Abstract
Starch synthesis and degradation require the participation of many enzymes, occur in both photosynthetic and nonphotosynthetic tissues, and are subject to environmental and developmental regulation. We examine the distribution of starch in vegetative tissues of Arabidopsis (Arabidopsis thaliana) and the expression of genes encoding core enzymes for starch synthesis. Starch is accumulated in plastids of epidermal, mesophyll, vascular, and root cap cells but not in root proper cells. We also identify cells that can synthesize starch heterotrophically in albino mutants. Starch synthesis in leaves is regulated by developmental stage and light. Expression of gene promoter-β-glucuronidase fusion constructs in transgenic seedlings shows that starch synthesis genes are transcriptionally active in cells with starch synthesis and are inactive in root proper cells except the plastidial phosphoglucose isomerase. In addition, ADG2 (for ADPG PYROPHOSPHORYLASE2) is not required for starch synthesis in root cap cells. Expression profile analysis reveals that starch metabolism genes can be clustered into two sets based on their tissue-specific expression patterns. Starch distribution and expression pattern of core starch synthesis genes are common in Arabidopsis and rice (Oryza sativa), suggesting that the regulatory mechanism for starch metabolism genes may be conserved evolutionarily. We conclude that starch synthesis in Arabidopsis is achieved by spatial coexpression of core starch metabolism genes regulated by their promoter activities and is fine-tuned by cell-specific endogenous and environmental controls.
Plants can synthesize starch in chloroplasts of photosynthetic cells and in amyloplasts of nonphotosynthetic cells. In spite of the importance of these processes, our understanding of the nature and regulation of the pathways for starch synthesis and degradation is incomplete (Smith et al., 2005; Zeeman et al., 2007; Deschamps et al., 2008). In previous studies on starch synthesis, several Arabidopsis (Arabidopsis thaliana) mutants with low or aberrant starch in their leaves have been characterized. They are nucleus-encoded recessive mutants (e.g. pgi1, pgm1, adg1, adg2, isa1 isa2, ss1, ss4, be2, and be3), indicating that plastidial phosphoglucose isomerase (PGI), phosphoglucose mutase (PGM), ADP-Glc pyrophosphorylase (AGPase), debranching enzymes (ISA), members of the starch synthase (SSs) family, and branching enzymes (BE) are required for normal starch synthesis in photosynthetic cells (Caspar et al., 1985; Lin et al., 1988; Wang et al., 1997, 1998; Zeeman et al., 1998; Yu et al., 2000; Delvalle et al., 2005; Dumez et al., 2006; Roldán et al., 2007). These nucleus-encoded enzymes are synthesized in the cytosol and transported into plastids for their functions.
Along with the plastidial forms of PGI and PGM, there are cytosolic isozymes of PGI and PGM in Arabidopsis. Both cytosolic and plastidial PGI and PGM enzymes can transform Fru-6-P to Glc-6-P (G6P) and G6P to Glc-1-P, respectively. The Arabidopsis pgi1 mutant is deficient in plastidial PGI and lacks starch in mesophyll cells, but the starch synthesis in root cap cells and guard cells is not affected (Yu et al., 2000). This phenotype suggests that in the absence of plastidial PGI, cytosolic G6P can be transported into chloroplasts of guard cells and amyloplasts of root cap cells for starch synthesis, while chloroplasts of mesophyll cells cannot import G6P efficiently. In Arabidopsis, there are two G6P/phosphate (Pi) translocators (GPT1 [At5g54800] and GPT2 [At1g61800]). While GPT1 is expressed ubiquitously during plant development, GPT2 expression is restricted to a few tissues. It has been demonstrated that ectopic expression of either G6P/Pi translocator can import cytosolic G6P to chloroplasts of mesophyll cells and can rescue the starch deficiency in mesophyll cells of the pgi1 mutant (Niewiadomski et al., 2005).
The deficiency of starch in the pgm1 mutant (Caspar et al., 1985), which has no detectable plastidial PGM activity, indicates that cytosolic Glc-1-P is not efficiently transported into plastids as the substrate for AGPase to produce ADP-Glc (ADPG). AGPase is a tetrameric enzyme containing either four small subunits or two large and two small subunits (Li and Preiss, 1992). In Arabidopsis, two genes encode for small subunits (APS1 and a pseudogene APS2) and four genes encode for large subunits (APL1–APL4; Crevillen et al., 2005). The adg1 mutant has a mutation in the small subunit gene (designated as APS1), and the adg2 mutant has a mutation in the large subunit gene (designated as APL1). The starchless phenotype of the adg1 mutant, which lacks AGPase activity, suggests that ADPG produced in plastids is the substrate for starch synthesis (Lin et al., 1988; Wang et al., 1998). The adg2 mutant, in which AGPase is suggested to be a homotetramer with four small subunits (Li and Preiss, 1992), has decreased starch content in leaves. While there is evidence that homotetrameric AGPases of Arabidopsis APS1 (Crevillen et al., 2003) can function in Escherichia coli, the APS1 homotetramer was much less sensitive to 3-phosphoglycerate activation and more sensitive to Pi inhibition. It has been suggested that APL1 homologs (APL2, APL3, and APL4) may also form heterotetramers with APS1 based on the presence of specific isoforms and play roles in starch metabolism in response to various metabolic states of tissues (Crevillen et al., 2005).
The enzymes mentioned above provide the substrate, ADPG, for starch synthesis. Many other enzymes (e.g. SSs, BEs, and ISAs) are involved in the biosynthesis of the starch granule itself. Mutants lacking these genes frequently have decreased starch content in leaves but also altered starch structure (Zeeman et al., 1998; Satoh et al., 2003; Delvalle et al., 2005; Zhang et al., 2005; Dumez et al., 2006; Roldán et al., 2007).
Genetic evidence indicates that plant cells need cooperative actions of multiple genes to accomplish starch synthesis and degradation. Starch synthesis occurs in both photosynthetic and nonphotosynthetic tissues, yet not every plant cell accumulates starch. For instance, starch granules are present in root cap cells (Yu et al., 2000; Siedlecka et al., 2003) but are absent in other cells of the root (i.e. the part of the capless root designated as root proper). This tissue-specific distribution of starch must be associated with the starch synthesis machinery, suggesting that starch synthesis genes may be coexpressed in a tissue-specific manner. Coordinated expression of starch synthesis genes during the endosperm development of maize (Zea mays) and rice (Oryza sativa) has been shown (Giroux et al., 1994; Zhu et al., 2003; Ohdan et al., 2005). By Pearson correlation coefficient analysis, 30 genes whose expression is highly correlated with starch debranching enzyme genes in Arabidopsis were identified (Li et al., 2007). Of these 30 genes, 12 are involved in starch metabolism and 10 are known for starch degradation. Some starch metabolism genes of Arabidopsis have a similar diurnal expression pattern in leaves (Harmer et al., 2000; Smith et al., 2004), suggesting that there is a temporal coordinated regulation. In Arabidopsis, high-resolution microarray expression profiles of various types of sorted root cells and root cells dissected from different development stages demonstrate not only developmental but also spatial coregulation of many clusters of genes (Brady et al., 2007). However, only the steady-state level of mRNA was analyzed in these previous studies, and the mechanism for the coordinated expression of these genes was not investigated. To address the regulation of starch synthesis genes, we investigated the pattern of spatial expression of several starch synthesis genes. In this study, we demonstrate that PGM1, APS1, APL1, and SS1 promoters are transcriptionally inactive in cells without starch synthesis (e.g. root proper cells). Our results suggest that starch synthesis in Arabidopsis requires spatial coexpression of core starch metabolism genes, which are regulated by their promoter activities. Furthermore, starch synthesis can be fine-tuned by the accessibility of carbon sources under cell-specific and environmental controls.
RESULTS
Starch Distribution Is Similar in Arabidopsis and Rice Seedlings
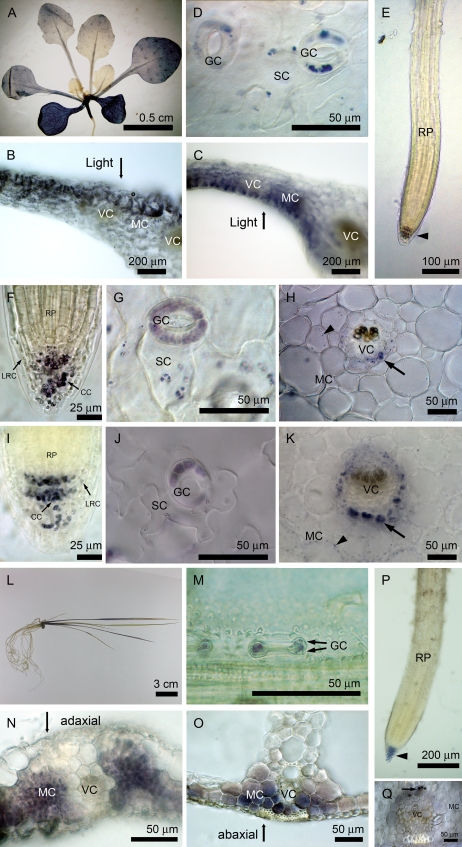
To examine the distribution of starch in vegetative tissues at the end of the light period, we used Lugol's solution to stain depigmented plants of Arabidopsis and rice. As shown in Figure 1, starch stained with iodine, as the blue-purple color was found mainly in mesophyll cells of Arabidopsis. The staining of starch was more intense in older leaves than in younger leaves (Fig. 1A). Within a cross-section of Arabidopsis leaf, mesophyll cells at the adaxial side had more starch than cells at the abaxial side at the end of the day (Fig. 1B). However, when we inverted the leaves at the end of the night and started the illumination for 12 h, the pattern of starch accumulation was reversed (Fig. 1C). Thus, mesophyll cells toward the light could accumulate more starch than those underneath cells sheltered from light. In epidermis, starch was found in guard cells and could be detected in subsidiary cells neighboring the guard cells (Fig. 1D). The root cap comprises columella cells and the outer lateral root cap cells. Starch granules were present mainly in columella cells and some lateral root cap cells but were absent in other cells of the root (i.e. root proper; Fig. 1E). Because root cells do not contain chloroplasts normally, starch synthesis in root cap cells would be heterotrophic. To identify cells with heterotrophic starch synthesis, we examined starch distribution in an albino mutant, ispH (Hsieh and Goodman, 2005), grown on Murashige and Skoog (MS) medium with Suc. Because the ispH mutant has no chlorophyll in plastids to support photosynthesis, starch synthesis must rely on an exogenous carbon source. We found that starch was present in root cap cells of the ispH mutant as expected (Fig. 1F). In addition, starch was synthesized in epidermal cells (Fig. 1G), vascular cells (including vascular bundle cells and bundle sheath cells), and some mesophyll cells neighboring vascular bundles of the ispH mutant (Fig. 1H). Our result indicated that the majority of mesophyll cells synthesize starch phototrophically and the other types of cells can synthesize starch heterotrophically. This interpretation was further substantiated with an identical starch distribution in the pgi1 mutant to that in the ispH mutant (Fig. 1, I–K), suggesting that starch synthesis was possible in the plastids of these cells, presumably through the uptake of G6P from the cytosol (see “Discussion”).
Figure 1.
Starch distribution in Arabidopsis and rice seedlings. Fourteen-day-old seedlings were depigmented and stained with Lugol's solution for starch (Arabidopsis wild type, A, D, and E; pgi1, I–K; rice wild type, l–P; rice albino mutant, Q). Seedlings of the 42-d-old ispH mutant were harvested and examined (F–H). Cross-sections (B and C) were from wild-type rosette leaves of 28-d-old plants. In Arabidopsis wild type (A), mesophyll cells of leaves contained starch in a gradient pattern in which more starch accumulated in mesophyll cells toward the light (B and C). Starch was detected in guard cells and subsidiary cells of epidermis (D). Starch could be detected in root cap cells (arrowhead) but not in the root proper (E). In Arabidopsis ispH and pgi1 mutants, starch accumulation could be found in the root cap (including columella cells and lateral root cap cells; F and I), epidermal cells (G and J), vascular cells of leaf veins (arrows) and much less in mesophyll cells (arrowhead; H and K, which show representative cross sections from petioles). In rice wild type (L), starch was synthesized in bone-shaped guard cells (arrows; M), mesophyll cells of leaf blade (N) and leaf sheath (O), and root cap cells (arrowhead) but not in the root proper (P). In rice albino mutant, starch can be detected in vascular cells (arrow) but not in mesophyll cells (Q, which shows a cross-section from a leaf blade). CC, Columella cell; GC, guard cell; LRC, lateral root cap cell; MC, mesophyll cell; RP, root proper; SC, subsidiary cell; VC, vascular cell.
In rice, starch was found in guard cells, mesophyll cells, and root cap cells but not in root proper cells (Fig. 1, l–P), with a starch distribution pattern similar to that of Arabidopsis. Interestingly, the starch accumulation level was higher in the abaxial side of leaf sheath, reflecting the orientation of cells toward the light (Fig. 1, N and O). Besides, we also found starch in vascular cells (Fig. 1Q), guard cells, and root cap cells in an albino rice mutant (T58504) similar to that in the Arabidopsis ispH mutant.
Enzyme Activities of Specific Starch Synthesis Genes Are Absent in Root
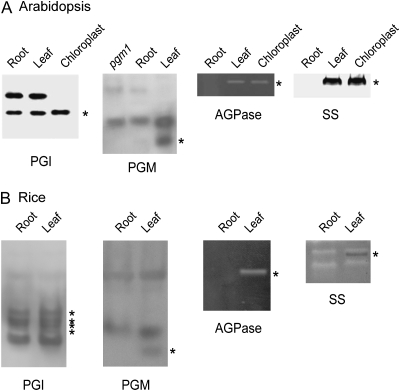
Genetic studies of starch-deficient mutants of Arabidopsis and other species have shown that the cooperation of PGI, PGM, AGPase, and SS enzyme activities is required for normal starch biosynthesis. Cells without starch could potentially lack either a set of starch synthesis enzymes or a single key enzyme. To correlate the distribution of starch and enzymes required for starch synthesis, we analyzed the enzyme activities in extracts from leaves and roots by native activity gels. Leaf extract of wild-type Arabidopsis contained plastidial PGI, PGM, AGPase, and SS activities, which was expected for normal starch synthesis (Fig. 2A). Root extract of wild-type Arabidopsis did not have significant plastidial PGM, AGPase, and SS activities but unexpectedly did have plastidial PGI activity (Fig. 2A). Root cap cells synthesize starch and should contain enzyme activities for starch synthesis. The root extracts analyzed in our experiments included root cap cells; however, enzyme activities of root cap cells in the root tissue was proportionally very low and therefore barely detectable (e.g. PGM in Fig. 2A). Both root and leaf extracts contained cytosolic isozymes of PGI and PGM. The activities of cytosolic isozymes were not correlated with the difference in starch accumulation between leaves and roots, suggesting that cytosolic PGI and PGM do not contribute directly to starch synthesis in chloroplasts. We further examined these enzyme activities present in leaf and root extracts of rice seedlings with zymograms. Based on rice genomic sequence data, there are four genes encoding plastidial PGI and two genes for cytosolic PGI. Six activity bands for PGI were present in both extracts from leaves and roots (Fig. 2B). Specific enzyme activity bands for PGM, AGPase, and SS were present in leaf extracts but were not detected in root extracts of rice seedlings (Fig. 2B). We confirmed that bands specific to leaf extracts were localized in plastids by comparison with zymograms of isolated rice chloroplasts (data not shown). Thus, the pattern of enzyme activities examined in rice was similar to that of Arabidopsis (i.e. a set of plastidial starch synthesis enzymes was present in leaf but not detected in root, with the exception of PGI).
Figure 2.
Zymograms of several enzymes required for starch synthesis. Arabidopsis (A) and rice (B) extracts from roots, leaves, and isolated chloroplasts as indicated were separated on 7.5% polyacrylamide gels and assayed for enzyme activities with specific substrates. Enzyme activities localized in plastids are labeled with asterisks.
Message RNA of Specific Starch Synthesis Genes Is Absent in Root
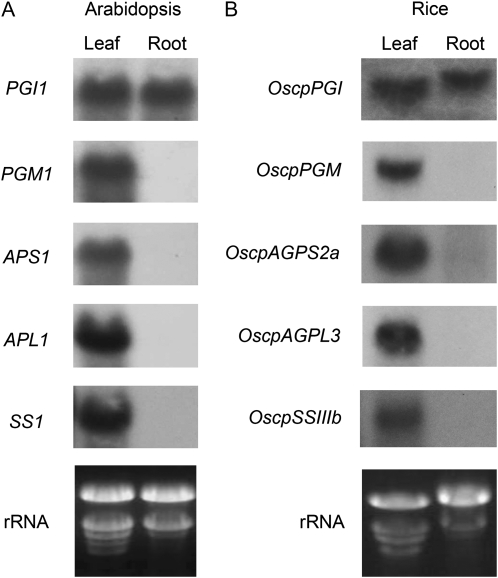
The regulation of gene expression can be achieved at many levels, including transcription, translation, processing of mRNA, and through protein turnover. To investigate the mechanism underlying the lack of starch biosynthetic enzyme activities in roots, we examined mRNA in leaves and roots of Arabidopsis by northern-blot analysis with gene-specific probes. As shown in Figure 3A, PGI1 was expressed in both leaves and roots and the other starch synthesis genes examined (PGM1, APS1, APL1, and SS1) were only expressed in leaves. Our results suggest that these enzymes are not synthesized in roots because the corresponding mRNAs are absent. To test if this gene coexpression pattern in leaves and roots is conserved among different plant species, we also examined the expression of genes encoding enzymes for starch synthesis in rice. By sequence comparison, starch synthesis genes of rice with homology to those of Arabidopsis were selected for the analysis of their expression in leaves and roots (Table I). Northern-blot analysis showed that the transcripts of the genes were present specifically in leaves but not in roots (Fig. 3B), with the exception of OscpPGI mRNA, which was detected in both leaves and roots. Therefore, the coexpression pattern of these rice genes was very similar to that observed in Arabidopsis.
Figure 3.
Northern-blot analysis of starch synthesis genes expressed in leaves or roots. Total RNA (20 μg) isolated from Arabidopsis (A) or rice (B) leaves and roots was separated on 1% agarose gels containing formaldehyde. RNA was transferred to Nytran membranes and probed with radioactively labeled probes as indicated. A portion of ethidium bromide-stained gels is shown for the rRNA bands.
Table I.
Primers used to amplify rice cDNA encoding for starch synthesis genes
| Gene | Enzyme | Accession No. | Primer Sequence (5′ to 3′) | Targeta |
|---|---|---|---|---|
| OscpPGIb | Phosphoglucoisomerase | AK107494 | [F]AGGATCCTACTTGTTCTTAACGAAGCCAG | P |
| [R]TTAAGAATTCATATCATCATCCACGTTGC | ||||
| OscpPGM | Phosphoglucomutase | AK064893 | [F]TGGAAACTATACCCTTCAGTTTGCCGATG | P |
| [R]TTATGTTATGACAGTAGGCTTATCTCTTC | ||||
| OsAGPS2a | ADP-Glc pyrophosphorylase small subunit | AK103906 | [F]TTGATGCTGATGTGACAGACAGTGTCATT | P |
| [R]TCATATAACTGTTCCGCTAGGGAGCAAAG | ||||
| OsAGPL3 | ADP-Glc pyrophosphorylase large subunit | AK069296 | [F]TGTTGTTGGAATCCGTTCTAGGATAGGTT | P |
| [R]TCAGATGACTAATCCATCTGCAATTATCG | ||||
| OsSSIIIb | Starch synthase III | AF432915 | [F]GAGCCACTATCTCATTTGATTTATGCTGG | P |
| [R]TCAGTTCTTGCGAGCGGAATGGTACAATT |
Target indicates the subcellular localization that is predicted by the TargetP Web-based program. P, Plastidial.
Except for OscpPGI, the other cDNA probes are gene specific.
Promoter Activities of Specific Starch Synthesis Genes Are Inactive in Cells without Starch
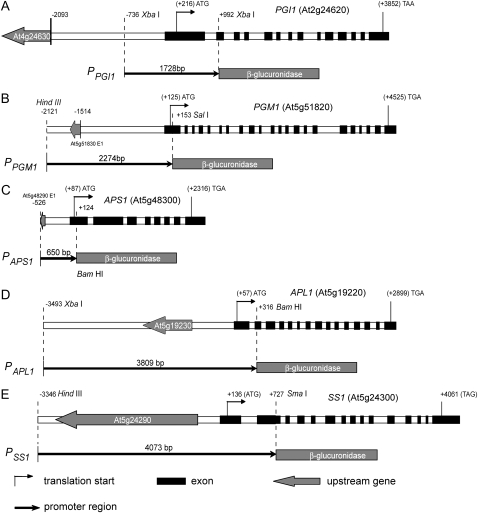
The absence of mRNA of specific starch synthesis genes in roots could be due to the absence of transcription or the instability of transcripts. It is possible to distinguish between these two alternatives by examining gene promoter-reporter fusion expression in transgenic plants. In addition, the expression of reporter genes can be resolved to the individual cell level, thus compensating for the resolution of our analysis of enzyme activity and mRNA levels to the whole organ level. We generated transgenic Arabidopsis plants carrying the promoter region of PGI1 (1,728 bp), PGM1 (2,274 bp), APL1 (3,809 bp), APS1 (650 bp), and SS1 promoter (4,073 bp) fused to the GUS reporter (Fig. 4).
Figure 4.
Diagrams of promoter-GUS fusion constructs used for Arabidopsis transformation. The top line in each panel shows the upstream gene, intragenic region, and coding region of each gene with the transcription start site as +1. The bottom line shows the fragment and length of DNA fused to the GUS of pPZP212GUS vector in each construct. The diagrams were drawn according to the sequence data deposited in The Arabidopsis Information Resource database proportionally (http://www.arabidopsis.org).
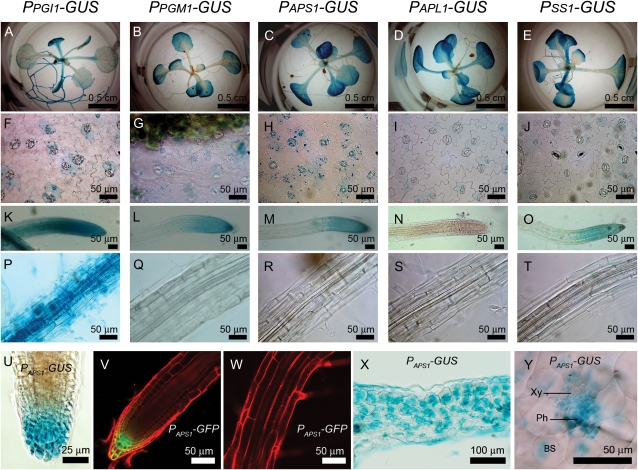
The GUS expression in transgenic seedlings with various promoter-GUS fusion constructs indicated that PGI1, PGM1, APS1, APL1, and SS1 promoters were active in mesophyll cells of leaves, with higher activity in older leaves than younger leaves (Fig. 5, A–E). These GUS staining patterns were similar to that of iodine staining of leaf starch (Fig. 1A). The GUS activity did not show distinct differences among mesophyll cells within a cross-section of leaves (Fig. 5X); however, starch was accumulated to a higher level in cells toward the light (Fig. 1, C and D). GUS staining of epidermal peels from the PGI1, PGM1, APS1, APL1, and SS1 promoter-GUS fusion transgenic plants showed that GUS activity was expressed mainly in guard cells and scantly in subsidiary cells neighboring the guard cells (Fig. 5, F–J). We also observed GUS activities present in bundle sheath cells and vascular bundle cells except dead tracheids of leaf veins in transgenic plants (Fig. 5Y). It has been shown that distinct small starch granules are present in plastids within subsidiary cells, bundle sheath cells, and phloem cells (Delatte et al., 2005; Streb et al., 2008). The starch-containing plastids in these cells with few membrane stacks compared with chloroplasts of mesophyll cells (Delatte et al., 2005; Streb et al., 2008).
Figure 5.
GUS activity and GFP fluorescence in Arabidopsis transgenic lines with various promoter fusion constructs. GUS activities present in seedlings (A–E), epidermal peels (F–J), roots (K–O), and root proper (P–T) of transgenic lines carrying various promoter-GUS fusion constructs (as indicated at the top of each column) are shown. For each construct, more than 10 independent transgenic lines were examined. Identical expression patterns for each construct were observed in independent transgenic lines. At the bottom, expression of GUS (U, X, and Y) driven by the APS1 promoter in various tissues is shown. BS, Bundle sheath cell; Ph, phloem; Xy, xylem. Longitudinal confocal sections of a representative root tip (V) and the segment of the nearby root proper (W) of PAPS1-GFP transgenic plants show GFP fluorescence (green) and cells counterstained with propidium iodide (red).
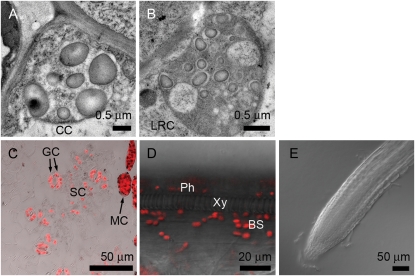
GUS activity was present in every root cell of PPGI1-GUS transgenic lines (Fig. 5, K and P). For transgenic lines carrying APS1, PGM1, and SS1 promoter-GUS fusion constructs, GUS activity was present in columella cells and lateral root cap cells (Fig. 5, L, M, O, and U) but not in root proper cells, including root vascular bundle cells (Fig. 5, Q–T). Similar expression patterns in roots were observed in transgenic plants with ISA gene promoter-GUS fusion constructs (Li et al., 2007). To confirm that the expression of GUS is limited to root cap cells, we also examined the GFP expression in transgenic lines carrying a PAPS1-GFP fusion construct. This analysis also showed that GFP expression was present in columella cells and lateral root cap cells (Fig. 5V) but not in root proper cells (Fig. 5W). The intensity of green fluorescence present in columella cells was higher than that in lateral root cap cells. We compared the starch granules present in columella cells and lateral root cap cells by electron microscopy. The number of plastids and the size of starch granules are more and larger in columella cells than in lateral root cap cells (Fig. 6, A and B). To find out whether these plastids contain chlorophylls, we further examined these cells with a confocal microscope for autofluorescence emitted from chlorophyll upon the excitation of 488-nm laser light. In contrast with plastids of leaf cells, no red fluorescence was observed in root cells, confirming that plastids in root cells did not contain chlorophylls (Fig. 6, C–E).
Figure 6.
Plastids in various cell types of Arabidopsis examined by electron and confocal microscopy. Electron micrographs of Arabidopsis columella cell (CC) and lateral root cap cell (LRC; A and B) showed starch granules present in amyloplasts. Confocal micrographs of epidermal peel (C), longitudinal section of leaf vascular bundle (D), and root (E) showed that red fluorescence from chlorophylls was present in guard cells (GC) and subsidiary cells (SC), was present in phloem cells (Ph) and bundle sheath cells (BS), and was absent in xylem vessel (Xy) and root cells. Some mesophyll cells (MC) attached to the epidermal peel are shown in C.
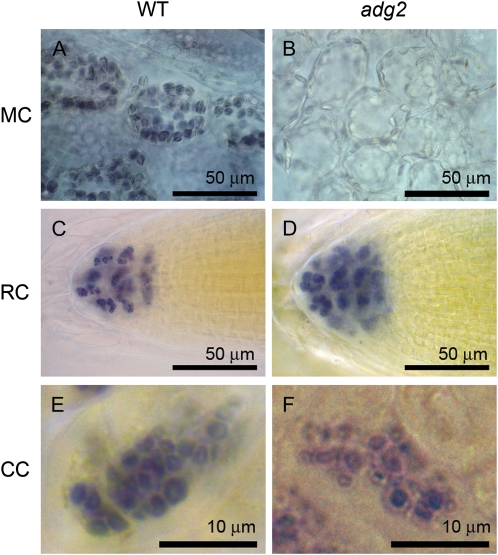
Surprisingly, APL1 promoter activity was not detected in root, including root cap cells (Fig. 5N). Because of this unexpected expression pattern of APL1, we examined starch granules in root cap cells of the wild type and the adg2 mutant that lacks a functional large subunit AGPase (Wang et al., 1997). While the appearance of starch granules of mesophyll cells in the wild type and the adg2 mutant was apparently different (Fig. 7, A and B), starch granules in columella cells of adg2 were indistinguishable from those in the wild type (Fig. 7, C–F), suggesting that APL1 is not required for starch synthesis in root cap cells.
Figure 7.
Starch granules in root cells and mesophyll cells of Arabidopsis wild type (WT) and adg2 mutant. Roots and leaves of Arabidopsis wild type (A, C, and E) and adg2 mutant (B, D, and F) were stained with iodine for starch granules. Starch granules were clearly observed in mesophyll cells (MC) of wild-type leaves (A) but barely found in adg2 mutant (B). On the contrary, starch granules present in root cap (RC) and columella cells (CC) of the wild type (C and E) and the adg2 mutant (D and F) were similar in size and number.
Expression Profile of Arabidopsis Starch Metabolism Genes
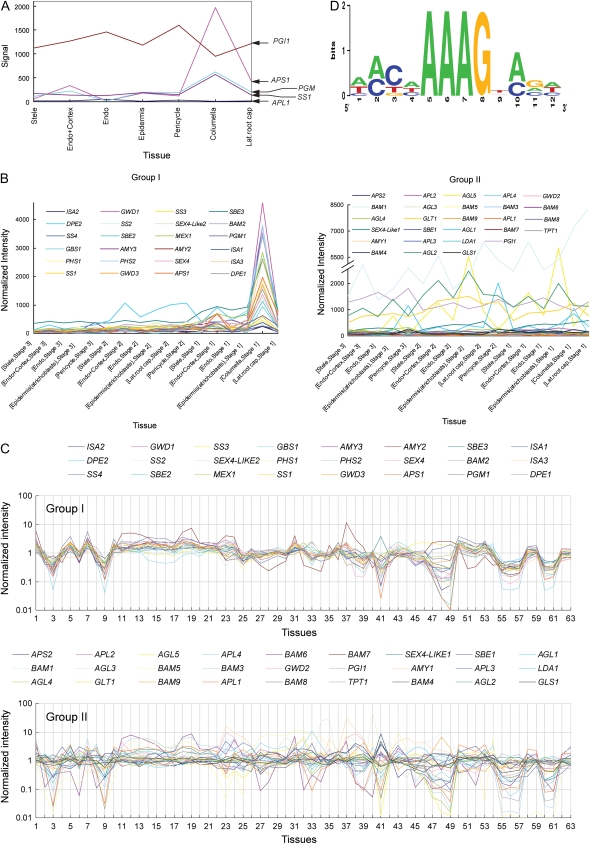
Cooperative actions of multiple enzymes are required for starch metabolism in plant cells. Tissue-specific distribution of starch in plants must be associated with the starch metabolism machinery, implicating that starch metabolism genes may be coexpressed in a tissue-specific manner. Because we only examined the expression pattern of a limited number of starch synthesis genes, we extended our analysis to other starch metabolism-related genes (Supplemental Table S1; defined by Smith et al., 2004) using existing bioinformatic data. We extracted the transcriptome data for starch metabolism genes from the root expression map database (www.arexdb.org; Birnbaum et al., 2003; Brady et al., 2007). The expression profiles of starch metabolism genes in root (Fig. 8A) are similar to the GUS staining patterns of PGM1, APS1, and SS1 promoter-GUS transgenic lines (i.e. the mRNAs were expressed mainly in the columella and extended to the lateral root cap). Similarly, our results on the expression pattern of PPGI1-GUS and PAPL1-GUS also agreed with microarray data (Fig. 8A), indicating that the promoter-driven GUS expression correlated with the location of mRNA of starch synthesis genes in root. Thus, microarray data not only supported our observation of coexpression of some starch synthesis genes in roots but could be extended to discover tissue-specific coexpression of other starch metabolism genes. As shown in Figure 8B, a set of starch metabolism genes were clustered into two groups: groups I and II. Group I genes had high signal in root cap cells and low or no signal in root proper cells (Supplemental Fig. S1). Genes involved in both starch synthesis and degradation pathways were found in group I. We further examined the expression profiles of group I and group II genes in other tissues (Fig. 8C; Supplemental Table S2; Schmid et al., 2005). We found that their expression also showed similar coordinated expression patterns for group I genes but not for group II genes. Expression of group I genes had a direct correlation with the spatial distribution of starch in plants, suggesting that at least some of these genes play important roles as core enzymes in starch metabolism.
Figure 8.
Expression profiles of starch metabolism-related genes of Arabidopsis. A, Expression profiles of starch synthesis genes in root showed a similar expression pattern for APS1, PGM1, and SS1 but not for PGI1 and APL1. B, Normalized expression profiling data for starch metabolism genes in roots were analyzed. Based on the expression patterns, starch metabolism-related genes could be clustered into two groups (Supplemental Fig. S1): group I genes were mainly expressed in columella cells, but not group II genes. C, In addition, group I genes coexpressed in various tissues (for conditions of tissues, see Supplemental Table S2) but not group II genes. D, Sequence logo of a motif statistically overrepresented in promoter regions (Supplemental Table S3) of coregulated group I genes.
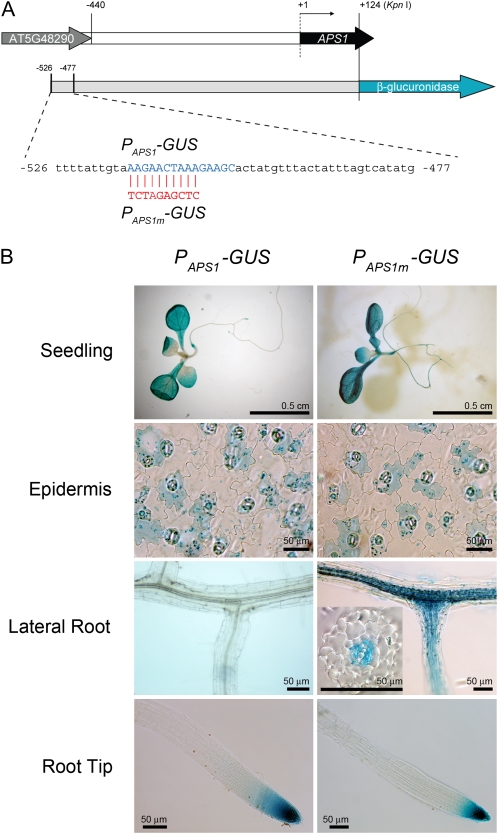
Group I genes showed a similar coordinated expression pattern but not for group II genes, suggesting that a general transcriptional coregulation might exist for the group I genes. cis-Acting motifs in group I genes were evaluated using Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/) for statistically overrepresented motifs in the promoter region of coregulated group I genes. A cis-acting motif with a signature of [ATC][AC][CTG][ATC]AAAGN[AC][GCA][ATC] (Fig. 8D; degenerate bases at a defined position are bracketed) was found in 83.3% (20 of 24) of group I genes (Supplemental Table S3) but only 48.1% (13 of 27) of group II genes and 31.2% ± 6.7% (n = 10) of 50 random sequences generated with Arabidopsis promoter sequence model. This motif is present in the promoter region (−516 to −502) of the APS1 gene. To verify the identified motif as a transcriptional control element, we modified this motif in the PAPS1-GUS construct (as PAPS1m-GUS; Fig. 9A) and examined the expression of GUS in PAPS1m-GUS transgenic plants. The GUS expression patterns in transgenic plants carrying either PAPS1-GUS or PAPS1m-GUS were similar in leaves and roots (Fig. 9B), except that GUS activities were observed at the site of lateral root initiation in PAPS1m-GUS lines but not in PAPS1-GUS lines. The GUS activities were found in stele cells of the lateral root base in PAPS1m-GUS lines (Fig. 9B). Mutation of this cis-element switched on APS1 promoter activity in stele cells of the lateral root base, indicating that this cis-element could be a target for transcription repression.
Figure 9.
Mutation of a cis-acting motif in the APS1 promoter switched on transcriptional activity at the sites of lateral root initiation. A, A cis-acting motif (−516 to −507) in the PAPS1-GUS construct was modified by PCR-directed mutagenesis. The APS1 promoter-GUS fusion constructs of the wild type (PAPS1-GUS) and mutant (PAPS1m-GUS) are illustrated. B, GUS activities present in seedlings, epidermal peels, lateral roots, and root tips of 9-d-old transgenic lines carrying PAPS1-GUS or PAPS1m-GUS are shown. The inset in the PAPS1m-GUS lateral root panel shows that GUS activities were present in stele cells at the lateral root base.
DISCUSSION
Starch metabolism is a complex biological process requiring the function of multiple enzymes not only for synthesis but also for degradation. For cells containing chloroplasts (e.g. mesophyll cells), starch is synthesized from the carbon source fixed during photosynthesis. Enzymes required for starch metabolism, therefore, must be expressed in phototrophic cells. The representative enzymes for starch synthesis (PGI, PGM, AGPase, and SSI) examined in our study were indeed expressed in mesophyll cells. Furthermore, data from expression profiles of starch metabolism genes, including genes with roles in starch degradation, also support this prediction (Fig. 8C).
The transcription activities of starch synthesis genes showed a developmental expression pattern, as the older leaf cells had higher expression levels than the younger leaf cells (Fig. 5, A–E). This pattern coincided with the starch accumulation level detected by iodine staining (Fig. 1A), suggesting that the starch synthesis enzymes may be the limiting factors. On the contrary, the GUS activity did not show distinct differences within a cross-section of leaves (Fig. 5X); however, starch was accumulated to a higher level in cells toward the light (Fig. 1, B and C). This pattern suggested that limited substrates derived from photosynthesis are available for starch synthesis in those cells shielded from light under a normal growth condition. The relative GUS activities in guard cells versus subsidiary cells (Fig. 5, F–J) and columella cells versus lateral root cap cells (Fig. 5, L, M, O, and V) are correlated with the starch accumulation level in these cell types (Fig. 1, D, F, G, I, and J), suggesting the existence of a cell-specific endogenous control for starch gene expression.
In heterotrophic cells, the ability to synthesize starch would not only rely on the presence of required starch metabolism enzymes but also on the availability of a carbon source exported from source cells. We found that plastids of root cap cells, epidermal cells, vascular cells, and some mesophyll cells neighboring vascular bundles could synthesize starch in the albino ispH mutant and the pgi1 mutant (Fig. 1, F–K). The ispH mutant has no chlorophyll in plastids to support photosynthesis; starch synthesis in these cells must rely on an exogenous carbon source. The presence of starch in these cells of the pgi1 mutant suggests that the starch synthesis in them would be plastidial PGI independent. Otherwise, these cells contain additional plastidial PGI activities that are not affected by the pgi1 mutation. However, only two genes in the Arabidopsis genome are identified as encoding for the cytosolic and plastidial PGI enzymes, which is consistent with our native gel analysis (Supplemental Fig. S2). In addition, we analyzed PGI activities present in extracts of whole leaf or epidermis of the wild type and the pgi1 mutant upon heat inactivation treatment (Supplemental Table S4). We found that heat inactivation percentage was higher for extracts from the wild-type leaves (37.4%) than from the pgi1 mutant (2.2%), indicating that the pgi1 mutant is deficient in the plastidial PGI, which is heat labile (Supplemental Fig. S2). If there are additional PGI enzymes specifically expressed in those cells with starch in the pgi1 mutant (e.g. epidermis), the percentage of heat inaction of PGI activity would be changed notably. The heat inactivation percentage for PGI activity remained similar for the extract from the epidermal cells of the wild type (38.8%) and whole leaf extract (37.4%) and for the extract from epidermal cells of the pgi1 mutant (2.7%) and whole leaf extract (2.2%). These results indicated that epidermal cells of the pgi1 mutant did not contain additional PGI activities.
Our data suggested that heterotrophic starch synthesis in the ispH and pgi1 mutants relied on cytosolic G6P imported to their plastids as the substrate. The expression of G6P/Pi translocators in root cap cells, guard cells, and mesophyll cells for importing cytosolic G6P to plastids has been demonstrated (Niewiadomski et al., 2005). The expression level of GPT genes among mesophyll cells is similar and is at least 10-fold less than that in guard cells (Niewiadomski et al., 2005), suggesting that mesophyll cells cannot efficiently import G6P from cytosol to chloroplasts for starch synthesis. Most mesophyll cells would synthesize starch with assimilated carbon during photosynthesis. Only those mesophyll cells near the vascular bundle have imported carbon available for starch synthesis in the pgi1 mutant and the albino ispH mutant. The morphology of membrane stacking in chloroplasts of leaf vascular cells and epidermal cells was distinct from that of mesophyll cells (Delatte et al., 2005; Streb et al., 2008). We suggest that plastids in vascular cells and epidermal cells can use either assimilated carbon or imported carbon for starch synthesis. Therefore, we could differentiate at least three types of plastids active in starch synthesis: (1) the chloroplasts of mesophyll cells, (2) the chloroplasts of epidermal cells and vascular cells of leaves, and (3) amyloplasts of root cap cells. Future studies on novel distinctions in metabolism and gene expression among these cell types would be worthwhile.
For root proper cells, which are heterotrophic and have no apparent starch accumulation, we showed that key starch metabolism enzymes are absent, including PGM1, APS1, APL1, and SS1; the other group I genes are not expressed as well (Fig. 8B). These genes were transcriptionally inactive in the root proper cells based on the results of promoter-reporter analyses (Fig. 5, Q–T) and so may be regulated coordinately by transcription factors. However, PGI1 was expressed in root proper cells, regardless of their inability to synthesize starch in the absence of PGM, AGPase, and SSI. The expression of PGI1 in starch-free cells may reflect its biological role in pathways other than starch synthesis (e.g. the production of NADPH through the oxidative pentose phosphate pathway; ap Rees, 1980, 1985).
Unexpectedly, APL1 was not expressed in root cap cells, which could synthesize starch. It has been shown that the adg2-1 mutant, which has a mutation in APL1, has AGPase formed as a homotetramer with four small subunits (Li and Preiss, 1992). The starch granules accumulated in root cap cells of the adg2-1 mutant were indistinguishable from those of the wild type, suggesting that the expression of APL1 is not essential for starch synthesis in root cap cells. It is possible that in root cap cells the AGPase is composed of small subunits only. However, the levels of allosteric effectors required for optimal homotetrameric APS1 activity are not favored in cells. Alternatively, APL1 homologs in Arabidopsis may replace APL1 to form heterotetrameric AGPase in root cap cells. There are homologs to APL1 genes in the Arabidopsis genome (i.e. APL2, APL3, and APL4). Microarray data indicate that APL2 and APL4 are expressed in root cap cells (Birnbaum et al., 2003). Recently, it has been shown that APL2 and APL4 may replace APL1 to form functional heterotetrameric AGPase (Ventriglia et al., 2008).
We showed that starch distribution and gene expression pattern in rice were similar to those of Arabidopsis. In fact, we observed similar starch distribution in seedlings of other species (e.g. tomato [Solanum lycopersicum], pea [Pisum sativum], onion [Allium cepa], and maize; i.e. starch present in mesophyll, epidermal, and root cap cells but not in root proper cells). Moreover, the ability of heterotrophic starch synthesis, in plastids of epidermal cells, vascular cells, and root cap cells, was found in albino mutants of rice and Arabidopsis. Our results suggest that the spatial regulation and expression of starch metabolism genes may be conserved among plants. In this study, we showed that the starch was absent in primary roots. However, in some plants, starch accumulation appears in secondary roots, such as in Cape Erica (Erica species; Verdaguer and Ojeda, 2002) and buttercup (Ranunculus asiaticus; Kamenetsky et al., 2005). In addition, in some plants (e.g. sweet potato [Ipomoea batatas]), the root can synthesize and store starch. We hypothesize that in those roots with starch, starch metabolism genes would be activated coordinately during developmental differentiation (Li and Zhang, 2003).
Correlation analyses of transcript profiles could help identify genes that are functionally related. By bioinformatics analysis, we found that starch metabolism genes could be clustered into two groups based on their spatial expression patterns. Expression of group I genes, involved in both starch synthesis and degradation pathways, had a direct correlation with the spatial distribution of starch in plants (Fig. 8B), suggesting that at least some of these genes play important roles as core enzymes in starch metabolism. For example, in addition to APS1, PGM, and SS1, the spatial patterns of coexpressed ISA1 and ISA2 during plant development (Li et al., 2007) are fully consistent with their functions in starch metabolism, as the proteins encoded by these two genes form a heteromultimeric complex. It has been shown that SBEI, SBEIIb, and starch phosphorylase form a protein complex involved in starch synthesis of wheat (Triticum aestivum) plastids (Tetlow et al., 2004), suggesting that proteins derived from coexpressed group I genes may form high-Mr starch synthesis/degradation complexes in Arabidopsis.
On the contrary, expression of group II genes does not correlate well with starch distribution. Many starch metabolism genes are in multigene families. We noticed that most starch metabolism genes clustered in the group II are duplicated genes related to group I genes. Group II genes may acquire different transcriptional regulation from that of group I genes. For example, APL family genes (APL1–APL4) can be formed as heterotetramers with APS1 in different tissues of Arabidopsis based on the expression of specific APL isoforms. The various forms of heterotetramers would respond to substrates and allosteric effectors for starch synthesis and carbon partition in a tissue-specific manner (Crevillen et al., 2003). On the other hand, group II genes could exert functions other than the basic starch metabolism. Presumably, these genes enable the plants to adjust starch metabolism in response to different environments (e.g. response to stress conditions; Sparla et al., 2006).
Our promoter-reporter analysis of several starch synthesis genes (Fig. 5) indicated that these genes were coregluated spatially by transcriptional activity of the promoters. Both genes related to starch synthesis and degradation pathways were found in group I. We suggest that a common mechanism for the transcriptional coregulation of group I genes exists, presumably by activation and/or repression, via specific cis-elements in the gene promoters. A motif with a signature of [ATC][AC][CGT][ATC]AAAGN[AC][GCA][ATC] was identified in promoter regions of group I genes (Fig. 8D). Modification of this conserved sequence located upstream of the APS1 gene ectopically switched on its promoter activity in some root cells where starch biosynthetic genes are not normally active. Although more analyses are required, our preliminary results suggest that this conserved cis-element may be a target site for transcription repression. We also analyzed the genome-wide expression profile with clustering parameters shown in Supplemental Figure S1. The 373 genes (group I-associated genes; Supplemental Table S5) with similar expression patterns to group I starch metabolism genes were identified. The genes related to transcriptional regulation were found within group I-associated genes by gene ontology analysis. Some of them may be involved in the regulation of group I gene expression. Identification of transcription factors and other cis-elements conserved in group I genes would provide clues for future study of spatial coregulation and ultimately provide a strategy to turn on or increase starch synthesis in tissues that make little starch, to increase starch production for food or biofuels.
MATERIALS AND METHODS
Plant Materials and Molecular Analysis
Plants were grown in potting soil at 23°C under a 12-h/12-h day/night cycle with 100 μmol quanta m−2 s−1 fluorescent light. For root tissue, plants were grown hydroponically in water (rice [Oryza sativa]) or in B5 medium with 2% Suc (Arabidopsis [Arabidopsis thaliana]). The rice albino mutant (T58504) used in this study was isolated from a T-DNA insertion-mutagenized population. The albino mutants (ispH and T58504) were grown on MS medium with 2% Suc. Standard cloning and northern-blot analysis were performed as described (Sambrook and Russell, 2001). Total RNA of Arabidopsis (Columbia ecotype) and rice (TNG67) was extracted with Trizol reagent and separated by agarose gels containing formaldehyde. Probes for northern-blot analysis were isolated from cDNA clones and labeled with [α-32P]dCTP. Arabidopsis cDNA clones (PGI1, C105270; PGM1, U25342; APS1, U18261; APL1, U15657; and SS1, pSport1:SS) were obtained from Arabidopsis Biological Resource Center. Rice cDNA fragments were amplified by reverse transcription-PCR with primers shown in Table I. The amplified fragments were cloned into pZero2.1 and confirmed by DNA sequencing. Probes used for northern-blot analysis were gene specific, except that for OscpPGI.
For construction of promoter-reporter fusion clones, a 1.7-kb fragment of PGI1, a 2.2-kb fragment of PGM1, and a 0.65-kb fragment of APS1 containing the promoter region were amplified by PCR with designed primers and subcloned in front of the GUS reporter of a binary T-DNA vector, pPZP212GUS. A 3.9-kb fragment of APL1 promoter and a 4.3-kb fragment of SS1 promoter were subcloned from genomic clones to pPZP212GUS digested with XbaI/BamHI and HindIII/SmaI, respectively. A 1.8-kb fragment of APS1 containing the promoter region was subcloned from a genomic clone in front of the GFP reporter of pPZP212GFP. PAPS1m-GUS was constructed from PAPS1-GUS by a PCR-directed mutagenesis with primer 5′-ATTGTATCTAGAGCTCGAAGCACTATGTTTAC-3′. The promoter-reporter constructs were transformed into Arabidopsis by vacuum infiltration with Agrobacterium tumefaciens (GV3101) carrying the binary constructs. Transgenic plants were selected for kanamycin resistance on MS medium, then transferred to pots, and confirmed by Southern-blot analysis. For each transgenic construct, more than 10 independent transgenic lines were examined.
Native Gel Assay for Starch Synthesis Enzymes
Native gel assays for PGI and PGM (Caspar et al., 1985), AGPase (Yu et al., 2000), and SS1 (Fontaine et al., 1993) activities were carried out according to previously described methods with 7.5% (w/v) polyacrylamide gels. Leaf and root samples were extracted with extraction buffer (100 mm Tris-HCl, pH 7.0, 40 mm β-mercaptoethanol, 10 mm MgCl2, 100 mm KCl, and 15% glycerol; 2 mL g−1 fresh tissue). The crude extract was cleared by centrifugation, and the supernatant was assayed for enzyme activities. For heat inactivation, the extracts were treated at 50°C for 10 min in a thermocycler. PGI activity was assessed by following the enzyme-linked reduction of NADP+ at 340 nm in a Powerwave 340 microplate reader (Bio-Tek) at 25°C. The reaction mixture (0.1 mL) consisted of 0.1 m Tris-HCl (pH 7.0), 4.2 mm Fru-6-P, 1.3 mm NADP+, and 1 unit mL−1 G6P dehydrogenase (Sigma). PGI activity was expressed as units per μg protein (one unit converting 1 pmol of Fru-6-P to G6P per min).
Histological Detection of Starch and GUS Activity
Plants were depigmented with ethanol to remove chlorophylls and stained with Lugol's iodine solution (6 mm iodine, 43 mm KI, and 0.2 n HCl) to detect starch granules. GUS staining of transgenic seedlings was performed as described. Stained specimens were cleared and mounted with chloral hydrate clearing solution. Stained seedlings were examined with a Nikon SMZ 1500 stereomicroscope. To peel epidermis from mesophyll cells, leaf samples were glued between two transparent tapes and the tapes were pulled apart; an epidermal layer would adhere to the separated tapes. The epidermis on the tape was covered by GUS staining buffer and incubated in a chamber assembled from a microscope slide and a coverslip and incubated at room temperature overnight. Epidermis was examined with an Olympus BHT microscope. Images were captured with a Nikon DS-5Mc digital camera.
Confocal and Transmission Electron Microscopic Analyses
A confocal laser-scanning microscope (Zeiss LSM-510) using the 488-nm laser light for excitation was used to examine autofluorescence of chlorophyll (emission filter was long-pass 650 nm), GFP fluorescence (emission filter was band-pass 505–530 nm), and propidium iodide fluorescence (emission filter was long-pass 560 nm) in cells. Roots of the transgenic plants carrying the PAPS1-GFP fusion construct were stained with 10 μg mL−1 propidium iodide for 1 min and mounted in water, then examined for GFP and propidium iodide fluorescence. For electron microscopic analysis, Arabidopsis tissues were fixed in a solution of 4% paraformaldehyde and 2.5% glutaraldehyde in 100 mm sodium phosphate, pH 7.3, at room temperature for 4 h. Tissues were washed with 100 mm sodium phosphate, pH 7.3, three times and postfixed with 1% osmium tetraoxide overnight. The postfixed tissues were dehydrated with an ethanol series, infiltrated, and embedded in LR White acrylic resin (London Resin Company). Ultrathin sections stained with uranyl acetate and Reynold's lead citrate were examined and photographed with a Tecnai G2 Spirit TWIN electron microscope (FEI).
Microarray Data Analysis
The cell type-specific expression data of roots (Birnbaum et al., 2003) were obtained from the AREX databases (www.arexdb.org). The expression data of Arabidopsis development (Schmid et al., 2005) were downloaded from PLEXdb (http://www.plexdb.org/index.php). Expression profiles were analyzed with the Gene Spring program. Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/) were used to analyze motifs present in the promoter (including 1,000 bp from the upstream region of the transcription start site). Sequence Logo was drawn with WebLogo (http://weblogo.berkeley.edu/logo.cgi).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A flow chart diagram for steps and parameters used in the expression profile analysis of starch metabolism genes.
Supplemental Figure S2. Heat inactivation of PGI isozymes in leaf extracts of the wild type and the pgi1 mutant.
Supplemental Table S1. Starch metabolism genes defined by Smith et al. (2004).
Supplemental Table S2. Samples extracted from microarray data available at the Plant Expression Database under the accession number AT-40.
Supplemental Table S3. Motif-matching positions in upstream regions of group I genes.
Supplemental Table S4. Heat inactivation of PGI activity in extracts of leaf and epidermis from the wild type and the pgi1 mutant.
Supplemental Table S5. Genome-wide expression profiles were analyzed with the same clustering conditions for group I genes.
Supplementary Material
Acknowledgments
We thank Steve Smith, Samuel Zeeman, Hsou-min Li, and Tien-Shin Yu for reading the manuscript prior to publication.
This work was supported by the National Science Council, Taiwan, Republic of China (grant nos. NSC91–2311–B–002–068 to S.-M.W. and NSC96–2311–B–001–004 to J.C.) and Academia Sinica, Taipei, Taiwan (to J.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jychian Chen (mbjchen@gate.sinica.edu.tw).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- ap Rees T (1980) Assessment of the contributions of metabolic pathways to plant respiration. In DD Davis, ed, The Biochemistry of Plants, Vol 2. Academic Press, New York, pp 1–29
- ap Rees T (1985) The organization of glycolysis and the oxidative pentose phosphate pathway in plants. In R Douce, DA Day, eds, Encyclopedia of Plant Physiology, New Series, Vol 18. Springer, Berlin, pp 391–417
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillen P, Ballicora MA, Merida A, Preiss J, Romero JM (2003) The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme. J Biol Chem 278 28508–28515 [DOI] [PubMed] [Google Scholar]
- Crevillen P, Ventriglia T, Pinto F, Orea A, Merida A, Romero JM (2005) Differential pattern of expression and sugar regulation of Arabidopsis thaliana ADP-glucose pyrophosphorylase-encoding genes. J Biol Chem 280 8143–8149 [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41 815–830 [DOI] [PubMed] [Google Scholar]
- Delvalle D, Dumez S, Wattebled F, Roldan I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, et al (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43 398–412 [DOI] [PubMed] [Google Scholar]
- Deschamps P, Haferkamp I, d'Hulst C, Neuhaus HE, Ball SG (2008) The relocation of starch metabolism to chloroplasts: when, why and how. Trends Plant Sci 13 574–582 [DOI] [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvallé D, Planchot V, Ball SG, D'Hulst C (2006) Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell 18 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein ML, Routier F, Pepin TM, Decq A, Wieruszeski JM, Delrue B, Van den Koornhuyse N, et al (1993) Toward an understanding of the biogenesis of the starch granule: evidence that Chlamydomonas soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem 268 16223–16230 [PubMed] [Google Scholar]
- Giroux MJ, Boyer C, Feix G, Hannah LC (1994) Coordinated transcriptional regulation of storage product genes in the maize endosperm. Plant Physiol 106 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Goodman HM (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetsky R, Peterson RL, Melville LH, Machado CF, Bewley JD (2005) Seasonal adaptations of the tuberous roots of Ranunculus asiaticus to desiccation and resurrection by changes in cell structure and protein content. New Phytol 166 193–204 [DOI] [PubMed] [Google Scholar]
- Li L, Ilarslan H, James MG, Myers AM, Wurtele ES (2007) Genome wide co-expression among the starch debranching enzyme genes AtISA1, AtISA2, and AtISA3 in Arabidopsis thaliana. J Exp Bot 58 3323–3342 [DOI] [PubMed] [Google Scholar]
- Li L, Preiss J (1992) Characterization of ADPglucose pyrophosphorylase from a starch-deficient mutant of Arabidopsis thaliana (L.). Carbohydr Res 227 227–239 [Google Scholar]
- Li XQ, Zhang D (2003) Gene expression activity and pathway selection for sucrose metabolism in developing storage root of sweet potato. Plant Cell Physiol 44 630–636 [DOI] [PubMed] [Google Scholar]
- Lin TP, Caspar T, Somerville C, Preiss J (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADP-glucose pyrophosphorylase activity. Plant Physiol 86 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flugge UI, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56 3229–3244 [DOI] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Lucas MM, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D'Hulst C, Mérida Á (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 49 492–504 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Siedlecka A, Ciereszko I, Mellerowicz E, Martz F, Chen J, Kleczkowski LA (2003) The small subunit ADP-glucose pyrophosphorylase (ApS) promoter mediates okadaic acid-sensitive uidA expression in starch-synthesizing tissues and cells in Arabidopsis. Planta 217 184–192 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56 73–98 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol 141 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventriglia T, Kuhn ML, Ruiz MT, Ribeiro-Pedro M, Valverde F, Ballicora MA, Preiss J, Romero JM (2008) Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol 148 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer D, Ojeda F (2002) Root starch storage and allocation patterns in seeder and resprouter seedlings of two Cape Erica (Ericaceae) species. Am J Bot 89 1189–1196 [DOI] [PubMed] [Google Scholar]
- Wang SM, Chu B, Lue WL, Yu TS, Eimert K, Chen J (1997) adg2-1 represents a missense mutation in the ADPG pyrophosphorylase large subunit gene of Arabidopsis thaliana. Plant J 11 1121–1126 [DOI] [PubMed] [Google Scholar]
- Wang SM, Lue WL, Yu TS, Long JH, Wang CN, Eimert K, Chen J (1998) Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 13 63–70 [DOI] [PubMed] [Google Scholar]
- Yu TS, Lue WL, Wang SM, Chen J (2000) Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol 123 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, ap Rees T (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15 357–365 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2007) The diurnal metabolism of leaf starch. Biochem J 401 13–28 [DOI] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Provart N, Chang HS, Guimil S, Su W, Estes B, Zou G, Wang X (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1 59–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.