Abstract
Photosynthetic organisms experience changes in light quantity and light quality in their natural habitat. In response to changes in light quality, these organisms redistribute excitation energy and adjust photosystem stoichiometry to maximize the utilization of available light energy. However, the response of other cellular processes to changes in light quality is mostly unknown. Here, we report a systematic investigation into the adaptation of cellular processes in Synechocystis species PCC 6803 to light that preferentially excites either photosystem II or photosystem I. We find that preferential excitation of photosystem II and photosystem I induces massive reprogramming of the Synechocystis transcriptome. The rewiring of cellular processes begins as soon as Synechocystis senses the imbalance in the excitation of reaction centers. We find that Synechocystis utilizes the cyclic photosynthetic electron transport chain for ATP generation and a major part of the respiratory pathway to generate reducing equivalents and carbon skeletons during preferential excitation of photosystem I. In contrast, cytochrome c oxidase and photosystem I act as terminal components of the photosynthetic electron transport chain to produce sufficient ATP and limited amounts of NADPH and reduced ferredoxin during preferential excitation of photosystem II. To overcome the shortage of NADPH and reduced ferredoxin, Synechocystis preferentially activates transporters and acquisition pathways to assimilate ammonia, urea, and arginine over nitrate as a nitrogen source. This study provides a systematic analysis of cellular processes in cyanobacteria in response to preferential excitation and shows that the cyanobacterial cell undergoes significant adjustment of cellular processes, many of which were previously unknown.
Light is one of the most important environmental factors for photosynthetic organisms. It is used to drive photosynthesis and to regulate growth and development. The primary reactions of oxygenic photosynthesis are catalyzed by two large pigment-protein complexes, PSII and PSI (for review, see Pakrasi, 1995). These two complexes act in series to drive several light-dependent electrochemical reactions. PSII catalyzes light-dependent oxidation of water and reduction of plastoquinone (PQ). The electrons from reduced PQ are transferred to PSI via a membrane-bound cytochrome b6f complex and a diffusible pool of plastocyanin or cytochrome c553, leading to reduction of ferredoxin and NADP+. The ability of both PSII and PSI to catalyze electrochemical reactions is dependent on the presence of unique light-harvesting antennae structures that are used for the efficient absorption of light. These antennae optimize the capture of light by absorption at totally different wavelengths. Cyanobacteria utilize two main antenna pigments, chlorophyll (Chl) and bilin, to absorb light energy (Glazer, 1977; MacColl, 1998). Chls are mostly associated with PSI and absorb light of maximum absorbance wavelengths (λmax) 435 and 680 nm. Bilins are covalently attached to light-harvesting proteins called phycobiliproteins and are mostly associated with PSII. The specific combination of apoproteins and bilins present in a phycobilisome (PBS) determines its light absorption profile. Two major phycobiliproteins commonly present in the PBS of cyanobacteria are the red light-absorbing allophycocyanin (AP) with λmax ∼ 650 nm and phycocyanin (PC) with λmax ∼ 620 nm.
Light is the only source for the generation of energy and reduced carbon (C) for cyanobacteria under photoautotrophic conditions. It is critically important that these organisms balance the excitation of PSII and PSI to maximize the quantum yield of photosynthetic light reactions. Changes in the spectral composition of light can affect the rate of quanta transfer to one reaction center over the other, which in turn leads to a decreased photosynthetic efficiency and damages the photosynthetic apparatus (Anderson et al., 1995; Dietzel et al., 2008). To counteract such imbalance, cyanobacteria, like plants and algae, have developed short-term and long-term adaptation mechanisms (Manodori and Melis, 1986; Chow et al., 1990; Anderson et al., 1995; Fujita, 1997; Allen, 2003; Dietzel et al., 2008). As a short-term mechanism that occurs in the time scale of seconds to minutes, cyanobacteria utilize a process known as state transition: the adaptive, complementary transfer of light energy from the overexcited photosystem to the underexcited photosystem (Bonaventura and Myers, 1969; Murata, 1969). Under light conditions that predominantly excite Chl associated with PSI (PSI light), cyanobacteria maintain balance by decreasing energy transfer to PSI and increasing energy transfer to PSII. Similarly, preferential excitation of PSII (PSII light) leads to an increase in energy transfer to PSI and decreases energy transfer to PSII in order to maintain balance. In contrast, the long-term mechanism requires the adjustment of the photosystem stoichiometry to regulate the balance of electron flow between two reaction centers (Chow et al., 1990; Anderson et al., 1995; Dietzel et al., 2008). Thus, by modulating the composition, structure, and functions of the photosynthetic apparatus, cyanobacteria ensure an effective balance of light-driven electron transfer between the two photosystems under changing light quality.
The physiological and molecular basis of short-term and long-term adaptations under changing light quality have been well characterized (Anderson et al., 1995; Fujita, 1997; Allen, 2003; Dietzel et al., 2008). However, the response of cellular functions other than photosynthesis to changing light quality is largely unexplored. This is particularly striking given the wealth of recent data suggesting that the photosynthetic process is tightly coupled to other principal metabolic pathways (Ma et al., 2001; Palenchar et al., 2004; Aurora et al., 2007; Singh et al., 2008b). In addition, photosynthetic organisms have evolved complex transcriptional networks that dictate the acclimation process in response to environmental cues (Jiao et al., 2007). Thus, it can be speculated that the acclimation process of cyanobacteria to changing light quality is not limited to the photosynthetic apparatus only but also involves other cellular processes. Indeed, a few physiological results have previously shown that a change in light quality affects other cellular processes. For example, respiration in higher plants and algae is activated by PSI light (Kowallik, 1982). In contrast, cytochrome c oxidase activity, the terminal component of respiration, in Synechocystis species PCC 6714 is reported to be preferentially activated by PSII light (Gu et al., 1994). PII (a small nitrogen [N] regulatory protein) has been shown to be covalently modified by PSII light (Tsinoremas et al., 1991). It has been suggested that PII links the state of central C and energy metabolism to the control of N assimilation in cyanobacteria (Forchhammer, 2004). In this work, we have utilized time series DNA microarrays to investigate the response of Synechocystis species PCC 6803 (hereafter Synechocystis) to PSI and PSII light. Our results show that preferential excitation of PSII and PSI leads to the massive reprogramming of the Synechocystis transcriptome. This reprogramming allows certain cellular processes to be preferentially utilized under a given light condition. These adjustments could enable Synechocystis to overcome the limited production of energy and reducing equivalents and, in the process, allow growth and development under changing light quality.
RESULTS
Physiological Response of Synechocystis to PSI Light and PSII Light
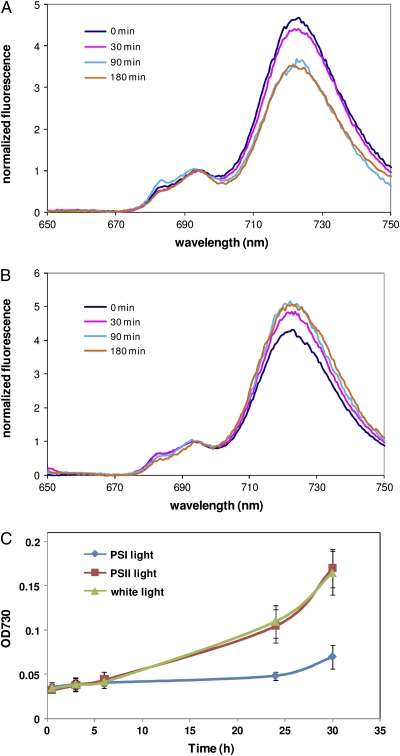
The physiological response of white light-grown Synechocystis to PSI and PSII light was monitored by measuring the growth and 77 K emission spectra of Chl fluorescence (Fig. 1). The fluorescence emission spectra at 77 K can be used to estimate the efficiency of energy transfer from pigments to PSI and PSII reaction centers. This measurement has been extensively used to study state transitions in cyanobacteria (Bonaventura and Myers, 1969; Murata, 1969; Huang et al., 2003; Fujimori et al., 2005; Kondo et al., 2009). Figure 1 shows typical emission spectra obtained at 77 K following excitation of Chl at 435 nm. The peaks at 685 and 695 nm correspond to the core antennae of PSII, CP43 and CP47, whereas the 725-nm peak arises from PSI (Van Dorssen et al., 1987; Siefermann-Harms, 1988). The relative intensity of PSI fluorescence gradually decreased following illumination of Synechocystis with PSI light (Fig. 1A). This gradual decrease was observed until 90 min and remained at the same level thereafter. The decrease of PSI fluorescence indicates a decreased energy transfer to PSI and increased energy supply to PSII during illumination with PSI light. In contrast, the relative intensity of PSI fluorescence gradually increased in Synechocystis illuminated with PSII light (Fig. 1B). This gradual increase in PSI fluorescence was observed until 90 min and was maintained at this level thereafter. The increase of PSI fluorescence indicates a decreased energy transfer to PSII and increased energy supply to PSI during illumination with PSII light. We also observed that Synechocystis grew at a significantly slower rate under PSI light compared with cells grown under white light (Fig. 1C). In contrast, cells under PSII light grew at a rate that was comparable to growth of cells under white light.
Figure 1.
Physiological responses of Synechocystis to PSI and PSII light. A and B, Chl fluorescence emission spectra of cells illuminated with PSI light (A) and PSII light (B) were measured following excitation of Chl at 435 nm. Curves were normalized to the fluorescence intensity at 695 nm. C, Cell growth was monitored by measuring absorption (OD [optical density]) at 730 nm under PSI light or PSII light. Error bars represent sd based on mean values of three independent growth experiments.
Synechocystis Genes Responsive to PSI Light and PSII Light
Synechocystis cells sampled at 15 min, 45 min, 1.5 h, 2 h, 3 h, and 6 h following illumination with either PSI light or PSII light were used to determine changes in transcript abundance using DNA microarray. We calculated the fold change of a gene by dividing the normalized intensity obtained from PSII light-illuminated cells by that obtained with PSI light-illuminated cells. A positive fold change suggests greater transcript abundance under PSII light, whereas a negative fold change suggests greater transcript abundance under PSI light. Using these criteria, we find that illumination of Synechocystis with a specific light has a significant impact on transcript levels of genes involved in various processes (Supplemental Table S1). In total, 1,202 genes showed changes in transcript abundance in response to illumination with PSI light and PSII light of at least 1.3-fold (P < 0.01). Of these, 467 genes (224 genes with greater transcript abundance under PSI light and 243 genes with greater transcript abundance under PSII light) were regulated only at one given time point, whereas the remaining genes showed significant changes in transcript levels at two time points at least. Further analysis led to the identification of several functional categories, where for a given process transcript levels of some genes were greater under PSI light while transcript levels of other genes involved in the same process were greater under PSII light. Additionally, ribosomal genes, whose expression is typically linked to an organism's growth, responded to both PSI and PSII light (Supplemental Fig. S1; Supplemental Table S2). These results clearly show that the differential growth rate observed in Figure 1C during illumination with PSI and PSII light has minimal impact on the differential regulation of genes. The complete list of differentially regulated genes identified in this study is provided in Supplemental Table S2.
Complementary Regulation of Photosystem Genes by PSI and PSII Light Establishes Proof of Concept
Transcript levels of PSII genes including those required for PSII biogenesis were greater in Synechocystis illuminated with PSI light compared with PSII light (Fig. 2). Alteration in the transcript abundance of PSII genes under PSI light appears to be immediate, as significant changes could be observed within 15 min of the onset illumination. Similarly, transcript levels of PSI genes were greater in Synechocystis illuminated with PSII light compared with PSI light (Fig. 2). However, changes in transcript levels of PSI genes under PSII light were observed only after 15 min of illumination. Regulation of the psbA gene encoding the D1 protein of PSII by PSI light and that of the psaE gene encoding the PsaE protein of PSI by PSII light has been reported previously (Tsinoremas et al., 1994; Alfonso et al., 2000; El Bissati and Kirilovsky, 2001). The same results were observed in this study, supporting the conclusion that preferential illumination of Synechocystis with PSI light or PSII light led to the complementary regulation of photosystem genes.
Figure 2.
A hierarchical cluster display of photosystem genes regulated by PSI and PSII light. Expression ratios [log2(PSII light/PSI light)] of all PSI and PSII genes were used to generate the cluster using Spotfire Decisionsite version 8.0. Euclidean distance was used as a measure of similarity between various time points, and genes were clustered using the weighted pair gene method with arithmetic means. The color scale used to define the regulation pattern of a gene is provided at the bottom. The fold changes of genes are provided in Supplemental Table S2.
Cellular Processes Responsive to PSII Light
Our data show that transcript abundance of a large number of genes encoding proteins for various cellular processes is intimately linked to illumination with PSI and PSII light (Fig. 3). In addition to PSI genes, transcript levels of genes coding for heat shock proteins (HSPs), cytochrome c oxidase, and those required for Arg catabolism were greater under PSII light (Table I). Most HSP gene expression could be linked to illumination with PSII light. However, transcript levels of a few HSPs, including the sll1384 gene encoding an HSP, were greater under PSI light (Table II). Changes in transcript levels of the sll1384 gene by PSI light could be seen within 15 min of the onset of illumination and remained high throughout the duration of illumination. Induction of expression of several HSP genes during heat shock in cyanobacteria is known to be modulated by light (Glatz et al., 1997; Asadulghani et al., 2003); however, to our knowledge, expression of HSP genes has not been previously shown to be regulated by illumination with a specific spectrum of light.
Figure 3.
A schematic representation of major cellular processes controlled by PSI and PSII light. Blue and orange circles represent responses associated with PSI light and PSII light, respectively. Circles containing both blue and orange represent cellular processes controlled by both PSI and PSII light. The list of regulated genes belonging to these processes is provided in Supplemental Table S2. CHEM, Chemotaxis; CO2, CO2 fixation; COX, cytochrome oxidase; Cytbf, cytochrome b6f complex; EM, energy metabolism; EM-AA, energy metabolism amino acids; PIGM, pigment biosynthesis; PR, photoreceptor(s); PROT, proteases.
Table I.
List of genes belonging to cellular processes whose transcript levels were greater under PSII light
The fold changes are the log2 ratios of transcript abundance in cells illuminated with PSII light versus PSI light at various time points. The complete list of genes with fold changes and respective P values is provided in Supplemental Table S2.
| Open Reading Frame | Open Reading Frame Product | 15 min | 45 min | 90 min | 2 h | 3 h | 6 h |
|---|---|---|---|---|---|---|---|
| Heat shock proteins | |||||||
| sll0170 | DnaK2 | 3.62 | 2.01 | 0.41 | 0.19 | 0.3 | −0.55 |
| sll0416 | GroEL2 | 2.07 | 0.94 | 0.45 | 0.28 | 0.29 | −0.07 |
| sll0430 | HtpG | 3.94 | 0.95 | 0.32 | 0.05 | −0.02 | −0.48 |
| sll0897 | DnaJ | 1.44 | 0.38 | 0.28 | 0.25 | 0.22 | 0.17 |
| sll1514 | HspA | 2.11 | 1.87 | 1.03 | 0.7 | 0.36 | −0.34 |
| slr0093 | DnaJ | 3.34 | 0.19 | 0.01 | −0.13 | −0.28 | −0.35 |
| slr2075 | GroES | 0.97 | 2.68 | 1.88 | 1.27 | 1 | 0.32 |
| slr2076 | GroEL1 | 0.25 | 2.47 | 1.7 | 1.18 | 0.9 | 0.35 |
| Glycolate cycle | |||||||
| sll1831 | Glycolate oxidase subunit | 1.42 | 0.88 | 0.71 | 0.78 | 0.51 | 0.08 |
| Cytochrome oxidase | |||||||
| sll1899 | Cytochrome c oxidase folding protein (CtaB) | 0.09 | 0.12 | 0.21 | 0.35 | 0.51 | 0.82 |
| slr1136 | Cytochrome c oxidase subunit II (CtaC1) | −0.13 | −0.14 | 0.29 | 0.39 | 0.63 | 1 |
| slr1137 | Cytochrome c oxidase subunit I (CtaD1) | −0.23 | −0.04 | 0.17 | 0.32 | 0.35 | 0.71 |
| slr1138 | Cytochrome c oxidase subunit III (CtaE1) | −0.13 | −0.03 | 0.14 | 0.24 | 0.4 | 0.5 |
| sll1245 | Cytochrome cM (CytM) | 0.04 | 0.46 | 0.37 | 0.52 | 0.51 | 0.54 |
| N assimilation | |||||||
| sll0108 | Ammonium permease (Amt1) | 1.31 | 1.14 | 1.43 | 1.69 | 1.61 | 1.86 |
| sll0537 | Ammonium permease (Amt3) | 0.45 | 0.21 | 0.2 | 0.2 | 0.15 | 0.37 |
| sll1017 | Ammonium permease (Amt2) | 1.52 | 0.5 | 0.72 | 0.73 | 0.48 | 0.8 |
| sll0374 | Urea transport system (UrtE) | −0.15 | 0.12 | 0.16 | 0.35 | 0.49 | 0.54 |
| slr1200 | Urea transport system permease | 0.69 | 0.12 | 0.32 | 0.51 | 0.66 | 0.97 |
| slr1201 | Urea transport system permease | 0.62 | 0.14 | 0.47 | 0.53 | 0.51 | 0.99 |
| sll0420 | Urease β-subunit (UreB) | −0.13 | 0.26 | 0.24 | 0.33 | 0.35 | 0.4 |
| slr1219 | Urease accessory protein E (UreE) | 0.04 | 0.08 | 0.06 | 0.12 | 0.13 | 0.38 |
| slr1899 | Urease accessory protein F (UreF) | 0.09 | 0.41 | 0.51 | 0.35 | 0.3 | 0.39 |
| sll1883 | Arg biosynthesis (ArgJ) | 0.02 | 0.48 | 0.49 | 0.62 | 0.75 | 0.98 |
| slr0288 | Glu-ammonia ligase (GlnN) | 1.11 | 0.37 | 0.24 | 0.39 | 0.46 | 0.93 |
| slr0898 | Ferredoxin-nitrite reductase (NirA) | 0.74 | −0.09 | 0.07 | 0.19 | 0.3 | 1.11 |
| slr1756 | Glu-ammonia ligase (GlnA) | −0.42 | 0.01 | 0.25 | 0.4 | 0.36 | 0.72 |
| ssl0707 | N regulatory protein P-II (GlnB) | 0.32 | −0.21 | 0.28 | 0.47 | 0.68 | 0.8 |
| sll1423 | Global N regulator (NtcA) | 0.29 | 0.43 | 0.28 | 0.49 | 0.43 | 0.37 |
| sll1102 | Glu transport (GtrA) | 0.44 | 0.24 | 0.13 | 0.36 | 0.45 | 0.51 |
| Arg catabolism | |||||||
| sll0370 | Carbamoyl phosphate synthase (PyrA) | 0.11 | −0.1 | 0.15 | 0.37 | 0.56 | 0.57 |
| sll1077 | Agmatinase (SpeB2) | 0.18 | 0.15 | 0.41 | 0.66 | 0.94 | 1.06 |
| sll1498 | Carbamoyl phosphate synthase small chain | 0.11 | 0.38 | 0.52 | 0.55 | 0.53 | 0.67 |
| slr0585 | Argininosuccinate synthetase (ArgG) | 0.15 | 0.3 | 0.53 | 0.73 | 0.96 | 1.1 |
| slr1022 | N-Acetyl-Orn aminotransferase (ArgD) | −0.13 | 0.14 | 0.19 | 0.36 | 0.39 | 0.52 |
| sll0422 |
Asparaginase |
−0.14 |
−0.05 |
0.04 |
0.19 |
0.28 |
0.5 |
Table II.
List of genes belonging to cellular processes whose transcript levels were greater under PSI light
The fold changes are the log2 ratios of transcript abundance in cells illuminated with PSII light versus PSI light at various time points. The complete list of genes with fold changes and respective P values is provided in Supplemental Table S2.
| Open Reading Frame | Open Reading Frame Product | 15 min | 45 min | 90 min | 2 h | 3 h | 6 h |
|---|---|---|---|---|---|---|---|
| Energy metabolism | |||||||
| sll0018 | Fru-bisphosphate aldolase (CI-Fba) | −1.18 | −0.98 | −0.73 | −0.89 | −0.67 | −0.61 |
| sll1275 | Pyruvate kinase 2 (Pyk2) | 0.13 | −0.37 | −0.38 | −0.31 | −0.27 | −0.15 |
| slr0394 | Phosphoglycerate kinase (Pgk) | −0.09 | −0.65 | −0.17 | −0.16 | 0.2 | 0.43 |
| slr0884 | Glyceraldehyde 3-phosphate dehydrogenase (Gap1) | 0.08 | 0.18 | 0.24 | 0.46 | 0.74 | 1.17 |
| slr0943 | Fru-bisphosphate aldolase (CII-Fba) | −0.29 | −0.28 | −0.22 | −0.47 | −0.46 | −0.51 |
| sll0329 | 6-Phosphogluconate dehydrogenase (Gnd) | −0.53 | −0.27 | −0.04 | 0.05 | 0.25 | 0.65 |
| slr1843 | Glc-6-P dehydrogenase (Zwf) | −0.49 | −0.46 | −0.38 | −0.31 | −0.21 | 0.08 |
| slr0301 | Phosphoenolpyruvate synthase (Pps) | 0.43 | 0.54 | −0.57 | −1 | −0.83 | −1.34 |
| slr0194 | Ribose 5-phosphate isomerase (RpiA) | −0.74 | −0.81 | −0.6 | −0.69 | −0.55 | −0.26 |
| slr0942 | Alcohol dehydrogenase | −0.3 | −0.38 | −0.42 | −0.65 | −0.71 | −0.7 |
| sll0823 | Probable succinate dehydrogenase | −0.41 | −0.47 | −0.3 | −0.2 | 0.12 | 0.26 |
| sll0891 | Malate dehydrogenase | 0.07 | 0.33 | −0.16 | −0.42 | −0.56 | −0.96 |
| slr1233 | Succinate dehydrogenase | −0.41 | −0.09 | −0.3 | −0.35 | −0.57 | −0.5 |
| sll0771 | Glc transport protein (Gtr) | 0.14 | −0.67 | −0.65 | −0.41 | −0.37 | −0.79 |
| C fixation | |||||||
| sll0934 | Carboxysome formation protein (CcmA) | −0.48 | 0.39 | 0.11 | 0.12 | −0.03 | −0.13 |
| sll1028 | CO2-concentrating mechanism protein (CcmK) | −0.02 | −0.62 | −0.37 | −0.18 | 0.19 | 0.43 |
| sll1029 | CO2-concentrating mechanism protein (CcmK) | −0.13 | −0.62 | −0.32 | −0.08 | 0.28 | 0.68 |
| sll1030 | CO2-concentrating mechanism protein (CcmL) | −0.2 | −0.68 | −0.36 | −0.05 | 0.32 | 0.76 |
| sll1031 | CO2-concentrating mechanism protein (CcmM) | −0.58 | −0.7 | −0.42 | −0.18 | 0.18 | 0.67 |
| sll1032 | CO2-concentrating mechanism protein (CcmN) | −0.08 | −0.63 | −0.42 | −0.24 | −0.05 | 0.32 |
| sll1525 | Phosphoribulokinase (Prk) | −0.39 | −0.79 | −0.49 | −0.23 | −0.03 | −0.06 |
| slr0009 | Ribulose bisphosphate carboxylase (RbcL) | −0.83 | −1.04 | −0.75 | −0.64 | −0.29 | 0.49 |
| slr0012 | Ribulose bisphosphate carboxylase (RbcS) | −1.17 | −1.02 | −0.81 | −0.68 | −0.28 | 0.56 |
| Cytochrome b6f complex | |||||||
| sll1317 | Apocytochrome f (PetA) | −0.83 | −0.52 | −0.39 | −0.35 | −0.21 | −0.18 |
| slr0342 | Cytochrome b6 (PetB) | −1.23 | −0.83 | −0.66 | −0.92 | −0.87 | −0.8 |
| slr0343 | Cytochrome b6f complex (PetD) | −1.32 | −0.92 | −0.84 | −0.79 | −0.79 | −0.85 |
| sml0004 | Cytochrome b6f complex (PetN) | 0.07 | −0.74 | −0.42 | −0.02 | 0.19 | 0.11 |
| smr0003 | Cytochrome b6f complex (PetM) | −0.06 | −0.8 | −0.42 | −0.1 | 0.27 | 0.02 |
| N assimilation | |||||||
| sll1450 | Nitrate/nitrite transport system (NrtA) | −0.05 | −0.17 | −0.16 | −0.19 | −0.24 | 0.6 |
| sll1452 | Nitrate/nitrite transport system (NrtC) | −0.73 | −0.24 | −0.21 | −0.21 | −0.31 | 0.21 |
| sll1453 | Nitrate/nitrite transport system (NrtD) | −0.66 | −0.03 | −0.03 | −0.07 | −0.2 | 0 |
| sll1454 | Ferredoxin-nitrate reductase (NarB) | −0.57 | 0.04 | 0.02 | 0.04 | −0.06 | 0.27 |
| sll1499 | Glu synthase (GlsF) | −0.55 | −0.74 | −0.57 | −0.58 | −0.48 | −0.25 |
| slr0710 | Glu dehydrogenase (GdhA) | 0.19 | −0.32 | −0.44 | −0.44 | −0.44 | −0.44 |
| ssl1911 | Gln synthetase-inactivating factor (IF7) | −1.36 | −1.11 | −1.65 | −2.08 | −2.21 | −2.11 |
| sll1515 | Gln synthetase-inactivating factor (IF17) | −0.24 | 0.04 | −0.35 | −0.71 | −0.91 | −1.4 |
| Heat shock proteins | |||||||
| sll0058 | DnaK1 | −0.41 | −0.3 | −0.19 | −0.14 | −0.15 | −0.12 |
| sll1384 |
Similar to DnaJ protein |
−0.64 |
−0.52 |
−0.63 |
−0.89 |
−0.94 |
−1.01 |
Transcript levels of genes encoding aa3-type cytochrome c oxidase were greater under PSII light (Table I). These included all three genes present in the ctaCDE operon encoding aa3-type cytochrome c oxidase subunits C (Slr1136), D (Slr1137), and E (Slr1138). Neither of the other known terminal oxidases in Synechocystis, cytochrome bd-quinol oxidase and the alternative respiratory terminal oxidase (Hart et al., 2005), responded to specific light. Additionally, transcript levels of genes encoding cytochrome c oxidase folding protein (Sll1899) and Cyt cM (Sll1245), a small c-type cytochrome, were also greater under PSII light. It has been reported that cytochrome oxidase activity increases in Synechocystis species PCC 6714 under PSII light (Gu et al., 1994). Our results suggest that a similar regulation of cytochrome c oxidase under PSII light occurs in Synechocystis and that its increased activity is due to greater transcript abundance of genes encoding aa3-type cytochrome oxidase.
Transcript levels of the sll1831 gene encoding a subunit of glycolate oxidase was greater under PSII light (Table I). Glycolate oxidase is the key enzyme involved in the salvage of 2-phosphoglycolate (2PG) to phosphoglycerate and O2 (Douce and Neuburger, 1999). 2-Phosphoglycolate is produced by the oxidase activity of Rubisco when the O2-CO2 ratio increases. Preferential excitation of PSII is expected to increase the O2-CO2 ratio; therefore, positive regulation of glycolate oxidase may constitute an important adaptation under these conditions. It must be noted that the sll0404 gene encoding glycolate dehydrogenase, identified by Eisenhut et al. (2006) and annotated as glycolate oxidase subunit in Cyanobase, is not regulated by either PSI light or PSII light in our study.
Cellular Processes Responsive to PSI Light
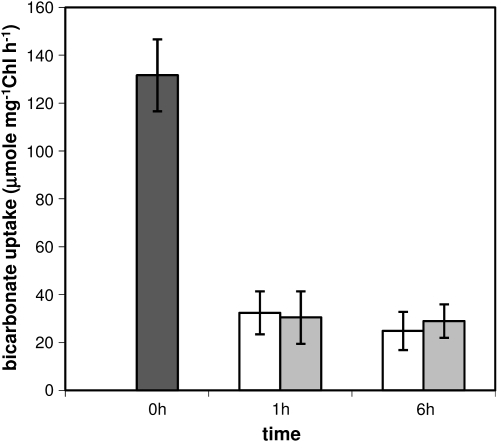
In addition to PSII genes, transcript levels of genes involved in a number of processes, including CO2 fixation, energy metabolism, and cytochrome b6f complex, were greater under PSI light (Table II). We found that transcript levels of genes encoding Rubisco subunits (Slr0009 and Slr0012), carbon-concentrating mechanism proteins (Sll0934 and Sll1028–Sll1032), and phosphoribulokinase (Sll1525) were greater under PSI light. In contrast, genes encoding transporters for the uptake of CO2 and HCO3− were not preferentially regulated by either PSI light or PSII light. These contrasting changes in transcript abundance of genes involved in transport and CO2 fixation have also been observed during high-light treatment (Singh et al., 2008b). It was found that genes encoding Rubisco and carbon-concentrating mechanism proteins were down-regulated, whereas those coding for transporters required for active uptake of CO2 and bicarbonate were up-regulated during high light. The apparent inconsistency between the regulation of genes involved in transport and CO2 fixation was explored by measuring the effect of PSI and PSII light on the uptake of [14C]bicarbonate by Synechocystis. Uptake of bicarbonate in white light-grown Synechocystis was at the rate of 130 μmol mg−1 Chl h−1 under our growth condition (Fig. 4). The high uptake rate of bicarbonate in Synechocystis is due to the growth of cells at ambient CO2 concentration, which is known to induce transporters involved in uptake of bicarbonate (Omata et al., 1999). Illumination of Synechocystis with either PSI light or PSII light led to a significant but similar decrease in the uptake of bicarbonate (Fig. 4). These results show that illumination of Synechocystis with either PSI light or PSII light does not preferentially affect the uptake of bicarbonate, which is in accordance with the transcriptional pattern of bicarbonate transporters observed in this study.
Figure 4.
Uptake of [14C]bicarbonate in air-grown Synechocystis under PSI and PSII light. Synechocystis cells grown under white light (dark gray bar) were illuminated with either PSII light (white bars) or PSI light (light gray bars) for 1 and 6 h, and the rate of bicarbonate uptake was measured as described in “Materials and Methods.” Error bars represent sd based on mean values of four independent measurements.
Transcript levels of genes encoding cytochrome b6f complex (Table II) and some NADH dehydrogenase subunits (Supplemental Table S2) were greater under PSI light. Additionally, transcript levels of genes encoding ATP synthase were transiently greater under PSI light (Supplemental Table S2). These complexes participate in both photosynthetic and respiratory electron transport chains in cyanobacteria (Hart et al., 2005). In addition, transcript levels of a number of genes coding for proteins involved in energy metabolism were greater under PSI light (Table II). These included genes involved in Glc transport, Glc catabolism via the glycolytic and oxidative pentose phosphate (OPP) pathways, and the tricarboxylic acid cycle. The OPP pathway is the major route of Glc catabolism in Synechocystis (Yang et al., 2002). Transcript levels of Glc-6-P dehydrogenase (Slr1843) and phosphogluconate dehydrogenase (Sll0329), the two rate-controlling enzymes of the OPP pathway, were more abundant under PSI light. Taken together, these results would suggest that illumination of Synechocystis with PSI light leads to an activation of respiration, a conclusion similar to that observed in plants and algae (Kowallik, 1982). However, a significant difference in the activation of respiration in Synechocystis and plants is in the regulation of cytochrome c oxidase, the terminal component of respiration. Our data showed that transcript levels of genes encoding cytochrome c oxidase in Synechocystis were greater under PSII light.
Cellular Processes Responsive to PSI and PSII Light
We identified several cellular processes, where for a given process transcript levels of some genes were greater under PSI light while transcript levels of other genes involved in the same process were greater under PSII light. Such processes include light harvesting, pigment biosynthesis, and protein degradation (Table III). PBS is the major light-harvesting complex in cyanobacteria. It consists of a core complex formed by AP and a rod complex formed by PC. The core and rod complexes are connected to each other via linker proteins. We found that transcript levels of genes encoding AP and core linker proteins were greater under PSI light, a response similar to PSII genes. In contrast, transcript levels of the cpc operon encoding PC and rod linker proteins were greater under PSII light, a response similar to PSI genes. In addition, transcript levels of the cpcG1 (slr2051) gene encoding a rod-core linker protein was greater under PSI light, whereas the cpcG2 gene encoding a second rod-core linker protein was not responsive to either light. It has been suggested that PBS containing the CpcG1 consists of both core and rod complexes and transfers light energy preferentially to PSII, whereas PBS containing the CpcG2 consists of only rod complex and transfers light energy preferentially to PSI (Kondo et al., 2007).
Table III.
List of genes belonging to cellular processes whose transcript levels were greater under both PSII and PSI light
The fold changes are the log2 ratios of transcript abundance in cells illuminated with PSII light versus PSI light at various time points. The complete list of genes with fold changes and respective P values is provided in Supplemental Table S2.
| Open Reading Frame | Open Reading Frame Product | 15 min | 45 min | 90 min | 2 h | 3 h | 6 h |
|---|---|---|---|---|---|---|---|
| Pigment biosynthesis | |||||||
| slr0056 | Chl a synthase | 0.11 | 0.33 | 0.28 | 0.43 | 0.58 | 0.46 |
| sll1875 | Heme oxygenase (HO2) | 0.21 | 0.49 | 0.57 | 0.87 | 0.97 | 0.36 |
| slr1030 | Magnesium protoporphyrin IX chelatase | 0.05 | 0.29 | 0.27 | 0.39 | 0.44 | 0.21 |
| sll1184 | Heme oxygenase (HO1) | −0.65 | −0.65 | −0.38 | −0.18 | 0 | −0.18 |
| sll1185 | Coproporphyrinogen III oxidase | −0.29 | −0.07 | 0.06 | 0.4 | 0.52 | 0.53 |
| sll1994 | Porphobilinogen synthase | −0.31 | −0.53 | −0.42 | −0.24 | −0.27 | −0.38 |
| slr0260 | Cob(I)alamin adenosyltransferase | 0.46 | −0.35 | −0.34 | −0.18 | −0.07 | −0.21 |
| slr0749 | Light-independent protochlorophyllide reductase (ChlL) | −0.6 | −0.63 | −0.59 | −0.64 | −0.48 | −0.62 |
| slr0772 | Light-independent protochlorophyllide reductase (ChlB) | −1.05 | −0.75 | −0.74 | −0.77 | −0.78 | −1 |
| Cellular protection | |||||||
| slr1738 | Transcription regulator (PerR) | 1.26 | 0.56 | 0.54 | 0.64 | 0.55 | 0.42 |
| sll1621 | AhpC/TSA family protein (PrxII) | 0.28 | 0.47 | 0.64 | 0.69 | 0.63 | 0 |
| ssr2061 | Glutaredoxin | 0.13 | 0.39 | 0.52 | 0.7 | 0.85 | 0.92 |
| sll1159 | Probable bacterioferritin comigratory protein | −0.02 | 0.26 | 0.73 | 0.97 | 1.18 | 1.18 |
| slr1238 | Glutathione synthetase | −0.2 | 0.26 | 0.43 | 0.49 | 0.64 | 0.87 |
| slr0233 | Thioredoxin M (TrxM) | −0.12 | −0.14 | −0.3 | −0.3 | −0.41 | −0.63 |
| slr0600 | NADP-thioredoxin reductase (NTR) | 0.24 | 0.06 | −0.11 | −0.3 | −0.5 | −0.76 |
| ssr1789 | CAB/ELIP/HLIP-related protein (HliD) | 0.04 | −0.2 | −0.27 | −0.33 | −0.39 | −0.43 |
| ssr2595 | High-light-inducible polypeptide (HliB) | −1.25 | −0.64 | −0.67 | −0.54 | −0.66 | −0.02 |
| Phycobilisome | |||||||
| sll1577 | Phycocyanin β-subunit (CpcB) | −0.15 | −0.22 | 0.59 | 0.65 | 0.61 | 0.63 |
| sll1578 | Phycocyanin α-subunit (CpcA) | −0.1 | −0.2 | 0.5 | 0.5 | 0.54 | 0.55 |
| sll1579 | Phycobilisome rod linker polypeptide (CpcC2) | 0.18 | −0.18 | 0.38 | 0.38 | 0.43 | 0.21 |
| sll1580 | Phycobilisome rod linker polypeptide (CpcC1) | 0.12 | −0.19 | 0.47 | 0.57 | 0.63 | 0.31 |
| ssl0452 | Phycobilisome degradation protein (NblA1) | −0.18 | 0.64 | 0.48 | 0.26 | 0.24 | 0.31 |
| ssl0453 | Phycobilisome degradation protein (NblA2) | −0.39 | 0.48 | 0.33 | 0.05 | 0.07 | 0.33 |
| sll0928 | Allophycocyanin B (ApcD) | −0.49 | −0.32 | −0.08 | 0.04 | 0.07 | −0.01 |
| slr0335 | Phycobilisome core membrane linker (ApcE) | −0.61 | −0.74 | −0.29 | −0.21 | −0.05 | −0.16 |
| slr1459 | Phycobilisome core component (ApcF) | −0.88 | −0.73 | −0.38 | −0.33 | −0.18 | −0.34 |
| slr1986 | Allophycocyanin β-subunit (ApcB) | −1.11 | −0.65 | −0.23 | −0.14 | 0.12 | 0.09 |
| slr2051 | Phycobilisome rod-core linker (CpcG1) | −0.45 | −0.39 | −0.09 | −0.09 | 0.09 | −0.07 |
| slr2067 | Allophycocyanin α-subunit (ApcA) | −0.75 | −0.61 | −0.17 | −0.1 | 0.15 | 0.11 |
| ssr3383 | Phycobilisome small core linker (ApcC) | −1.28 | −0.94 | −0.35 | 0.43 | 0.58 | 0.34 |
| Protein degradation | |||||||
| sll0020 | Clp protease ATPase | 0.14 | 0.52 | 0.4 | 0.18 | −0.07 | −0.37 |
| sll1679 | Periplasmic protease (HhoA) | 0.49 | 0.38 | 0.31 | 0.33 | 0.18 | −0.09 |
| sll2008 | Processing protease | 0.28 | 0.61 | 0.81 | 0.97 | 0.92 | 1.29 |
| sll2009 | Processing protease | 0.2 | 0.55 | 0.83 | 0.87 | 0.82 | 0.95 |
| slr1204 | Protease (HtrA) | 1.43 | 0.44 | 0.29 | 0.11 | 0.15 | 0.53 |
| slr1641 | ClpB protein (ClpB1) | 3.31 | 0.45 | −0.01 | −0.32 | −0.43 | −0.63 |
| slr0008 | C-terminal processing protease (CtpA) | 2.06 | −0.05 | −0.15 | −0.14 | −0.17 | −0.18 |
| slr0164 | ATP-dependent Clp protease (ClpP4) | −0.57 | −0.06 | −0.1 | −0.38 | −0.45 | −0.37 |
| slr0165 | ATP-dependent Clp protease (ClpP3) | −0.59 | −0.11 | −0.12 | −0.33 | −0.45 | −0.43 |
| slr1751 |
Periplasmic C-terminal protease |
−0.54 |
−0.05 |
−0.25 |
−0.33 |
−0.59 |
−0.58 |
Not many genes involved in pigment biosynthesis showed changes in transcript levels during preferential illumination of Synechocystis (Table III). For example, transcript levels of most genes encoding proteins required for the conversion of Glu to protoporphyrin IX did not change in response to changes in light quality. However, we found that transcript levels of slr0056 and slr1030 genes involved in the conversion of protoporphyrin IX to Chl were greater under PSII light, whereas transcript levels of the sll1184 gene involved in the conversion of protoporphyrin IX to bilin were greater under PSI light. Interestingly, transcript levels of the ho2 (sll1875) gene were greater under PSII light. Both ho1 and ho2 genes have been suggested to encode heme oxygenase; however, only the ho1 gene has been found to be involved in the multistep monooxygenase reaction to produce biliverdin IXα and CO from protoheme (Cornejo et al., 1998). We also found that transcript levels of chlB (slr0772) and chlL (slr0749) genes encoding the light-independent protochlorophyllide reductase were greater under PSI light, whereas the por gene encoding light-dependent protochlorophyllide reductase was not affected by either light.
Genes encoding proteins with significant function in cellular protection and redox homeostasis also showed light quality-dependent changes in transcript levels (Table III). Transcript levels of genes encoding ClpB1 (Slr1641), HtrA (Slr1204), HhoA (Sll1679), CtpA (Slr0008), and two processing proteases (Sll2008 and Sll2009) were greater under PSII light, whereas transcript levels of genes encoding ClpP3 (Slr0165), ClpP4 (Slr0164), and C-terminal protease (Slr1751) were greater under PSI light. Similarly, transcript levels of trxM (slr0233) and ntr (slr0600) genes encoding thioredoxin M (TrxM) and NADP-thioredoxin reductase (NTR), respectively, were greater under PSI light. These two genes were also up-regulated during high light (Singh et al., 2008b). Transcript levels of hliB (ssr2595) and hliD (ssr1789) genes encoding high light-inducible proteins (HLIPs) were greater under PSI light. Transcript levels of the sll1621 gene encoding a type II peroxiredoxin (PrxII) and the ssr2061 gene encoding a glutaredoxin (Grx) were greater under PSII light. Transcript levels of the slr1738 gene encoding PerR, which controls the expression of the sll1621 and ssr2061 genes (Li et al., 2004), was also greater under PSII light. Transcript levels of glutathione synthase (Slr1238) were increased coordinately with the sll1159 gene under PSII light. Sll1159 contains a Cys-X-X-Cys motif and has a typical Grx domain.
Opportunistic Utilization of N Assimilation Pathways under PSII Light
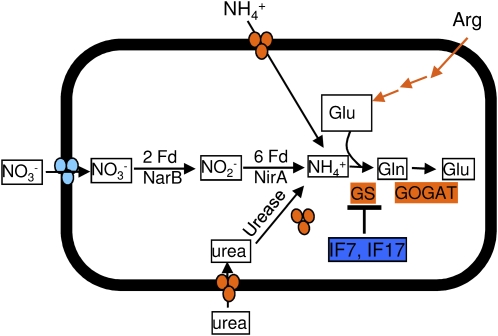
A major adaptation in Synechocystis under PSII light appears to be the preferential utilization of N substrates other than nitrate. The transcript levels of the nrt operon involved in transport and assimilation of nitrate were transiently greater within 15 min of illumination with PSI light (Table II). In contrast, transcript levels of genes coding for proteins involved in the transport and assimilation of ammonium, urea, Glu, and Arg were greater under PSII light (Table I; Fig. 5). These included genes encoding ammonium permease, urea permease, and urease. In addition, transcript levels of the gtrA (sll1102) gene encoding a sodium-dependent Glu transporter were greater under PSII light. The imported N substrates are first converted to ammonia and then combined with Glu to produce Gln by the GS-GOGAT cycle. Glutamine synthase (GS) is the first enzyme of the GS-GOGAT cycle and combines Glu and ammonia to produce Gln. Transcript levels of the glnA (slr1756) and glnN (slr0288) genes encoding GS were greater under PSII light (Table I). Commensurate with this finding, transcript levels of genes encoding IF7 (Ssl1911) and IF17 (Sll1515) were greater under PSI light (Table II). These two proteins inhibit the activity of GS (Garcia-Dominguez et al., 1999). Transcript levels of ntcA (sll1423) and glnB (ssl0707) genes, involved in the regulation of N assimilation in cyanobacteria (Forchhammer, 2004), were also greater under PSII light. Additional genes involved in N assimilation and positively regulated by PSII light included an asparaginase (Sll0422) that converts Asn to ammonia and subunits of carbamoyl phosphate synthase (Sll0370 and Sll1498) involved in the conversion of l-Gln to carbamoyl l-phosphate (Table I). Transcript levels of the glsF (sll1499) gene encoding ferredoxin-dependent Glu synthase and the gdhA (slr0710) gene encoding Glu dehydrogenase were greater under PSI light (Table II). We also found that transcript levels of several genes encoding proteins involved in Arg catabolism (Quintero et al., 2000) were greater under PSII light (Table II).
Figure 5.
Effects of PSI and PSII light on N assimilation pathways. Pathways controlled by PSII light are colored orange, whereas those controlled by PSI light are colored blue. Transcript levels of genes encoding nitrate transporter are transiently greater under PSI light and are colored light blue. The lists of regulated genes belonging to these processes are provided in Tables I and II.
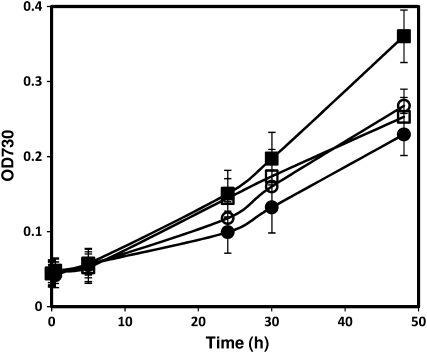
The DNA microarray data presented in this work suggest that Synechocystis cells preferentially transport and assimilate ammonia, urea, and Arg under PSII light over nitrate, a common N substrate present in BG11 medium. To study the physiological effect of this adaptation, we measured the growth of Synechocystis in BG11 medium in the presence or absence of 2 mm NH4NO3 under illumination with either white light or PSII light. Cells grew at a similar rate under illumination with white light and PSII light in BG11 medium (Fig. 6). Addition of 2 mm NH4NO3 into BG11 medium had no effect on the growth rate of Synechocystis under illumination with white light. However, the growth rate of Synechocystis increased significantly under PSII light in the presence of 2 mm NH4NO3 (Fig. 6). The growth of Synechocystis in the presence and absence of NH4NO3 confirmed results obtained from microarray data and showed that assimilation of N substrates other than nitrate is beneficial to the growth of Synechocystis under PSII light.
Figure 6.
Effects of NH4NO3 and light quality on the growth of Synechocystis. Synechocystis cells were grown under 30 μE m−2 s−1 white light (circles) and 10 μE m−2 s−1 PSII light (squares) in BG11 with (black) or without (white) 2 mm NH4NO3. Cell growth was monitored by measuring absorption (OD [optical density]) at 730 nm. Error bars represent sd based on mean values of three independent growth experiments.
DISCUSSION
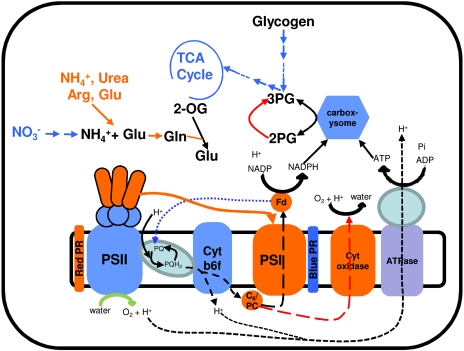
In this work, we have systematically investigated the adaptation of cellular processes in Synechocystis in response to changes in light quality. A custom-designed light-emitting diode (LED) source with a narrow bandwidth of lights tailored to excite either Chl or PBS was used to preferentially stimulate energy flow through PSI and PSII. Measurement of transcript abundance by DNA microarray shows that approximately 33% of genes in Synechocystis are regulated in response to changes in light quality. Analysis of these genes has led to the identification of cellular processes that enable Synechocystis to circumvent the reduced production of energy and reducing power and that protect cells from reactive oxygen species during changes in light quality. Figure 7 provides an overview of major adaptations revealed by this work. Overall, our results suggest that state transitions and adjustments of photosystem stoichiometry alone are insufficient to reverse the effects of excitation imbalance. Most cellular processes respond immediately to the imbalance in the excitation of reaction centers.
Figure 7.
A summary of key cellular adaptations during changes in light quality. Blue and orange colors represent responses associated with PSI light and PSII light, respectively. The photosynthetic linear electron transfer chain is represented by the dotted black line. The dotted blue line (from ferredoxin [Fd] to PQ) denotes the cyclic electron transfer chain under PSI light. The dotted red lines show the electron transfer to cytochrome c oxidase under PSII light. The solid orange line denotes the movement of rod complex to PSI under PSII light. 2-OG, 2-Oxoglutarate; 2PG, 2-phosphoglycolate; 3PG, 3-phosphoglycerate.
Illumination of Synechocystis with PSII light leads to the accumulation of reduced PQ, which must pass electrons to acceptor molecules in order to avoid photooxidative damage. Our results indicate that Synechocystis utilizes at least two routes to transfer electrons from reduced PQ under PSII light (Fig. 7). In the first route, the electrons are received by PSI. This is accomplished by the preferential energy transfer from PBS to PSI. Despite an extensive study of the structure and function of PBS, little is known about the regulation of PBS interaction with PSII and PSI (Adir, 2005; Mullineaux, 2008). Recently, it was found that Synechocystis possesses two types of PBS that differ by the presence of rod-core linker proteins (CpcG1 and CpcG2; Kondo et al., 2007). CpcG1-PBS contains both core and rod complexes and transfers light energy preferentially to PSII. In contrast, CpcG2-PBS contains only rod complex and transfers light energy preferentially to PSI. Our results show that transcript levels of PC genes but not of AP genes are greater under PSII light. The regulation of transcript abundance of PC genes is in accordance with the role of CpcG2-PBS in preferential energy transfer to PSI, as suggested by Kondo et al. (2007). In the second route, electrons from reduced PQ are received by cytochrome c oxidase via cytochrome cM, the terminal component of respiration. It should be noted that several components of the electron transport chain including PQ are shared by both respiration and photosynthesis in cyanobacteria (Hart et al., 2005). Utilization of cytochrome c oxidase as a terminal acceptor of electrons originating from PSII suggests that preferential transfer of light energy by CpcG2-PBS to PSI is not sufficient to accept all of the electrons from reduced PQ. The use of two terminal electron acceptors during preferential excitation of PSII enables similar production of ATP as under normal light conditions. However, it leads to the limited production of reduced ferredoxin and NADPH, as the electrons received by cytochrome c oxidase are used to reduce O2. The limiting NADPH and reduced ferredoxin can significantly affect the assimilation of nutrients, especially C and N. The conversion of 1 mol of nitrate (present in BG11 medium) to nitrite and then to NH4+ by nitrate reductase and nitrite reductase, respectively, requires 8 mol of reduced ferredoxin (Flores et al., 2005). Our data show that cells overcome the shortage of reduced ferredoxin by the activation of pathways involved in the transport and assimilation of ammonia, urea, Arg, and Glu. This adjustment not only overcomes the shortage of ferredoxin but also ensures that the electrons from ferredoxin will be maximally used to produce NADPH that can be used to fix C. Further support for the importance of these adjustments of cellular processes comes from the measurement of physiological growth in the presence of different N substrates under PSII light. We find that growth of Synechocystis increases significantly in the presence of ammonium under PSII light compared with white light (Fig. 6).
Synechocystis grew poorly under PSI light in our growth conditions (Fig. 1C). We propose that poor growth of Synechocystis under PSI light is related to a lack of NADPH production. Photosynthesis provides at least two modes for electron flow. In the first mode, a linear electron flow from PSII to PSI allows the production of ATP and reducing power in the form of reduced ferredoxin or NADPH. In the second mode, a cyclic electron flow driven solely by PSI allows the production of ATP without the accumulation of reduced species. We suggest that Synechocystis can only generate ATP via the cyclic electron flow under PSI light. To overcome the limited production of NADPH (or lack thereof), our data show that Synechocystis activates pathways involved in Glc catabolism, including the OPP pathway and the tricarboxylic acid cycle. However, as reported in “Results,” cytochrome c oxidase, the terminal component of respiration, was responsive to PSII light in Synechocystis. These results together suggest that Synechocystis utilizes the C metabolism pathways only to generate NADPH and C skeletons for biosynthesis under PSI light. These adaptations will allow Synechocystis to grow optimally under PSI light in the presence of reduced C.
Our results have also led to the identification of key factors involved in cellular protection during changes in light quality. We find that Synechocystis utilizes the glutathione/Grx system in combination with Prxs for protection under PSII light. The glutathione/Grx system is known to protect cells during oxidative stress by reducing peroxides and Prxs and by protecting thiol groups of enzymes via glutathionylation/deglutathionylation mechanisms (Holmgren et al., 2005). Additional cellular protection under PSII light is provided by HSPs. A role for HSPs in cellular protection under PSII light is consistent with earlier results obtained in Synechocystis suggesting that the transcription of HSP genes is induced in response to a reduced state of PQ and to oxidative stress (Glatz et al., 1997; Asadulghani et al., 2003; Singh et al., 2003; Li et al., 2004). Similarly, the TrxM/NTR system and HLIPs appear to protect cellular functions under PSI light (Fig. 7). Recent studies have also suggested a role for TrxM/NTR in the antioxidant network involved in mitigating reactive oxygen species (Hishiya et al., 2008; Singh et al., 2008b). HLIPs have been suggested to be involved in the protection of PSI during stress (Jantaro et al., 2006).
In summary, our analyses provide significant insights into the adaptations of Synechocystis to changes in light quality. We show that a significant adjustment of cellular processes in addition to previously well-studied adaptation mechanisms (e.g. state transitions and adjustment of photosystem stoichiometry) is necessary to reverse the effects of excitation imbalance. A significant finding relates to the growth of Synechocystis under PSII light to a rate typically observed under white light. Our results have led to the identification of necessary cellular adaptations that could enable the growth of Synechocystis during illumination with PSII light (Fig. 7). In contrast, Synechocystis under PSI light grows poorly due to insufficient production of reducing equivalents. In this situation, our results show that cells depend exclusively on metabolic pathways involved in Glc catabolism and respiration to fulfill the requirements of reducing power for the assimilation of nutrients and C skeletons for biosynthesis (Fig. 7). Taken together, our results underline the importance of preferential adjustments in several cellular processes, including photosynthesis, for the survival of photosynthetic organisms during changes in light quality.
MATERIALS AND METHODS
Custom Design of a LED Panel
A LED panel (26 × 26 cm2) was custom designed and constructed at Photon Systems Instruments (www.psi.cz). The panel consists of 14 rows of alternating royal blue- and red-emitting diodes separated by approximately 1.8 cm. Each row consists of seven LEDs. The royal blue-emitting diodes have a wavelength range from 440 to 460 nm with a typical light emission at 455 nm. The red-emitting diodes have a wavelength range from 620.5 to 645 nm with a typical light emission at 627 nm. The LED panel is connected to a programmable box that allows the control of light output intensity from 0 to 200 μE m−2 s−1 for both royal blue- and red-emitting diodes.
Illumination of Synechocystis with PSI Light and PSII Light
Synechocystis species PCC 6803 cells were grown at 30°C in BG11 medium buffered with 10 mm TES-KOH, pH 8.2 (Singh et al., 2008a), and bubbled with air. Illumination was at 30 μE m−2 s−1 provided by fluorescent cool-white light. Growth of Synechocystis was spectrophotometrically monitored by measuring optical density at 730 nm on a DW2000 spectrometer (SLM-Aminco). Unless otherwise noted, white light-grown Synechocystis cells (approximately 1 × 108 cells mL−1) were harvested by centrifugation at 6,000g and resuspended in fresh BG11 medium to a cell density of approximately 5 × 107 cells mL−1. These cells were then transferred in a long test tube (3 cm in diameter) and used for illumination with PSI light (provided by royal blue-emitting diodes) or PSII light (provided by red-emitting diodes). The tubes containing cells were transferred in a thermostat water bath maintained at 30°C and illuminated with either PSI light or PSII light provided by the custom-designed LED panel. Light intensity was maintained at 10 μE m−2 s−1. Cells were air bubbled during light illumination. Cells were collected after 15 min, 45 min, 1.5 h, 2 h, 3 h, and 6 h following illumination with light. Cells were spun down by centrifugation at 6,000g, frozen in liquid N, and stored at −80°C.
RNA Isolation and DNA Microarray Hybridization
Total RNA from Synechocystis was isolated using the RNAwiz Kit (Ambion) as described (Singh et al., 2008b). The quantity and quality of extracted RNA were determined spectrophotometrically in a Nanodrop ND-1000 (Thermo Scientific) at 260 and 280 nm and by the Bio-analyzer (Agilent) as described by the manufacturers. Total RNA isolated from PSI light- or PSII light-illuminated cells was fluorescently labeled with either Cy3 or Cy5 using the Micromax ASAP RNA Labeling Kit (Perkin-Elmer Life Sciences) as described (Singh et al., 2008b). The specific activity of the labeled RNA was determined in a Nanodrop. Fluorescently labeled probes (700 ng of Cy3- and Cy5-labeled RNA to a final specific activity of 50 pmol μg−1 RNA) were hybridized to Synechocystis 11 K custom oligonucleotide DNA microarrays. Hybridization, scanning, and data extraction were performed by MOgene essentially as described (Singh et al., 2008b).
Statistical Analysis
The experimental design used to identify regulated genes in response to PSI light and PSII light is essentially as described (Singh et al., 2008b). For each time point, we used two biological replicates, and each biological replicate consisted of three process replicates including a dye swap. Microarray data were processed using Matlab (MathWorks). Determination of the coefficient of variations of individual spots showed that the pixel intensity variation within the spots was quite low. We used a Lowess-based data normalization procedure with a window size of 25% for removing the intensity-based trends observed in the microarray data. The standard t test was used to quantify the consistency of measurements across different replicates. Transcript levels of a gene were considered as greater under PSI light or PSII light if the absolute value of its log ratio value exceeded a threshold of 0.3785 (i.e. 1.3-fold change) and the P value was less than 0.01 at any of the time points studied. We have previously shown that genes satisfying these criteria can be confidently identified as differentially regulated (Singh et al., 2008b). The transcriptome data generated in this work have been submitted to the ArrayExpress database at the European Bioinformatics Institute (accession no. E-TABM-339).
Assay for Uptake of Bicarbonate
The uptake of bicarbonate in white light-grown Synechocystis illuminated with PSI light or PSII light was measured using NaH14CO3 (Amersham). One milliliter of Synechocystis was mixed with 1 μCi of NaH14CO3 in a clear colorless Eppendorf tube. Following illumination with white light (500 μE m−2 s−1) for 30 s, reaction was terminated by rapid filtration of the cells onto a glass filter (GF/B; Whatman) by suction, followed by immediate washing of the filter with 5 mL of BG11. The filter was subjected to the measurement of radioactivity.
Fluorescence Measurements
Fluorescence emission spectra of Synechocystis at 77 K were measured on a Fluoromax-2 fluorometer with excitation at 435 nm (Jobin Yvon).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression patterns of genes responding to changes in light quality.
Supplemental Table S1. Summary of differentially regulated genes.
Supplemental Table S2. List of genes regulated by PSI light and PSII light.
Supplementary Material
Acknowledgments
We thank S. Rangwala (MOgene) for his help in the DNA microarray experiments and the members of the Pakrasi laboratory for collegial discussions.
This work was supported by the National Science Foundation Frontiers in Integrative Biological Research program (grant no. EF0425749).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Himadri B. Pakrasi (pakrasi@wustl.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adir N (2005) Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res 85 15–32 [DOI] [PubMed] [Google Scholar]
- Alfonso M, Perewoska I, Kirilovsky D (2000) Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803: involvement of the cytochrome b6/f complex. Plant Physiol 122 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF (2003) State transitions: a question of balance. Science 299 1530–1532 [DOI] [PubMed] [Google Scholar]
- Anderson JM, Chow WS, Park YI (1995) The grand design of photosynthesis: acclimation of the photosynthetic apparatus to environmental cues. Photosynth Res 46 129–139 [DOI] [PubMed] [Google Scholar]
- Asadulghani, Suzuki Y, Nakamoto H (2003) Light plays a key role in the modulation of heat shock response in the cyanobacterium Synechocystis sp PCC 6803. Biochem Biophys Res Commun 306 872–879 [DOI] [PubMed] [Google Scholar]
- Aurora R, Hihara Y, Singh AK, Pakrasi HB (2007) A network of genes regulated by light in cyanobacteria. OMICS 11 166–185 [DOI] [PubMed] [Google Scholar]
- Bonaventura C, Myers J (1969) Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim Biophys Acta 189 366–383 [DOI] [PubMed] [Google Scholar]
- Chow WS, Melis A, Anderson JM (1990) Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA 87 7502–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo J, Willows RD, Beale SI (1998) Phycobilin biosynthesis: cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J 15 99–107 [DOI] [PubMed] [Google Scholar]
- Dietzel L, Brautigam K, Pfannschmidt T (2008) Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry. Functional relationships between short-term and long-term light quality acclimation in plants. FEBS J 275 1080–1088 [DOI] [PubMed] [Google Scholar]
- Douce R, Neuburger M (1999) Biochemical dissection of photorespiration. Curr Opin Plant Biol 2 214–222 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Kahlon S, Hasse D, Ewald R, Lieman-Hurwitz J, Ogawa T, Ruth W, Bauwe H, Kaplan A, Hagemann M (2006) The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol 142 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bissati K, Kirilovsky D (2001) Regulation of psbA and psaE expression by light quality in Synechocystis sp PCC 6803: a redox control mechanism. Plant Physiol 125 1988–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E, Frias JE, Rubio LM, Herrero A (2005) Photosynthetic nitrate assimilation in cyanobacteria. Photosynth Res 83 117–133 [DOI] [PubMed] [Google Scholar]
- Forchhammer K (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev 28 319–333 [DOI] [PubMed] [Google Scholar]
- Fujimori T, Hihara Y, Sonoike K (2005) PsaK2 subunit in photosystem I is involved in state transition under high light condition in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 280 22191–22197 [DOI] [PubMed] [Google Scholar]
- Fujita Y (1997) A study on the dynamic features of photosystem stoichiometry: accomplishments and problems for future studies. Photosynth Res 53 83–93 [Google Scholar]
- Garcia-Dominguez M, Reyes JC, Florencio FJ (1999) Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci USA 96 7161–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz A, Horvath I, Varvasovszki V, Kovacs E, Torok Z, Vigh L (1997) Chaperonin genes of the Synechocystis PCC 6803 are differentially regulated under light-dark transition during heat stress. Biochem Biophys Res Commun 239 291–297 [DOI] [PubMed] [Google Scholar]
- Glazer AN (1977) Structure and molecular organization of the photosynthetic accessory pigments of cyanobacteria and red algae. Mol Cell Biochem 18 125–140 [DOI] [PubMed] [Google Scholar]
- Gu TQ, Iwama Y, Murakami A, Adhikary SP, Fujita Y (1994) Changes in the cytochrome c oxidase activity in response to light regime for photosynthesis observed with the cyanophyte Synechocystis PCC 6714. Plant Cell Physiol 35 1135–1140 [Google Scholar]
- Hart SE, Schlarb-Ridley BG, Bendall DS, Howe CJ (2005) Terminal oxidases of cyanobacteria. Biochem Soc Trans 33 832–835 [DOI] [PubMed] [Google Scholar]
- Hishiya S, Hatakeyama W, Mizota Y, Hosoya-Matsuda N, Motohashi K, Ikeuchi M, Hisabori T (2008) Binary reducing equivalent pathways using NADPH-thioredoxin reductase and ferredoxin-thioredoxin reductase in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol 49 11–18 [DOI] [PubMed] [Google Scholar]
- Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH (2005) Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33 1375–1377 [DOI] [PubMed] [Google Scholar]
- Huang C, Yuan X, Zhao J, Bryant DA (2003) Kinetic analyses of state transitions of the cyanobacterium Synechococcus sp. PCC 7002 and its mutant strains impaired in electron transport. Biochim Biophys Acta 1607 121–130 [DOI] [PubMed] [Google Scholar]
- Jantaro S, Ali Q, Lone S, He Q (2006) Suppression of the lethality of high light to a quadruple HLI mutant by the inactivation of the regulatory protein PfsR in Synechocystis PCC 6803. J Biol Chem 281 30865–30874 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8 217–230 [DOI] [PubMed] [Google Scholar]
- Kondo K, Mullineaux CW, Ikeuchi M (2009) Distinct roles of CpcG1-phycobilisome and CpcG2-phycobilisome in state transitions in a cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 99 217–225 [DOI] [PubMed] [Google Scholar]
- Kondo K, Ochiai Y, Katayama M, Ikeuchi M (2007) The membrane-associated CpcG2-phycobilisome in Synechocystis: a new photosystem I antenna. Plant Physiol 144 1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowallik W (1982) Blue light effects on respiration. Annu Rev Plant Physiol 33 51–72 [Google Scholar]
- Li H, Singh AK, McIntyre LM, Sherman LA (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacColl R (1998) Cyanobacterial phycobilisomes. J Struct Biol 124 311–334 [DOI] [PubMed] [Google Scholar]
- Manodori A, Melis A (1986) Cyanobacterial acclimation to photosystem I or photosystem II light. Plant Physiol 82 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW (2008) Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res 95 175–182 [DOI] [PubMed] [Google Scholar]
- Murata N (1969) Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim Biophys Acta 172 242–251 [DOI] [PubMed] [Google Scholar]
- Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T (1999) Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc Natl Acad Sci USA 96 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakrasi HB (1995) Genetic analysis of the form and function of photosystem I and photosystem II. Annu Rev Genet 29 755–776 [DOI] [PubMed] [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM (2004) Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol 5 R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero MJ, Muro-Pastor AM, Herrero A, Flores E (2000) Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and arginase pathway. J Bacteriol 182 1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefermann-Harms D (1988) Fluorescence properties of isolated chlorophyll-protein complexes. In HK Lichtenthaler, ed, Applications of Chlorophyll Fluorescence. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 45–54
- Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB (2008. a) Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem 283 15762–15770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Elvitigala T, Bhattacharyya-Pakrasi M, Aurora R, Ghosh B, Pakrasi HB (2008. b) Integration of carbon and nitrogen metabolism with energy production is crucial to light acclimation in the cyanobacterium Synechocystis. Plant Physiol 148 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA (2003) Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132 1825–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Castets AM, Harrison MA, Allen JF, Tandeau de Marsac N (1991) Photosynthetic electron transport controls nitrogen assimilation in cyanobacteria by means of posttranslational modification of the glnB gene product. Proc Natl Acad Sci USA 88 4565–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Schaefer MR, Golden SS (1994) Blue and red light reversibly control psbA expression in the cyanobacterium Synechococcus sp. strain PCC 7942. J Biol Chem 269 16143–16147 [PubMed] [Google Scholar]
- Van Dorssen RJ, Breton J, Plijter JJ, Satoh K, Van Gorkom HJ, Amesz J (1987) Spectroscopic properties of the reaction center and of the 47 kDa chlorophyll protein of photosystem II. Biochim Biophys Acta 893 267–274 [Google Scholar]
- Yang C, Hua Q, Shimizu K (2002) Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng 4 202–216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.