Abstract
Phosphorus (P) deficiency is widespread in regions where the common bean (Phaseolus vulgaris), the most important legume for human consumption, is produced, and it is perhaps the factor that most limits nitrogen fixation. Global gene expression and metabolome approaches were used to investigate the responses of nodules from common bean plants inoculated with Rhizobium tropici CIAT899 grown under P-deficient and P-sufficient conditions. P-deficient inoculated plants showed drastic reduction in nodulation and nitrogenase activity as determined by acetylene reduction assay. Nodule transcript profiling was performed through hybridization of nylon filter arrays spotted with cDNAs, approximately 4,000 unigene set, from the nodule and P-deficient root library. A total of 459 genes, representing different biological processes according to updated annotation using the UniProt Knowledgebase database, showed significant differential expression in response to P: 59% of these were induced in P-deficient nodules. The expression platform for transcription factor genes based in quantitative reverse transcriptase-polymerase chain reaction revealed that 37 transcription factor genes were differentially expressed in P-deficient nodules and only one gene was repressed. Data from nontargeted metabolic profiles indicated that amino acids and other nitrogen metabolites were decreased, while organic and polyhydroxy acids were accumulated, in P-deficient nodules. Bioinformatics analyses using MapMan and PathExpress software tools, customized to common bean, were utilized for the analysis of global changes in gene expression that affected overall metabolism. Glycolysis and glycerolipid metabolism, and starch and Suc metabolism, were identified among the pathways significantly induced or repressed in P-deficient nodules, respectively.
A key to the success of the legume family, which comprises approximately 700 genera with more than 18,000 species (Doyle and Luckow, 2003), was the evolution of mutualistic symbioses with nitrogen (N)-fixing bacteria of the family Rhizobiaceae to directly capture atmospheric dinitrogen (N2) to support plant growth. Symbiotic nitrogen fixation (SNF) takes place in specialized rhizobia-induced legume root nodules and involves a tight association between the two symbionts. SNF and legume crop production might be affected by disease and insect pressures but also by edaphic constraints that include climatic conditions, nutrient deficiency, soil acidity, and metal toxicity.
Phosphorus (P) is an essential macronutrient for plant growth and development, with P concentration ranging from 0.05% to 0.5% plant dry weight. P is taken by the plants as phosphate (Pi), but Pi is unevenly distributed and relatively immobile in soils. As a result, crop yield in 30% to 40% of arable land is limited by P availability (Vance et al., 2003). Widespread P deficiency is a major restriction for SNF and legume crop productivity (Andrew, 1978).
N2-fixing legumes require more P than legumes growing on mineral N, but little is known about the basis for the higher P requirement. Growing root nodules are strong P sinks in legumes. For example, P concentration in the nodules of soybean (Glycine max; Sa and Israel, 1991) and white lupin (Lupinus albus; Schulze et al., 2006) from P-deficient plants can reach up to 3-fold that of other plant organs. The deleterious effect of P deficiency on SNF and plant growth has been evidenced through the evaluation of physiological and biochemical parameters, such as nodule number and mass, nitrogenase and carbon/N assimilatory enzyme activities, CO2 fixation, photosynthesis, energy status, organic acid synthesis, release of protons or organic acids, and nodule O2 diffusion. Such studies have been performed in different legume species such as lupin, soybean, alfalfa (Medicago sativa), white clover (Trifolium repens), Medicago truncatula, pea (Pisum sativum), and common bean (Phaseolus vulgaris; Jakobsen, 1985; Israel, 1987; Sa and Israel, 1991; Ribet and Drevon, 1995a, 1995b; Al-Niemi et al., 1997; Vadez et al., 1997; Almeida et al., 2000; Tang et al., 2001, 2004; Hogh-Jensen et al., 2002; Olivera et al., 2004; Schulze and Drevon, 2005; Schulze et al., 2006; Le Roux et al., 2008).
Common bean is the world's most important grain legume for direct human consumption. P deficiency is widespread in the bean-producing regions of the Third World and is perhaps the factor that most limits N2 fixation on small farms. Bean genotypes differ in N2 fixation ability and P use efficiency under P deficiency. Considering the greater P need of nodulated legumes, P-tolerant cultivars that in addition partition a significant percentage of their P uptake to nodules will be a prerequisite for improved bean N2 fixation (Graham, 1981; Broughton et al., 2003; Graham et al., 2003; Tang et al., 2004).
Plant response to P deficiency and stress tolerance involves multiple genes and intricate regulatory mechanisms. In the case of common bean, two reports discuss gene expression analyses in the roots of P-deficient plants. Hernández et al. (2007) identified 126 P-responsive genes, through transcript profile analysis, and Tian et al. (2007) identified 240 P stress-induced genes, through the analysis of a suppressive subtractive cDNA library. In addition, using our platform for transcription factor (TF) profiling, based on quantitative reverse transcription (qRT)-PCR technology, we identified 17 TF genes that were differentially expressed in P-deficient roots of the common bean (Hernández et al., 2007). We demonstrated that the PvPHR1 (for P. vulgaris Phosphate Starvation Response 1) TF, which is induced in P-deficient bean roots, and the PvmiR399 microRNA both play essential roles in the P-starvation signaling of the common bean (Valdés-López et al., 2008).
Microarray and macroarray approaches enabled the identification of a large number of genes that are differentially expressed in legume nodules of M. truncatula, Lotus japonicus, soybean, and bean (Colebatch et al., 2002, 2004; El Yahyaoui et al., 2004; Kouchi et al., 2004; Lee et al., 2004; Asamizu et al., 2005; Ramírez et al., 2005; Starker et al., 2006; Brechenmacher et al., 2008). However, to our knowledge, there are no reports on the transcriptome of P-deficient legume nodules.
The understanding of the mechanisms for the adaptation to P deficiency of common bean plants under SNF conditions will become useful for future crop improvement. In an attempt to contribute to such efforts, we performed research focused on the identification of genes, gene networks, and signaling pathways that are relevant for P-deficient bean nodules. We undertook a macroarray-based transcript profiling screen of P-deficient bean nodules elicited by Rhizobium tropici. Furthermore, we used qRT-PCR to assess nodule gene expression of the whole set of proposed bean TFs (Hernández et al., 2007) in order to identify TFs that may regulate gene expression in P-stressed nodules. Third, we performed a nontargeted metabolite profiling of bean nodules using gas chromatography-mass spectrometry (GC-MS) and correlated metabolic changes to the orchestrated global changes of gene transcription as a response to P starvation.
In order to interpret the gene expression data, we used the MapMan (Thimm et al., 2004) and PathExpress (Goffard and Weiller, 2007b; Goffard et al., 2009) bioinformatics tools, which were adapted to the common bean. PathExpress allowed identification of the differentially expressed genes that were assigned EC numbers and thus associated to the relevant metabolic pathways operating in P-deficient bean nodules. Such metabolic pathways inferred by transcriptome analysis were additionally associated to some of the discovered P deficiency-responsive metabolites. The overall goal of our research was to identify candidate genes that may be useful to bean improvement and that will contribute to the understanding of the acclimation to P deficiency of the N2-fixing common bean.
RESULTS
Phenotypic Characterization
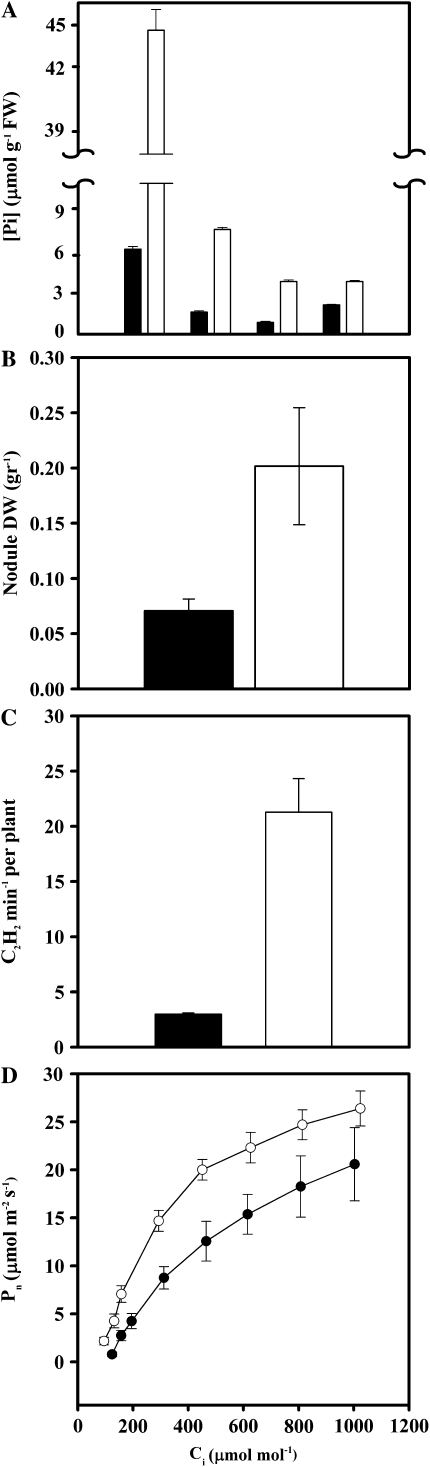
Germinated common bean seedlings were inoculated with R. tropici CIAT899 and then subjected to long-term P deficiency (–P) under an otherwise controlled environment using a 200-fold lower Pi concentration as compared with P-sufficient (+P) control plants. The performance of the plants was assessed 21 d post inoculation (dpi) and exposure to the –P condition. Control plants accumulated higher concentrations of soluble Pi in leaves (7-fold), stems (4-fold), and roots (4-fold) but only 1.5-fold in nodules as compared with –P plants (Fig. 1A). As expected, nodulation and SNF were affected in –P bean plants. These plants showed 3.5-fold less nodule mass (Fig. 1B) and 85% reduction in nitrogenase-specific activity (Fig. 1C).
Figure 1.
Effect of P deficiency on bean in symbiosis with R. tropici. A, Soluble Pi in different plant organs. FW, Fresh weight. B, Nodule dry weight (DW). C, Nitrogenase activity determined by acetylene reduction assay. D, Net photosynthesis (Pn) rate as a function of changing internal CO2 (Ci). Plants inoculated with R. tropici CIAT899 were grown for 21 d under P deficiency (black bars or circles) or P-sufficient conditions (white bars or circles). Values are means ± se of 12 determinations from three independent experiments with four replicates per experiment.
The content of photosynthetic pigments such as chlorophyll a and b and carotenes was similar in plants under –P and +P treatments (data not shown). However, P-deficient plants exhibited an inhibition of the net photosynthetic rate (Pn). Pn was 40% lower at ambient CO2 concentrations (350 μmol mL−1) and reflected the lower carboxylation efficiency under –P conditions (Fig. 1D). The maximum Pn was not significantly affected in –P plants, indicating that the Rubisco and ribulose 1,5-bisphosphate regeneration was maintained. The latter observation suggests that symbiotic P-deficient bean plants were capable of regulating photosynthetic activity.
Macroarray Analysis for Nodule Response to P Deficiency
Global gene expression in P-deficient bean nodules as compared with control P-sufficient nodules was determined by macroarray analyses. Two different macroarrays were prepared by spotting nylon filters with ESTs from the common bean –P root and mature nodule cDNA libraries (Ramírez et al., 2005; Graham et al., 2006). The root and nodule macroarrays included 2,212 and 1,786 unigene sets, respectively, as reported (Ramírez et al., 2005; Hernández et al., 2007).
Total RNA was isolated from plants grown under similar conditions as described for each treatment (−P or +P). Eight nylon filter root arrays and eight nodule arrays were hybridized with radiolabeled first-strand cDNA synthesized from four independent sources of total RNA. From the eight hybridizations, four replicates of each array and of each treatment, with high determination coefficients (r2 ≥ 0.8), were chosen for the analysis of differential gene expression. A total of 459 genes (tentative consensus sequences [TCs] or singletons) showed significant (P ≤ 0.05) differential expression in P-deficient nodules (Supplemental Tables S1 and S2).
In order to aid gene annotation, cDNAs were assigned to TCs (Dana Farber Cancer Institute [DFCI] Phaseolus vulgaris Gene Index [PhvGI], version 2.0). The annotation of all ESTs from the nodule and root cDNA library ESTs was updated by comparison (BLASTX; E value < 10−4) with the UniProt Knowledgebase (UniProtKB) database (release 14.1; UniProt Consortium, 2008; Supplemental Table S3). From the 7,129 total EST sequences, 5,102 ESTs had significant best matches to UniProtKB/Swiss-Prot, 621 ESTs had significant best matches to UniProtKB/trEMBL but not to Swiss-Prot, while 1,406 ESTs did not have significant matches to UniProtKB. A UniProt keyword was assigned to each EST. The biological process was the preferred keyword; ESTs were classified in 39 different biological processes (Supplemental Table S4). If this keyword was not available, other keywords such as the molecular function or the cellular component were assigned (Supplemental Table S4).
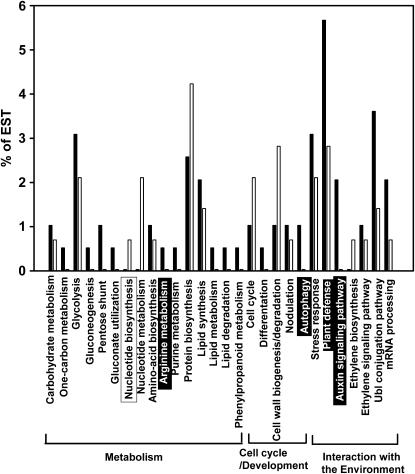
Supplemental Tables S1 and S2 show the genes that were induced (263) or repressed (196) 2-fold or more in P-deficient nodules. These genes were initially grouped in four main categories: metabolism, cell cycle and development, interaction with the environment, and unknown function. The latter includes genes with similarity to a hypothetical protein or DNA sequences with unknown function and those for which no BLAST hit was found. Figure 2 shows the more relevant biological processes that group the genes differentially expressed in P-deficient nodules.
Figure 2.
Distribution of selected bean ESTs into biological processes according to UniProtKB keywords (UniProt Consortium, 2008). Black bars, ESTs induced in –P nodules; white bars, ESTs repressed in –P nodules. The percentage represents the proportion of submitted ESTs that have been assigned in the corresponding category. The biological processes overrepresented in the set of induced or repressed ESTs, compared with the remaining ESTs (Supplemental Table S4), are highlighted with black or white boxes, respectively. Main functional categories that group the different biological processes are indicated.
The induced genes (Supplemental Table S1) were classified into the categories metabolism (30%), cell cycle and development (6%), interaction with the environment (34%), and unknown function (30%). The biological processes statistically overrepresented in the set of induced ESTs, compared with the remaining ESTs, were Arg metabolism, autophagy, auxin signaling pathway, and plant defense (Fig. 2; Supplemental Table S1). Several biological processes from the carbon (one-carbon metabolism, glycolysis, gluconeogenesis, pentose shunt, gluconate utilization), N (Arg and purine metabolism), and lipid (lipid synthesis, lipid metabolism, lipid degradation) metabolisms showed high proportions of induced ESTs (Fig. 2).
The most abundant category among the repressed genes (Supplemental Table S2) was interaction with the environment (41%), followed by metabolism (25%), unknown function (24%), and cell cycle and development (10%). “Nucleotide metabolism” was the only biological process that was statistically overrepresented in the set of repressed ESTs (Fig. 2; Supplemental Table S2). In contrast to the main induced biological processes, several processes from N metabolism (nucleotide metabolism and biosynthesis, protein biosynthesis) showed a high proportion of repressed ESTs, similar to processes like cell cycle and cell wall biosynthesis and degradation (Fig. 2).
Expression Analyses of Selected Genes by RT-PCR
Nine –P nodule-induced ESTs and nine repressed ESTs were randomly selected from Supplemental Tables S1 and S2 in order to confirm the macroarray expression data by semiquantitative RT-PCR (sRT-PCR). The selected genes corresponded to different functional categories and biological processes and showed high –P/+P expression ratios in macroarray analysis (≥4 or ≤−4). In addition, 15 genes assigned as enzymes that participate in significantly induced or repressed metabolic pathways (see below) were selected to confirm their macroarray expression data by real-time qRT-PCR. These included the gene (NOD_210_H10) annotated as malate dehydrogenase (EC 1.1.1.7), which was not detected as significantly induced through macroarray analyses but participated in the glycolysis/gluconeogenesis significantly induced pathway (see below). Primers and conditions for RT-PCR analyses are given in Supplemental Table S5.
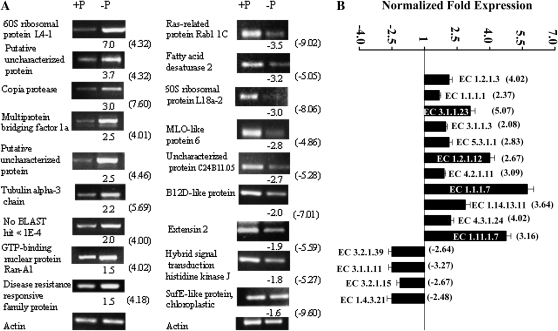
As shown in Figure 3, all of the genes that were tested for expression responses using sRT-PCR or qRT-PCR gave results that confirmed the expression results obtained with the macroarray experiment regarding the induction or repression of each gene in P-deficient nodules. However, there was a variation of –P/+P expression ratios for each tested gene when comparing the values obtained from macroarray with those from RT-PCR; in general, the values obtained from macroarray analysis were higher (Fig. 3). The latter may be related to the different sensitivity of the technologies used.
Figure 3.
Verification of macroarray results by sRT-PCR and qRT-PCR analyses. A, Selected genes identified as induced (left panel) or repressed (right panel) in P-deficient nodules were evaluated by sRT-PCR. The actin gene was included as a control for uniform RT-PCR conditions (bottom). The intensity of the bands was quantified densitometrically, and the –P/+P normalized expression ratios are shown below each gel image. B, Selected genes assigned to represent enzymes (with indicated EC numbers) induced or repressed in P-deficient nodules. Enzyme names corresponding to each EC number are indicated in Supplemental Tables S1 and S2 and Figures 6 and 7. Values represent the normalized –P/+P fold expression as the average of three biological replicates ± sd. For ratios lower than 1 (genes repressed in P deficiency), the inverse of the ratio was estimated and the sign was changed. The –P/+P expression ratios obtained from the macroarray analyses (Supplemental Tables S1 and S2) are shown in parentheses. The primer sequences and reaction conditions for sRT-PCR and qRT-PCR analyses are presented in Supplemental Table S5.
TF Transcript Profiling by qRT-PCR
We identified a set of 372 TF genes from common bean that were selected from DFCI PhvGI (version 1.0) and had been included into the reported qRT-PCR platform of TF expression profiling (Hernández et al., 2007). We used this platform to determine the differential expression of bean TF genes that might be involved in regulating the gene expression response of P-deficient nodules. Three biological replicates of –P- and +P-treated nodules were analyzed. In each qRT-PCR run, the phosphatase gene (TC3168), known to be induced in –P bean roots (Ramírez et al., 2005; Hernández et al., 2007), was included as a P-deficiency control of the –P response in bean nodules. This gene was highly induced in nodules and had an average expression ratio comparing –P with +P of 30.12 (P = 1E-6), as determined by qRT-PCR. Thus, the P-deficient status of the nodules under investigation was confirmed. Table I shows the 37 TF genes that were differentially expressed, 2-fold or more (P ≤ 0.05), in P-deficient nodules. These genes were classified into 17 different TF families according to the Arabidopsis (Arabidopsis thaliana) TF classification (Riechmann, 2002). Only one TF gene from the C3H-type 1(Zn) family was repressed; all others were induced. None of the differentially expressed TF genes detected by qRT-PCR analysis was detected by macroarray analysis, indicating the much higher sensitivity and accuracy of the qRT-PCR platform for TF expression profiles (Table I; Supplemental Tables S1 and S2). Most of the induced TF genes (10) belong to the C2C2(Zn) or the C2H2(Zn) family, and three belong to the MYB superfamily. The participation of MYB TFs in P-starvation signaling is known for Arabidopsis (Rubio et al., 2001; Todd et al., 2004). We have demonstrated earlier the respective role of a MYB TF in bean roots: PvPHR1 was reported to be relevant for the P-starvation signaling by Valdés-López et al. (2008). A TF from the ZIM/TIFY family, TC7761, was the only TF gene found to be induced both in nodules (Table I) and in roots (Hernández et al., 2007) of P-deficient bean plants.
Table I.
TF genes significantly expressed in nodules of P-deficient plants identified by real-time RT-PCR
| GenBank Accession No. (TC No.) | Annotation | BLASTX E Value | TF Family/Domain | Expression Ratio –P/+P | P |
|---|---|---|---|---|---|
| Induced in –P | |||||
| CV531158 | O23379 Putative zinc finger protein CONSTANS-LIKE 11 | 1.00 E-8 | C2C2(Zn) | 5.55 | 0.00130 |
| CB542189 | Q96502 Zinc finger protein CONSTANS-LIKE 2 | 1.00 E-8 | C2C2(Zn) | 2.71 | 0.04174 |
| TC3558 | Q96288 Salt tolerance protein | 1.00 E-76 | C2C2(Zn) | 2.33 | 0.00672 |
| CB540841 | Q9SSE5 Zinc finger protein CONSTANS-LIKE 9 | 3.00 E-7 | C2C2(Zn) | 2.23 | 0.01626 |
| BQ481766 (TC4839)a | Q96502 Zinc finger protein CONSTANS-LIKE 2 | 1.00 E-9 | C2C2(Zn) | 2.00 | 0.00360 |
| TC3525 | Q42430 Zinc finger protein 1 (WZF1) | 2.00 E-13 | C2H2(Zn) | 3.37 | 0.00122 |
| CB541538 | B9IEY1 Predicted protein | 6.00 E-40 | C2H2(Zn) | 3.31 | 0.01795 |
| TC4594 | Q42430 Zinc finger protein 1 (WZF1) | 4.00 E-149 | C2H2(Zn) | 2.57 | 0.03902 |
| CV542423 | Q39266 Zinc finger protein 7 | 4.00 E-07 | C2H2(Zn) | 2.25 | 0.01362 |
| TC6164 | Q8L6Y4 Polycomb group protein EMBRYONIC FLOWER 2 | 1.00 E-149 | C2H2(Zn) | 2.06 | 0.01031 |
| CV533267 | Q9FJ93 Dehydration-responsive element-binding protein 1D | 2.00 E-41 | AP2/EREBP | 4.70 | 0.01197 |
| BQ481785 | Q84QC2 Ethylene-responsive transcription factor ERF017 | 3.00 E-34 | AP2/EREBP | 2.71 | 0.05405 |
| CB540147 (TC6825)a | Q8LC30 Ethylene-responsive transcription factor RAP2-1 | 2.00 E-39 | AP2/EREBP | 2.55 | 0.05779 |
| TC6676 | O80337 Ethylene-responsive transcription factor 1A | 3.00 E-63 | AP2/EREBP | 2.15 | 0.02622 |
| TC3112 | Q9SAH7 Probable WRKY transcription factor 40 | 4.00 E-74 | WRKY(Zn) | 2.76 | 0.01113 |
| CX129652 | Q8S8P5 Probable WRKY transcription factor 33 | 3.00 E-52 | WRKY(Zn) | 2.50 | 0.03801 |
| TC3738 | Q9SUP6 Probable WRKY transcription factor 53 | 3.00 E-57 | WRKY(Zn) | 2.02 | 0.04625 |
| TC4946 | Q9LX82 Transcription factor MYB48 | 6.00 E-25 | MYB superfamily | 2.36 | 0.01668 |
| TC2999 | Q6ZNW5 UPF0580 protein C15orf58 | 6.00 E-40 | MYB superfamily | 2.32 | 0.01079 |
| DN153793 | Q9M2Y9 Transcription factor RAX3 | 1.00 E-20 | MYB superfamily | 2.04 | 0.05997 |
| CV536452 | Q9FIW5 Putative NAC domain-containing protein 94 | 8.00 E-27 | NAC | 2.34 | 0.03376 |
| CV534675 | Q5CD17 NAC domain-containing protein 77 | 3.00 E-30 | NAC | 2.26 | 0.03562 |
| CV530141 | B9RH27 Transcription factor, putative | 4.00 E-11 | NAC | 2.04 | 0.02866 |
| TC7761 | Q9LMA8 Protein TIFY 10A | 1.00 E-30 | ZIM/TIFY | 3.40 | 0.01489 |
| TC5396 | Q9LMA8 Protein TIFY 10A | 1.00 E-39 | ZIM/TIFY | 2.48 | 0.00604 |
| TC7287 | Q9FXG8 BEL1-like homeodomain protein 10 | 9.00 E-48 | Homeobox | 2.69 | 0.04828 |
| TC3707 | Q6YWR4 Homeobox-Leu zipper protein HOX16 | 4.00 E-24 | Homeobox | 2.18 | 0.00083 |
| CV538920 | P24068 Ocs element-binding factor 1 | 7.00 E-21 | bZIP | 2.27 | 0.00289 |
| TC3839 | Q69IL4 Transcription factor RF2a | 3.00 E-39 | bZIP | 2.21 | 0.02188 |
| BQ481439 | Q0PJI4 MYB transcription factor MYB138 | 1.00 E-52 | Homeodomain-like | 6.83 | 0.04473 |
| CV536165 | Q9SMX9 Squamosa promoter-binding-like protein 1 | 1.00 E-54 | SBP | 4.38 | 0.00422 |
| TC6556 | P13089 Auxin-induced protein AUX28 | 2.00 E-99 | AUX/IAA | 3.37 | 0.00049 |
| TC6822 | Q8VY21 Tubby-like F-box protein 3 | 1.00 E-153 | TUB | 2.86 | 0.03833 |
| TC5176 | Q69VG1 Chitin-inducible gibberellin-responsive protein 1 | 1.00 E-153 | GRAS | 2.58 | 0.02628 |
| CV529418 | O49403 Heat stress transcription factor A-4a | 2.00 E-27 | HSF | 2.13 | 0.01078 |
| CV529563 (TC3533)a | Q9CAA4 Transcription factor BIM2 | 8.00 E-50 | bHLH | 2.00 | 0.03141 |
| Repressed in –P | |||||
| CV530991 |
Q0D3J9 C3H53_ORYSJ Zinc finger CCCH domain-containing protein 53 |
2.00 E-12 |
C3H-type 1(Zn) |
−2.56 |
0.00581 |
Previous singletons (in PhvGI version 1.0) now correspond to the indicated TC according to PhvGI version 2.0.
Metabolome Analyses
Nontargeted metabolite profiling of bean roots using GC-MS was performed in order to assess the degree to which changes in plant gene expression in P-deficient bean nodules affect metabolism. The complete information of 81 covered mostly primary metabolites and nonidentified mass spectral metabolite tags (MSTs) detected in bean nodules when subjected to –P and +P treatments is provided as Supplemental Table S6. Thirty-nine of the identified metabolites and MSTs showed a response ratio higher than 1, indicating an increase in P-deficient nodules, while 31 showed a decrease in –P nodules. Eleven of the detected metabolites and MSTs were not affected by the nutrient stress (response ratio = 1; Supplemental Table S6).
Table II shows those metabolites and MSTs (45) included in significantly induced or repressed pathways (see below), those with –P/+P response ratios higher than 1.5-fold, and those with lower but significant (P ≤ 0.05) ratios. In agreement with previous analyses (Desbrosses et al., 2005; Hernández et al., 2007), the identified metabolites were mostly primary metabolites belonging to the following compound classes: amino acids, N compounds, organic acids, polyhydroxy acids, sugar phosphates, polyols, and sugars. Most of the carbon metabolites, such as organic and polyhydroxy acids, sugars, and polyols were increased significantly in P-stressed nodules, while most of the amino acids and other N compounds showed a decrease in P-stressed nodules. As expected, phosphates such as Fru-6-P, Glc-6-P, and glycerate-6-phosphate were also decreased in P-starved nodules (Table II).
Table II.
Selected metabolites identified by GC-MS in bean nodules from –P- and +P-treated plants
| Metabolite | RI, Expected | RI, sd | Response Ratio −P/+Pa |
|---|---|---|---|
| Amino acids | |||
| Gly | 1,304.5 | −0.17 | −1.4 |
| β-Ala | 1,424.7 | −0.12 | 3.5 |
| Ser | 1,252.9 | 0.09 | −1.2 |
| Asn | 1,665.8 | −0.02 | 2.0 |
| Thr | 1,290.9 | −0.02 | −2.0 |
| 4-Hyp | 1,518.0 | 0.21 | −1.4 |
| Gln | 1,767.6 | −0.05 | −1.5 |
| Leu | 1,151.0 | 0.52 | 1.6 |
| Lys | 1,847.3 | −0.02 | 1.2 |
| Phe | 1,553.1 | 0.15 | 2.4 |
| N compounds | |||
| Putrescine (agmatine)b | 1,737.2 | −0.24 | −1.4 |
| Pipecolic acid | 1,366.6 | −0.16 | −1.6 |
| Picolinic acid | 1,327.2 | 0.20 | 1.5 |
| Spermidine | 2,251.1 | −0.22 | −1.6 |
| Urea | 1,235.6 | 0.06 | −1.4 |
| Organic acids | |||
| Malonic acid | 1,195.0 | 0.30 | 1.4 |
| Tartaric acid | 1,625.9 | 0.03 | −1.7 |
| Malic acid | 1,477.3 | 0.11 | 1.5 |
| 2,4-Dihydroxybutanoic acid | 1,403.6 | −0.08 | −2.1 |
| cis-Aconitic acid | 1,740.5 | −0.02 | 1.6 |
| Citric acid | 1,804.6 | −0.04 | 1.2 |
| Vanillic acid | 1,761.6 | 0.10 | 1.7 |
| Polyhydroxy acids | |||
| Glyceric acid | 1,321.7 | −0.02 | −1.1 |
| Threonic acid | 1,546.4 | −0.04 | 1.4 |
| Galactonic acid-1,4-lactone | 1,877.4 | −0.31 | 1.3 |
| Galactaric acid | 2,032.5 | −0.22 | 1.5 |
| Galactonic acid | 1,984.7 | −0.29 | 1.8 |
| Gulonic acid | 1,951.4 | −0.21 | 2.0 |
| Phosphates | |||
| Glyceric acid-3-phosphate | 1,790.2 | 0.06 | −1.1 |
| Fru-6-P | 2,292.8 | −0.03 | −2.2 |
| Glu-6-P | 2,329.6 | −0.13 | −1.3 |
| Polyols | |||
| Threitol | 1,485.1 | −0.12 | 3.3 |
| Sugars | |||
| Fru | 1,856.2 | −0.24 | −1.1 |
| Man | 1,869.0 | −0.17 | 1.4 |
| Suc | 2,629.6 | −0.28 | 2.2 |
| α,α′-Trehalose | 2,730.1 | −0.26 | 1.6 |
| MSTscd | |||
| [516; 1H-Indole-2,3-dione, 1-(tert-butyldimethylsilyl)-5-isopropyl-, 3-(O-methyloxime)] | 1,691.2 | −0.04 | −2.4 |
| [771; α-d-Methylfructofuranoside (4TMS)] | 1,760.9 | −0.02 | 1.5 |
| [802; Methylcitric acid (4TMS)] | 1,909.57 | −0.06 | 1.2 |
| [965; Gluconic acid, 2,3,4,5,6-pentakis-O-(trimethylsilyl)-trimethylsilyl ester] | 2,002.9 | −0.27 | 1.6 |
| [834; 2-O-Glycerol-β-d-galactopyranoside (6TMS)] | 2,160.3 | −0.04 | −1.5 |
| [926; Galactosylglycerol (6TMS)] | 2,297.2 | −0.12 | 2.4 |
| [802; Gulose (5TMS)] | 2,424.3 | 0.26 | −1.9 |
| [533; Lactose methoxyamine (8TMS)] | 2,878.2 | −0.10 | 1.3 |
| [882; Melibiose (8TMS)] |
3,092.9 |
−0.27 |
2.6 |
The response ratio of average –P nodule response compared with average +P nodule response is listed (t test significance of P < 0.05 is indicated by boldface numbers for the response ratio). For ratios lower than 1, the inverse of the ratio was estimated and the sign was changed.
Represents the sum of two or more metabolites.
Reference substance not yet available.
MSTs are characterized by match factor and mass spectral hit.
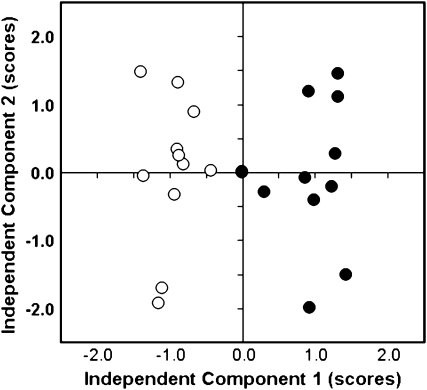
The quantitative data on the relative pool size changes of the metabolites listed in Supplemental Table S6 were subjected to independent component analysis (ICA). A major difference of the metabolic phenotype between P-deficient and P-sufficient nodules was revealed using an ICA scores plot (Fig. 4). This analysis of the metabolite response ratios of all observed metabolites in 12 samples from P-deficient nodules and 12 samples of P-sufficient nodules allowed unambiguous partitioning into two sample groups, showing the clear metabolic differentiation of –P-stressed individual plants from the P-sufficient metabolite phenotype (Fig. 4).
Figure 4.
ICA of the major metabolic variances in bean nodules determined by GC-MS-based metabolite profiling. Bean plants grown in P-sufficient (white circles) and P-deficient (black circles) conditions were used. Score analysis demonstrated the clear difference between the P-sufficient and P-deficient metabolic phenotypes.
Transcriptome and Metabolome Data Analyses
The data of differentially expressed genes from P-stressed nodules, generated in this work through macroarray analyses and TF gene profiling, were analyzed using the MapMan (Thimm et al., 2004) and PathExpress (Goffard and Weiller, 2007b; Goffard et al., 2009) software tools, which allow visualization and interpretation of the data in the context of known biological networks. For this task, both software tools were customized to the common bean as described (see “Materials and Methods”).
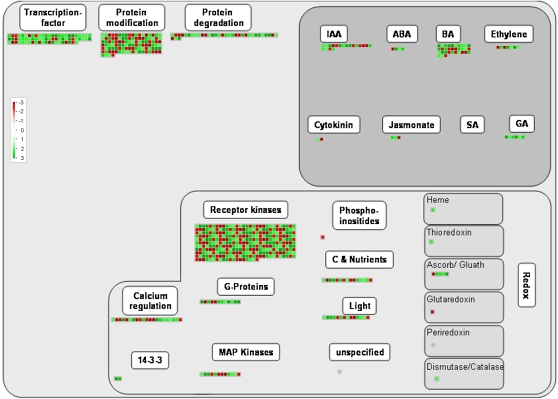
For MapMan data analyses, a recently created soybean mapping (S. Yang, unpublished data) was the basis for a common bean mapping file containing the differentially expressed genes resulting from the current macroarray and TF profiling approaches (Supplemental Table S7). After submitting the –P/+P expression ratios of the determined bean genes, different graphical representations were obtained for visual analysis from MapMan. To avoid an overlap with the PathExpress investigation, the MapMan analysis focused on the maps describing pathways other than the metabolic. Figure 5 shows the bean nodule MapMan graph representation of the regulation overview map. As was expected from our manual gene expression results, the majority of the genes assigned to the different categories in the regulation overview map were induced. Evident abundant categories, which included most of the induced regulatory genes, were TFs, receptor kinases, and protein degradation. In addition, several genes from the overrepresented induced biological processes, auxin signaling pathway and autophagy (Fig. 2), are included in the regulatory categories from Figure 5.
Figure 5.
MapMan regulation overview map showing differences in transcript levels between P-deficient and P-sufficient bean nodules. In the color scale, green represents higher gene expression and red represents lower gene expression in P-deficient nodules as compared with control (+P) nodules. The lists of normalized expression values are given in Table I and Supplemental Tables S1 and S2. The complete sets of genes submitted to MapMan analysis are given in Supplemental Table S7. IAA, Indole-3-acetic acid; ABA, abscisic acid; BA, brassinosteroid; SA, salicylic acid; MAP, mitogen-activated protein.
The input files for the PathExpress analysis comprised the list of genes that were differentially expressed in P-deficient bean nodules (Supplemental Tables S1 and S2). PathExpress uses the subset of submitted genes that can be assigned EC numbers and reports all metabolic networks that include these EC numbers as well as the enzymes in these networks that correspond to submitted identifiers. Table III shows the list of significant (P < 0.05) pathways or subpathways that were induced or repressed in P-stressed bean nodules. The enzymes assigned to the significantly induced or repressed pathways from Table III are highlighted in Supplemental Tables S1 and S2, respectively. Since PathExpress graphical representations of metabolic pathways contain two types of nodes, enzymes labeled with EC numbers and metabolites labeled with Kyoto Encyclopedia of Genes and Genomes (KEGG) identifiers (Kanehisa et al., 2004), we analyzed the graph representations to link increased or decreased metabolites, as shown in Table II, to the pathways revealed by gene expression analysis (Table III). Thus, we integrated both transcriptomic and metabolomic data with the known map of metabolic networks. However, such integration was limited to 13 metabolites detected in our analysis that were included in significant metabolic pathways (Tables II and III), while the rest of the detected metabolites (68) belong to other compound classes not included in these pathways (Supplemental Table S6). Figures 6 and 7 show graphical representations of selected induced and repressed pathways, respectively. These visualizations are based on the PathExpress output and include the significant P-responsive enzymes (Fig. 3; Supplemental Tables S1 and S2) and the respective metabolites (Table II), highlighted according to up-regulation (green) or down-regulation (red) or pool concentration. In addition, Figures 6 and 7 include the EC numbers of those enzymes from each pathway that are included in the Phaseolus vulgaris Gene Index and other metabolites from the pathways, albeit undetected in our analysis.
Table III.
Significant pathways or subpathways identified by PathExpress analysis
Pathways that are significantly associated with the list of submitted sequence identifiers with a P value threshold of 0.05.
| Pathway/Subpathway | No. of Enzymes Included in Bean EST Sequences | No. of Differentially Expressed Enzymes | P |
|---|---|---|---|
| Induced | |||
| Glycerolipid metabolism | 9 | 5 | 0.00222 |
| Fatty acid metabolism | 2 | 2 | 0.01580 |
| Glycolysis-gluconeogenesis | 19 | 7 | 0.00496 |
| Carbon fixation | 4 | 4 | 0.00023 |
| Isoflavonoid biosynthesis | 2 | 2 | 0.01580 |
| 3-Chloroacrylic acid degradation | 2 | 2 | 0.01580 |
| Metabolism of xenobiotics by cytochrome P450 | 2 | 2 | 0.01580 |
| Phe metabolism | 6 | 3 | 0.02900 |
| Selenoamino acid metabolism | 6 | 3 | 0.02900 |
| Bile acid biosynthesis | 3 | 2 | 0.04350 |
| Drug metabolism, cytochrome P450 | 3 | 2 | 0.04350 |
| Phenylpropanoid biosynthesis | 7 | 3 | 0.04630 |
| Repressed | |||
| Starch and Suc metabolism | 22 | 4 | 0.00730 |
| Pentose and glucuronate interconversions | 7 | 2 | 0.00730 |
| β-Ala metabolism | 7 | 3 | 0.03920 |
| Aminoacyl-tRNA biosynthesis | 9 | 3 | 0.03460 |
| Lys biosynthesis |
4 |
2 |
0.03920 |
Figure 6.
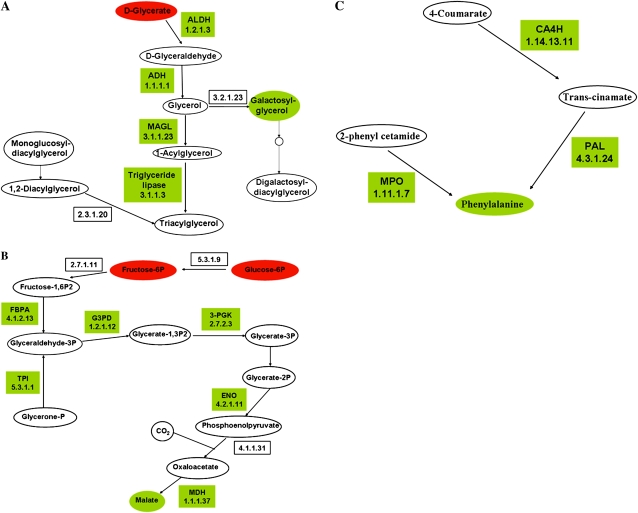
Representation of selected metabolic pathways induced in –P-stressed nodules. Drawings are based on PathExpress outputs (Goffard and Weiller, 2007b; Goffard et al., 2009) and contain two types of nodes: metabolites represented by ellipses, and enzymes (boxes) labeled with the enzyme name or abbreviation and/or the EC number. Enzymes coded by genes included in the PhvGI are shown. The color code indicates the induced gene expression of enzymes (Fig. 3; Supplemental Table S1) or increased metabolite pools (Table II; green) or respective decrease (red). Other metabolites shown in each pathway (white ellipses) were not detected in our analysis. A, Glycerolipid metabolism. ADH, Alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; MAGL, monoglyceride lipase. B, Glycolysis-gluconeogenesis-carbon fixation. ENO, Enolase 2; FBPA, Fru-bisphosphate aldolase; G3PD, glyceraldehyde-3-phosphate dehydrogenase; MDH, malate dehydrogenase; 3-PGK, phosphoglycerate kinase; TPI, triosephosphate isomerase. C, Phe metabolism. CA4H, trans-Cinnamate 4-monooxygenase; MPO, myeloperoxidase; PAL, Phe ammonia lyase.
Figure 7.
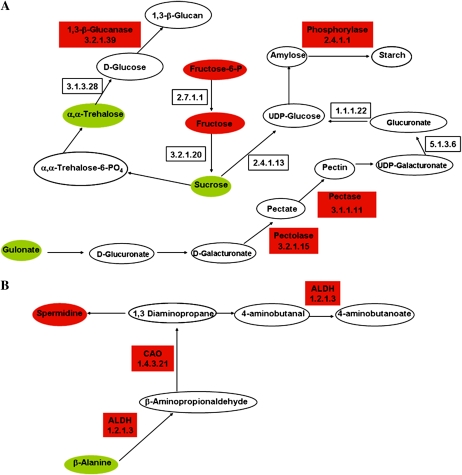
Representation of selected metabolic pathways repressed in –P-stressed nodules (Fig. 3; Tables II and III; Supplemental Table S2). For color-coded representation of the relative changes of metabolite pools and enzyme gene expression, compare with Figure 6. A, Starch and Suc metabolism. B, β-Ala metabolism. ALDH, Aldehyde dehydrogenase; CAO, primary amine oxidase.
The significantly induced pathway of glycerolipid metabolism is depicted in Figure 6A. This pathway includes four induced enzymes, slightly decreased glycerate, and increased galactosyl-glycerol content in P-deficient nodules. The gene products take part in the biosynthesis of galactolipids such as digalactosyl-diacylglycerol, which has been reported as an important component of plasma membranes from P-deficient plants (Andersson et al., 2003; Tjellström et al., 2008). A common plant response to P starvation is the modification of membrane lipid composition by increasing polar lipid production with low Pi content, such as galacto- and sulfo-lipids (Essigmann et al., 1998; Hartel et al., 2000; Andersson et al., 2003, 2005; Tjellström et al., 2008).
Symbiotic carbon supply is a key plant process of nodule metabolism that is facilitated mainly by a high production of organic acids that are offered to the bacteroid symbiont for enabling efficient N2 fixation. Figure 6B depicts the induced glycolysis/gluconeogenesis/carbon fixation pathway, which includes six induced enzymes, slightly decreased Glc-6-P, decreased Fru-6-P, and slightly increased malate contents in P-stressed nodules. This pathway is in agreement with what has been demonstrated for malate synthesis in legume nodules, involving mainly CO2 fixation through phosphoenolpyruvate carboxylase and malate dehydrogenase, rather than through the tricarboxylic acid cycle (Vance and Heichel, 1991).
Although the content of several amino acids was reduced in –P nodules, Phe was increased more than 2-fold (Table II) and the metabolic pathway for this amino acid was accordingly induced (Table III). Figure 6C shows the details of the Phe pathway with three –P-induced enzymes.
Figure 7 depicts two significantly repressed metabolic pathways. The starch and Suc pathway includes four down-regulated enzymes, indicating the repression of starch and pectin biosynthesis and a rechanneling of carbon toward synthesis of soluble sugars, such as the increasing Suc and α,α-trehalose pools, as well as increased gulonate in P-stressed nodules (Fig. 7A). The subpathway of β-Ala metabolism, depicted in Figure 7B, was significantly repressed in –P nodules. The repression of two enzymes from this pathway correlates with the increase in β-Ala and the decreased spermidine content observed (Table II). From the significantly repressed Lys biosynthesis pathway (Table III), Lys was found increased (Table II) and the enzyme diaminopimelate decarboxylase, which converts this amino acid to meso-2,6-diaminoheptane dioate, was down-regulated (Supplemental Table S2).
DISCUSSION
A low P level in the soil is an important constraint for bean production, especially in Latin America and Africa (Graham, 1981). In order to understand the molecular responses of bean for adaptation to P deficiency, we have analyzed the root transcriptomic and metabolomic profiles of P-stressed bean plants (Hernández et al., 2007). Considering that P deficiency is one of the most limiting factors for SNF (Andrew, 1978; Graham, 1981; Graham et al., 2003), in this work we undertook functional genomic approaches to advance the understanding of the adaptation of R. tropici-inoculated bean plants to P stress. Transcript and metabolic responses were analyzed from mature bean nodules of P-deficient plants with evident deleterious effects on nodulation and SNF (Fig. 1).
The P-deficient inoculated bean plants analyzed showed much lower soluble Pi concentration in different plant organs as compared with control (P-sufficient) plant organs (Fig. 1). However, Pi was higher in nodules than in stems or roots of P-stressed bean plants (Fig. 1). This observation is in agreement with previous reports indicating that, particularly under P deficiency, nodules are strong sinks for P and show higher P concentration in nodules than other organs (Sa and Israel, 1991; Al-Niemi et al., 1997; Vadez et al., 1997; Tang et al., 2001; Hogh-Jensen et al., 2002; Schulze et al., 2006). It has been reported that N2 fixation tolerance to P deficiency varies among different common bean genotypes (Vadez et al., 1997; Tang et al., 2001). The common bean cv Negro Jamapa 81 used in this work showed a dramatic decrease in nodule mass and in N2 fixation capacity, as determined by acetylene reduction assay nitrogenase activity per plant (Fig. 1). The latter is in agreement with numerous studies that have reported the negative effects of P starvation on N2-fixing capacity of legumes (Jakobsen, 1985; Israel, 1987; Sa and Israel, 1991; Al-Niemi et al., 1997; Vadez et al., 1997; Tang et al., 2001, 2004; Olivera et al., 2004; Le Roux et al., 2008). Israel (1987) has postulated that P has specific roles in nodule initiation, growth, and functioning in addition to its involvement in host plant growth processes.
Transcript expression patterns revealed by hybridization of nylon filter arrays spotted with ESTs from bean –P roots and mature nodule cDNA libraries (approximately 4,000 unigene set) resulted in 459 differentially expressed genes with 2-fold or more induction (59% genes) or repression (41% genes) in –P nodules (Supplemental Tables S1 and S2). Most of the significantly up-regulated genes derived from the P-stressed root cDNA library, while the significantly down-regulated genes derived from both libraries (Supplemental Tables S1 and S2). This may be related with a probable biased overrepresentation of genes expressed in this nutrient deficiency. However, RT-PCR of selected induced and repressed genes confirmed their differential expression (Fig. 3). Furthermore, several of the induced genes revealed by macroarray analysis (Supplemental Table S1) have been predicted by Graham et al. (2006) as bean candidate P stress-induced genes through clustering analysis across four legume species and Arabidopsis. The bioinformatically predicted (Graham et al., 2006) and experimentally up-regulated genes detected in this work include glyceraldehyde-3-phosphate dehydrogenase, alcohol dehydrogenase, oxidoreductases, wound-induced and pathogenesis-related proteins, Ser/Thr kinases, peroxidases, and MYB and WRKY transcription factors.
The transcript profile of P-deficient noncolonized bean roots revealed 126 differentially expressed genes (Hernández et al., 2007). A comparative analysis between the P-responsive genes from roots (Hernández et al., 2007) and from Rhizobium-elicited nodules (Supplemental Tables S1 and S2) showed that only 24 genes, out of 585, have a common response in the two organs. Supplemental Figure S1 shows the results of such comparative analysis, including a flower diagram and the list of common responsive genes. Twelve genes are up-regulated in roots and nodules, and only two genes are down-regulated in both organs (Supplemental Fig. S1). The rest (10 genes) are differentially regulated in roots as compared with nodules (Supplemental Fig. S1). The main functional categories of the genes up-regulated in both –P bean roots and nodules include proteins from regulation/signal transduction processes (i.e. steroid-binding protein, translationally controlled tumor protein, RNA-binding protein), a pathogenesis-related protein, and proteins related to carbon/sugar metabolism or sensing (i.e. monosaccharide-sensing protein, aldehyde dehydrogenase; Supplemental Fig. S1). Specifically, we found that the gene coding for S-adenosylmethionine synthase 2 (TC2965) is up-regulated, while the gene coding for S-adenosylmethionine decarboxylase proenzyme (TC7398) is down-regulated, in both roots and nodules (Supplemental Fig. S1), pointing to a relevant role of S-adenosylmethionine or polyamine metabolism in P-deficiency response of common bean. The very small proportion of common P-responsive genes in bean roots and nodules suggests a rather different response of each organ to the same nutrient stress. From our comparative analysis of transcript and metabolite profiles, we can conclude that the main response of P-deficient roots is addressed to maintain P homeostasis and root architecture modification, while responses of P-stressed nodules are mainly oriented to maintain adequate carbon/N flux between the symbionts and to avoid oxidative stress.
Nontargeted metabolite analysis, based on GC-MS technology, led to the identification of 81 metabolites and MSTs from bean nodules (Supplemental Table S6). Some of the detected metabolites were increased in –P nodules, some were decreased, and some metabolite pools did not change in sufficient versus deficient conditions (response ratio –P/+P = 1; Supplemental Table S6). ICA analysis from the identified metabolites indicated major differences among phenotypes of P-deficient and P-sufficient nodules (Fig. 4). The PathExpress software tool (Goffard and Weiller, 2007b; Goffard et al., 2009) was used in an attempt to provide comprehensive and integrative analyses of the transcript and metabolic responses found in P-stressed bean nodules. We identified relevant metabolic pathways associated both with enzymes coded by a subset of induced or repressed nodule genes and with responsive nodule metabolites (Fig. 3; Tables II and III). From the detected metabolites, 13 responsive metabolites could be associated with significantly induced or repressed pathways (Figs. 6 and 7); the rest of the metabolites from these pathways were not detected in our analysis.
Our integrated analyses indicated that the reduction of SNF in P-stressed bean plants led to a reduction of general N metabolism. A decreased –P/+P response ratio was observed in several N metabolites, including the N compounds spermidine, putrescine, and urea, and most of the detected amino acids (Table II). The latter correlates with the diminished expression of three aminoacyl-tRNA enzymes and significant repression of this biosynthesis pathway (Table III; Supplemental Table S2). In addition, the nucleotide metabolism was overrepresented among the repressed biological processes of –P nodules (Fig. 2). These findings contrast with the metabolic response of P-stressed bean noncolonized roots, where a significant increase of amino acid concentration was reported (Hernández et al., 2007). Morcuende et al. (2007) also reported a higher amino acid concentration for P-deprived Arabidopsis seedling cultures, as compared with full-nutrition control cultures. Such a contrasting response supports the observations of particularly high sensitivity of inoculated legumes, depending solely on fixed N2, to environmental limitations such as P starvation, which result in diminished nodulation and SNF, as compared with nonsymbiotic plants, which may have N sufficiency in P-deficient soils.
P deficiency in plants alter carbon metabolism in shoot; higher levels of carbon are allocated to the root and thereby increase the root-shoot biomass ratio and alter the root morphology. Some P-starved plant species accumulate sugars in the root and reduce photosynthesis, because sugars exert metabolite feedback regulation, allowing changes in gene expression and excreting organic acids to the rhizosphere as responses for adaptation to stress (for review, see Vance et al., 2003; Hermans et al., 2006). Roots from P-deficient common bean plants showed a decreased concentration of organic acids, which was interpreted as resulting from their exudation to the rhizosphere. The same roots showed accumulation of sugars. This enhanced carbohydrate allocation can be interpreted as the root demand of photosynthate under conditions of decreased net photosynthesis (Hernández et al., 2007). Legume nodules are strong carbon sinks; photosynthate is highly required for symbiotic N2 fixation and for assimilation of fixed N into amino acids. Carbon and N metabolisms and their interaction/regulation are key processes of nodule function, and the regulation of the gene expression in response to the C:N status has been widely investigated for several years. Data from this work show that P-stressed bean nodules fixed N2, albeit at a reduced level (Fig. 1). Therefore, we assume that photosynthate is still demanded by –P bean nodules, resulting in low sugar accumulation and high organic acid accumulation (Table II) and maintenance of net photosynthesis (Fig. 1). This –P nodule phenotype contrasts with the reported phenotype of –P bean roots (Hernández et al., 2007). Only Suc, Man, and α,α-trehalose showed a modest increase in the –P/+P response ratio in bean nodules (Table II), while we found six sugars increased in bean roots (Hernández et al., 2007) and several sugars were found accumulated in Arabidopsis under P stress (Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007). The accumulation of α,α-trehalose in bean nodules might also be related to its role as an osmoprotector or to its function as a signal molecule activating stress tolerance pathways in plants (Paul, 2007). Although some P-stressed plant species accumulate starch in roots (Hermans et al., 2006; Morcuende et al., 2007), we observed that several enzymes from the starch and Suc metabolic pathways were repressed in –P bean nodules and did not detect accumulation of starch or other carbon polymers (Figs. 3 and 7), suggesting that in stressed nodules, sugars are channeled into glycolysis and organic acid synthesis rather than toward carbon polymer synthesis.
Photosynthate provided to nodules as Suc is metabolized to supply respiratory substrates, mainly malate, to the bacteroids and to provide carbon skeletons for the incorporation of fixed N to amino acids (Vance and Heichel, 1991). Our data show that the glycolysis/carbon-fixation pathway is significantly induced in P-stressed nodules (Fig. 6). This pathway includes the sequential action of phosphoenolpyruvate carboxylase and malate dehydrogenase, resulting in malate synthesis; this and other organic acids showed an increased –P/+P response ratio (Table II). The point of divergence of glycolysis at phosphoenolpyruvate serves to circumvent the conventional adenylate-requiring pyruvate kinase and has been interpreted as an adaptive response to P stress for roots and nodules of several plant species (Olivera et al., 2004; Misson et al., 2005; Hermans et al., 2006; Morcuende et al., 2007; Müller et al., 2007; Le Roux et al., 2008). A potential drawback of this branch point would be the competition of organic acids for the tricarboxylic acid cycle and for amino acid synthesis; accordingly, we observed low amino acid concentrations in –P nodules (Table II). Le Roux et al. (2008) reported that excessive malate accumulation in P-deficient lupin nodules may inhibit N2 fixation and N assimilation, an interpretation that might also hold true for bean P-deficient nodules.
Under P deficiency conditions, plants can remobilize P from internal resources, such as nucleic acids and phospholipids. In this regard, the induction of genes involved in the membrane-phospholipid degradation has been reported in different plant species (Hartel et al., 2000). Some of these genes participate in the galacto- and sulfo-lipid synthesis, which, under P deficiency, are the principal membrane components (Andersson et al., 2003; Tjellström et al., 2008). In Arabidopsis, lipid composition is more sensitive to P deficiency in leaves than in roots (Misson et al., 2005; Morcuende et al., 2007). We detected that monoglyceride lipase and triglyceride lipase genes, involved in galactolipid synthesis, were induced in Pi-deficient root nodules (Figs. 3 and 6; Supplemental Table S1). Although we do not have information about nodule lipid composition under –P conditions, there is evidence indicating that diacylglyceryl, N,N,N-trimethylhomoserine is the principal component of the bacteroid membranes from bean nodules under P deficiency (C. Sohlenkamp, personal communication).
Plant responses to abiotic stress are regulated at different levels, transcriptional and posttranscriptional, with both routes involving intricate signaling pathways. Our bioinformatic analysis based on the MapMan software tool (Thimm et al., 2004) revealed several cellular signaling and regulatory processes that involve a number of bean nodule P-deficient response genes (Fig. 5). Most of these genes, induced in –P nodules, included receptor and mitogen-activated protein kinases, genes involved in protein modification/degradation, in calcium regulation, and in phytohormone regulation, as well as TF genes (Fig. 5). Similar types of regulatory genes are induced in P-stressed Arabidopsis (Wu et al., 2003; Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007).
In this work, we found that 37 of the 372 identified bean TF genes (Hernández et al., 2007) were differentially expressed, and only one was repressed, in P-deficient nodules, as revealed by the TF expression platform based on qRT-PCR (Table I). TFs are master regulators of gene expression. In P deficiency, around 100 TFs were differentially expressed in Arabidopsis plants, while in bean roots, only 17 of 372 analyzed TFs showed differential expression (Wu et al., 2003; Misson et al., 2005; Hernández et al., 2007). The P-deficient root-responsive TFs belong to different gene families, comprising MYB, SCARECROW, AP2, F-box, HOMEOBOX, WRKY, NAC, ERF/AP2, NAM, and C2H2 Zinc-finger (Wu et al., 2003; Misson et al., 2005; Hernández et al., 2007; Müller et al., 2007). Some of the bean nodule P-responsive TFs have been implicated in specific responses to P deficiency in other plant species (i.e. members of the WRKY and C2H2 ZFP TF families) that are involved in the root architecture modification and in the regulation of some –P-responsive genes (Devaiah et al., 2007a, 2007b). Also, several TFs that respond in –P conditions in different species and organs, including bean roots and nodules, are additionally implicated in other stresses, such as drought (NAC, AP2/EREBP), pathogenesis (WRKY, TIFY), and salinity (C2C2 ZFP). These data revealed the cross talk of different signaling pathways in the adaptation to P deficiency.
Our data showed the induction of members of the AP2/EREBP and TIFY TF families in P-stressed bean nodules (Table I). The role of these TFs in legumes might be related to root and nodule developmental processes, since AP2/EREBP and TIFY TFs have been implicated in ethylene and jasmonic acid phytohormone signaling pathways, respectively (Kizis et al., 2001; Chini et al., 2007; Thines et al., 2007), and AP2/EREBP has been implicated in the root and nodule development in L. japonicus (Asamizu et al., 2008). There are no reports on the involvement of TIFY TFs in P-deficiency plant response. We are using reverse genetic approaches to investigate the function of a TIFY TF in the response of bean roots and nodules to P deficiency; preliminary results show that modification of TIFY gene expression affects the nodulation of bean plants (G. Hernández, unpublished data).
This work presents integrative analyses of transcript and metabolic expression data from stressed bean nodules in an attempt to provide important insight into the P-starvation response. However, the integration of transcriptomics with metabolomics, proteomics, and enzyme biochemistry will be needed to achieve a thorough understanding of the intricate mechanisms by which plant metabolism adapts to nutritional P deficiency. Our results provide an abundance of candidate regulatory genes and candidate metabolic pathways that are postulated to play important roles in the adaptation of symbiotic bean plants to P deficiency and that may be used for marker-assisted selection of P-efficient bean genotypes. To make relevant contributions to develop better N2-fixing bean genotypes, it is imperative to consider the improvement in both N use and P use. Information generated here combined with future studies, including direct and reverse genetic analyses, might lead to the long elusive goal of improving N2 fixation in agronomically important grain legumes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The common bean (Phaseolus vulgaris) Mesoamerican cv Negro Jamapa 81 was used in this study. Plants were grown during spring in controlled-environment greenhouses (26°C–28°C, 16-h photoperiod) at the Centro de Ciencias Genómicas/Universidad Nacional Autónoma de México (Cuernavaca, México) and the Max Planck Institute of Plant Molecular Physiology (Golm, Germany) or in growth chambers at the University of Minnesota (St. Paul). Surface-sterilized seeds were germinated at 25°C over sterile, wet filter paper. Three days postimbibition, seeds were sown in pots with vermiculite or coarse quartz sand and inoculated with Rhizobium tropici CIAT899 as reported (Ramírez et al., 2005). Pots were watered 3 d per week with Summerfield plant nutrient solution without N (Summerfield et al., 1977). For –P conditions, K2HPO4 concentration of the plant nutrient solution was reduced from 1 mm to 5 μm. In –P conditions, cotyledons from each plant were cut 5 d after planting. Plants were grown for 21 dpi before harvesting. Nodules for RNA isolation were harvested directly into liquid N and stored at –80°C.
Soluble Pi Concentration, Nitrogenase Activity, and Photosynthesis
Soluble Pi content was determined at 21 dpi in different organs of plants grown in –P or +P conditions as reported (Taussky and Shorr, 1953; Hernández et al., 2007). Nitrogenase activity was determined in detached, 21-dpi nodulated roots by the acetylene reduction assay. The relationships between CO2 assimilation rate (net photosynthetic rate) and increasing internal CO2, stomatal conductance, and resistance were determined using a portable photosynthesis system (LI-6200 Primer; LI-COR) in –P- versus +P-treated plants as reported (Hernández et al., 2007). Each value represents the average of 12 determinations from three independent experiments with plants grown in similar conditions and with four replicate assays from each treatment (–P or +P) per experiment.
EST Sequencing and Annotation
Because the macroarrays used in this study were spotted prior to sequencing, 82 of the spotted clones had poor-quality sequence and were not included in sequence-based analyses (Ramírez et al., 2005; DFCI PhvGI) or submitted to GenBank. In order to include these clones in our analyses, the clones were resequenced. DNA sequencing was performed at the Advanced Genetic Analysis Center (University of Minnesota). The new sequences were submitted to GenBank (accession nos. GO355314–GO355395).
The annotation of all EST sequences from the nodule and P-deficient root common bean cDNA libraries (DFCI PhvGI), including the newly sequenced ESTs (7,129 sequences), was updated by comparing with proteins from the UniProtKB database (http://www.uniprot.org, release 14.1; UniProt Consortium, 2008) using BLASTX. The best match, with a threshold E value of 1.00E-4, was selected and UniProtKB keywords were extracted; both were assigned to each EST (Supplemental Table S3). The sequences described in Supplemental Tables S1 and S2 were cross-referenced with the DFCI PhvGI (version 2.0) to find the corresponding TCs or singletons.
Nylon Filter Arrays and Hybridization
The preparation of cDNA libraries from P-deficient roots and from mature nodules from Negro Jamapa 81 bean plants and the sequences of ESTs have been reported (Ramírez et al., 2005; Graham et al., 2006). Two different macroarrays, with the ESTs from each library (root macroarray and nodule macroarray), were prepared as reported (Ramírez et al., 2005; Hernández et al., 2007).
Total RNA was isolated from 0.5 g of mature (21-dpi) nodules from inoculated bean plants grown under similar –P or +P conditions in four independent experiments. Synthesis of radiolabeled cDNA probes from 30 μg of total RNA and hybridization and washing conditions of nylon filters were as reported (Ramírez et al., 2005). Eight independent nylon filter root macroarrays and eight independent nodule macroarrays were hybridized with cDNA from each treatment: –P nodules and +P nodules.
Hybridized filters were exposed to phosphor screens for 5 d for root macroarray and for 2 d for nodule macroarray, and the fluorescent intensity of each spot was quantified as reported (Ramírez et al., 2005). The signal intensity of each spot was determined automatically using the software Array-Pro Analyzer (Media Cybernetics). To work with highly reproducible experiments, linear regression analysis was performed for each pair of membrane replicas; only those replicas for which the linear model could explain at least 80% of the variation (r2 ≥ 0.8) were considered. This process yielded four well-correlated replicas for each macroarray (root or nodule macroarray) and for each treatment (Supplemental Table S8). The housekeeping genes ubiquitin-conjugating enzyme (TC8137) and ubiquitin (TC5422) served as internal normalization controls for calculating expression ratios between the treatments from the root macroarray and the nodule macroarray, respectively. Each TC included more than one EST spotted in each array; the chosen housekeeping genes showed constant intensity values for all of the ESTs from each TC. The average intensity value from the ESTs of each TC was used for normalization for each macroarray. Student's t test for paired observations was applied to determine whether genes showed significant differential expression values (P value cutoff at 0.05) from each treatment. sRT-PCR and qRT-PCR approaches were used to verify macroarray expression data. Total RNA for RT-PCR was isolated from 0.5 g of frozen 21-dpi nodules. Quantification of transcripts by sRT-PCR was performed using two-step RT-PCR following the manufacturer's directions (Ambion) using a polythymine deoxynucleotide (dT) primer. Amplified sRT-PCR products were resolved on 2% (w/v) agarose gels in Tris-acetate-EDTA buffer. Amplification of the actin gene was used as a control for uniform PCR conditions. The intensity of the bands from sRT-PCR amplification was quantified by densitometry using ImageQuant 5.2 software (Molecular Dynamics), and normalized –P/+P expression ratios were obtained.
Quantification of transcripts by qRT-PCR was done by the one-step assay using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad). Assays were done in 25 μL of reaction volume, which contained 12.5 μL of 2× Master Mix, 100 nm forward primer, 100 nm reverse primer, 100 ng of RNA template, and 0.5 mL of iScript reverse transcriptase for one-step RT-PCR. DNase-RNase-free water was used to adjust the volume to 25 μL. Real-time one-step RT-PCR was performed in a 96-well format using the iQ5 Real-Time PCR Detection System and iQ5 Optical System Software (Bio-Rad). The thermal cycler settings for real-time one-step RT-PCR were as follows: 10 min at 50°C (cDNA synthesis), 5 min at 95°C (iScript reverse transcriptase inactivation), followed by 40 cycles for PCR cycling and detection of 30 s at 59.5°C. Each real-time one-step RT-PCR assay had a melt curve analysis consisting of 80 cycles of 1 min at 95°C, 1 min at 55°C, and 10 s at 55°C, increasing each by 0.5°C per cycle. For each reaction, a product between 100 and 280 bp could be visualized on an agarose gel. Each assay included at least two no-template controls, in which RNA was substituted by DNase-RNase-free water; no amplification was obtained for no-template controls. Quantification was based on a cycle threshold value, with the expression level of each gene in –P nodules as compared with +P nodules normalized by the ubiquitin gene calculated. The sequences of oligonucleotide primers and conditions used in sRT-PCR and qRT-PCR are shown in Supplemental Table S5.
TF profiling, based on real-time qRT-PCR, was performed at the Max Planck Institute of Molecular Plant Physiology to determine nodule differential expression of TF genes. The identification of a set of 372 bean TF genes, and the design and synthesis of RT-PCR primers for each gene, have been reported (Hernández et al., 2007). Total RNA for qRT-PCR was isolated from 200 mg of frozen nodules as reported (Hernández et al., 2007). Three biological replicas were isolated for each treatment (–P and +P nodules), extracting RNA from different sets of plants grown in similar conditions. RNA concentration was measured in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and 10 μg of total RNA was used for qRT-PCR analysis. Genomic DNA degradation, cDNA synthesis, and quality verification for qRT-PCR were done as reported (Hernández et al., 2007; Supplemental Table S9). Quantitative determinations of relative transcript levels of TF genes using RT-PCR were carried out according to Czechowski et al. (2004) and Hernández et al. (2007). The bean phosphatase gene (TC3168) was included as a marker for P deficiency in every qRT-PCR run, using the reported primers (Hernández et al., 2007). TF expression was normalized to that of UBC9, which was the more constant of the four housekeeping genes included in each PCR run. –P/+P average expression ratios were calculated as reported (Hernández et al., 2007). Student's t test was performed with a P value cutoff of 0.05. Data from statistically differentially expressed ESTs with a –P/+P expression ratio of 2 or more were analyzed as mentioned below.
Plant Metabolite Extraction
Plant metabolite extraction of nodule samples from –P- and +P-treated bean plants and GC-MS metabolite profiling were done as reported previously (Colebatch et al., 2004; Desbrosses et al., 2005; Hernández et al., 2007). Twelve replicate samples for each condition, namely nodules from plants grown under +P and –P conditions, were harvested at 21 dpi from pods, rinsed with tap water, dried on filter paper, and shock frozen in liquid N. Frozen samples of 35 to 70 mg fresh weight were ground by mortar and pestle under liquid N in order to keep samples metabolically deactivated. Frozen powder was extracted with hot methanol/CHCl3, and the fraction of polar metabolites was prepared by liquid partitioning into water. Further processing was as described by Desbrosses et al. (2005).
GC-Time of Flight-MS Metabolite Profiling
GC-time of flight (TOF)-MS profiling was performed using a FactorFour VF-5ms capillary column (30 m length, 0.25 mm i.d., 0.25 μm film thickness) with a 10 m EZ-guard precolumn (Varian) and an Agilent 6890N gas chromatograph with splitless injection and electronic pressure control mounted to a Pegasus III TOF mass spectrometer (LECO Instrumente). Details of the GC-TOF-MS adaptation of the original profiling method (Desbrosses et al., 2005) are described by Wagner et al. (2003) and Erban et al. (2006). Metabolites were quantified as relative changes of pool sizes after mass spectral deconvolution (ChromaTOF software version 1.00, Pegasus driver 1.61; LECO) of at least three mass fragments representing each analyte. Peak height representing arbitrary mass spectral ion currents of each mass fragment was normalized using the amount of the sample fresh weight, and ribitol was added as an internal standardization to correct for volume variations. Normalized responses (g−1 fresh weight) and response ratios were calculated as described (Colebatch et al., 2004; Desbrosses et al., 2005).
Identification of Metabolites within GC-MS Metabolite Profiles
Metabolites were identified using the NIST05 mass spectral search and comparison software (National Institute of Standards and Technology; http://www.nist.gov/srd/mslist.htm) and the mass spectral and retention time index (RI) collection (Schauer et al., 2005) of the Golm Metabolome Database (GMD; Kopka et al., 2005). Mass spectral matching was manually supervised, and matches were accepted with thresholds of match > 650 (with maximum match equal to 1,000) and RI deviation < 1.0% (for details, see Table II; Supplemental Table S6). Information on the polar metabolites, using the corresponding mass spectral identifiers, can be found at http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_smq.html Metabolites are characterized by Chemical Abstracts System identifiers and compound codes issued by KEGG (Kanehisa et al., 2004). Metabolites were identified by standard substances or by as yet unidentified MSTs of GMD. The term MST is used for repeatedly occurring but nonidentified compounds, which can be recognized by mass spectrum and RI as defined earlier (Colebatch et al., 2004; Desbrosses et al., 2005). MSTs are tentatively characterized and named by best mass spectral match to compounds identified by NIST05 or GMD using match value and hit name (Table II; Supplemental Table S6). The response ratio –P/+P for each metabolite/MST was calculated by dividing the average metabolite concentration from 12 nodule samples of P-deficient plants by the average metabolite concentration from 12 nodule samples from roots of control plants (Table II; Supplemental Table S6).
ICA and Statistical Analysis
ICA (Scholz et al., 2004) was applied to metabolite profiles (as compiled in “Supplemental Data”). Data were normalized by calculation of response ratios using the median of each metabolite as the denominator and subsequently subjected to logarithmic transformation (log10). Missing value substitution was as described earlier (Scholz et al., 2005). Statistical testing was performed using Student's t test. Logarithmic transformation of response ratios was applied to better approximate the Gaussian normal distribution of metabolite profiling data required for statistical analyses.
Data Analyses
Three bioinformatics-based approaches were used for analyses aimed to interpret the biological significance of gene expression data in combination with metabolome data.
First, we aimed to detect whether a certain category, as defined by the UniProt keywords, was statistically overrepresented in the differentially expressed sets of ESTs (induced or repressed in –P) compared with the rest of the ESTs. For this, the P value for all UniProt keywords was calculated using the hypergeometric distribution, as described in GeneBins (Goffard and Weiller, 2007a; Supplemental Table S4).
A second approach for expression data analysis was based on MapMan software version 2.2.0 (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/). In order to extend MapMan to common bean, a soybean (Glycine max) mapping developed by S. Yang (University of Minnesota; unpublished data) was uploaded to MapMan. Soybean genes homologous to common bean differentially expressed genes were manually identified by BLASTN comparisons with the soybean consensus sequences (http://www.affymetrix.com/products_services/arrays/specific/soybean.affx), and the Affymetrix-Gm identifier for each homologous gene was retrieved. To verify if the putative bean and soybean homologous genes indeed had the same gene annotation, each retrieved Affymetrix-Gm identifier was submitted to the soybase Affymetrix-Soybean Genome Array Annotation Version 2 Page (http://www.soybase.org/AffyChip/), and only the genes with similar annotation in bean and soybean were considered for MapMan submission. The expression ratio –P/+P of the induced or repressed bean genes, expressed in log2, in combination with the list of Affymetrix-Gm identifiers were used to visualized the common bean gene expression data. Supplemental Table S7 shows the complete list of bean genes submitted to MapMan with their homologous soybean gene identifiers and the expression ratios.
The third type of analysis used the PathExpress Web-based tool (Goffard and Weiller, 2007b; Goffard et al., 2009) in order to identify the most relevant metabolic pathways associated with the subsets of differentially expressed genes. PathExpress was extended to bean. The data used to build the bean metabolic network were derived from the current release of the KEGG LIGAND database (release 42.0; Kanehisa et al., 2004). If the best match in UniProtKB for all ESTs from the bean nodule and root cDNA libraries had been annotated as enzymes, each EST was assigned to the corresponding EC number. The two sets of ESTs corresponding to nodule –P-induced or -repressed genes were separately submitted to PathExpress and were compared with the list of all bean enzymes involved in the annotated pathways. The results allowed detection of those ESTs associated with metabolic pathways or subpathways that were statistically overrepresented (P ≤ 0.05) in the differentially expressed sets of ESTs. The graphs of significant metabolic pathways generated by PathExpress were manually checked in order to identify induced or repressed nodule metabolites participating in those pathways.
Sequence data for this article can be found in the GenBank/EMBL data libraries under accession numbers GO355314 to GO355395.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Differentially expressed genes common to roots and nodules from P-deficient bean plants.
Supplemental Table S1. Genes induced in nodules of P-deficient plants identified by macroarray analysis.
Supplemental Table S2. Genes repressed in nodules of P-deficient plants identified by macroarray analysis.
Supplemental Table S3. Annotation of ESTs from the root and nodule cDNA libraries (DFCI PhvGI) from common bean.
Supplemental Table S4. Total ESTs from bean nodule and root cDNA libraries and ESTs from differentially expressed sets assigned to a certain UniProtKB keyword.
Supplemental Table S5. Primers and conditions used for sRT-PCR and qRT-PCR.
Supplemental Table S6. Complete metabolic profile response from common bean roots.
Supplemental Table S7. Soybean genes homologous to differentially expressed common bean ESTs used for MapMan analysis.
Supplemental Table S8. Nodule transcript levels of all of the genes in the common bean root and nodule macoarrays.
Supplemental Table S9. Nodule transcript levels of all common bean TF genes determined by qRT-PCR.
Supplementary Material
Acknowledgments
We are grateful to Victor M. Bustos for plant maintenance. We acknowledge the advice and help of Mesfin Tesfaye, Michelle A. Graham, Tomasz Czechowski, Armin Schlereth, and Maren Wandrey at initial stages of this work.
This work was supported by the Dirección General de Asuntos del Personal Académico/Universidad Nacional Autónoma de México (grant no. PAPIIT: IN211607 and sabbatical fellowship to G.H.), by the U.S. Department of Agriculture, Agricultural Research Service (grant no. USDA–FAS MX161 to the University of Minnesota), by the German Academic Exchange Service (research stay fellowship to G.H.), and by the Consejo Nacional de Ciencia y Tecnología, México (studentship no. 200048 to O.V.-L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Georgina Hernández (gina@ccg.unam.mx).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Almeida JPF, Hartwig UA, Frehner M, Nösberger J, Lüscher A (2000) Evidence that P deficiency induces N feedback regulation of symbiotic N2 fixation in white clover (Trifolium repens L.). J Exp Bot 51 1289–1297 [PubMed] [Google Scholar]
- Al-Niemi TS, Kahn ML, McDermott TR (1997) P metabolism in the bean-Rhizobium tropici symbiosis. Plant Physiol 113 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS (2005) Phosphate-limited oat: the plasma membrane and the tonoplast as major targets for phospholipid to glycerolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylclygerol. FEBS Lett 537 128–132 [DOI] [PubMed] [Google Scholar]
- Andrew CS (1978) Nutritional restraints on legume symbiosis. In J Dobereiner RH Burris, A Hollaender, eds, Limitations and Potentials for Biological Nitrogen Fixation in the Tropics. Plenum Press, New York, pp 135–160
- Asamizu E, Nakamura Y, Sato S, Tabata S (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant Microbe Interact 18 487–498 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Shimoda Y, Kouchi H, Tabata S, Sato S (2008) A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol 147 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, et al (2008) Transcription profiling of bean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact 21 631–645 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.): model food legume. Plant Soil 252 55–128 [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al (2007) The JAZ family repressors is the missing link in jasmonate signalling. Nature 448 666–671 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15 411–420 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Desbrosses GC, Kopka J, Udvardi MK (2005) Lotus japonicus metabolic profiling: development of gas chromatography-mass spectrometry resources for the study of plant microbe interactions. Plant Physiol 137 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B, Karthikeyan AS, Raghothama KG (2007. a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B, Nagarajna VK, Raghothama KG (2007. b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg: legume diversity and evolution in a phylogenetic context. Plant Physiol 131 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J (2006) Non-supervised construction and application of mass spectral and retention time index libraries from time-of-flight GC-MS metabolite profiles. In W Weckwerth, ed, Metabolomics: Methods and Protocols. Humana Press, Totowa, NJ, pp 19–38 [DOI] [PubMed]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Frickey T, Weiller G (2009) PathExpress update: the enzyme neighbourhood method of associating gene-expression data with metabolic pathways. Nucleic Acids Res 37 W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Weiller G (2007. a) GeneBins: a database for classifying gene expression data, with application to plant genome arrays. BMC Bioinformatics 8 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffard N, Weiller G (2007. b) PathExpress: a Web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res 35 W176–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MA, Ramírez M, Valdés-López O, Lara M, Tesfaye M, Vance CP, Hernández G (2006) Identification of candidate phosphorus stress induced genes in Phaseolus vulgaris L. through clustering analysis across several plant species. Funct Plant Biol 33 789–797 [DOI] [PubMed] [Google Scholar]
- Graham PH (1981) Some problems of nodulation and symbiotic nitrogen fixation in Phaseolus vulgaris L.: a review. Field Crops Res 4 93–112 [Google Scholar]
- Graham PH, Rosas JC, Estevez de Jensen C, Peralta E, Tlusty B, Acosta-Gallegos J, Arraes Pereira PA (2003) Addressing edaphic constraints to bean production: the bean/cowpea CRSP project in perspective. Field Crops Res 82 179–192 [Google Scholar]
- Hartel H, Dörmann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White JP, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11 610–617 [DOI] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogh-Jensen H, Schjoerring JK, Soussana JF (2002) The influence of phosphorus deficiency on growth and nitrogen fixation of white clover plants. Ann Bot (Lond) 90 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DW (1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen I (1985) The role of phosphorus in nitrogen fixation by young pea plants (Pisum sativum). Physiol Plant 64 190–196 [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32 D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D, Lumbreras V, Pages M (2001) Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett 498 187–189 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSB.DB: the Golm Metabolome Database.. Bioinformatics 21 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Lee H, Hur CG, Oh CJ, Kim HB, Park SY, An CS (2004) Analysis of the root nodule-enhanced transcriptome in soybean. Mol Cells 18 53–62 [PubMed] [Google Scholar]
- Le Roux MR, Khan S, Valentine AJ (2008) Organic acid accumulation may inhibit N2 fixation in phosphorus-stressed lupin nodules. New Phytol 177 956–964 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al (2007) Genomic-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30 85–112 [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera M, Tejera N, Iribarne C, Ocaña A, Lluch C (2004) Growth, nitrogen fixation and ammonium assimilation in common bean (Phaseolus vulgaris): effect of phosphorus. Physiol Plant 121 498–505 [Google Scholar]
- Paul M (2007) Trehalose-6-phosphate. Curr Opin Plant Biol 10 303–309 [DOI] [PubMed] [Google Scholar]
- Ramírez M, Graham MA, Blanco-López L, Silvente S, Medrano-Soto A, Blair MW, Hernández G, Vance CP, Lara M (2005) Sequencing analysis of common bean ESTs: building a foundation for functional genomics. Plant Physiol 137 1211–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet J, Drevon JJ (1995. a) Phosphorus deficiency increases the acetylene induced decline of nitrogenase activity in soybean (Glycine max L. Merr.). J Exp Bot 46 1479–1486 [Google Scholar]
- Ribet J, Drevon JJ (1995. b) Increase in permeability to oxygen and in oxygen uptake of soybean nodules under deficient phosphorus nutrition. Physiol Plant 94 298–304 [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, http://www.asp.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Rubio V, Linhares F, Solano R, Martín A-C, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plant and unicellular algae. Genes Dev 15 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa T, Israel DW (1991) Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol 97 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J (2004) Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20 2447–2454 [DOI] [PubMed] [Google Scholar]
- Scholz M, Kaplan F, Guy CL, Kopka J, Selbig J (2005) Non-linear PCA: a missing data approach. Bioinformatics 21 3887–3895 [DOI] [PubMed] [Google Scholar]
- Schulze J, Drevon JJ (2005) P-deficiency increases the O2 uptake per N2 reduced in alfalfa. J Exp Bot 56 1779–1784 [DOI] [PubMed] [Google Scholar]
- Schulze J, Temple G, Temple S, Beschow H, Vance CP (2006) Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot (Lond) 98 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield RJ, Huxley PA, Minchin FR (1977) Plant husbandry and management techniques for growing grain legumes under simulated tropical conditions in controlled environments. Exp Agric 13 113–121 [Google Scholar]
- Tang C, Drevon JJ, Jaillard B, Souche G, Hinsinger P (2004) Proton release of two genotypes of bean (Phaseolus vulgaris L.) as affected by N nutrition and P deficiency. Plant Soil 260 59–68 [Google Scholar]
- Tang C, Hinsinger P, Drevon JJ, Jaillard B (2001) Phosphorus deficiency impairs early nodule functioning and enhances proton release in roots of Medicago truncatula L. Ann Bot (Lond) 88 131–138 [Google Scholar]
- Taussky HH, Shorr E (1953) A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem 202 675–685 [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MapMan: a user-driven tool to display genomic data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- Tian J, Venkatachalam P, Liao H, Yan X, Raghothama K (2007) Molecular cloning and characterization of phosphorus starvation responsive genes in common bean (Phaseolus vulgaris L.). Planta 227 151–165 [DOI] [PubMed] [Google Scholar]
- Tjellström H, Andersson MX, Larsson KE, Sandelius AS (2008) Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ 31 1388–1389 [DOI] [PubMed] [Google Scholar]
- Todd CD, Zeng P, Rodríguez AM, Hoyos ME, Polacco JC (2004) Transcripts of MYB-like genes respond to phosphorus and nitrogen deprivation in Arabidopsis. Planta 219 1003–1009 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2008) The universal protein resource (UniProt). Nucleic Acids Res (Suppl 1) 36: D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadez V, Beck DP, Lasso JH, Drevon JJ (1997) Utilization of the acetylene reduction assay to screen for tolerance of symbiotic N2 fixation to limiting P nutrition in common bean. Physiol Plant 99 227–232 [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Reyes JL, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA:PvmiR399 in phosphorus deficiency signaling in common bean roots. Plant Cell Environ 31 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vance CP, Heichel GH (1991) Carbon in N2 fixation: limitation or exquisite adaptation. Annu Rev Plant Physiol Plant Mol Biol 42 373–392 [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157 423–447 [DOI] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62 887–900 [DOI] [PubMed] [Google Scholar]
- Wu P, Ligeng M, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.