Abstract
A novel alliinase (EC 4.4.1.4) was detected and purified from the roots of the Amazonian medicinal plant Petiveria alliacea. The isolated enzyme is a heteropentameric glycoprotein composed of two α-subunits (68.1 kD each), one β-subunit (56.0 kD), one γ-subunit (24.8 kD), and one δ-subunit (13.9 kD). The two α-subunits are connected by a disulfide bridge, and both α- and β-subunits are glycosylated. The enzyme has an isoelectric point of 4.78 and pH and temperature optima of 8.0 and approximately 52°C, respectively. Its activation energy with its natural substrate S-benzyl-l-cysteine sulfoxide is 64.6 kJ mol−1. Kinetic studies showed that both Km and Vmax vary as a function of substrate structure, with the most preferred substrates being the naturally occurring P. alliacea compounds S-benzyl-l-cysteine sulfoxide and S-2-hydroxyethyl-l-cysteine sulfoxide. The alliinase reacts with these substrates to produce S-benzyl phenylmethanethiosulfinate and S-(2-hydroxyethyl) 2-hydroxyethanethiosulfinate, respectively.
Alliinases (EC 4.4.1.4) are Cys sulfoxide lyases found most often in genus Allium plants such as garlic (Allium sativum) and onion (Allium cepa), among others. The enzyme is compartmentalized in plant cell vacuoles, while its substrates, S-alk(en)yl-l-Cys sulfoxides (ACSOs), are located in the cytoplasm (Lancaster and Collin, 1981). Exposure of the alliinase to cytoplasmic ACSOs, such as occurs on plant tissue disruption, results in alliinase-mediated cleavage of the ACSOs via β-elimination to form pyruvate, ammonia, and sulfenic acids. The sulfenic acids rapidly condense with loss of water to afford the corresponding thiosulfinates (Scheme 1; Block, 1992, 2010; Shimon et al., 2007). The thiosulfinates are often further transformed into other organosulfur derivatives, including but not limited to sulfides, polysulfides, dithiins, cepaenes, and zwiebelanes (Block, 2010). The characteristic flavors and odors, as well as the broad spectrum of therapeutic activities of onion, garlic, and related Allium species, are attributed to these sulfurous compounds (Ramirez, 2003).
Scheme 1.
Proposed general mechanism for the catalysis of C-S bond cleavage in Cys sulfoxide derivatives by alliinase (Block, 1992; Shimon et al., 2007). Alliin, S-Allyl-l-Cys sulfoxide; 2-hydroxyethiin, S-2-hydroxyethyl-l-Cys sulfoxide; isoalliin, (E)-S-(1-propenyl)-l-Cys sulfoxide; methiin, S-methyl-l-Cys sulfoxide; petiveriin, S-benzyl-l-Cys sulfoxide; propiin, S-propyl-l-Cys sulfoxide.
Genus Allium alliinases that have been purified to homogeneity and characterized include those from garlic cloves (Mazelis and Crews, 1968), onion bulbs (Tobkin and Mazelis, 1979), leek (Allium porrum; Lohmuller et al., 1994), Chinese chive (Allium tuberosum; Manabe et al., 1998), and wild garlic/ramson (Allium ursinum; Landshuter et al., 1994). Previous reports have shown that garlic (Nock and Mazelis, 1986), leek (Won and Mazelis, 1989), onion (Tobkin and Mazelis, 1979), and Welsh onion (Allium fistulosum; Fujita et al., 1990) alliinases are glycoproteins, and all alliinases have been shown to be pyridoxal 5′-P (PLP) dependent. Isolated alliinases vary in molecular mass depending on the source of the enzyme, ranging from 67 kD in Chinese chive (Manabe et al., 1998) to 580 kD in leek (Won and Mazelis, 1989). The number of enzyme subunits has been shown to differ depending on the source of the alliinase, ranging from one for Chinese chive to 12 for leek. The affinity of the enzyme for substrate, expressed as Km and typically determined using S-ethyl-l-Cys sulfoxide as the substrate, also varies depending on the alliinase source, ranging from 2.7 mm for Chinese chive (Manabe et al., 1998) to 13 mm for leek (Won and Mazelis, 1989). The reported pH optima also vary depending on the species and the buffer conditions used, ranging from 6.5 (garlic in phosphate buffer; Nock and Mazelis, 1986) to 8.5 (Chinese chive in Tris-HCl buffer; Manabe et al., 1998). There have been no reports on the temperature optima for alliinases from various sources, although ambient temperature has been universally used in published activity assays (Ramirez, 2003).
Petiveria alliacea is a non-Allium herbaceous perennial of the family Phytolaccaceae. It is indigenous to the Amazon rainforest and tropical areas of Central and South America, the Caribbean, and sub-Saharan Africa (Kubec and Musah, 2001; Kubec et al., 2002). It has a long history of use in herbal medicine. Additionally, in a plant-screening program conducted at the University of Illinois at Chicago that evaluated more than 1,400 plant extracts as potentially novel therapies for the prevention and treatment of cancer, P. alliacea was one of 34 promising plants identified as having activity against cancer (Taylor, 2005). The analyses revealed that P. alliacea had a broad range of therapeutic properties, including antileukemic, antitumor, anticancer, immunostimulant, antiinflammatory, antimicrobial, and hypoglycemic activities (Taylor, 2005; Kim et al., 2006). Several organosulfur small molecules, many of which have been shown to have biological activity, have been isolated from P. alliacea, including benzyl 2-hydroxyethyl trisulfide (Szczepanski et al., 1972), cis-3,5-diphenyl-1,2,4-trithiolan (Adesogan, 1974), dibenzyl trisulfide (Sousa et al., 1990), dibenzyl disulfide (Ayedoun et al., 1998), S-benzyl-l-Cys sulfoxide (petiveriin), S-2-hydroxyethyl-l-Cys sulfoxide (2-hydroxyethiin), (Z)-phenylmethanethial S-oxide (PMTSO), and S-benzyl phenylmethanethiosulfinate (petivericin; Kubec and Musah, 2001; Kubec et al., 2002, 2003), among others.
In previous work, we isolated from P. alliacea roots four Cys sulfoxide derivatives, namely petiveriins A and B and 2-hydroxyethiins A and B, which are constitutively present (Scheme 1; Kubec and Musah, 2001; Kubec et al., 2002). We subsequently showed that when the root tissue is macerated, thiosulfinate structures derivative of the aforementioned petiveriins and 2-hydroxyethiins are formed (Kubec et al., 2002). The similarity of this chemistry to that which has been shown to occur in Allium plants, namely, the formation of thiosulfinates mediated by alliinase action on precursor Cys sulfoxides, alerted us to the possibility that P. alliacea might contain an alliinase. The purpose of the work described here was to determine whether the non-Allium plant P. alliacea contains an alliinase, to characterize it if present, and to compare and contrast it to alliinases found in genus Allium plants.
RESULTS
Confirmation of an Alliinase in P. alliacea
Our suspicion of the presence of an alliinase in P. alliacea roots was confirmed with our successful isolation of a protein that reliably resulted in the production of the thiosulfinate petivericin on exposure to its corresponding Cys sulfoxide precursor petiveriin. During the development of the chromatographic separation protocol for the isolation and purification of this protein, it became necessary for us to design a facile and efficient means by which to confirm and track the presence of alliinase activity. We have developed such a method based on modification of a procedure previously reported by Ukai and Sekiya (1997) for the determination of lyases that cleave C-S bonds in sulfur-containing compounds after PAGE. This method is based on the reduction of 3-(4′,5′-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) by thiol compounds in the presence of phenazine methosulfate (PMS), which results in the formation of deeply colored formazans. Thus, alliinase enzymes that are embedded in a polyacrylamide gel, when exposed to an appropriate substrate, will cleave the C-S bond to yield compounds that reduce MTT to form a formazan, which stains the gel purple and indicates the presence of a C-S bond-cleaving protein. The substrate used in these reactions was petiveriin. We have extended this method for use in a convenient 96-well-plate format in which active fractions derived chromatographically from each purification step can be identified visually (colorimetrically).
Purification and Biochemical Characterization of P. alliacea Alliinase
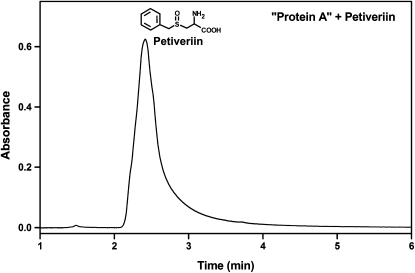
Using the aforementioned active staining method and extensive chromatographic purification procedures modeled after published protocols for the isolation of alliinases from garlic (Nock and Mazelis, 1986; Rabinkov et al., 1994), onion (Tobkin and Mazelis, 1979), and shiitake mushroom (Lentinus edodes; Kumagai et al., 2002), we have developed a purification procedure the steps of which are summarized in Table I. The alliinase was purified from P. alliacea roots using ion-exchange, hydroxyapatite, and gel-filtration chromatographies in sequence, yielding a single band of the alliinase as observed by native PAGE (Fig. 1A). Specifically, alliinase from P. alliacea roots that were pulverized in phosphate buffer was precipitated using 60% (w/v) ammonium sulfate and eluted between 220 and 265 mm NaCl by anion-exchange chromatography (first column). The alliinase eluent was subjected to hydroxyapatite chromatography (second column), where it eluted between 80 and 160 mm phosphate. Further purification of this eluent by gel-filtration chromatography yielded alliinase that eluted at 7.4 min. The protein was subjected to further purification by a second run through the gel-filtration column, which furnished purified alliinase. This 65-fold purified alliinase exhibited a specific activity of 107 nkat mg−1 (1 nkat = 1 nmol pyruvate s−1), with an activity recovery of 4.5% (nkat/nkat) and a protein recovery of 0.07% (w/w; Table I). The protein isolated by this procedure tested positive in our active staining protocol (Fig. 1B). Alliinase bands observed by native PAGE were consistently diffuse, implying that the protein might be glycosylated. This conjecture was confirmed by the “in-gel” detection of carbohydrate by oxidation of the carbohydrates in the gel to aldehydes, followed by reaction of the aldehyde with a hydrazide, which produced an easily detectable fluorescent conjugate (Fig. 1C). We used reverse-phase C-18 HPLC to determine the product formed on exposure of the protein to petiveriin and confirmed the production of petivericin (Fig. 2). The molecular mass of the alliinase was observed to be 145.1 kD by gel-filtration chromatography (Supplemental Fig. S1), a result corroborated by Ferguson plot analysis (FPA; 145.4 kD; Supplemental Fig. S2). By the FPA method, we also estimated the molecular net charge of the alliinase to be −3.44 × 10−12 Coulombs per molecule, which corresponds to −2.15 net protons per molecule. The protein's pI was determined to be 4.78 by chromatofocusing (Supplemental Fig. S3). Similar to what has been observed for other PLP-dependent alliinases, exposure of P. alliacea alliinase to hydroxylamine resulted in complete loss of enzyme activity.
Table I.
Purification of alliinase from 150 g of P. alliacea roots
| Fraction | Volume | Total Protein | Total Activity | Specific Activity | Recovery | Purification |
|---|---|---|---|---|---|---|
| mL | mg | nkat | nkat mg−1 | % | fold | |
| Homogenate | 230 | 289 | 482 | 1.67 | 100 | 1.0 |
| 60% (w/v) (NH4)2SO4 | 20 | 152 | 457 | 3.00 | 95 | 1.8 |
| Anion exchange | 4 | 25 | 333 | 13.3 | 69 | 8.0 |
| Hydroxyapatite | 0.8 | 5.0 | 208 | 41.6 | 43 | 25 |
| Gel filtration | 0.8 | 1.5 | 103 | 68.7 | 21 | 41 |
| Gel filtration |
0.7 |
0.2 |
21.7 |
109 |
4.5 |
65 |
Figure 1.
Physical characterization of P. alliacea alliinase. A, Native PAGE with Coomassie Brilliant Blue staining showing the molecular mass markers (left lane) and the alliinase (right lane) purified to homogeneity. B, Native PAGE showing the molecular mass marker (left lane) and a positive test for C-S cleavage activity based on reduction of MTT by thiol compounds in the presence of PMS to form a deeply colored formazan (right lane). C, Native PAGE of alliinase after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate (i.e. a positive test for the presence of sugars).
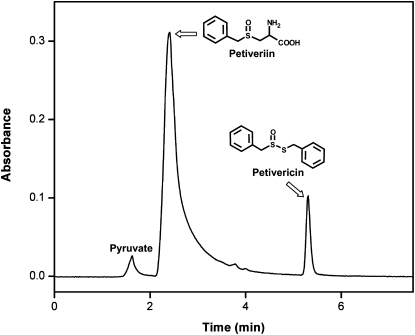
Figure 2.
Confirmation of the production of petivericin by P. alliacea alliinase-mediated breakdown of petiveriin using RP C-18 HPLC. Twenty microliters of a reaction mixture composed of 1.0 mL of 20 mm phosphate buffer, pH 8.0, 25 μm PLP, 1.5 mm petiveriin, and P. alliacea-derived alliinase that had been incubated for 10 min at room temperature was subjected to HPLC analysis under the following conditions: flow rate, 1.0 mL min−1; mobile phase, water:acetonitrile (30:70).
SDS-PAGE analysis of the protein was conducted both with and without β-mercaptoethanol (BME) in order to reveal the presence of disulfide linkages. The results of these experiments are shown in Figure 3. By SDS-PAGE in the absence of BME, four bands appeared, termed α′ (130.0 kD), β (56.0 kD), γ (24.8 kD), and δ (13.9 kD; Fig. 3C). However, in the presence of BME, the protein represented by the α′ band in Figure 3C collapses into a single band of lower molecular mass, termed α (68.1 kD), whereas the bands represented by subunits β, γ, and δ are retained (Fig. 3A). Thus, the protein is apparently composed of five subunits, two of which are similar and possibly identical and connected by a disulfide linkage (i.e. the two α-subunits). The two α-subunits and the β-subunit were observed to be glycosylated (Fig. 3B). Since band α′ in Figure 3C apparently consists of two monomers connected by a disulfide bond, the molecular mass estimate based on SDS-PAGE of the entire protein is 218.7 kD. This is approximately 75 kD higher than the molecular mass estimates of the alliinase molecule as measured by gel-filtration chromatography and FPA (i.e. about 145 kD; Supplemental Figs. S1 and S2). However, this discrepancy was not unexpected, given that it is well documented that glycoproteins as well as proteins with rigid disulfide linkages often exhibit anomalous migration profiles in SDS-PAGE, resulting in larger than normal molecular mass estimates (Segrest et al., 1971; Marciani and Papamatheakis, 1978). Thus, we believe the molecular mass estimates determined by gel-filtration chromatography and FPA to be more reliable than those determined by SDS-PAGE, since size-exclusion chromatography and FPA are more immune to this anomalous behavior. Limited availability of the protein prevented us from investigating this further.
Figure 3.
Characterization of P. alliacea alliinase subunits by SDS-PAGE. A, SDS-PAGE in the presence of BME, with Coomassie Brilliant Blue staining showing the molecular mass markers (left lane) and the four bands representing the subunits of which P. alliacea alliinase is composed (right lane). The band labeled “α” contains two subunits of approximately equal size. B, SDS-PAGE of alliinase in the presence of BME after oxidation of carbohydrates bound to the protein, followed by treatment with a hydrazide dye to yield a highly fluorescent conjugate (i.e. a positive test for the presence of sugars). The results show that the two largest subunits are glycosylated. C, SDS-PAGE of alliinase in the absence of BME showing the α′-band that collapses into the α-band in A.
Determination of the Importance of the Disulfide Bond in the P. alliacea Alliinase α′-Subunit to Alliinase Activity
In order to determine whether the disulfide bond in the P. alliacea alliinase α′-subunit is necessary for catalytic activity, the alliinase after reduction with BME was exposed to petiveriin. The reaction was then followed by C-18 HPLC in order to track the formation of products. The chromatogram showed that no products were formed (Supplemental Fig. S4A), indicating that with the α′-subunit reduced, the P. alliacea alliinase no longer exhibited alliinase activity.
Determination of the Ability of P. alliacea Alliinase to Precipitate Polysaccharides
Lectins are known to bind alliinases in garlic and ramson and to be copurified with them during chromatographic separation (Rabinkov et al., 1995; Smeets et al., 1997). In these cases, the sugar-binding behavior of the lectin is retained even while the lectin is complexed to the alliinase. In order to determine whether any of the subunits of the P. alliacea alliinase might have lectin activity, the ability of the alliinase to precipitate four polysaccharides (chitin, glycogen, dextran, and mannan) was tested by the method of Goldstein (1972). No precipitate was observed for any of the polysaccharides tested.
P. alliacea Alliinase Enzyme Kinetics and Substrate Specificity
Alliinases react with S-substituted Cys sulfoxide derivatives to produce sulfenic acids and α-aminoacrylic acid (Scheme 1). Both the sulfenic acids and α-aminoacrylic acid are fleeting intermediates whose formation cannot easily be tracked. The sulfenic acids condense with loss of water to form thiosulfinates. The α-aminoacrylic acid subsequently breaks down into pyruvate, the stable product formed in all reactions of Cys sulfoxide lyases with S-substituted Cys sulfoxide derivatives. Thus, the kinetics of the reactions of alliinases are determined by monitoring the formation of either the thiosulfinate(s) or pyruvate.
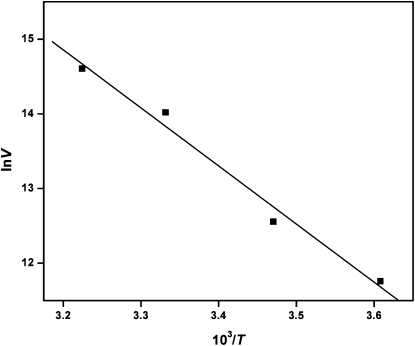
Using petiveriin as the substrate and monitoring pyruvate formation, P. alliacea alliinase showed detectable activity over a wide pH range of 5.0 to 10.0, with a pH optimum of 8.0 (Supplemental Fig. S5). Alliinase activity also increased with increasing temperature over a range of 4.0°C to 52°C. At 67°C, enzyme activity falls precipitously, and at 84°C, alliinase activity is completely lost (Supplemental Fig. S6). The relationship between the reaction velocity and temperature obeyed the Arrhenius equation, and thus by plotting the inverse of the reaction rate against the inverse of the absolute temperature over a temperature range of 4.0°C to 37°C (Fig. 4), the activation energy for the enzyme's reaction with petiveriin was calculated to be 64.6 kJ mol−1. We also observed that in a control system composed of the P. alliacea alliinase and its natural substrate petiveriin in buffer at ambient temperature, alliinase activity was retained for over 4.0 h (Fig. 5).
Figure 4.
Arrhenius plot for the cleavage of the C-S bond in petiveriin by P. alliacea alliinase. The reaction rate was monitored by following the production of petivericin by UV-visible analysis at 210 nm. The activation energy calculated from the Arrhenius equation was 64.6 kJ mol−1.
Figure 5.
Determination of the time frame during which P. alliacea alliinase remains active. Measurements were conducted at 25°C and were based on UV-visible detection of the amount of petivericin product produced during a period of 255 min when 1.0 μg of purified alliinase was suspended in 1.0 mL of 20 mm phosphate buffer at pH 8.0 containing 25 μm PLP and 1.5 mm petiveriin.
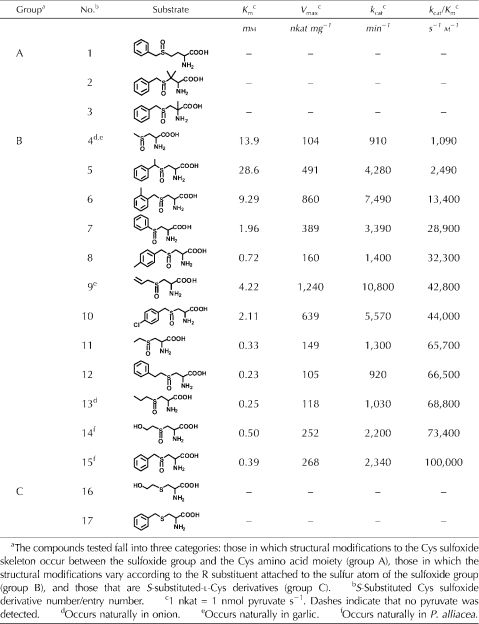
The efficiency and specificity of the P. alliacea alliinase reaction with a total of 15 natural and unnatural S-substituted Cys sulfoxides and two S-substituted-l-Cys derivatives, expressed in order of increasing kcat/Km (i.e. specificity constant), were evaluated. Purified alliinase in 20 mm phosphate buffer (pH 8.0) in the presence of 25 μm PLP was reacted with each of the substrates, and the Km, Vmax, turnover number (kcat), and kcat/Km were determined (Table II).
Table II.
Kinetics study of the reaction of P. alliacea alliinase with S-substituted-l-Cys sulfoxides and S-substituted-l-Cys derivatives
Within experimental error, both Km and Vmax (i.e. kcat) varied as a function of substrate structure. The compounds tested fall into three categories: those in which structural modifications to the Cys sulfoxide skeleton occur between the sulfoxide group and the Cys amino acid moiety (Table II, group A, entries 1−3), those in which the structural modifications vary according to the R substituent attached to the sulfur atom of the sulfoxide group (Table II, group B, entries 4−15), and those that were S-substituted-l-Cys derivatives (Table II, group C, entries 16 and 17). None of the tested compounds from groups A and C reacted with the alliinase, whereas all of the compounds in group B, which included compounds with S-substituted alkyl, alkenyl, aromatic, and polar side chains, did. A few of the tested compounds with which P. alliacea alliinase reacted occur in genus Allium species, such as compound 4 from both onion and garlic, compound 9 from garlic, and compound 13 from onion.
DISCUSSION
In this work, we have shown that the medicinal plant P. alliacea contains an alliinase that catalyzes reactions similar to those observed for the alliinases in garlic and onion. However, in many respects, P. alliacea alliinase exhibits important differences from those of the alliinases reported so far. For instance, we observed the P. alliacea alliinase to be composed of five protein subunits (Fig. 3), two of which are similar and possibly identical, and connected via a disulfide bridge, and the other three being structurally distinct protein subunits. To our knowledge, an alliinase composed of different protein subunits has never before been reported. The heteromultimeric nature of the P. alliacea alliinase is in sharp contrast to the alliinases found in Allium plants and in shiitake mushroom, which although varying greatly in size (from 67 kD in Chinese chive to 84 kD in shiitake to 580 kD in leek) have heretofore been reported to be multimers composed of identical subunits or a single monomeric unit showing only a single band by SDS-PAGE (Lohmuller et al., 1994; Manabe et al., 1998; Kumagai et al., 2002; Ramirez, 2003).
Since all previously isolated and characterized alliinases have been shown to be either monomers or homomultimers, the observation of a heteromeric protein exhibiting alliinase activity is surprising and begs the question of whether the observed nonidentical subunits are really parts of the alliinase enzyme required for alliinase activity or simply lectin contaminants that are complexed to the alliinase. Precedents for the formation of alliinase-lectin complexes include the observation in garlic and ramson of stable alliinase-lectin complexes (Rabinkov et al., 1995; Smeets et al., 1997). The lectins to which the garlic and ramson alliinases were complexed (i.e. A. sativum agglutinin I [ASAI] and A. ursinum agglutinin I [AUAI], respectively) are Man-binding heterodimers, and their complexation to the alliinases does not inhibit alliinase activity. It has also been observed that less than 5% of alliinase molecules are associated with the ASAI and AUAI lectins (Smeets et al., 1997). Thus, in both garlic and ramson, two alliinase-active protein fractions are observed: one in which alliinase is uncomplexed, and the other in which alliinase is complexed to a lectin. Of the two, the former contributes greater than 95% of the total alliinase activity observed in isolated alliinase-active samples (Smeets et al., 1997). Alliinase-lectin complex formation is mediated by the interaction of Man residues on the alliinase with the lectin (Rabinkov et al., 1995; Smeets et al., 1997).
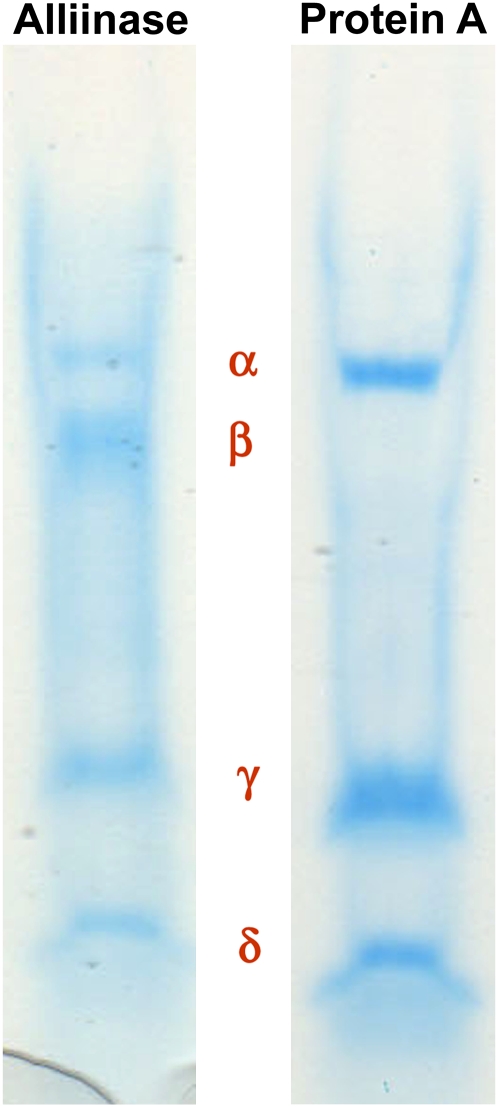
If any of the subunits of the P. alliacea alliinase reported here are complexed lectins, then by analogy with the alliinases in garlic and ramson, P. alliacea alliinase chromatographic isolation might be expected to yield two or more active fractions, at least one of which would remain uncomplexed with any lectins present in the plant extract. During the chromatographic isolation and purification of the P. alliacea alliinase, only a single active protein fraction showing five subunits by SDS-PAGE was observed. However, the first chromatography step of the purification procedure (Table I) yielded a second protein fraction (termed protein A in Fig. 6) that by SDS-PAGE appeared similar to that of the alliinase except that it did not possess the β-subunit. This observation afforded us the opportunity to test the importance of the β-subunit of the isolated P. alliacea alliinase to the observed alliinase activity. The first petiveriin-derived organosulfur compounds formed in freshly isolated P. alliacea root protein extracts are petivericin (Fig. 2) and the potent lachrymator PMTSO (Fig. 7). Petivericin is formed from alliinase-catalyzed cleavage of petiveriin, whereas PMTSO formation has been shown to be mediated by a lachrymatory factor synthase (LFS; see companion paper by Musah et al., 2009). Incubation of the isolated five-subunit alliinase with petiveriin resulted in the formation of petivericin as a single product (Fig. 2), whereas exposure of protein A to petiveriin did not yield any products (Fig. 8). In neither case was PMTSO observed.
Figure 6.
Characterization of the subunits of P. alliacea alliinase and protein A by SDS-PAGE. Protein A was acquired from the first chromatography step of the purification procedure outlined in Table I.
Figure 7.
Lachrymator isolated from P. alliacea (Kubec et al., 2003).
Figure 8.
Characterization of P. alliacea root-derived protein A. C-18 HPLC analysis of the reaction of protein A with petiveriin showing no detectable products.
If the α′-subunit by itself is an alliinase and the γ- and δ-subunits are lectins that are complexed to it, then by analogy to what has been observed in alliinase-lectin complexes in garlic and ramson (Rabinkov et al., 1995; Smeets et al., 1997) alliinase activity would be retained in protein A. Yet, we observed protein A to be devoid of alliinase activity. We interpreted this result to mean that the α′-subunit alone is not sufficient for alliinase activity. The same is true of the γ- and δ-subunits. However, the combination of the β-subunit of the alliinase with the α′-, γ-, and δ-subunits results in C-S lyase activity that accounts for all of the alliinase activity observed in P. alliacea roots. No other chromatographically derived protein fractions with alliinase activity were found. This observation demonstrates that the β-subunit is required for alliinase activity. That no other chromatographically derived protein fraction with alliinase activity was seen contrasts with what has been observed in garlic and ramson, where alliinase-active fractions both with and without complexed lectin have been observed (Smeets et al., 1997).
When the disulfide bond of either the alliinase or protein A was reduced with BME and the resulting protein was exposed to petiveriin, no products were formed (Supplemental Fig. S4, A and B). We concluded from this result that (1) the disulfide link of the α′-subunit is essential for alliinase activity and (2) the β-subunit, which itself does not have a disulfide link, does not on its own have alliinase activity. Therefore, both the α′- and β-subunits are necessary for P. alliacea alliinase activity, and the P. alliacea alliinase is heteromeric, which to our knowledge is a first for reported alliinases.
It has been observed that the Man-specific binding ability of ASAI and AUAI lectins is retained even when they are complexed to alliinases in garlic and ramson, respectively (Smeets et al., 1997). Therefore, we sought to determine whether the γ- and δ-subunits of the isolated P. alliacea alliinase might be lectins by assessing the ability of the alliinase to precipitate various saccharides. Incubation of the alliinase with mannan did not result in polysaccharide precipitation. That the alliinase did not precipitate mannan is significant because Man is the sugar shown to be associated with the alliinases from Allium plants. If the γ- and δ-subunits of the P. alliacea alliinase had Man-binding ability, the alliinase would be expected to precipitate mannan.
Since P. alliacea is not from the Allium family but rather from the pokeweed (Phytolacca americana) family, we also considered the possibility that the P. alliacea alliinase might contain a lectin with sugar-binding specificity different from that of the lectins that bind alliinases in Allium plants. For example, the lectin isolated from pokeweed is specific for GlcNAc sugars (Ken-ichi et al., 1996; Minoru et al., 2003; Tomomi et al., 2004). Therefore, we tested the ability of the P. alliacea alliinase to precipitate chitin, a polysaccharide of GlcNAc. Similar to what we observed for incubation of the P. alliacea alliinase with mannan, no precipitation of chitin was observed. We obtained the same results with dextran and glycogen. We concluded from these experiments that the subunits of the P. alliacea alliinase are not able to bind sugars that are known to be associated with alliinases or sugars known to bind lectins in plants from the same family of which P. alliacea is a member (i.e. pokeweed). Although the data we have acquired thus far imply that the P. alliacea subunits are not lectins, the possibility that these proteins may have agglutinating activity is the subject of continuing investigations in our laboratory.
A further fascinating observation was that whereas neither the alliinase nor protein A produced the P. alliacea sulfine lachrymator PMTSO when exposed to petiveriin, the combination of the two proteins in the presence of petiveriin immediately produced the lachrymator, with the lachrymator amounts produced depending upon the ratio of the alliinase to protein A. This observation indicates that protein A, with its constituent α′-, γ-, and δ-subunits, is far from an innocent bystander but rather exhibits LFS activity, since its presence led directly to the formation of the P. alliacea PMTSO lachrymator. The characterization of protein A, termed a LFS, is described in a companion article (Musah et al., 2009). That the PMTSO is only formed when both the alliinase and the LFS are combined in the presence of the alliinase substrate petiveriin is analogous to what has been observed in onion, where the lachrymator is only formed when both its alliinase and LFS are combined in the presence of the substrate isoalliin (Imai et al., 2002). However, in onion, based upon what is known of its LFS primary sequence, there are no similarities between the structure of the onion alliinase and the onion LFS. This contrasts with our observations in P. alliacea, where the LFS appears to share several subunits with the alliinase. In this regard, it is intriguing that although the P. alliacea alliinase that we isolated appears to contain the γ- and δ-subunits of the LFS, it does not itself possess LFS activity. The importance of the γ- and δ-subunits to the P. alliacea alliinase activity is the subject of further investigations in our laboratory.
The pI of P. alliacea alliinase was determined to be 4.78 by chromatofocusing (Supplemental Fig. S3), a result significantly different from that usually observed for alliinases, including those from leek (7.5; Landshuter et al., 1994), onion (7.6−9.3; Nock and Mazelis, 1987; Lancaster et al., 2000), and garlic (6.35; Rabinkov et al., 1994). The P. alliacea alliinase pI aligns most closely with that of shiitake mushroom (4−5; Kumagai et al., 2002).
By monitoring its activity over a temperature range of 0°C to 84°C (Supplemental Fig. S6), we observed that with its most preferred substrate petiveriin, P. alliacea alliinase showed 80% higher activity at 37°C and 100% higher activity at 52°C than at ambient temperature (25°C). Even at 67°C, alliinase activity was still 19% higher than that observed at room temperature, indicating that the alliinase retains significant functionality over a broad range of temperature and possesses a high overall intrinsic thermal stability. This feature may help the plant to survive under harsh climatic conditions in tropical regions where temperatures are known to climb well over 40°C. The activation energy for the enzyme's reaction with petiveriin was determined from an Arrhenius plot (Fig. 4) to be 64.6 kJ mol−1, which is relatively high and implies that substantial energy is required to form the sulfenic acid intermediate. The activation energies for leek alliinase have been reported to be 31, 54, and 28 kJ mol−1 for the substrates S-methyl-l-Cys sulfoxide (methiin), S-propyl-l-Cys sulfoxide (propiin), and S-allyl-l-Cys sulfoxide (alliin), respectively (Lohmuller et al., 1994).
The results of our kinetics study of P. alliacea alliinase on a total of 17 natural and unnatural substrates are consistent with the previously proposed catalytic mechanism in which the Cys sulfoxide precursor reacts with PLP at the alliinase active site to form a Schiff base (Block, 1992; Shimon et al., 2007; Scheme 1). A basic residue at the active site then abstracts the α-proton, which furnishes the sulfenic acid. The group A unnatural substrates (entries 1–3) failed to react. In the case of compounds 1 and 2, assuming that they bind to the active site, the inclusion of the additional carbon between the amino acid α-carbon and the sulfoxide moiety may create enough of a perturbation in the Cys sulfoxide configuration to prevent abstraction of the α-proton. This would result in the Cys sulfoxide amino group no longer being oriented in a way that permits it to be anchored to the active site through Schiff base formation with PLP. On the other hand, assuming successful formation of the Schiff base, it may be that the active site basic residue is no longer proximal enough to the α-hydrogen to abstract it. However, the possibility that the structural perturbations result in neither molecule binding to the active site, a fact that would also preclude the formation of product, cannot be ruled out. Compound 3, which does not possess an α-hydrogen that can be abstracted, was unreactive, as expected.
Like the group A substrates, those from group C also failed to react. This observation reveals that the P. alliacea alliinase is devoid of S-substituted-l-Cys lyase activity, at least for the two substrates tested (i.e. entries 16 and 17).
All of the group B Cys sulfoxide derivatives (Table II, entries 4−15), which differ from one another by the identity of the sulfur-bound R group, were observed to be viable substrates for P. alliacea alliinase. The alliinase reacted with its constitutively present substrates most efficiently (Table II, compounds 14 and 15). The enzyme demonstrated broad substrate specificity, reacting with Cys sulfoxides bearing aromatic (Table II, compounds 5−8, 10, 12, and 15), aliphatic (Table II, compounds 4, 11, and 13), alkenyl (Table II, compound 9), and polar (Table II, compound 14) substituents. Among the congener groups tested, no clear trends in catalytic efficiency were observed. For example, compounds 12, 15, and 7 are congeners that differ from one another by a benzene ring and sulfoxide group separated by two, one, or no methylene groups, respectively. As the number of methylene groups decreases, no trend in the value of kcat/Km is observed. Interestingly, however, an upward trend in turnover number (kcat) was observed as the number of methylene groups decreased (i.e. kcat was 920, 2,340 and 3,390 min−1 for compounds 12, 15, and 7, respectively). That the enzyme exhibited a higher catalytic efficiency for compound 15, even though the turnover number for compound 7 is 1.5 times higher than that of compound 15, is a consequence of the fact that the Km of compound 7 makes a substantial negative contribution to the catalytic efficiency (i.e. the Km of compound 7 is five times less than that of compound 15). Additionally, the binding efficiency, as reflected in the Km values, tended downward as the number of methylenes decreased, with compounds 12, 15, and 7 having Km values of 0.23, 0.39, and 1.96 mm, respectively.
Among the congeners that differ by the length of the alkyl chain attached to the sulfoxide moiety (compounds 4, 11, and 13), there is a trend toward decreasing Km values as the size of the alkyl group increases from methyl (compound 4, Km = 13.9) to ethyl (compound 11, Km = 0.33) to propyl (compound 13, Km = 0.25). This trend is similar to that observed for compounds 12, 15, and 7 and implies that among congeners, the larger the R group, the stronger the protein-substrate binding. Compound 4, the substrate possessing the smallest sulfur-bound R group, has minimal hydrophobic surface area to bind within the active site, and this is reflected in its extremely poor binding affinity compared with the other substrates (i.e. Km = 13.9 mm). This relatively high Km negatively impacts the specificity constant, yielding a kcat/Km value lower than that of all the compounds tested (i.e. 1,090 s−1 m−1).
Comparing compounds 5, 6, 8, and 10, which differ from the constitutively present compound 15 in having either an additional methyl group or a Cl atom, it is clear that the placement of a methyl group on the carbon that is alpha to sulfur has a dramatic negative impact on binding affinity (Km = 28.6 mm for compound 5 versus Km = 0.39 for compound 15). This results in compound 5 having the highest Km of all the compounds tested. Steric factors may prevent compound 5 from optimal binding in the active site. The data also show that, in terms of Km values, para substituents are better accommodated in the active site than ortho substituents. For example, compounds 8 and 10, both of which possess para substituents, have Km values of 0.72 and 2.11 mm for CH3 and Cl, respectively, versus compound 6, which has a methyl group in the ortho position and a Km value of 9.29 mm.
Unlike 4, 5, and 6, the other group B compounds have neither R groups that are too small/large nor branching. Thus, compared with the kcat/Km values corresponding to compounds 4 to 6, the catalytic efficiency with which the enzyme reacts with the remaining substrates is substantially higher, varying from 28,000 to 100,000 s−1 m−1. Of the substrates tested, P. alliacea alliinase reacted most efficiently with its two constitutively present S-substituted Cys sulfoxides petiveriin (Table II, compound 15, aromatic side chain; kcat/Km = 100,000 s−1 m−1) and 2-hydroxyethiin (Table II, compound 14, polar side chain; kcat/Km = 73,400 s−1 m−1). The relatively low Km value of 0.5 mm corresponding to 2-hydroxyethiin (compound 14) implies that the hydrophobic alliinase active site possesses a polar group with which its hydroxyl group interacts. The kinetics of P. alliacea alliinase action on naturally occurring alliin from garlic (compound 9) indicates that although alliin affinity for the enzyme is relatively weak (Km = 4.22 mm), the enzyme's turnover number with this substrate is higher than that of any of the other compounds tested. The balance between the relatively weak binding of alliin and the extremely high kcat value exhibited by the enzyme for this compound makes a favorable enough contribution to the specificity constant to place alliin in the middle of the specificity constant profile for the compounds tested.
It is clear that although very dissimilar in structure from the alliinases heretofore isolated and characterized, P. alliacea alliinase performs chemistry analogous to that observed for genus Allium-derived alliinases. This is quite remarkable given that all previously characterized alliinases, including that from shiitake mushroom, have been shown to be composed of a single monomer unit or are multimers of a single protein. Although the P. alliacea alliinase mediates the formation of thiosulfinates, just as do previously identified Cys sulfoxide lyases, P. alliacea's unique constitutively present precursor Cys sulfoxides (petiveriin and 2-hydroxyethiin) result in its having a thiosulfinate profile that is totally different from that commonly observed in Allium plants. Comparison with representative examples of alliinases from garlic and onion illustrates this point. Both contain S-alk(en)yl-Cys sulfoxides, with the sulfur-bound R groups most commonly observed in significant amounts being methyl (garlic and onion), propyl (onion), (E)-1-propenyl (onion), and 2-propenyl (garlic). Thus, the symmetrical and mixed thiosulfinate derivatives of these compounds possess only aliphatic substituents. Similar thiosulfinate profiles have been observed in leek, Chinese chive, and wild garlic. P. alliacea contains S-alk(en)yl-Cys sulfoxides only in trace amounts, whereas the two pairs of diastereomeric Cys sulfoxides, petiveriins A and B and 2-hydroxyethiins A and B, are present in abundance (Kubec and Musah, 2001; Kubec et al., 2002, 2003). The result is that alliinase-mediated breakdown of these compounds in P. alliacea yields a profile of symmetrical and mixed thiosulfinates that is totally different from what has heretofore been observed in Allium species. P. alliacea alliinase does display the ability to catalyze the transformation of S-alk(en)yl-Cys sulfoxides into thiosulfinates. This is similar to what has been observed for garlic alliinase, which acts on propiin found naturally in onion to form S-propyl propanethiosulfinate, even though garlic does not itself contain this precursor Cys sulfoxide. Whether any of the Allium alliinases exhibit broad enough substrate specificity to catalyze the transformation of the petiveriins or 2-hydroxyethiins into their corresponding thiosulfinates is the subject of ongoing investigations.
MATERIALS AND METHODS
Plants and Materials
Unless otherwise noted, all chemicals were obtained from Sigma-Aldrich. Whole fresh plants of Petiveria alliacea were obtained from Native Habitat Landscaping. They were collected in Vero Beach, Indian River County, Florida, and stored at −30°C until analysis. A voucher specimen is deposited in the herbarium PIHG at the Florida Department of Agriculture and Consumer Services, Division of Plant Industry, under accession number 7801.
Reference Compounds
S-Substituted-l-Cys derivatives and the corresponding diastereomeric S-substituted-l-Cys sulfoxides were synthesized according to the method of Kubec and Musah (2001).
Analytical Methods
HPLC separations were performed on a Dynamax SD-200 binary pump system employing a PDA 330 detector (Varian).
Native PAGE Analysis
Native PAGE was performed according to the method of Davis (1964) with 10% Tris-HCl Ready Gels (Bio-Rad Laboratories). Prestained SDS-PAGE Broad Range Standards (Bio-Rad Laboratories) were used as molecular mass markers. One part protein sample was mixed with two parts sample buffer. The sample buffer was composed of 62.5 mm Tris, 40% (v/v) glycerol, and 0.01% (w/v) bromphenol blue at pH 6.8. The running buffer was composed of 0.1 m Tris and 0.1 m Tricine at pH 8.3. The gel was run at a constant voltage of 120 V, with a starting current of 63 mA and a final current of 32 mA. The total run time was 45 min.
SDS-PAGE Analysis
SDS-PAGE was carried out by the method of Laemmli (1970) using 10% Tris-HCl Ready Gels (Bio-Rad Laboratories). Prestained SDS-PAGE Broad Range Standards (Bio-Rad Laboratories) were used as molecular mass markers. One part protein sample was mixed with two parts sample buffer, and the resulting solution was heated at 100°C for 15 min. The sample buffer was made from 980 μL of Tricine sample buffer (Bio-Rad Laboratories) and 20 μL of BME. The running buffer was composed of 0.1 m Tris, 0.1 m Tricine, and 0.1% (w/v) SDS at pH 8.3. The gel was run at a constant voltage of 120 V, with a starting current of 63 mA and a final current of 32 mA. The total run time was 45 min.
In-Gel Staining of Proteins
To detect the proteins, Bio-Safe Coomassie Brilliant Blue G-250 Stain (Bio-Rad Laboratories) was used after native PAGE and SDS-PAGE. To detect the presence of active enzymes, the gels were stained based on the method of Ukai and Sekiya (1997) with a staining solution composed of 50 mm Tris-HCl buffer at pH 8.0, 4.0 mm petiveriin, 100 μm PLP, 800 μm PMS, and 600 μm MTT. Gels were developed with gentle rocking at room temperature for 10 to 20 min and then rinsed with 7.5% (v/v) acetic acid.
Protein Extraction and Purification
P. alliacea alliinase was purified following a procedure modeled after published protocols for the isolation of alliinases from garlic (Allium sativum; Nock and Mazelis, 1986; Rabinkov et al., 1994), onion (Allium cepa; Tobkin and Mazelis, 1979), and shiitake mushroom (Lentinus edodes; Kumagai et al., 2002). All steps were carried out at 4°C. Fresh P. alliacea roots (150 g) were carefully cleaned in water and homogenized with a blender in 400 mL of 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP, 10% (v/v) glycerol, 5% (w/v) NaCl, 5% (w/v) polyvinylpolypyrrolidone, 5 mm EDTA, and 0.05% (v/v) BME. The homogenate was filtered through four layers of gauze, and the filtrate was centrifuged at 15,000g for 30 min. The 250 mL of supernatant obtained was brought to 60% (w/v) saturation with ammonium sulfate. The precipitate obtained after centrifuging at 15,000g for 30 min was resuspended in 15 mL of 20 mm Tris-HCl, pH 7.6, containing 20 μm PLP and 10% (v/v) glycerol and dialyzed overnight against the same buffer. The dialysate obtained (20 mL) was centrifuged at 10,000g for 10 min and then applied by HPLC in 5.0-mL aliquots to a HiPrep 16/10 DEAE FF column (16 mm i.d. × 100 mm; Amersham Biosciences) that had been preequilibrated with 20 mm Tris-HCl buffer at pH 7.6 containing 10% (v/v) glycerol. The protein sample was washed first with 65 mL of 20 mm Tris-HCl buffer at pH 7.6 containing 10% (v/v) glycerol. Then, the column was washed again using a linear gradient, over 100 min, as follows: solvent A, 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP and 10% (v/v) glycerol; solvent B, 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP, 10% (v/v) glycerol, and 280 mm NaCl. Finally, the column was washed with 30 mL of 20 mm Tris-HCl buffer at pH 7.6 containing 20 μm PLP, 10% (v/v) glycerol, and 280 mm NaCl. The flow rate was 2.0 mL min−1. Active fractions detected by PAGE active staining were pooled and concentrated to 4.0 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD) and then dialyzed overnight against 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The dialysate obtained was applied in 2-mL aliquots to a hydroxyapatite column (15 mm i.d. × 113 mm; Bio-scale CHT20-I; Bio-Rad Laboratories) that had been preequilibrated with 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The column was washed first with 60 mL of 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol, and then with the following solvent combination over a linear gradient lasting 100 min: solvent A, 20 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol; solvent B, 200 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol; and finally with 30 mL of 200 mm phosphate buffer at pH 7.0 containing 20 μm PLP and 10% (v/v) glycerol. The flow rate for this column was 1.0 mL min−1. Active fractions detected by PAGE active staining were pooled and concentrated to 0.8 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD). The resulting sample was applied to a gel-filtration column (7.8 mm i.d. × 300 mm; Bio-Sil SEC-250; Bio-Rad Laboratories) that had been preequilibrated with 20 mm phosphate buffer at pH 6.8 containing 0.15 m NaCl and 10% (v/v) glycerol. The loaded protein was washed with the same phosphate buffer at a flow rate of 1.0 mL min−1. Active eluents were pooled and concentrated to 0.8 mL using a Millipore centrifugal filter (molecular mass cutoff of 30 kD). The sample obtained was again applied to and eluted from the gel-filtration column under conditions identical to those described above. The active eluents were pooled and concentrated to yield 0.7 mL of sample. Protein concentrations were estimated based on the sample's A260 and A280 at a 1.0-cm path length using the following equation: protein concentration (mg mL−1) = 1.55 × A280 − 0.75 × A260 (Simpson, 2004).
Molecular Mass Determination
Two methods were used to determine the molecule mass of P. alliacea alliinase. (1) Gel-filtration chromatography with a Bio-Sil SEC-250 gel-filtration column (7.8 mm i.d. × 300 mm; Bio-Rad Laboratories). The sample was run according to the manufacturer's specifications. The gel-filtration standard was a lyophilized mixture of five molecular mass markers ranging from 1.35 to 670 kD (Bio-Rad Laboratories). The experimental conditions were as follows: mobile phase, 0.1 m phosphate buffer and 0.15 m NaCl, pH 6.8; flow rate, 1.0 mL min−1; monitoring wavelength, 280 nm. (2) FPA (Ferguson, 1964). Bio-Rad's β-galactosidase (124 kD), bovine serum albumin (80 kD), ovalbumin (49.1 kD), and carbonic anhydrase (34.8 kD) were used as the molecular mass markers. Native PAGE, as described above, was run at three different gel concentrations (5%, 10%, and 15%). The molecular mass of the protein was estimated from plotting the logarithm of the protein's mobility as a function of the gel concentration. The slope of the resulting line (i.e. the retardation coefficient) is proportional to the protein's molecular mass. The subunit molecular masses were estimated by SDS-PAGE as previously described.
pI Measurement
The pI was determined using a chromatofocusing column (5.0 mm i.d. × 200 mm Mono P 5/200 GL; Amersham Biosciences) according to the manufacturer's specifications. The pH interval was 5.7 to 3.5. The flow rate was 0.5 mL min−1. The UV monitor wavelength was set at 280 nm.
Carbohydrate Detection
Protein glycosylation was detected using a high-sensitivity fluorescent glycoprotein detection kit (Sigma-Aldrich) according to the manufacturer's specifications. Following PAGE, the proteins were fixed on the gel with an acetic acid:methanol:water (3:50:47, v/v/v) solution. Protein-bound carbohydrates were oxidized to aldehydes with periodic acid. A hydrazide dye was reacted with the aldehydes, forming a stable fluorescent conjugate that was viewed using a standard fluorescent UV-transilluminator with emission at 312 nm.
Detection of P. alliacea Alliinase Activity and Substrate Specificity
Alliinase activity of crude protein fractions was detected in vitro using petiveriin as the substrate. The reaction mixture in a total volume of 1.0 mL of 20 mm phosphate buffer at pH 8.0 contained 1.5 mm petiveriin, 25 μm PLP, and 2 to 50 μL of the isolated protein fraction in the concentration range of 0.3 to 7.6 mg mL−1, depending on the stage of purification (see “Protein Extraction and Purification” above). The mixture was incubated for 10 to 15 min at room temperature and then the reaction was quenched by the addition of 1.0 mL of 10% (w/v) TCA. The generated pyruvate and thiosulfinate in the reaction were observed by RP C-18 HPLC using 20-μL injections of the reaction solution under the following conditions: flow rate, 1.0 mL min−1; mobile phase, water:acetonitrile (30:70, v/v); detection wavelength, 210 nm; column, Microsorb-MV 100Å, 250 × 4.6 mm, 5 μm (Varian). When alliinase was present in a protein fraction, pyruvate and petivericin eluted at 1.79 and 5.30 min, respectively. Similar experiments were conducted to determine substrate specificity. Sample solutions identical to that described above were made, except that the petiveriin substrate was replaced with the Cys sulfoxide derivative being tested. P. alliacea alliinase ability to cleave S-benzyl-l-Cys and S-2-hydroxyethyl-l-Cys was tested similarly. After incubation for 15 min, the resulting solution was analyzed by HPLC as described above to detect the formation of pyruvate and the corresponding thiosulfinate. The activity of alliinase purified to homogeneity was also detected using a pyruvate assay kit (BioVision) according to the manufacturer's specifications. To reaction solutions prepared as described above and incubated for 10 to 15 min were added 3 mL of boiling water to quench the reaction by inactivating the alliinase. The sample tube was then suspended in boiling water for an additional 3 min. The amount of pyruvate formed was assayed in 80-μL aliquots of the reaction mixture using the pyruvate assay kit. To determine the time frame during which the alliinase was active at room temperature, reaction solutions prepared as described above and containing 1.0 μg of purified alliinase (6.9 nm) were incubated with 1.5 mm petiveriin, and petivericin formation was monitored by HPLC at 5, 15, 25, 45, 65, 95, 125, 165, 205, and 255 min.
Determination of Enzyme Kinetics
The kinetics data of P. alliacea alliinase (Km, Vmax, kcat, and kcat/Km) were determined from Lineweaver-Burk plots and were based on the formation of pyruvate during catalytic turnover. Analyses were conducted in vitro with the substrates outlined in Table II at concentrations between 0 and 4.0 mm. The reaction mixture in a total volume of 200 μL was composed of substrate, 25 μm PLP, and 0.29 μg of purified alliinase (11.5 nm) in 20 mm phosphate buffer at pH 8.0. At various time points over a 20-min period at 25°C, the reaction was quenched by adding 600 μL of boiling water, followed by immersion of the reaction tube in a boiling-water solution for 3 min. The pyruvate formed over the time course of each reaction was oxidized by pyruvate oxidase to generate a colored adduct that was monitored by UV-visible (λ = 570 nm) or fluorescence (Ex/Em = 535/587 nm) spectroscopy.
Determination of the Importance of the Disulfide Bond in the P. alliacea Alliinase α′-Subunit to Alliinase Activity
In order to determine whether the disulfide bond in the P. alliacea alliinase α′-subunit is necessary for catalytic activity, the alliinase after reduction with BME was exposed to petiveriin according to the following protocol. A solution composed of 1.0 μg of purified alliinase, 20 μL of 20 mm phosphate buffer at pH 8.0, 2% (v/v) BME, and 25 μm PLP was prepared. After a 20-min incubation period, the solution containing the reduced alliinase was added to a second solution composed of 1.0 mL of 20 mm phosphate buffer at pH 8.0, 1.5 mm petiveriin, 2% (v/v) BME, and 25 μm PLP. After a 20-min incubation period, 20 μL of the reaction mixture was subjected to C-18 HPLC analysis to observe the production of any reaction product. The HPLC conditions were as follows: flow rate, 1.0 mL min−1; mobile phase, water:acetonitrile (30:70, v/v); column, Microsorb-MV 100Å, 250 × 4.6 mm, 5 μm (Varian).
Determination of the Ability of P. alliacea Alliinase to Precipitate Polysaccharides
The ability of P. alliacea alliinase to precipitate polysaccharides was investigated according to the method of Goldstein (1972): 23 μg of P. alliacea alliinase was incubated for 24 h with 30 μg of various polysaccharides (chitin, glycogen, dextran, and mannan) in 150 μL of 20 mm phosphate buffer at pH 7.2 containing 1.0 m NaCl. The turbidity of the solution was observed visually.
Protein Inactivation by Hydroxylamine
PLP-dependent enzymes are known to be inhibited by hydroxylamine. To assess the susceptibility of P. alliinase activity to hydroxylamine, reaction solutions similar to those described above (see “Determination of Enzyme Kinetics”) containing petiveriin were exposed to 0, 10, and 50 μm hydroxylamine. The effects of hydroxylamine were determined based on the formation of petivericin, as monitored by HPLC peak areas as a function of hydroxylamine concentration.
Determination of P. alliacea Alliinase pH Optimum
The pH optimum of the alliinase was determined by monitoring the production of petivericin produced from 1.0 μg of purified alliinase (6.9 nm) and 1.5 mm petiveriin as a function of varying pH in the following buffers: 20 mm 1-methylpiperazine (pH 4.0−6.0), 20 mm phosphate (pH 6.0−8.5), and 20 mm piperazine (pH 8.5−11.0). The peak areas of petivericin detected by HPLC with UV-diode array detection were integrated and plotted against pH in the range of 4.0 to 11.0.
Determination of P. alliacea Alliinase Temperature of Optimum Activity
The temperature optimum of alliinase was determined using a procedure similar to that described above (“Determination of P. alliacea Alliinase pH Optimum”). The buffer used was 20 mm phosphate, pH 8.0. Petivericin peak areas (using the highest product peak area as the benchmark) were plotted against temperature in the range of 4°C to 84°C over a period of 10 min. By plotting the inverse of the reaction rate against the inverse of the absolute temperature, the activation energy for the enzyme's reaction with petiveriin was estimated according to the Arrhenius equation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular mass determination of P. alliacea alliinase by gel-filtration chromatography.
Supplemental Figure S2. Molecular mass determination of P. alliacea alliinase by FPA.
Supplemental Figure S3. P. alliacea alliinase pI determination by chromatofocusing.
Supplemental Figure S4. Determination of the importance of the disulfide bond in the P. alliacea alliinase α′-subunit to alliinase activity.
Supplemental Figure S5. pH optimum determination of P. alliacea alliinase.
Supplemental Figure S6. Determination of temperature of optimum activity for P. alliacea alliinase.
Supplemental Materials and Methods S1. Supplemental Materials and Methods corresponding to Supplemental Figures S1 to S6.
Supplementary Material
Acknowledgments
We thank Distinguished Professor Dr. Eric Block for helpful discussions and critical reading of the manuscript.
This work was supported by the National Science Foundation (grant no. 0239755) and the Research Foundation of the State University of New York.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rabi A. Musah (musah@albany.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adesogan EK (1974) Trithiolaniacin, a novel trithiolan from Petiveria alliacea. J Chem Soc Chem Commun 21 906–907 [Google Scholar]
- Ayedoun MA, Moudachirou M, Sossou PV, Garneau FX, Gagnon H, Jean FI (1998) Volatile constituents of the root oil of Petiveria alliacea L. from Benin. J Essent Oil Res 10 645–646 [Google Scholar]
- Block E (1992) The organosulfur chemistry of the genus Allium: implications for the organic chemistry of sulfur. Angew Chem Int Ed Engl 31 1135–1178 [Google Scholar]
- Block E (2010) Garlic and Other Alliums: The Lore and the Science. Royal Society of Chemistry, Cambridge, UK
- Davis B (1964) Disk electrophoresis. II. Method and application to human serum proteins. Ann N Y Acad Sci 121 404–427 [DOI] [PubMed] [Google Scholar]
- Ferguson KA (1964) Starch-gel electrophoresis: application to the classification of pituitary proteins and polypeptides. Metabolism 13 985–1002 [DOI] [PubMed] [Google Scholar]
- Fujita M, Endo M, Sano M (1990) Purification and characterization of alliin lyase from Welsh onion, Allium fistulosum L. Agric Biol Chem 54 1077–1079 [Google Scholar]
- Goldstein IJ (1972) Use of concanavalin A for structural studies. Methods Carbohydr Chem 6 106–119 [Google Scholar]
- Imai S, Tsuge N, Tomotake M, Nagatome Y, Sawad H, Nagata T, Kumagai H (2002) An onion enzyme that makes the eyes water. Nature 419 685. [DOI] [PubMed] [Google Scholar]
- Ken-ichi Y, Ayumi M, Gunki F (1996) Amino acid sequence and some properties of lectin-D from the roots of pokeweed (Phytolacca americana). Biosci Biotechnol Biochem 60 1380–1382 [DOI] [PubMed] [Google Scholar]
- Kim S, Kubec R, Musah RA (2006) Antibacterial and antifungal activity of sulfur-containing compounds from Petiveria alliacea L. J Ethnopharm 104 188–192 [DOI] [PubMed] [Google Scholar]
- Kubec R, Kim S, Musah RA (2002) S-Substituted cysteine derivatives and thiosulfinate formation in Petiveria alliacea: part II. Phytochemistry 61 675–680 [DOI] [PubMed] [Google Scholar]
- Kubec R, Kim S, Musah RA (2003) The lachrymatory principle of Petiveria alliacea. Phytochemistry 63 37–40 [DOI] [PubMed] [Google Scholar]
- Kubec R, Musah RA (2001) Cysteine sulfoxide derivatives in Petiveria alliacea. Phytochemistry 58 981–985 [DOI] [PubMed] [Google Scholar]
- Kumagai H, Kono H, Sakurai H, Tokimoto K (2002) Comparison of C-S lyase in Lentinus edodes and Allium sativum. Biosci Biotechnol Biochem 66 2560–2566 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lancaster JE, Collin HA (1981) Presence of alliinase in isolated vacuoles and alkyl cysteine sulphoxides in the cytoplasm of bulbs in onion (Allium cepa). Plant Sci Lett 22 169–176 [Google Scholar]
- Lancaster JE, Shaw ML, Joyce MP, McCallum JA, McManus MT (2000) A novel alliinase from onion roots: biochemical characterization and cDNA cloning. Plant Physiol 122 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landshuter J, Lohmuller EM, Knobloch K (1994) Purification and characterization of a C-S-lyase from ramson, the wild garlic Allium ursinum. Planta Med 60 343–347 [DOI] [PubMed] [Google Scholar]
- Lohmuller EM, Landshuter J, Knobloch K (1994) On the isolation and characterisation of a C-S-lyase preparation from leek, Allium porrum. Planta Med 60 337–342 [DOI] [PubMed] [Google Scholar]
- Manabe T, Hasumi A, Sugiyama M, Yamazaki M, Saito K (1998) Alliinase (S-alk(en)yl-l-cysteine sulfoxide lyase) from Allium tuberosum (Chinese chive), purification, localization, cDNA cloning and heterologous functional expression. Eur J Biochem 257 21–30 [DOI] [PubMed] [Google Scholar]
- Marciani DJ, Papamatheakis JD (1978) Anomalous behavior of the major avian myeloblastosis virus glycoprotein in the presence of sodium dodecyl sulfate. J Virol 26 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazelis M, Crews L (1968) Purification of the alliin lyase of garlic, Allium sativum L. Biochem J 108 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoru H, Tomomi F, Mika H, Masatsune I, Yasuo H (2003) Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed. J Mol Biol 334 551–565 [DOI] [PubMed] [Google Scholar]
- Musah RA, He Q, Kubec R (2009) Discovery and characterization of a novel lachrymatory factor synthase in Petiveria alliacea and its influence on alliinase-mediated formation of biologically active organosulfur compounds. Plant Physiol 151 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock LP, Mazelis M (1986) The C-S lyase of higher plants: preparation and properties of homogeneous alliin lyase from garlic (Allium sativum). Arch Biochem Biophys 249 27–33 [DOI] [PubMed] [Google Scholar]
- Nock LP, Mazelis M (1987) The C-S lyases of higher plants: direct comparison of the physical properties of homogeneous alliin lyase of garlic (Allium sativum) and onion (Allium cepa). Plant Physiol 85 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinkov A, Wilchek M, Mirelman D (1995) Alliinase (alliin lyase) from garlic (Allium sativum) is glycosylated at ASN146 and forms a complex with a garlic mannose-specific lectin. Glycoconj J 12 690–698 [DOI] [PubMed] [Google Scholar]
- Rabinkov A, Zhu XZ, Grafi G, Galili G, Mirelman D (1994) Alliin lyase (alliinase) from garlic (Allium sativum): biochemical characterization and cDNA cloning. Appl Biochem Biotechnol 48 149–171 [DOI] [PubMed] [Google Scholar]
- Ramirez EC (2003) Alliinases. Food Sci Technol 122 1043–1050 [Google Scholar]
- Segrest JP, Jackson RL, Andrews EP, Marchesi VT (1971) Human erythrocyte membrane glycoprotein: a re-evaluation of the molecular weight as determined by SDS polyacrylamide gel electrophoresis. Biochem Biophys Res Commun 44 390–395 [DOI] [PubMed] [Google Scholar]
- Shimon LJW, Rabinkov A, Shin I, Miron T, Mirelman D, Wilchek M, Frolow F (2007) Two structures of alliinase from Allium sativum L.: apo form and ternary complex with aminoacrylate reaction intermediate covalently bound to the PLP cofactor. J Mol Biol 366 611–625 [DOI] [PubMed] [Google Scholar]
- Simpson RJ (2004) Purifying Proteins for Proteomics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Smeets K, Van Damme EJM, Van Leuven F, Peumans WJ (1997) Isolation and characterization of lectins and lectin-alliinase complexes from bulbs of garlic (Allium sativum) and ramsons (Allium ursinum). Glycoconj J 14 331–343 [DOI] [PubMed] [Google Scholar]
- Sousa JR, Demuner AJ, Pinheiro JA, Breitmaier E, Cassels BK (1990) Dibenzyl trisulphide and trans-N-methyl-4-methoxyproline from Petiveria alliacea. Phytochemistry 29 3653–3655 [Google Scholar]
- Szczepanski C, Zgorzelak P, Hoyer GA (1972) Isolierung, Strukturaufklarung und Synthese einer antimikrobiell wirksamen Substanz aus Petiveria alliacea L. Arzneimittelforschung 22 1975–1976 [PubMed] [Google Scholar]
- Taylor L (2005) The Healing Power of Rainforest Herbs. Square One Publishers, Garden City Park, NY, pp 166–170
- Tobkin HE Jr, Mazelis M (1979) Alliin lyase: preparation and characterization of the homogeneous enzyme from onion bulbs. Arch Biochem Biophys 193 150–157 [DOI] [PubMed] [Google Scholar]
- Tomomi F, Minoru H, Mika H, Masatsune I, Yasuo H (2004) Structures of two lectins from the roots of pokeweed (Phytolacca americana). Acta Crystallogr D Biol Crystallogr D60 665–673 [DOI] [PubMed] [Google Scholar]
- Ukai K, Sekiya J (1997) A new staining method for lyases catalyzing cleavage of a C-S bond in sulfur containing compounds after polyacrylamide gel electrophoresis. Biosci Biotechnol Biochem 61 124–126 [Google Scholar]
- Won T, Mazelis M (1989) The C-S lyase of higher plants: purification and characterization of homogeneous alliin lyase of leek (Allium porrum). Physiol Plant 77 87–92 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.