binding protein (PABP), symplekin, and RNA polymerase II C-terminal domain also belong to the pre-mRNA cleavage and polyadenylation machinery. All of the protein factors, except PABP, are required for in vitro cleavage reaction, while only CPSF, PAP, and PABP are required for in vitro polyadenylation (Zhao et al., 1999). Although the yeast 3′ end-processing complex was named differently from the mammalian complex, they share significant similarities. The subcomplexes of the yeast machinery include the cleavage and polyadenylation factor (CPF), CF-IA, and CF-IB. CPF can be further separated into CF-II and polyadenylation factor I. CF-II contains some subunits that are homologous to those in mammalian CPSF, and CF-IA contains some subunits that are homologous to those in mammalian CstF and cleavage factor II. The functions of some of these homologs can be different in yeast and in mammals (Zhao et al., 1999).
Among those 15 to 20 protein factors, only one subgroup of proteins is needed for both the cleavage and polyadenylation steps. This protein subgroup is CPSF in both mammals and plants and CPF in yeast (Zhao et al., 1999). Mammalian CPSFs consist of five subunits, four of which are closely associated. These four protein factors are named according to their molecular mass values (CPSF30, CPSF73, CPSF100, and CPSF160) and were purified as a large protein complex from HeLa cells (Bienroth et al., 1991; Murthy and Manley, 1992). Antibody against one of the protein factors, CPSF100, coimmunoprecipitated the whole complex, confirming that CPSFs exist as a complex in the cell. CPSF160 is the largest subunit that is conserved in mammals and yeast, and it is essential in both systems. CPSF160 interacts directly with pre-mRNA to define the cleavage site by specifically binding to the AAUAAA domain (Zhao et al., 1999). CPSF160 interacts with other CPSF factors, such as hFip1 [for factor interacting with poly(A) polymerase in human] and CPSF100 (Chanfreau et al., 1996; He et al., 2003). CPSF100 belongs to the β-CASP (for metallo-β-lactamase-associated CPSF Artemis SNM1/PSO2) superfamily that is involved in nucleic acid binding and processing (Dominski, 2007). However, CPSF100 is catalytically inactive in the absence of critical residues (Callebaut et al., 2002). Structurally similar to CPSF100, CPSF73 also belongs to the β-CASP superfamily (Dominski, 2007). It has been shown that CPSF73 possesses nuclease activity by both crystal structural analysis and in vitro activity assays (Mandel et al., 2006), and it is proposed as the endonuclease for pre-mRNA cleavage (Ryan et al., 2004; Dominski, 2007). The smallest CPSF subunit, CPSF30, is a zinc finger protein that interacts with hFip1. CPSF30 has not always been detected with active CPSF preparations, although it is frequently immunoprecipitated with other CPSF factors (Hirose and Manley, 2000). hFip1 is the fifth mammalian CPSF subunit and is less closely connected with other CPSF factors. However, hFip1 is tightly associated with CPSF30 as well as other polyadenylation-related protein factors such as PAP (Kaufmann et al., 2004).
The yeast CPF is functionally similar to mammalian CPSF. It contains approximately seven protein factors that were copurified with the tag-fused Fip1p from Saccharomyces cerevisiae (Preker et al., 1997). Yeast CPF can be further grouped into CF-II and polyadenylation factor I (PF-I) subcomplexes (Chen and Moore, 1992). In the CF-II complex, three (Yhh1p, Ydh1p, and Ysh1p) of the four factors are homologous to the mammalian CPSF160, -100, and -73, respectively. Yhh1p binds to sequences near the A-rich cleavage site (Dichtl et al., 2002) and interacts with other CPF factors such as Fip1p and Ydh1p, the depletion of which abolished in vitro cleavage and polyadenylation (Stumpf and Domdey, 1996). Ydh1p interacts with other CPF factors, such as Yhh1p, Ysh1p, and Pta1p, as well as Pfs2p (Kyburz et al., 2003). Ysh1p is the yeast homolog of CPSF73, which is essential for cell viability (Chanfreau et al., 1996). However, the fourth yeast CF-II factor, Pta1p, is the homolog of symplekin, which is currently not considered a CPSF factor in the mammalian system (Mandel et al., 2008). Pta1p interacts with Ysh1p, Ydh1p, and many other factors, indicating that it may play a scaffolding role (Kyburz et al., 2003). Similarly, two of the three PF-I factors, Yth1p and Fip1p, are homologous to the mammalian CPSF factors CPSF30 and hFip1, respectively. The third yeast PF-I factor, Pfs2p, has a homolog in mammals, but with unknown function (Mandel et al., 2008). In plants, Pfs2p belongs to a highly conserved group of eukaryotic proteins represented by the RNA 3′ end-processing factor AtFY (Simpson et al., 2003).
Compared with the relatively better understood mammalian CPSF and yeast CPF, the composition of CPSF and its in vivo interaction pattern in plants has not yet been elucidated. To date, Arabidopsis (Arabidopsis thaliana) polyadenylation machinery is the best-studied system in plants, largely owing to the extensively annotated complete genome sequences and the availability of genetic resources. The Arabidopsis genome encodes all of the protein factors analogous to the already defined mammalian CPSF (Xu et al., 2006; Hunt, 2007). However, it remains unclear whether these Arabidopsis CPSF (AtCPSF) factors form an in vivo protein complex similar to their mammalian counterparts.
Studies aimed at exploring the in vivo composition and interaction patterns among plant CPSF components have been limited by the low expression level of CPSF and the lack of an efficient purification approach able to isolate protein factors while maintaining their natural interacting complexes. Here, we report an investigation of Arabidopsis CPSF composition and its in vivo interactions using Arabidopsis suspension cell cultures. Using the tandem affinity purification (TAP) approach from Arabidopsis, cell cultures overexpressing six TAP tag-fused Arabidopsis polyadenylation-related proteins, CPSF factors, and other polyadenylation-related proteins were identified by mass spectrometry (MS) and western immunoblots. Thus, a plant CPSF model is proposed based on the identified protein factors. The similarities and differences between plant CPSF and its mammalian and yeast counterparts are discussed.
RESULTS
Expression of Arabidopsis CPSF by TAP Tagging
The identities of the subunits of plant CPSF have been previously predicted using bioinformatics approaches, and their pair-wise interactions have been shown by yeast two-hybrid (Y2H) and in vitro pull-down assays (Xu et al., 2006; Hunt et al., 2008). However, direct evidence that these proteins exist and interact as a complex in vivo would be best studied by isolating the protein complex through immunoprecipitation or affinity purification. Therefore, with the goal of isolating an intact Arabidopsis CPSF complex, we investigated the Arabidopsis CPSF composition and the interacting partners using the TAP methodology (Rohila et al., 2004).
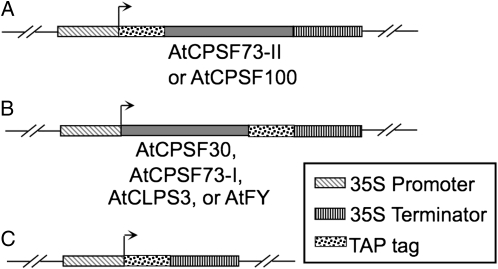
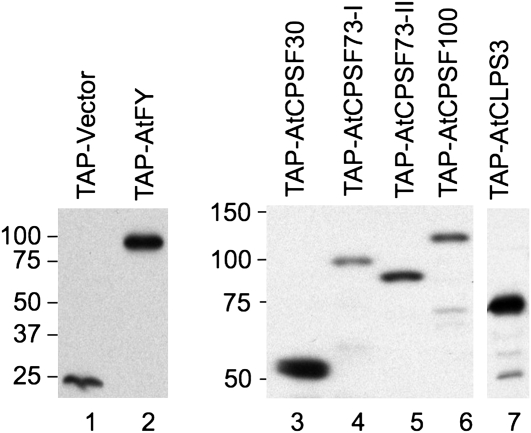
The cDNAs of four previously identified Arabidopsis CPSF subunits, AtCPSF30, AtCPSF73-I, AtCPSF73-II, and AtCPSF100 (Xu et al., 2006), as well as two potential interacting partners of CPSF, AtCLPS3 (Xing et al., 2008a) and AtFY (Herr et al., 2006), were fused with TAP tags (Fig. 1). The fusion proteins were expressed in Arabidopsis suspension cell cultures, as described in “Materials and Methods.” As a control, an empty vector was also transformed and expressed using the exact same procedures. While protein extracts were detected using an antibody (peroxidase-conjugated anti-peroxidase; Sigma) that specifically recognizes the protein A domain of the TAP tag, the cell culture that overexpressed the empty vector showed only one unique band of about 22 kD (Fig. 2, lane 1). The position of this protein band agreed with the predicted size of the TAP tag (Rohila et al., 2004). As shown in other lanes of Figure 2, cell cultures overexpressing TAP tag fusion proteins of AtFY, AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF100, and AtCLPS3 also produced predicted protein bands (about 22 kD larger than their original protein sizes). The expression levels among these fusion proteins were comparable when similar amounts of cells were used, with some variation where AtCPSF73-I was the lowest and AtCLPS3 was the highest.
Figure 1.
TAP fusion protein constructions of cDNAs encoding CPSF subunits or CPSF-associated factors. The genes were fused to TAP tags, either at the N terminus of the target genes, such as AtCPSF73-II (locus identifier is listed in Supplemental Table S1) and AtCPSF100 (A), or the C terminus of the target genes, such as AtCPSF30, AtCPSF73-I, AtCLPS3, and AtFY (B). The fusion genes were driven by the cauliflower mosaic virus 35S promoter. C, A vector control without any gene fused to the TAP tag. The transcript start site is marked by the rightward arrow. The engineered genes were hosted by a binary vector (Rohila et al., 2004), which was mobilized into agrobacteria for Arabidopsis cell transformation.
Figure 2.
TAP-fused proteins were detected by an antibody against the protein A portion of the TAP tag. Total proteins from cell cultures overexpressing the TAP-fused target proteins (marked on the top) were separated by SDS-PAGE and analyzed by western blots. The protein sizes are labeled to the left side of each gel (kD). The molecular masses (kD) of the target proteins are as follows: AtCPSF30, 29; AtCPSF73-I, 78; AtCPSF73-II, 69; AtCPSF100, 83; AtCLPS3, 48; and AtFY, 73.
Purification and Identification of Arabidopsis CPSF and Its Associated Proteins
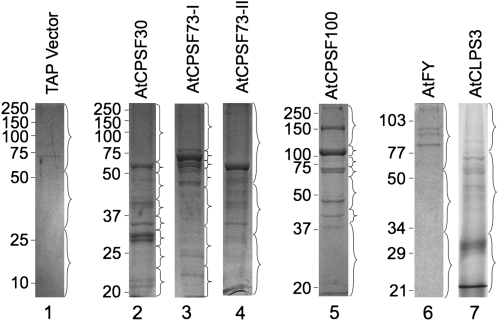
Knowing that the fusion proteins were expressed, we isolated the proteins that form complexes with these TAP-tagged proteins. From similar amounts of cell cultures, the tagged proteins were isolated using procedures as described in “Materials and Methods.” After TAP procedures (IgG and calmodulin-binding protein-conjugated beads), the proteins were then separated by SDS-PAGE and stained by Coomassie Brilliant Blue R-250. In most cases, multiple distinct protein bands were visible (Fig. 3, lanes 2–7), with the exception of the purification from the cell culture overexpressing the empty TAP vector (Fig. 3, lane 1). After staining, each gel lane was horizontally divided into sections so that the major bands were collected into different fractions (braces in Fig. 3). This was an effort to reduce the possibility of some abundant proteins masking the less abundant bands in the following identification steps. The resulting gel slices were then individually digested by trypsin, and the subsequent peptides were identified by HPLC with tandem MS (LC-MS/MS). The MS data were used to search protein databases using the search engine Mascot (Matrix Science). The identified proteins were listed according to their scores, which represent the probability that an observed match between the experimental data and the database sequence is a random event: the higher the scores are, the less likely it is that the proteins will be false positives. In our study, only protein hits with scores higher than 57 (less than 5% chance of getting a false-positive match [Pappin et al., 1993]) were considered as positively identified protein factors interacting with the TAP-fused target proteins. Accordingly, the accepted proteins are listed in Supplemental Table S2.
Figure 3.
Coomassie Brilliant Blue staining of proteins isolated by TAP purifications from Arabidopsis suspension cell cultures. The proteins were separated by 12% SDS-PAGE. Each gel lane was divided into a number of segments as indicated by the braces. The gel fragments were digested by trypsin overnight and subjected to LC-MS/MS for protein identification. Protein markers are as shown on the left (kD).
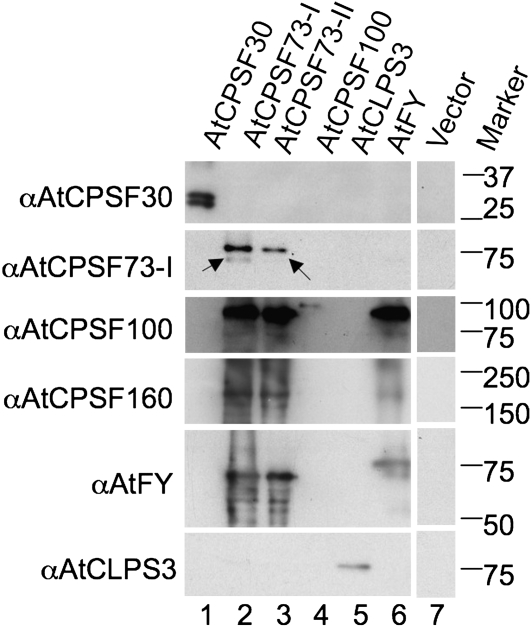
As a complement to the MS approach, we also performed western blotting to identify proteins that may be missed by MS. Antibodies against AtCPSF30, -73-I, -100, -160 (Xu et al., 2006), AtFY (Simpson et al., 2003), and AtCLPS3 (Xing et al., 2008a) were employed. As a result, many of the proteins that were originally identified by MS were also confirmed by their corresponding antibodies. Moreover, as it has been observed that, in most cases, western blots offer much higher sensitivity than MS (Copse and Fowler, 2002; Shevchenko et al., 2007), some protein factors that were not found by MS were identified by the former method (Fig. 4). For example, AtCPSF160 and AtFY, which were absent from the protein pool copurified with AtCPSF 73-I, were detected by western blots (Fig. 4, lane 2). Similar examples included AtCPSF100, AtCPSF160, and AtFY in the AtCPSF73-II purification (Fig. 4, lane 3) and AtCPSF160 in the AtFY purification (Fig. 4, lane 6). A complete list of RNA-processing/polyadenylation-related protein factors identified by LC-MS/MS and western blots is presented in Table I.
Figure 4.
TAP-purified proteins detected by antibodies in western blots. Proteins were extracted from Arabidopsis suspension cell cultures overexpressing a TAP-fused polyadenylation-related protein, as depicted at the top of each lane. After tandem purifications, the proteins were separated by 12% SDS-PAGE and then transferred to a PVDF membrane. The proteins on the membrane were detected by antibodies (α) raised against the proteins listed at the left side of the blot. Protein size markers (kD) are listed on the right.
Table I.
Arabidopsis CPSF components and their closely associated protein factors identified by TAP purifications and Y2H assays
Letters in brackets are symbols as used in “Discussion.”
| Coexisting in TAP Purification | Interaction in Y2H Assaysa |
|---|---|
| AtCPSF30-AtCPSF30[A]bc | AtCPSF30-AtCPSF30[a] |
| AtCPSF30-AtFIPS5[B]b | AtCPSF30-AtFIPS5[b] |
| N.D.d | AtCPSF30-AtCPSF100[c]e |
| N.D. | AtCPSF30-AtCPSF160[d] |
| N.D. | AtCPSF30-AtCLPS3[e] |
| AtCPSF73-I-AtCPSF73-I[C]bc | N.D. |
| AtCPSF73-I-AtCPSF100[D]bc | AtCPSF73-I-AtCPSF100[f]e |
| AtCPSF73-I-AtCPSF160[E]c | N.D. |
| AtCPSF73-I-AtCLPS3[F]b | N.D. |
| AtCPSF73-I-AtFY[G]c | N.D. |
| AtCPSF73-II-AtCPSF30[H]b | N.D. |
| AtCPSF73-II-AtCPSF73-II[J]b | N.D. |
| AtCPSF73-II-AtCPSF100[K]c | AtCPSF73-II-AtCPSF100[g]e |
| AtCPSF73-II-AtCPSF160[L]c | N.D. |
| AtCPSF73-II-AtFY[M]c | N.D. |
| N.D. | AtCPSF100-AtCPSF73-I[h] |
| N.D. | AtCPSF100-AtCPSF73-II[i] |
| AtCPSF100-AtCPSF100[O]bc | N.D. |
| AtCPSF100-AtCPSF160[P]b | AtCPSF100-AtCPSF160[j]e |
| N.D. | AtCPSF100-AtFY[k] |
| N.D. | AtCPSF100-AtCPSF30[l] |
| N.D. | AtCLPS3-AtCPSF30[m] |
| AtCLPS3-AtCLPS3[Q]bc | N.D. |
| AtCLPS3-AtSYM5[R]b | N.D. |
| AtCLPS3-AtPCFS4[S]b | AtCLPS3-AtPCFS4[n]f |
| AtFY-AtCPSF73-II[T]b | N.D. |
| AtFY-AtCPSF100[U]bc | AtFY-AtCPSF100[o] |
| AtFY-AtCPSF160[V]c | N.D. |
| AtFY-AtFY[W]bc |
N.D. |
Interactions are based on the Y2H results presented by Hunt et al. (2008), except as otherwise indicated.
Detected by MS.
Detected by immunoblots.
N.D., Not detected.
Although there were few bands visible on the gel from the fraction purified from cell culture overexpressing the empty TAP vector (Fig. 3, lane 1), the gel lane was still divided into three equal sections and digested by trypsin. This was done to faithfully exclude any potential nonspecific binding caused by the TAP tag. After LC-MS/MS, our database search revealed significant protein hits from human, pig, bovine, and other species (Supplemental Table S2). However, no recognizable Arabidopsis proteins were identified through this method or by antibodies (Fig. 4, lane 7). This indicates that the proteins identified from TAP purifications resulted from specific binding to the TAP-fused target proteins, instead of nonspecific binding or contaminants, to the TAP moiety of the fusion proteins.
Components of the Arabidopsis CPSF Complex Revealed by Proteomic Study
Proteins identified by LC-MS/MS could be categorized into three groups: (1) polyadenylation-related proteins (in red in Supplemental Table S2); (2) abundant cellular proteins that are involved in cell structure, protein translation, and metabolic processes (in black); and (3) chaperons, or proteins known to interact with unfolded polypeptides, such as heat shock proteins (in blue). Whether or not a protein was categorized as a plant polyadenylation-related protein was judged by two criteria: (1) this protein has known or predicted polyadenylation-related functions; and/or (2) this protein has an ortholog in mammals or yeast that has been identified as a polyadenylation factor.
Nineteen proteins were copurified with the TAP-tagged AtCPSF30 (Supplemental Table S2). Using the criteria above, two polyadenylation-related proteins, AtCPSF30 and AtFIPS5, were discovered via the TAP purification of AtCPSF30. In particular, AtCPSF30 was also detected by antibody specifically against it (Fig. 4, lane 1). As shown in lane 1, both the TAP-tagged and native AtCPSF30 were identified at similar strength, suggesting that AtCPSF30 may exist as a dimer in CPSF (Hunt et al., 2008). AtFIPS5 was not confirmed by immunoblotting because of the lack of a specific antibody. The other 17 of the 19 identified proteins from AtCPSF30 TAP purification were classified as non-polyadenylation-related proteins, which nonspecifically bind to the TAP-tagged AtCPSF30. Among these 17 proteins, five were cytoskeleton proteins/subunits of actins and tubulins, 10 were enzymes involved in photosynthesis/respiration processes, translation, and other metabolic processes, and two were involved in protein binding. As demonstrated in the following paragraphs, these proteins appeared at different TAP purifications, no matter what target proteins were used. These repeatedly identified proteins may come from nonspecific binding to the fusion proteins and thus would not be considered as proteins specifically interacting with the target polyadenylation factors. Therefore, these proteins will not be included in the results of TAP purifications hereafter. Such elimination of nonspecific purified proteins is supported by a recent report, where most of these proteins were not found to be associated with polyadenylation complex, except EF-α (Shi et al., 2009). A list of polyadenylation-related proteins identified by MS can be found in Supplemental Table S3, but more detailed information about these peptides is shown in Supplemental Table S4.
In the protein pool copurified with AtCPSF73-I, several Arabidopsis polyadenylation factors whose orthologs in yeast or mammals have known polyadenylation-related functions were discovered (Hunt et al., 2008). These proteins include AtCPSF73-I, AtCPSF100, and AtCLPS3 (Supplemental Table S3). The same affinity-purified proteins were also subjected to detection by available antibodies. Interestingly, besides AtCPSF73-I and AtCPSF100, which had already been detected by MS, AtCPSF160 and AtFY were also detected (Fig. 4, lane 2). However, AtCLPS3 was not detectable (Fig. 4, lane 2, bottom), despite the fact that in the same sample it was detectable by MS. This could be attributed to the low sensitivity of the antibody available to us: even in the cell line overexpressing AtCLPS3, the signal is relatively weak (Fig. 4, lane 5, bottom). Aside from the TAP-tagged AtCPSF73-I (Fig. 4, lane 2, second row, top band), its endogenous version (the bottom band marked by an arrow) was also detected with reduced signal.
Five plant polyadenylation proteins were discovered in the copurification of AtCPSF73-II (Supplemental Table S3). These proteins are AtCPSF30, AtCPSF73-II, AtCPSF100, AtCPSF160, and AtFY. Among them, AtCPSF30 and AtCPSF73-II were identified by MS, while AtCPSF100, AtCPSF160, and AtFY were detected by their antibodies (Fig. 4, lane 3). Surprisingly, AtCPSF73-I appeared to be one of the protein factors copurified with AtCPSF73-II (Fig. 4, lane 3, second row from top). However, two reasons suggest that it is rather a cross-reaction between AtCPSF73-I antibody and AtCPSF73-II protein than the recognition of AtCPSF73-I by AtCPSF73-I antibody: (1) AtCPSF73-I and AtCPSF73-II are highly similar to each other, with 55% similarity and 38% identity (Xu et al., 2004); and (2) the band is at the position equivalent to the TAP-tagged AtCPSF73-I, which is very close to TAP-tagged AtCPSF73-II. Detection of TAP-tagged AtCPSF73-I in the AtCPSF73-II TAP purification is very unlikely. Consequently, the band at the second row of lane 3 (Fig. 4) should not be interpreted as a copurification relationship between AtCPSF73-I and -II. As side evidence, a barely visible faint band can be seen about 3 kD lower than the first band (marked by an arrow). Three kilodaltons happens to be the molecular mass of the remaining CBP portion of the tobacco etch viral protease (TEV)-cleaved TAP tag (Rohila et al., 2004). Therefore, this weak band should correspond to the endogenous version of AtCPSF73-II (Xu et al., 2004), a trace amount of which was copurified with the TAP-tagged AtCPSF73-II.
Surprisingly, only three plant polyadenylation factors were copurified with AtCPSF100: AtCPSF100 itself, AtCPSF160, and AtFY (Supplemental Table S3), among which only the TAP-tagged AtCPSF100 could be detected by its antibody (Fig. 4, lane 4, third row from top). This was unexpected, since a similar MS study found that Arabidopsis AtCPSF73-I and AtSYM5 were associated with the AtCPSF100 fused to a Flag (Herr et al., 2006). To account for this, we hypothesize that CPSF may be a dynamic complex such that AtCPSF100, under different developmental stages or in different tissues, may associate with different partners (Manzano et al., 2009). For example, in the whole plants used by Herr et al. (2006), the CPSF may form a variant that possesses some components different from the suspension-cultured cells used in this research. Another possible factor contributing to this discrepancy is the different sizes of the tags used in the two assays: the eight-amino acid Flag tag and the 22-kD TAP tag.
A TAP purification using tagged AtCLPS3 as the bait was employed after we found that AtCLPS3 was one of the proteins copurified with AtCPSF73-I (Supplemental Table S3). Three plant polyadenylation factors, AtCLPS3, AtPCFS4, and AtSYM5, were recovered from the proteins copurified with TAP-fused AtCLPS3, confirming its functional involvement in RNA processing/polyadenylation (Supplemental Table S3). AtCLPS3 was successfully confirmed by its antibody (Fig. 4, lane 5, bottom row). AtPCFS4 and AtSYM5 were not tested because of the lack of appropriate antibodies.
AtFY is an ortholog of yeast Pfs2p and mammalian hPfs2 (unknown function; Mandel et al., 2008). We observed that AtFY was copurified with AtCPSF73-I and AtCPSF73-II (Supplemental Table S3). When this finding is added to a previous study showing the association of AtFY with AtCPSF73-I, AtCPSF100, AtCPSF160, and AtSYMS5 (Herr et al., 2006), we conclude that AtFY may be an important polyadenylation factor in Arabidopsis. Therefore, a TAP-tagged AtFY was constructed for affinity purification, and the proteins copurified with AtFY, AtCPSF73-II, AtCPSF100, AtCLPS3, and AtFY, were identified (Supplemental Table S3). As shown in Figure 4, a tagged AtFY as well as AtCPSF100 and AtCPSF160 were detected by their respective antibodies (Fig. 4, lane 6).
Taken together, using the TAP-tagging approach, 10 proteins belonging to, or associating with, Arabidopsis CPSF were identified with confidence by this study. These identified CPSF-related proteins are AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF100, AtCPSF160, AtFIPS3, AtFY, AtCLPS3, AtSYM5, and AtPCFS4.
DISCUSSION
Study of Plant CPSF Using TAP Methodology
While playing a central role in pre-mRNA 3′ end formation, the structure of plant CPSF has not been well understood. It has been observed that some Arabidopsis CPSF orthologs, such as AtCPSF160, AtCPSF100, AtCPSF73-I, AtCPSF73-II, and AtCPSF30, are all localized in the nucleus (Delaney et al., 2006; Xu et al., 2006), which indicates that they might coexist in the same complex. Y2H assay data show some pair-wise interactions between CPSF factors as well (Hunt et al., 2008). However, whether or not these proteins form a protein complex in vivo is not clear.
Our early attempts to isolate Arabidopsis CPSF in vivo complex using TAP tagging in transgenic plants encountered issues with abnormal plant phenotypes. The resulting transgenic plants were either dead or defective in some developmental steps that caused limited seed production (Xu et al., 2006). Even though a few plants survived, the expression pattern of the overexpressed proteins might have been severely disrupted. This phenomenon was also observed by others when using TAP tagging (Van Leene et al., 2007). To circumvent this problem, Arabidopsis suspension cell culture was successfully employed owing to the advantages it offers. First, when using transgenic plants, developmental problems are avoided. Second, it can provide almost unlimited materials by scaling up cultures. Third, the reduced levels of proteins related to photosynthesis and stress resistance help to boost the relative contents of proteins related to essential metabolism, such as mRNA processing (Baerenfaller et al., 2008).
Application of the TAP approach in plants only recently emerged as an efficient tool for isolating protein complexes and studying in vivo protein interactions. As shown in this study, when Arabidopsis polyadenylation-related proteins were fused with the TAP tag, many proteins were copurified repeatedly, no matter what target protein was fused to the TAP tag (Supplemental Table S2). The fact that these abundant cellular proteins were copurified may simply be a function of their overwhelming presence in cells. On the other hand, when protein expression was elevated in cells, the levels of proteasome and heat shock proteins were also increased (Voellmy and Boellmann, 2007). In this study, subunits of cytoskeleton, such as actin and tubulin, and several heat shock proteins and protein folding-related proteins, such as DNA-J, were repeatedly found in many TAP purifications. Therefore, these proteins were classified as contaminants, as they were also observed and treated as such in other studies (Séraphin et al., 2002). The reason these contaminating proteins were not purified by the empty TAP might be attributed to the relatively smaller size of the TAP tag. In other words, compared with the fused target proteins, whose average molecular mass is about 80 kD, the 22-kD tag was much smaller. As size decreases, the chance of interacting or forming aggregates with other proteins (Séraphin et al., 2002) also lessens. However, given that these proteins were not equally associated with all of the tested target proteins, a further experimental approach would need to be implemented in order to completely rule out their potential specific association.
In this study, immunoblotting was also used in protein identification, since it has been reported that it is more sensitive than LC-MS/MS (Copse and Fowler, 2002; Shevchenko et al., 2007). However, in reality, we observed that, in two cases, proteins were identified by MS, but surprisingly, not by their antibodies. The first example was AtCLPS3, which was detected as a copurified protein with AtCPSF73-I by MS but not by western blots. The second example was AtCPSF30 in the AtCPSF73-II purification, which was detectable by MS but not by western blots. The discrepancy observed in the behavior of AtCLPS3 can be explained by the low sensitivity of AtCLPS3 antibody. Even in the TAP purification from an overexpressing cell line (Fig. 2), AtCLPS3 was detected as a weak band by its antibody (Fig. 4). In contrast, the AtCPSF30 antibody seems to be both specific and sensitive (Fig. 4). However, it was so observed only when AtCPSF30 was overexpressed. Therefore, in the TAP purification from AtCPSF73-II, where AtCPSF30 was not overexpressed, its antibody might not be sensitive enough to detect the signal.
An Arabidopsis CPSF Model
Based on the protein interaction patterns derived from TAP-purified Arabidopsis CPSF factors (Supplemental Table S3) and Y2H data from the literature (Table I), a CPSF model is proposed largely in accordance with the physical closeness of each factor. This is, to our knowledge, the first plant CPSF model that is built on a complete set of data derived from the study of in vivo coexistence and interaction of each CPSF subunit.
In this study, the polyadenylation factors isolated from TAP purification can be classified into three subgroups according to their concurrency in the same TAP-purified protein pools: (1) AtFIPS5 and AtCPSF30; (2) AtCPSF30, -73-I, -73-II, -100, -160, AtFY, and AtCLPS3; and (3) AtCPSF100, -160, AtFY, AtCLPS3, AtPCF4, and AtSYM5 (Fig. 5). These subgroups may then be assembled into a bigger complex according to the recurrence of some common factors among different subgroups (e.g. AtCPSF30 and AtCLPS3). However, direct or indirect interaction between factors cannot be distinguished by the very nature of affinity purification. Therefore, evidence from assays such as Y2H or phage display is required. Fortunately, a couple of studies have been carried out in recent years, and some protein-protein interaction data are available, as summarized in Table I.
Figure 5.
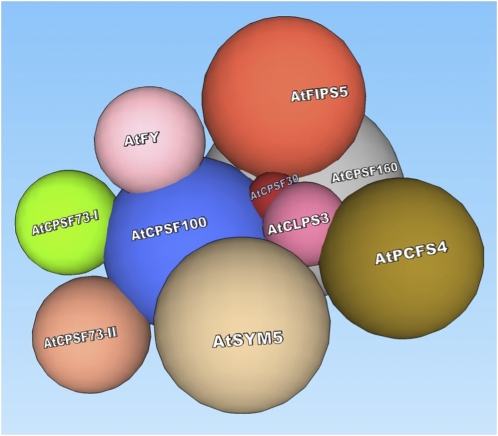
A proposed model of in vivo Arabidopsis CPSF structure. A plant CPSF complex consists of AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF100, AtCPSF160, AtFY, and AtFIPS5. AtCPSF100 serves as a core factor onto which other factors assemble. AtCPSF30 mediates interactions between CPSF and protein factors from other complexes, such as AtCLPS3 and AtPCFS4. Immediate contact indicates direct interaction, while relative remote distance means indirect interaction. The names of the factors are marked on the surfaces. The sizes of the factors are drawn to scale relative to their molecular masses. [See online article for color version of this figure.]
In this model, AtCPSF100 is proposed as the core factor of CPSF for the following reasons. (1) In this study, AtCPSF100 is one of the most frequently occurring proteins from different TAP purifications: AtCPSF100 appeared in all of the TAP purifications, except that of AtCPSF30 (Table I; see the interacting pairs labeled as [D], [K], [O], [P], and [U]). (2) AtCPSF100 is the most pair-wise active protein in Y2H (Table I, right column): AtCPSF100 interacts with AtCPSF30 (Table I, [c]), AtCPSF73-I [f], AtCPSF73-II [g], AtCPSF160 [j], and AtFY [k] (Xu et al., 2006; Hunt et al., 2008). (3) AtCPSF100 has been observed interacting with other Arabidopsis CPSF factors, such as AtCPSF160, AtCPSF73-I, and AtCPSF73-II in vitro pull-down assays (Xu et al., 2006). (4) In another study where a Flag tag was used as an affinity epitope, AtCPSF100 was shown to associate with AtCPSF160, AtFY, AtSYM5, and AtCstF64 (Herr et al., 2006). Therefore, AtCPSF100 is the best candidate for the CPSF core.
Other factors, such as AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF160, and AtFY, were either not observed in other TAP purifications (e.g. AtCPSF30 only with AtFIPS5) or did not interact with factors other than AtCPSF100 (i.e. AtCPSF73-I, -73-II, and -160). However, those factors have been observed as associated proteins of CPSF complexes in plants (Herr et al., 2006) or their orthologs in mammalian and yeast systems (Mandel et al., 2008). Therefore, they are proposed as CPSF factors that form the CPSF complex through their association with AtCPSF100.
Other remaining factors frequently did not copurify or interact with CPSF factors. These should be considered as factors located at the periphery of the CPSF complex or factors belonging to other polyadenylation complexes but that still closely interact with CPSF. Among them, AtFIPS5 only exists in the protein pool copurified with AtCPSF30 (Table I, [B]), and its only interacting partner identified by Y2H study was also AtCPSF30 (Table I, [b]; Forbes et al., 2006). Moreover, in mammals, CPSF30 and hFip1 belong to the same CPSF complex (Zhao et al., 1999), and their yeast homologs, Yth1p and Fip1p, exist in the same polyadenylation complex, PF-I (Mandel et al., 2008). These observations, therefore, support the idea that AtFIPS5 is a CPSF subunit and that it incorporates into Arabidopsis CPSF via its interaction with AtCPSF30.
AtCLPS3 may associate with CPSF through a mechanism similar to AtFIPS5, since AtCPSF30 was also observed interacting with AtCLPS3 (Table I, [e]). AtCLPS3 is an Arabidopsis ortholog of hClp1 in mammals and Clp1p in yeast (Xing et al., 2008a). Research has shown that AtCLPS3 might be involved in the processing of pre-mRNAs encoded by a distinct subset of genes important in plant development (Xing et al., 2008a). However, there is not enough valid evidence to support AtCLPS3 as a CPSF member. Thus, AtCLPS3 is listed as a polyadenylation-related factor that interacts with CPSF through AtCPSF30. For the same reason, AtPCFS4 is not considered a CPSF factor in this study. Research has shown that AtPCFS4 is in the same protein complex as AtCLPS3 and interacts directly with it (Table I, [n]; Xing et al., 2008b). AtPCFS4 has an ortholog in both mammals (hPcf11) and yeast (Pcf11p; Xing et al., 2008b). It was shown that hPcf11 and Pcf11p play a role in RNA polymerase II transcription, especially in termination (Sadowski et al., 2003; Zhang et al., 2007). In Arabidopsis, AtPCFS4 promotes flowering by regulating FCA alternative polyadenylation (Xing et al., 2008b). Similar to these two factors, AtSYM5 is an ortholog of mammalian symplekin and Pta1p in yeast, which may play a scaffolding role in vivo (Kyburz et al., 2003). The function of AtSYMS5 is unclear, yet its mutation impacts gene silencing (Herr et al., 2006).
The association of AtCLPS3 and AtPCFS4 to the CPSF complex underscores the importance of AtCPSF30 in the CPSF structure. AtCPSF30 might serve as a local pivot bridging CPSF factors (such as AtFIPS5) or non-CPSF factors (such as AtCLPS3) to the CPSF complex (AtCPSF100) through its intermediate interaction to those factors. This is evident by its multiple Y2H interacting partners: AtCPSF30 interacts with itself (Table I, [a]), AtFIPS5 (Table I, [b]), AtCPSF100 (Table I, [c]), AtCPSF160 (Table I, [d]), and AtCLPS3 (Table I, [e]).
By compiling the TAP purification and Y2H data, we proposed an in vivo structural model of Arabidopsis CPSF, as shown in Figure 5. According to this model, AtCPSF100 is the foundation of CPSF, onto which AtCPSF160, AtCPSF73-I, AtCPSF73-II, AtFY, and AtCPSF30 are assembled. AtCPSF30 acts as a connecting point through which another CPSF factor, AtFIPS5, is recruited. Also, factors, such as AtCLPS3, AtSYM5, and AtPCFS4, interact with CPSF via AtCPSF30.
It is surprising that so few polyadenylation-related proteins, like AtCstF subunits and AtPAP, have been identified by the proteomic approach. This might be explained by a recent plant polyadenylation machinery model based on pair-wise protein-protein interactions, in which 28 currently identified protein factors were organized into three hubs centering around AtCPSF100, AtCLPS3, and AtFIPS5 (Hunt et al., 2008). Concurring with this model, most of the protein factors identified in this study belong to a subset of the above-mentioned polyadenylation-related protein network (the CPSF complex), which is centered on AtCPSF100. Therefore, it is no surprise that protein factors at the center of other hubs were not copurified. Affinity purification using tagged proteins that belong to other hubs should be helpful in generating a full picture of these other plant polyadenylation factors.
Unique Features Distinguish Plant CPSF from Its Yeast and Mammalian Counterparts
The general similarities between yeast CPF and mammalian CPSF complexes have been reviewed in detail by Zhao et al. (1999) and Mandel et al. (2008), as summarized in Supplemental Table S1. The results in this study showed that plant CPSF is evolutionarily conserved as well. Among the seven proposed plant CPSF factors, five can find their corresponding orthologs in both mammals and yeast. However, results also showed that plant CPSF possesses two unique features: AtCPSF73-II and AtFY.
The first plant-unique feature is AtCPSF73-II, which has only recently been identified (Xu et al., 2004). Compared with AtCPSF73-I, it is more distantly related to the mammalian CPSF73, the endonuclease in pre-mRNA processing (Mandel et al., 2006). AtCPSF73-II has no homolog in yeast. In mammals, AtCPSF73-II is homologous to RC-68, which is not a mammalian CPSF (Shi et al., 2009). Given that AtCPSF73-I and AtCPSF73-II were both identified as CPSF factors in plants but were exclusive of each other's copurified protein pool (Table I; Fig. 4), it is reasonable to propose that they may compete to form a functional CPSF complex, in which AtCPSF73-I and AtCPSF73-II play roles in different subgroups of substrates. Alternatively, AtCPSF73-I and AtCPSF73-II may even form two distinct CPSF complexes. These two CPSF variants may recruit some common basal factors, such as AtCPSF100, AtCPSF160, and AtCPSF30, which provide general polyadenylation/RNA 3′ processing activity. For example, in contrast to the housekeeping roles of AtCPSF73-I, AtCPSF73-II may be involved in processes such as female gamete transmission and development (Xu et al., 2004). Thus, the presence of such a unique component may be essential for plant-specific biological processes.
This dynamic CPSF scenario is supported by the reminiscent circumstances of RC-68 in mammals. RC-68 is required for cell cycle progression, the depletion of which arrested HeLa cells in G1 phase. RC-68 has 40% identity and 60% similarity in the first 450 amino acids with CPSF-73. RC-68 interacts with RC-74, whose N-terminal region has about 20% identity to CPSF-100 (Dominski et al., 2005). Since RC-68 and RC-74 interact with each other but do not associate with subunits of the CPSF complex in HeLa and mouse cells, it is proposed that RC-68/RC-74 may form a distinct complex involved in the 3′ end processing of a subset of pre-mRNAs, with at least some of them encoding proteins that play a role in cell cycle regulation (Dominski et al., 2005).
The second plant-unique feature is AtFY. AtFY has a mammalian ortholog, hPfs2, the function of which is still unknown (Mandel et al., 2008). In contrast, the yeast ortholog of AtFY, Pfs2p, is a subunit of PF-I factor that encodes an RNA-binding protein with WD domains. Pfs2p interacts with proteins from PF-I (Fip1p and Yth1p) and CF-I (Rna14p), suggesting that Pfs2p plays a role in tethering these two factors together (Ohnacker et al., 2000). A similar close relationship among AtFY, AtCPSF30 (ortholog of Yth1p), and AtFIPS5 has not yet been observed in plants.
In fact, AtFY plays a role in flowering time control by regulating a floral repressor gene, FLC (Simpson et al., 2003). AtFY interacts with FCA, which regulates FLC through a transcriptional silencing mechanism via the histone demethylase activity of FLD. Additionally, FCA plays more general roles in silencing transgenes and transposons in the Arabidopsis genome (Bäurle et al., 2007), which presents a potential connection between the RNA-processing activities and chromatin regulation mediated by AtFY.
AtFY stably associates with AtCPSF100 and AtCPSF160 in vivo. However, this association is dynamically switched by the interaction of AtFY with FCA: loss of FCA function or loss of the FCA interaction domain in AtFY disrupted formation of the larger complexes containing AtCPSF160 but not those containing AtFY, suggesting that the Arabidopsis 3′ RNA-processing components AtFY/AtCPSF160 may exist in functionally distinct complexes in vivo (Manzano et al., 2009). Therefore, AtFY may function in both housekeeping posttranscriptional processes and chromatin regulation. AtFY can form a multiprotein complex with AtCPSF160 and AtCPSF100 for regular posttranscriptional processing. Upon the interaction of AtFY with FCA, for example, in the case of FLC, AtFY can form bigger complexes, recruiting other RNA-silencing factors such as FLD (a histone demethylase), through which the chromatin regulation pathway is turned on.
Dynamic Plant CPSF
As detailed above, the study of plant CPSF reveals two unique plant CPSF factors associated with a dynamic CPSF complex. These two factors, AtCPSF73-II and AtFY, happen to be involved in plant-specific processes, such as plant female development and flowering. On the basis of canonical eukaryotic CPSF, this may suggest that plants have evolved variants of CPSF to deal with specific developmental or environmental cues.
Interestingly, the concept of variants in the Arabidopsis CPSF complex has been proposed by other researchers as well. Herr et al. (2006) reported the identification of an AtCstF64-like homolog (ESP1) that encodes an Arabidopsis RNA-processing component. Affinity purification showed that, like its homolog AtCstF64, ESP1 coexists with canonical polyadenylation/RNA 3′ processing factors such as AtCPSF100 and AtSYMS5 (Herr et al., 2006). Based on their observation, the authors proposed that there are at least two complexes that contain an AtCstF64-like homolog. They could both be AtCstF-like complexes that bind a sequence downstream of the cleavage site. One of these complexes would be the standard AtCstF, which uses the RNA recognition motif of AtCstF64 to bind downstream of the mRNA3′ end formation sites. The other would use ESP1 and a separate RNA-binding protein to recognize alternative 3′ end formation sites. In another study, Rao et al. (2009) proposed that AtCPSF30 may be involved in the relocation of some plant CPSF factors, which potentially form different CPSF complexes. Taken together, the identification of AtCPSF73-II and AtFY as Arabidopsis CPSF components in this research supports the new concept that the polyadenylation/RNA 3′ processing apparatus can be dynamic in order to facilitate the gene regulation that best responds to developmental and environmental cues.
CONCLUSION
Based on their in vivo coexistence and pair-wise interacting data, we propose that the plant CPSF consists of seven factors: AtCPSF30, AtCPSF73-I, AtCPSF73-II, AtCPSF100, AtCPSF160, AtFIPS5, and AtFY. These factors form a complex where AtCPSF100 serves as the core. Through AtCPSF30, CPSF interacts with other polyadenylation-related factors, such as AtSYMS5, AtCLPS3, and AtPCF4. While this model is structurally similar to its mammalian and yeast counterparts, the plant CPSF features AtCPSF73-II and AtFY as unique components, as both have been associated with plant-specific biological processes. Our results also support a dynamic Arabidopsis CPSF model that may offer more flexibility in RNA processing adjusted according to developmental and environmental cues.
MATERIALS AND METHODS
Construction of Fusion Proteins
For cloning the Arabidopsis (Arabidopsis thaliana) CPSF cDNA sequences (Xu et al., 2006) into the Gateway-compatible binary vectors containing the TAP tag (TAPi [Rohila et al., 2004]; a gift from Dr. Michael Fromm, University of Nebraska), PCR-amplified CPSF cDNA sequences were first cloned into pENTR vector, which were then fused to the binary vectors by LR recombination reactions (Invitrogen). After fusion, the TAPi tag sequences were either located at the C terminus of the protein-coding sequences or the N terminus (Fig. 1). The cloning of AtCLPS3 and AtFY was described by Xing et al. (2008b). The fused constructs were confirmed by DNA sequencing. Plasmids containing correct constructs were transformed into the Agrobacterium tumefaciens strain GV3505 by electroporation (Xu and Li, 2008).
Cell Culture and Transformation
Arabidopsis cell culture (MM1 from Landsberg erecta; a gift from Dr. Chris Makaroff, Miami University) was maintained and transformed as described by Menges and Murray (2004). After the Agrobacterium-mediated transformation, cells were selected on Murashige and Skoog plates supplemented with cefotaxime (Bioplus; 100 μg mL−1 final concentration) and glufosinate ammonium (Sigma; 800 μg mL−1 final concentration) for resistant calli. A single callus (approximately 0.5 cm in diameter) from each transformation was selected, crunched by pipette tips, and cultured with 10 mL of Murashige and Skoog medium containing cefotaxime and glufosinate ammonium by rocking at 23°C (130 rpm) with a 16-h/8-h photoperiod. Detailed procedures can be found in Supplemental Methods S1.
TAP Purification
TAP purification was done basically as described by Rohila et al. (2004) with some modifications. Cells were ground in liquid nitrogen to fine powder, which was mixed with about 150 mL of extraction buffer (for recipes, see Supplemental Methods). The mixture was spun at 10,000g for 40 min at 4°C, and the supernatant was incubated with 100 μL (bed volume) of IgG beads (GE Health) at 4°C for 2 to 4 h. The IgG beads were then collected by passing through a mini column (Bio-Rad), where the beads were washed with 10 mL of IPP-150 (for immunoprecipitation-150) buffer three times and with 10 mL of TEV cleavage buffer once. After the washes, 1 mL of TEV cleavage buffer, 10 μL of 0.1 m dithiothreitol, and 10 μL (100 units) of TEV (Invitrogen) were added. The column was incubated at 4°C overnight with gentle rocking. Three milliliters of calmodulin-binding buffer and 3 μL of 1 m CaCl2 were added to the flow-through collected from the previous step (IgG binding). The column was incubated at 4°C for 1 h, and the beads were washed with 10 mL of IPP-150 three times. Finally, the proteins were eluted with 1 to 1.5 mL of elution buffer into the desired number of fractions, which were monitored by A280. Eluted proteins were precipitated by TCA and sodium deoxycholate before loading onto a SDS-PAGE gel. To each volume of proteins, a 1:100 volume of 2% sodium deoxycholate was added and incubated on ice for 30 min. One hundred percent TCA was added to 6% of the final volume, and the mixture was kept on ice for 1 h. The tubes were centrifuged at 2,500g for 45 min at 4°C, and the pellets were washed with cold 100% acetone and spun at 2,500g for another 45 min at 4°C. The pellets were dried by a SpecVac for 1 min and dissolved in SDS-PAGE loading buffer before being separated on 12% SDS-PAGE gels. The proteins on the gels were then analyzed by MS as described below or transferred to a polyvinylidene difluoride (PVDF) membrane and detected by antibodies.
Trypsin Digestion and MS Analysis
The in-gel digestion was done essentially as described by Séraphin et al. (2002). After separation by SDS-PAGE and staining by Coomassie Brilliant Blue R-250 (Sigma), the desired bands were excised and chopped into small fragments, washed twice with 50% methanol (v/v) by vortexing for 15 min, and then incubated with 50% acetonitrile (Sigma)/50 mm NH4HCO3 (v/v; pH 9.0) by vortexing for 30 min. After a serial treatment of acetonitrile, the gels were dried in a SpecVac. The dried gels were immersed with 55 mm iodoacetamide (Sigma), incubated in the dark for 45 min at room temperature, and then washed with 500 μL of 25 mm NH4HCO3. Finally, the gels were dehydrated by adding 100 μL of 100% acetonitrile. After drying in a SpecVac for 5 min, each sample was digested with 0.5 μg of sequencing-grade trypsin (Promega) in 250 μL of 10 mm NH4HCO3 (pH 9.0) at 37°C overnight. The next day, the solution containing the digested proteins was collected. The gel fragments were further extracted by 250 μL of 0.1% trifluoric acid/water, 250 μL of 0.1% trifluoric acid/30% acetonitrile (v/v), 250 μL of 0.1% trifluoric acid/60% acetonitrile, and finally 250 μL of 0.1% trifluoric acid/90% acetonitrile, each with 30 min of vortexing. The supernatants were pooled and dried to minimal volumes in a SpecVac and reconstituted into 10 μL of trifluoric acid/water in the case of overdrying. The digested proteins were analyzed by LC-MS/MS (Mass Spectrometry and Proteomics Facility, Ohio State University).
Western-Blot Analysis
After separation by SDS-PAGE, proteins were transferred to a PVDF membrane and visualized using chemiluminescence, as described previously (Xing et al., 2008b). Primary antibodies raised against the proteins (Xu et al., 2006; Xing et al., 2008a; AtFY antibody was a gift from Caroline Dean, John Innes Centre) were diluted at 1:1,000.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Conserved eukaryotic cleavage and polyadenylation-related factors.
Supplemental Table S2. LC-MS/MS-identified proteins from protein pools copurified with TAP-fused proteins.
Supplemental Table S3. Polyadenylation-related proteins purified via TAP-fused proteins.
Supplemental Table S4. Sequence information of MS-identified RNA processing/polyadenylation-related protein factors.
Supplemental Methods S1. A more detailed description of the methods.
Supplementary Material
Acknowledgments
We thank Chris Makaroff, Mike Fromm, and Kari Green-Church for materials and expert technical assistance and services. We are grateful to Dr. Arthur Hunt for his suggestions and the rest of Li laboratory members for helpful discussions, in particular to Ruqiang Xu for gene cloning and John Heffernan for cell culture characterization. We also appreciate Diana Kroll and David Martin for proofreading the manuscript.
This work was supported by the National Science Foundation (grant nos. MCB 0313472 and IOS–0817829 to Q.Q.L.), the National Institutes of Health (grant no. 1R15GM07719201A1 to Q.Q.L.), as well as Sigma Xi and Miami University Botany Academic Challenge Grants to H.Z.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Qingshun Quinn Li (liq@muohio.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941 [DOI] [PubMed] [Google Scholar]
- Bäurle I, Smith L, Baulcombe DC, Dean C (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318 109–112 [DOI] [PubMed] [Google Scholar]
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W (1991) Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem 266 19768–19776 [PubMed] [Google Scholar]
- Buratowski S (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17 257–261 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP (2002) Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res 30 3592–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Noble SM, Guthrie C (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science 274 1511–1514 [DOI] [PubMed] [Google Scholar]
- Chen J, Moore C (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol 12 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copse C, Fowler SJ (2002) Detection of proteins on western blots using chemifluorescence. In JM Walker, ed, The Protein Protocols Handbook. Humana Press, Totowa, NJ, pp 421–428
- Danckwardt S, Hentze MW, Kulozik AE (2008) 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney KJ, Xu RQ, Zhang JX, Li QQ, Yun KY, Falcone DL, Hunt AG (2006) Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol 140 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W (2002) Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z (2007) Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol 42 67–93 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF (2005) A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes KP, Addepalli B, Hunt AG (2006) An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J Biol Chem 281 176–186 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN (2002) Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol 22 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XY, Khan AU, Cheng HL, Pappas DL, Hampsey M, Moore CL (2003) Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Molnar A, Jones A, Baulcombe DC (2006) Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA 103 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14 1415–1429 [PubMed] [Google Scholar]
- Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D (2006) Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol 26 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG (2007) Messenger RNA 3′-end formation and the regulation of gene expression. In CL Bassett, ed, Regulation of Gene Expression in Plants: The Role of Transcript Structure and Processing. Springer, New York, pp 101–122
- Hunt AG, Xu R, Addepalli B, Rao S, Xing D, Zhao H, Mo M, Forbes KP, Meeks LR, Marion A, et al (2008) Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Sadowski M, Dichtl B, Keller W (2003) The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res 31 3936–3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L (2008) Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang HL, Gebauer D, Vethantham V, Manley JL, Tong L (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano D, Marquardt S, Jones AM, Baurle I, Liu F, Dean C (2009) Altered interactions within FY/AtCPSF complexes required for Arabidopsis FCA-mediated chromatin silencing. Proc Natl Acad Sci USA 106 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JAH (2004) Cryopreservation of transformed and wild-type Arabidopsis and tobacco cell suspension cultures. Plant J 37 635–644 [DOI] [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1992) Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem 267 14804–14811 [PubMed] [Google Scholar]
- Ohnacker M, Barabino SML, Preker PJ, Keller W (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J 19 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappin DJC, Hojrup P, Bleasby AJ (1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol 3 327–332 [DOI] [PubMed] [Google Scholar]
- Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W (1997) A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J 16 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16 272–278 [DOI] [PubMed] [Google Scholar]
- Rao S, Dinkins RD, Hunt AG (2009) Distinctive interactions of the Arabidopsis homolog of the 30 kD subunit of the cleavage and polyadenylation specificity factor (AtCPSF30) with other polyadenylation factor subunits. BMC Cell Biol 10 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Martinson HG (2008) Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 28 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Cerny R, Fromm ME (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J 38 172–181 [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL (2004) Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W (2003) Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J 22 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Puig O, Bouveret E, Rutz B, Caspary F (2002) Tandem affinity purification to enhance interacting protein identification. In Protein-Protein Interactions: A Molecular Cloning Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 313–328
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, Manley JL (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113 777–787 [DOI] [PubMed] [Google Scholar]
- Stumpf G, Domdey H (1996) Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science 274 1517–1520 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, et al (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6 1226–1238 [DOI] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F (2007) Chaperone regulation of the heat shock protein response. In P Csermely, L Vígh, eds, Molecular Aspects of the Stress Response: Chaperones, Membranes and Networks. Springer, New York, pp 89–99
- Xing D, Zhao H, Li QQ (2008. a) Arabidopsis CLP1-SIMILAR PROTEIN3, an ortholog of human polyadenylation factor CLP1, functions in gametophyte, embryo, and postembryonic development. Plant Physiol 148 2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Zhao H, Xu R, Li QQ (2008. b) Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J 54 899–910 [DOI] [PubMed] [Google Scholar]
- Xu R, Li QQ (2008) Streamline cloning of genes into binary vectors in Agrobacterium via the Gateway® TOPO vector system. Plant Methods 4 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Ye X, Li QQ (2004) AtCPSF73-II gene encoding an Arabidopsis homolog of CPSF 73 kDa subunit is critical for early embryo development. Gene 324 35–45 [DOI] [PubMed] [Google Scholar]
- Xu R, Zhao H, Dinkins RD, Cheng X, Carberry G, Li QQ (2006) The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol Biol 61 799–815 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Henderson AJ, Gilmour DS (2007) Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev 21 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.