Abstract
Abscisic acid (ABA) is a plant hormone that regulates plant growth as well as stress responses. In this study, we identified and characterized a new Arabidopsis (Arabidopsis thaliana) protein, Nuclear Protein X1 (NPX1), which was up-regulated by stress and treatment with exogenous ABA. Stomatal closure, seed germination, and primary root growth are well-known ABA responses that were less sensitive to ABA in NPX1-overexpressing plants. NPX1-overexpressing plants were more drought sensitive, and the changes in response to drought were due to the altered guard cell sensitivity to ABA in transgenic plants and not to a lack of ABA production. The nuclear localization of NPX1 correlated with changes in the expression of genes involved in ABA biosynthesis and ABA signal transduction. To understand the function of NPX1, we searched for interacting proteins and found that an ABA-inducible NAC transcription factor, TIP, interacted with NPX1. Based on the whole plant phenotypes, we hypothesized that NPX1 acts as a transcriptional repressor, and this was demonstrated in yeast, where we showed that TIP was repressed by NPX1. Our results indicate that the previously unknown protein NPX1 acts as a negative regulator in plant response to changes in environmental conditions through the control of ABA-regulated gene expression. The characterization of this factor enhances our understanding of guard cell function and the mechanisms that plants use to modulate water loss from leaves under drought conditions.
In plant and animal genomes, there is a relatively high percentage of unknown genes that currently lack defined motifs or domains (Gollery et al., 2006). Most of the differences in the composition of genes between species have been attributed to proteins with obscure features (POFs). Arabidopsis (Arabidopsis thaliana) contains 5,069 POFs, and 2,045 have no obvious homologs in rice (Oryza sativa) or poplar (Populus species). These POFs have been suggested to be involved in species- or phylogenesis-specific functions in Arabidopsis. Therefore, the study of unknown genes or POFs is an important task that should provide information that will enhance our understanding of what makes plant species unique (Gollery et al., 2007). A couple of approaches have been suggested for defining the functions of proteins of unknown function. Genome-wide clustering and the analysis of gene function in clusters allowed for the association of 1,541 proteins of unknown function with coexpressed genes for proteins of known function (Horan et al., 2008). Gene expression analysis was also used to identify a comprehensive set of abiotic stress-response genes (Horan et al., 2008). Coexpression and genome-wide transcript profiling data represent an important resource for guiding future functional characterization experiments of proteins or genes of unknown function. In this study, we were able to develop hypotheses regarding potential gene function based on available expression data in the AtGenExpress database (Arabidopsis eFP Browser; Winter et al., 2007).
Plants grow in environments where they encounter dynamic stresses such as cold, drought, and variations in nutrient concentrations. Molecular and cellular responses to abiotic stresses are becoming understood, which will allow for the engineering of how plants respond to such conditions (Zhu, 2002; Shinozaki and Yamaguchi-Shinozaki, 2007). The early events of plant adaptation to environmental stresses trigger specific and more general stress signal transduction networks, leading to specific changes in gene expression. The up-regulation of expression of several stress-regulated genes results in improved tolerance to drought, salt, and cold (Kasuga et al., 1999; Saijo et al., 2000).
The importance of potassium for plant growth and development is well documented. It plays a role in a wide range of functions in plants, including photosynthesis, enzyme activation, protein synthesis, osmotic adjustment (Marschner, 1995), and transpiration (Schroeder et al., 2001a). The protein of unknown function that we characterized in this work was identified as being regulated by potassium availability. Potassium is also known to be important under drought conditions. Stomatal closure occurs following K+ and anion efflux, resulting in the loss of water from the cell and leading to a reduction in cell turgor and pore closure (Schroeder et al., 2001b). Reducing water loss through stomatal closure is one strategy to minimize the adverse effects of drought (Riera et al., 2005); therefore, K+ homeostasis may be considered as a key factor in drought adaptation.
The phytohormone abscisic acid (ABA) is also a key factor in regulating developmental and physiological processes in plants, including seed dormancy and germination and seedling growth, as well as in controlling responses to many abiotic stresses such as drought (Schroeder et al., 2001b; Assmann, 2003; Yamaguchi-Shinozaki and Shinozaki, 2006). Under drought conditions, the ABA concentrations in leaves increase and this increased ABA acts as a signal (Christmann et al., 2007; Schachtman and Goodger, 2008). The production of ABA in roots and its transport to the leaves also provides the plant with a mechanism for transmitting a signal to report on the water status of the soil. ABA is also required to maintain root growth in addition to its role in reducing transpiration by triggering stomatal closure under water deficit (Schachtman and Goodger, 2008). Additionally, ABA may act as a signal under reduced nutrient supply (Peuke et al., 1994, 2002). Although increased ABA in grains from K+-deficient wheat (Triticum aestivum) plants and increased ABA concentrations in K+-deficient wheat flag leaves (Haeder and Beringer, 1981) have been measured, the physiological consequences of this increase in ABA are unknown.
To identify the initial cellular responses to K+ deprivation, we performed microarray experiments on Arabidopsis roots (Shin and Schachtman, 2004). From these experiments, the expression of the unknown gene, Nuclear Protein X1 (NPX1), was found to be up-regulated by potassium deprivation. In this study, we show that this protein of unknown function in Arabidopsis is a component in the ABA signaling pathway. We present evidence that NPX1 acts on the ABA signal transduction pathway by repressing at least one transcription factor, which results in multiple, well-known ABA-associated phenotypes, including increased drought tolerance.
RESULTS
Expression of NPX1 Was Enhanced under Stress Conditions
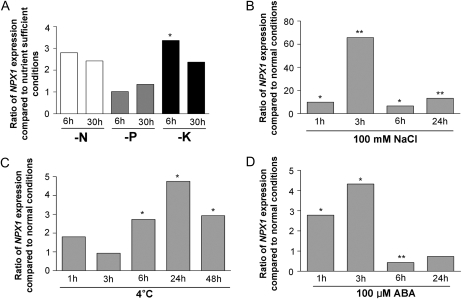
Previously, we found that the unknown Arabidopsis nuclear factor NPX1 was up-regulated in roots under K+ deprivation. To determine whether NPX1 is regulated by other nutrient stresses, real-time PCR was performed on root RNA isolated from plants that had been deprived of potassium, nitrogen, or phosphorus. The abundance of the NPX1 transcript increased significantly in roots starved of K+ for 6 h (Fig. 1A). NPX1 was widely expressed in different tissues (Supplemental Fig. S1) and was also found to be regulated by abiotic stresses, as determined by the AtGenExpress database (Arabidopsis eFP Browser; Winter et al., 2007). We verified that the expression of NPX1 increased in response to salt stress (Fig. 1B), increased under cold stress (Fig. 1C), and responded to ABA treatment (Fig. 1D). The NPX1 gene expression patterns suggested that NPX1 may be involved in ABA-dependent abiotic stress signaling.
Figure 1.
The expression of NPX1 was up-regulated by nutrient deprivation and abiotic stresses. Quantitative real-time PCR analysis of NPX1 under nutrient-deprived and abiotic stress conditions and ABA treatment is shown. A, Arabidopsis ecotype Columbia roots grown under nutrient-sufficient conditions, K+ deficiency (−K), N deficiency (−N), and P deficiency (−P) 6 or 30 h after incubation. B, Expression of NPX1 in the whole seedlings after treatment with 100 mm NaCl for 1, 3, 6, and 24 h. C, Expression of NPX1 in the whole seedlings after cold stress (4°C) for 1, 3, 6, 24, and 48 h. D, Expression of NPX1 in the whole seedlings after 100 μm ABA treatment for 1, 3, 6, and 24 h. Data represent means of three biological replicates. Asterisks indicate significant differences (* P < 0.05, ** P < 0.01, by Student's t test).
Overexpression of NPX1 Causes ABA Insensitivity
To further elucidate the function of NPX1, a homozygous T-DNA inactivation line, npx1-1 (SAIL_123), was isolated, and NPX1-overexpressing (NPX1-ox) lines under the control of the figwort mosaic virus promoter were created (for expression data on these lines, see Supplemental Fig S2). One NPX1-ox line with the highest expression level was chosen for further analysis. Multiple transgenic lines containing a 35S promoter-driven NPX1 cDNA with a yellow fluorescent protein at the N terminus (NYFP-NPX1-ox) showed the same phenotypes described below (Supplemental Fig. S2). The growth of npx1-1 was tested multiple times, and we found no significant differences in growth as compared with the wild type under control or K+-deprived conditions.
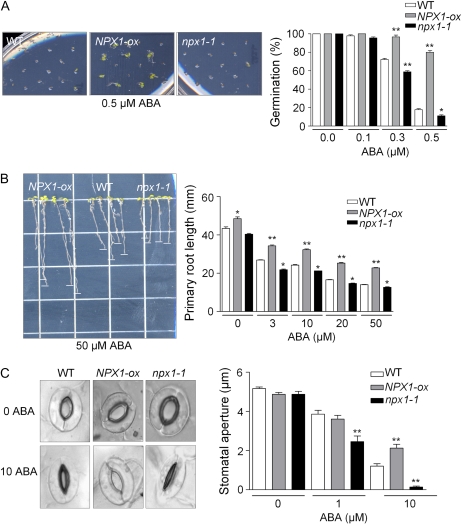
To elucidate the role of NPX1 in ABA signaling, we tested whether the disruption or overexpression of NPX1 affected ABA responses. Since ABA signaling is known to be an important component in seed germination, we determined the ABA sensitivity of germination (Fig. 2A). In the absence of exogenous ABA, NPX1-ox and npx1-1 mutant seeds germinated as well as wild-type seeds (Fig. 2A); in the presence of 0.3 and 0.5 μm ABA, NPX1-ox seed germination was less sensitive to ABA, whereas npx1-1 seed germination was more sensitive. NYFP-NPX1-ox seed germination was also less sensitive to ABA treatment (Supplemental Fig. S2B). After germination, ABA may still regulate seedling growth; therefore, we analyzed this aspect of ABA sensitivity by transferring 6-d-old seedlings on 0.25× Murashige and Skoog (MS) medium with sufficient potassium to plates containing 0, 3, 10, 20, and 50 μm ABA (Kuhn et al., 2006). The primary root of NPX1-ox and two NYFP-NPX1-ox lines was longer than the wild type under control conditions and also on medium containing 3 to 50 μm ABA (Fig. 2B; Supplemental Fig. S2C), whereas the primary root growth of npx1-1 was more sensitive to ABA. The complemented NPX1-transformed npx1-1 plants (npx1-1-NPX1) showed the same ABA sensitivity as the wild type in germination and root growth (Supplemental Fig. S2). When grown under K+-deprived conditions, the primary root length of NPX1-ox was significantly longer as compared with the wild type (Supplemental Fig. S3).
Figure 2.
Constitutive expression of NPX1 results in ABA insensitivity, while disruption of NPX1 causes ABA hypersensitivity during seed germination, inhibition of primary root growth, and stomatal closure. A, Germination of wild-type (WT), NPX1-ox, and npx1-1 seeds at 0.5 μm ABA after 5 d (left), and germination rates of seeds exposed to 0, 0.1, 0.3, and 0.5 μm ABA at 5 d (right). Data represent means ± sd of three independent experiments with 36 seeds per genotype and experiment. B, The photographs show plants of the wild type, NPX1-ox, and npx1-1 treated with 0.25× MS medium supplemented with 50 μm ABA 10 d after transfer of 6-d-old seedlings from medium without ABA (left). At right is a comparison of root elongation of wild-type, NPX1-ox, and npx1-1 seedlings; 6-d-old seedlings were transferred to plates supplemented with 0, 3, 10, 20, and 50 μm ABA, and root elongation was monitored after 6 d. Each data point represents the mean ± sd (n = 70). Asterisks indicate significant differences between the wild type and NPX1-ox or npx1-1 plants (* P < 0.05, ** P < 0.01). C, Photographs show the stomatal apertures in ABA-induced stomata closure assays with 0 (top row) and 10 μm ABA (bottom row) after 2 h of incubation. At right are stomatal aperture measurements of wild-type, NPX1-ox, and npx1-1 plants in response to 0, 1, and 10 μm ABA. Data represent means ± sd with 50 stomata per data point. Asterisks indicate significant differences between the wild type and NPX1-ox or npx1-1 plants (**P < 0.01).
Since it is well known that both ABA and potassium are critical components in stomatal movement and response to drought, we analyzed stomatal responses to ABA in loss- and gain-of-function NPX1 plants (Fig. 2C). Compared with the wild type, guard cells from NPX1-ox plants exhibited greater insensitivity to ABA-induced stomatal closure. Ten micromolar ABA (Fig. 2C) resulted in stomatal closure in wild-type plants but not in the NPX1-ox lines. In contrast, the disruption of NPX1 increased stomatal sensitivity and stomatal closure responses at 1 μm. These data show that NPX1 plays an important role in multiple processes that involve ABA signal transduction, including germination, root growth, and the regulation of stomatal aperture. Taken together, our data suggest an important role for NPX1 as a negative regulator of ABA signaling.
Plants Overexpressing NPX1 Show Hypersensitivity to Drought Stress
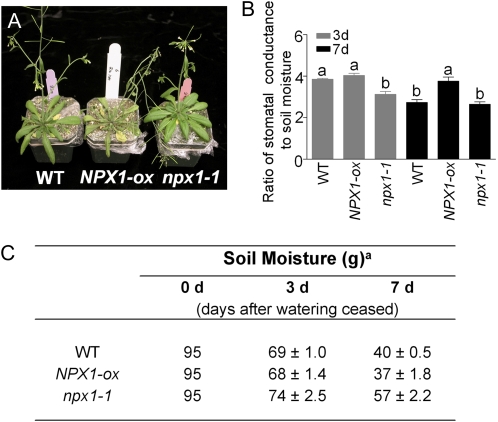
The control of water loss from the leaf surface triggered by ABA is a crucial survival mechanism for plants during drought periods. To investigate the role of NPX1 in regulating drought tolerance and water loss, the soil of 3-week-old plants was covered with plastic and water was withheld for 2 weeks. NPX1-ox plants wilted more quickly than the other lines, whereas npx1-1 remained turgid longer (Fig. 3A). Since survival depends on the rate at which water is depleted from pots, we also measured the stomatal conductance 3 and 7 d after watering was stopped and calculated the ratio of stomatal conductance to soil moisture. This provides a measure of the sensitivity of stomatal closure to soil moisture or stomatal response to soil water deficit. Three days after watering was stopped, the soil moisture levels in the pots were still relatively high (Fig. 3C), but the npx1-1 plants had significantly lower rates of water loss (Fig. 3B). In contrast, the transpiration rate of NPX1-ox plants was not significantly different from the wild type (Fig. 3B). However, after 7 d, NPX1-ox plants showed a 1.4-fold increase in water loss compared with the wild type and npx1-1 (Fig. 3, B and C). This result correlated with the different stomatal responses that we observed when adding ABA (Fig. 2C). From these results, we conclude that overexpression of NPX1 reduces sensitivity to ABA, so those plants lose water faster and are more drought sensitive than npx1-1 and the wild type.
Figure 3.
Plants overexpressing NPX1 wilted more quickly under terminal drought stress. A, Photographs of plants after drought stress treatment that involves withholding water. Three-week-old plants of wild-type (WT), NPX1-ox, and npx1-1 plants were gown for a period of 14 d without added water. B, Stomatal conductance and soil moisture after 3 and 7 d without added water. Different letters indicate significant differences between means at P < 0.05 (Tukey's honestly significant difference; n = 20 plants). C, Table shows the soil moisture during drought stress treatment (n = 20 for each group). aThe values are presented as means ± se.
NPX1 Is a Nuclear Protein
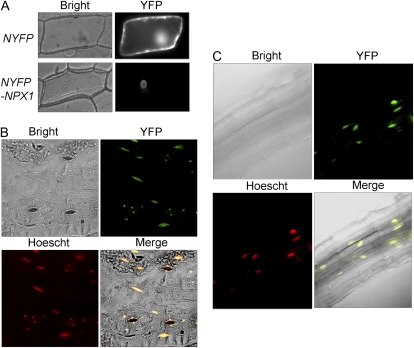
We identified three putative nuclear localization sequences (softberry ProtComp6.0; http://linux1.softberry.com/berry.phtml?topic=protcompan&group=help&subgroup=proloc) in the C-terminal region of NPX1, which suggested that NPX1 could be localized to the nucleus. Therefore, we tested the cellular localization of NPX1 fused to yellow fluorescent protein at the N terminus of NPX1 (NYFP-NPX1) by transient expression in onion (Allium cepa) epidermal cells. Fluorescence was observed in the nucleus of cells where the NYFP-NPX1 was expressed, whereas the NYFP empty vector control was mainly present in cytoplasm (Fig. 4A). To confirm the nuclear localization of NPX1 protein, NYFP-NPX1 was stably transformed into Arabidopsis and the fluorescence was measured in both leaves (Fig. 4B) and roots (Fig. 4C). Cells expressing the NYFP-NPX1 also showed that the YFP signal appeared only in the nucleus.
Figure 4.
NYFP-NPX1 protein was localized to the nucleus of root and guard cells. A, Fluorescent proteins were transiently expressed in onion epidermal cells: NYFP (top) and NYFP-NPX1 (bottom). Signals from bright field (bright) and NYFP (YFP) are shown. B and C, NPX1 localization in stable transgenic Arabidopsis leaves (B) and roots (C). Signals from bright field (bright) and NYFP (YFP; green) and Hoescht signal (red) indicate nuclear localization, and the merge of the three signals (merge; yellow) is shown.
NPX1 Regulates the Expression of ABA Signaling Genes
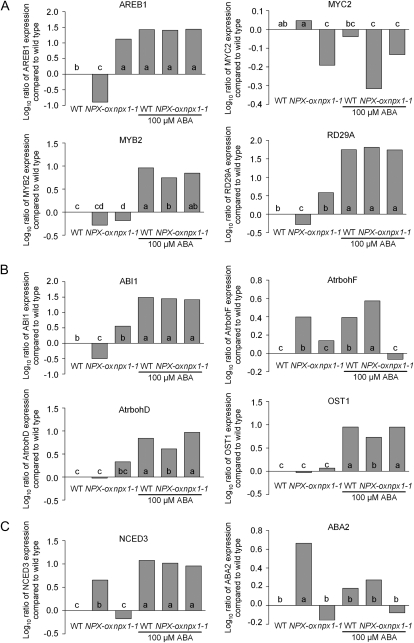
Since results suggested that NPX1 may be involved in ABA signaling and was localized to the nucleus, we tested whether NPX1 is involved in regulating the expression of genes important for ABA signaling in the wild type, NPX1-ox, and npx1-1 before and after treatment with 100 μm ABA. Our results clearly show that the expression of ABA-Responsive Element Binding1 (AREB1), RD29A, and ABI1 was down-regulated in NPX1-ox without ABA treatment (Fig. 5, A and B). In contrast, the expression of ABA biosynthesis genes NCED3 and ABA2 as well as AtrbohF was up-regulated in NPX1-ox without ABA treatment (Fig. 5C). In the wild type, the expression of MYB2, AtrbohD, and Open Stomata1 (OST1) was up-regulated after treatment with ABA (Fig. 5). However, the induction of these genes was greatly attenuated or abolished in NPX1-ox. The AtrbohF gene was expressed at higher levels in the NPX1-ox lines even without the application of exogenous ABA, and the expression pattern was opposite that of AtrbohD (Fig. 5).
Figure 5.
Constitutive expression of NPX1 or disruption of NPX1 alters the gene expression of ABA signaling genes. Seedlings growing on agar plates were untreated or treated with 100 μm ABA for 3 h. Gene expression before and after ABA treatment by real-time PCR analysis is shown. Real-time PCR for expression of genes involved in the ABA-dependent stress-responsive pathway (A), ABA-induced stomatal closing (B), or the ABA biosynthetic pathway (C). Different letters indicate significant differences between means at P < 0.05 (Student's t test). For full information on the genes represented in this figure, see Supplemental Table S2. WT, Wild type.
NPX1 Contributes to the Regulation of ABA Biosynthesis
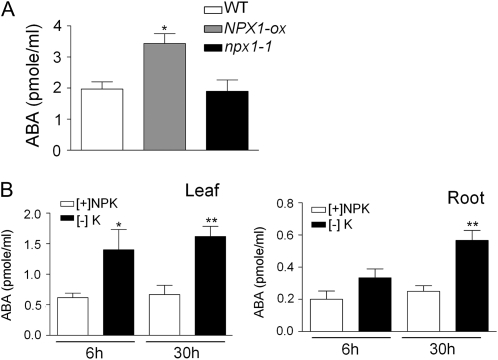
The fact that NPX1 is coexpressed with the ABA biosynthesis gene NCED3 (Ma and Bohnert, 2008) and that the expression of NCED3 and ABA2 is up-regulated in NPX1-ox without ABA treatment (Fig. 5C) suggest that NPX1 may be involved in the regulation of biosynthesis of ABA. To investigate this possibility, we measured ABA in the wild type, NPX1-ox, and npx1-1 using multiple plants in three sets of independent biological replicates. The ABA levels were significantly higher in NPX1-ox than in the other lines (Fig. 6A). We also measured ABA levels in leaves and roots after K+ starvation. The level of ABA in leaves increased 6 and 30 h after deprivation, whereas the production of ABA in roots increased only 30 h after deprivation (Fig. 6B), which confirms previous results in Ricinus communis (Peuke et al., 2002) and helps to explain why this gene was up-regulated under potassium-deprived conditions.
Figure 6.
Constitutive expression of NPX1 and potassium deprivation increased the biosynthesis of ABA. A, ABA concentrations in leaves of wild-type (WT), NPX1-ox, and npx1-1 plants grown on agar plates. Data represent means of three biological replicates each containing multiple plants. B, Comparison of ABA concentrations in leaves or roots of plants grown under full nutrient conditions (+NPK) or K+ deficiency (−K) for 6 or 30 h. Data represent means of six biological replicates. Asterisks indicate significant differences as compared with other means in each subfigure (* P < 0.05, ** P < 0.01, by Student's t test).
NPX1 Interacts with TIP and Represses Transcription in Yeast
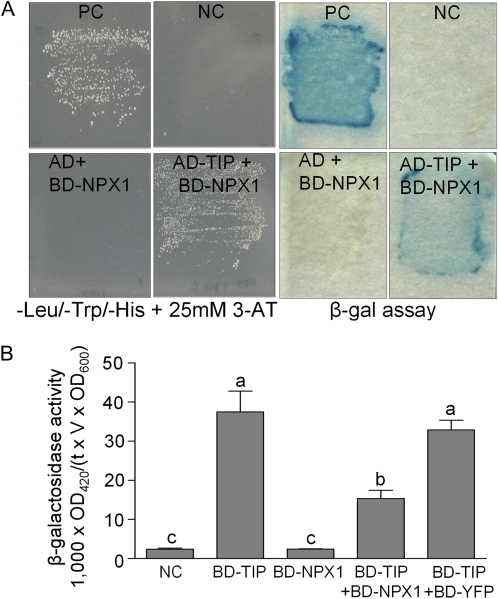
Because NPX1 was localized to the nucleus and the expression of several genes involved in ABA biosynthesis and signaling was altered in NPX1-ox plants, we tested the transcriptional activation of NPX1 even though it did not contain a classical DNA-binding domain (BD). For this experiment, NPX1 was fused with GAL4 BD and transformed with GAL4 activation domain (AD) containing empty vector. NPX1 did not show transcriptional activation activity in yeast (data not shown). Therefore, we tested whether NPX1 may regulate gene expression through interactions with other proteins such as transcription factors. To identify proteins that interact with NPX1, we used the yeast two-hybrid system with NPX1 fused in-frame to the GAL4 BD (BD-NPX1). First, we checked for interactions between BD-NPX1 and GAL4 AD fused to eight different transcription factors that were already known to be regulators of ABA signaling: AREB1, AREB2, AREB3, DREB2A, MYB2, MYC2, CBF4, and ATHB6 (Riera et al., 2005; Nilson and Assmann, 2007). However, none of these eight candidates interacted with NPX1 (data not shown). Therefore, we screened an Arabidopsis root library fused to the GAL4 AD. Of the 1.5 × 106 transformants screened, 44 colonies initially grew on plates without Leu, Trp, and His. When these 44 clones were retransformed and subjected to colony-lift filter assay, 20 clones exhibited β-galactosidase activity. The cDNAs were recovered from these 20 colonies and sequenced (Supplemental Table S1). One of the cDNAs encoded a NAC transcriptional factor previously named TIP (Ren et al., 2000). The interaction between NPX1 and TIP was confirmed using additional plasmid combinations and constructs to test for the following: autoactivation of the HIS3 and lacZ reporter genes, possible direct interactions with the GAL4 BD or AD, and potential artifacts caused by high expression of the encoded GAL4 fusion protein. BD-NPX1 alone did not activate the HIS3 or lacZ reporter gene, whereas the cotransformation of BD-NPX1 and AD-TIP activates HIS3 or lacZ reporter gene (Fig. 7A). This interaction was confirmed by quantitative β-galactosidase assays using o-nitrophenyl-β-d-galactopyranoside as a substrate. The β-galactosidase activity of NPX1- and TIP-cotransformed cells was about 34% of that observed for the positive control (BD-Krev1 and AD-RalGDS-wt; data not shown).
Figure 7.
NPX1 interacts with TIP and represses the transcription activity of TIP in yeast. A, Interaction analysis of NPX1 using the yeast two-hybrid system. The yeast strain MaV203 was cotransformed as indicated. PC, Positive control (BD-Krev1 and AD-RalGDS-wt were used); NC, negative control (BD-Krev1 and AD-RalGDS-m2 were used). Activation of HIS3 and lacZ is indicated by growth on −His medium and by the blue color, respectively. White colonies indicate a lack of reporter activation. B, NPX1 represses the activity of TIP. BD-NPX1 represses the activity of TIP in yeast, as indicated by the decreased β-galactosidase (β-gal) activity. BD-TIP but not NC (cotransformants of pGBKT7 and pDEST 32 vector) and BD-NPX1 activate lacZ expression. Data represent means ± sd of three independent experiments. Different letters indicate significant differences between means at P < 0.05 (Student's t test). OD, Optical density.
To reveal the underlying molecular mechanism of interaction between NPX1 and TIP, we tested whether this direct interaction could lead to activation or repression of the reporter gene expression in yeast. Yeast containing an integrated GAL4-lacZ reporter was transformed with BD-TIP in the presence or absence of BD-NPX1. BD-TIP was previously reported to activate a lacZ reporter (Ren et al., 2000), and we confirmed this activation (Fig. 7B). The activity of BD-TIP was significantly reduced by 54% when BD-NPX1 was present but not significantly reduced by BD-YFP, which was used as a control (Fig. 7B). Taken together, our results indicate that NPX1 interacts with TIP and functions as a transcriptional repressor.
DISCUSSION
NPX1 Is a Novel Protein Involved in ABA Responses
The previously unknown gene NPX1 was identified as a gene whose expression was up-regulated by potassium deficiency (Shin and Schachtman, 2004). To gain insight into the function of the Arabidopsis NPX1 gene, we searched databases for hormones and environmental conditions that regulated the transcription of this gene. NPX1 was found to be up-regulated by dehydration, low temperature, and high salt and coexpressed with the ABA biosynthetic gene NCED3 (Ma and Bohnert, 2008). The phytohormone ABA plays a central role in regulating plant responses to environmental stress, including water deficit, high salt, and low temperature (Leung and Giraudat, 1998). To confirm whether NPX1 played a role in ABA responses, we studied how exogenously applied ABA affected germination, stomatal conductance, drought tolerance, and root growth, which are all known to be physiological responses that are influenced by ABA (Nilson and Assmann, 2007). In germination and root growth assays, we showed that the overexpression of NPX1 decreased the sensitivity to exogenously applied ABA.
ABA plays a key role in reducing plant water loss and stimulating root growth under conditions of water deficit. When plants are drought stressed, ABA concentrations in leaves increase, which leads to stomatal closure ultimately controlling whole plant water loss (Nilson and Assmann, 2007). Guard cells, which make up each stomatal complex, shrink, which results in closure and swell, which results in opening. Stomatal apertures in response to ABA in isolated epidermal peels containing guard cells were compared between wild-type, overexpression, and knockout lines of NPX1. Those studies clearly showed that isolated guard cells from NPX1-ox plants were relatively insensitive to the application of low concentrations of ABA, which would normally close guard cells in wild-type plants (Pei et al., 1997). To demonstrate that the ABA insensitivity of guard cells was also evident in transpiring leaves on whole plants, we carefully measured soil moisture content and water loss as well as plant survival in NPX1-ox, wild-type, and npx1-1 lines. Soil moisture content plays a key role in stomatal response, and it is well known that transpiration declines as a result of soil drying due to increased leaf ABA concentrations (Schachtman and Goodger, 2008). At the whole plant level, we clearly showed that guard cells in plants overexpressing NPX1 did not close as rapidly upon soil drying, so NPX1-ox plants survived for a shorter time period as compared with other lines under water-limiting conditions, because they depleted the soil moisture more rapidly. This phenotype is reminiscent of other mutations such as abi1-1 and abi2-1 (Leung et al., 1997). Taken together, NPX1-ox plants were less sensitive to ABA at both the cellular and whole plant levels, indicating that NPX1 acts a negative regulator of ABA response or biosynthesis. Measurement of ABA concentrations showed that insensitivity in the NPX1-ox lines does not occur due to reduced ABA biosynthesis, because NPX1-ox plants had higher levels of ABA than wild-type plants or the npx1-1 knockout. Other molecular events, including the action of transcription factors and phosphorylation, also serve as positive and negative regulators in ABA signaling (Nilson and Assmann, 2007).
NPX1 Is a Negative Transcriptional Regulator of ABA Responses
To understand how NPX1 could modulate ABA responses, we scanned the encoded protein for known domains and found that NPX1 had some limited similarity to the global transcription factor group E8 that contains a bromodomain. Although NPX1 does not contain a functional DNA BD, we tested for transcriptional activity in yeast using β-galactosidase assays with GAL4 BD-fused NPX1 and found that the protein does not show activity that might be associated with a transcription factor (data not shown). Localization studies showed that a GFP fusion of NPX1 was localized to the nucleus and therefore may be involved in the regulation of transcription by binding to other proteins.
Previous studies aimed at revealing the molecular determinants of ABA signaling have highlighted regulators that act at the transcriptional and posttranscriptional levels (Riera et al., 2005; Nilson and Assmann, 2007). Since NPX1 was localized to the nucleus and acts as a negative regulator of ABA responses, we determined whether there was a link between NPX1 and transcription of many well-known genes involved in ABA signal transduction and biosynthesis. One SnRK2 kinase, OST1, plays a central role in ABA signaling events and the control of transpiration under abiotic stress (Mustilli et al., 2002; Yoshida et al., 2002). This kinase functions upstream of the NADPH oxidases AtrbohD and AtrbohF to activate reactive oxygen species production in guard cells and drive stomatal closure (Kwak et al., 2003). OST1 also phosphorylates proteins that contain AREB1, AREB2, and ABI5 (Furihata et al., 2006). The phosphorylation by OST1 activates transcription of these factors. In addition to changes in activity due to phosphorylation, expression studies revealed that transcript levels of OST1, AREB1, and ABI1 are sensitive to ABA (Leonhardt et al., 2004; Fujita et al., 2005), but the upstream factors that activate OST1 are unknown. In this study, AREB1 and ABI1 expression was reduced in the NPX1-ox plants without added ABA, and OST1 expression was lower when ABA was added (Fig. 5). We also showed that the overexpression of NPX1 inhibits the expression of ABA-induced OST1 and AREB1 (Fig. 5), which suggests that the decreased stomatal qsensitivity to ABA in NPX1-ox might be caused in part by the repression of drought stress-regulated genes such as OST1 and AREB1. Expression of AtbohD is also regulated by NPX1, but there appears to be some compensation in the form of the increased expression of AtbrohF. We have seen this type of compensation in the expression of NADPH oxidase family members in roots of the rhd2 mutant (Shin et al., 2005).
Other negative transcriptional regulators of ABA signaling have been identified, including ATHB6 and AtERF7 (Himmelbach et al., 2002; Song et al., 2005), which act to repress transcription through binding to DNA. Transgenic plants overexpressing ATHB6 or AtERF7 exhibited similar responses to NPX1-ox, such as increased water loss and reduced stomatal closure (Himmelbach et al., 2002; Song et al., 2005). However, these two transcriptional repressors act downstream of NPX1 and do not appear to be direct targets of NPX1. ATHB6 has been shown to target ABI1 (Himmelbach et al., 2002) and AtERF7 targets PKS3, which are Ser/Thr protein kinases in the pathway (Song et al., 2005). This is the first report, to our knowledge, of an upstream transcriptional regulator of the OST1-mediated signaling pathway.
ABA also induces the expression of stress-related genes (Shinozaki and Yamaguchi-Shinozaki, 2007) such as RD29A. More than half of drought-inducible genes such as RD29A are also induced by high salinity and/or ABA treatment, implicating significant cross talk between the drought, high salinity (Shinozaki and Yamaguchi-Shinozaki, 2007), and ABA response pathways. In our study, the ABA-inducible NPX1 was also up-regulated by NaCl and cold stress (Fig. 1), and NPX1-ox plants were hypersensitive to drought stress (Fig. 3). The regulation of RD29A is enhanced in npx1-1 plants without the addition of ABA, but upon addition of ABA RD29A expression increases in all lines tested. This suggests that either the application of exogenous ABA short circuits NPX1 effects or the effects of NPX1 are subtle and tightly regulated at endogenous levels of ABA.
The regulation of gene expression under drought stress is mediated by multiple transcriptional cascades (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). In each cascade, a set of transcription factors are induced by ABA, which in turn activates or represses downstream target genes important for drought resistance (Shinozaki and Yamaguchi-Shinozaki, 2007). From our results, we conclude that NPX1 negatively regulates ABA signaling in part by reducing the expression of ABA-inducible transcription factors such as AREB1 and MYB2 and stomata closure by regulating ABI1 and OST1 (Fig. 5). Thus, NPX1-ox plants are hypersensitive to drought, whereas npx1-1 plants are more tolerant (Fig. 3). ABI1 is known as a negative regulator in stomatal closing and has been shown to be a negative regulator of OST1 (Yoshida et al., 2006). The changes in expression of MYC2 with added ABA and the lower expression in the npx1-1 line are also consistent with the conclusion that NPX1 modulates sensitivity to ABA through transcriptional changes (Abe et al., 2003). Therefore our data on the expression of genes involved in ABA signal transduction indicate that NPX1 defines one new component of these stress-responsive transcriptional cascades.
NPX1 Modulates ABA Signaling, Which Feeds Back to ABA Biosynthesis
NPX1-ox lines have higher ABA content, which is presumably due to the enhanced expression of ABA biosynthesis genes such as ABA2 and NCED3, which are also strongly induced by drought (Iuchi et al., 2001; Figs. 5C and 6A). Even though levels of ABA are higher in NPX1-ox, these plants appear to be relatively less sensitive to ABA. Therefore, we suggest that NPX1 does not merely contribute to ABA signaling by increasing ABA content. Rather, we propose that the increased ABA concentrations are due to feedback that may be initially caused by insensitivity, which triggers increased ABA biosynthesis to overcome a perceived reduction in sensitivity. The idea of this type of feedback was suggested earlier (Xiong et al., 2001), and our study provides evidence that this occurs. The observation that NPX1-ox is less sensitive to but contains more ABA differs from other mutants such as ahg2, which accumulates 50% higher endogenous ABA but appears to be more hypersensitive to ABA (Nishimura et al., 2005). In contrast, the sad1 mutant shows enhanced sensitivity to ABA but does not produce more ABA under stress conditions as compared with the wild type (Xiong et al., 2001). In the case of sad1, the authors suggested that ABA production was reduced in sad1, which resulted in enhanced sensitivity to ABA and abiotic stress. Multiple results from different groups suggest that ABA responses are complex and regulated at different levels, including direct effects and feedback responses. Our results suggest that NPX1 modulates ABA signaling through effects on the signal transduction pathway, but the possibility still exists that it could act indirectly by inducing ABA biosynthesis. Thus, we conclude that NPX1 reduces ABA sensitivity through the repression of stress-inducible gene expression but activates the induction of ABA biosynthesis via positive feedback regulation.
A NAC Transcriptional Factor Is a Target of NPX1
To identify how NPX1 modulates ABA signaling, and since our hypothesis was that modulation occurred through protein-protein interactions rather than through direct binding to DNA, we employed yeast two-hybrid assays. We found that NPX1 interacted with multiple targets, including a NAC transcription factor. NACs are plant-specific transcription factors that participate in controlling plant development and are involved in pathogen attack and responses to various environmental stresses (Aida et al., 1997; Collinge and Boller, 2001; Hu et al., 2006). Several NAC transcriptional factors are induced by drought, high salinity, and ABA and regulate ABA- and stress-inducible gene expression (Hegedus et al., 2003; Fujita et al., 2004; Tran et al., 2004; Hu et al., 2006). Recently, a drought- and ABA-inducible NAC transcription factor (RD26) was identified, and RD26-ox plants were found to be hypersensitive to ABA whereas RD26 dominant negative transgenic plants were insensitive (Fujita et al., 2004). In our study, one of the NAC transcription factors, previously named TIP (Ren et al., 2000), was isolated as an interacting protein of NPX1. Previously, TIP was shown to act as an important participant in the turnip crinkle virus resistance response through direct activation of the transcription of genes involved in basal resistance (Ren et al., 2000).
Plants have evolved a wide range of mechanisms to cope with biotic and abiotic stresses, and there is cross talk between these biotic and abiotic stress responses (Fujita et al., 2006). For example, the importance of ABA in pathogen resistance was highlighted by OST1's involvement in part of the innate immunity defense against bacterial infection by regulating stomatal closure (Melotto et al., 2006), and ABA-responsive elements have also been identified in the promoters of defense-related genes (Adie et al., 2007). We found that TIP not only interacted with NPX1 but was also induced under potassium-deprived conditions and by ABA and NaCl treatments (Supplemental Fig. S4). Furthermore, we identified regions in the promoters of OST1 and AREB1 that contained a CATGTG motif (data not shown) that have been shown to be NAC-binding motifs (Tran et al., 2004). Since TIP activates reporter gene expression in yeast cells (Ren et al., 2000), we used a yeast repressor assay to test whether NPX1 may directly repress TIP activity. We showed that TIP activity was reduced when NPX1 was expressed in the same yeast cells (Fig. 7B). ABI1 expression was reduced in the tip plants without added ABA, and OST1 expression was lower when ABA was added (Supplemental Fig. S5B). The regulation of these two genes in the tip knockout is as we would predict based on our results in NPX1-ox plants (Fig. 5).
Based on the yeast two-hybrid interaction, the fact that NPX1 and TIP have overlapping expression patterns, and the repressor assay results, we suggest that NPX1 acts as a repressor of TIP and perhaps other proteins and that this interaction modulates ABA responses. Although NPX1 does not bind directly to DNA, it is up-regulated under abiotic stress conditions and may be targeted to relevant gene promoters via a complex with other proteins such as transcription factors.
NPX1 Stimulates ABA Biosynthesis in Nutrient-Deficient Plants
Our initial result showing that potassium deficiency induced the expression of NPX1 led to the finding that ABA biosynthesis is enhanced when Arabidopsis is deprived of potassium (Jeschke et al., 1997; Peuke et al., 2002). At this time, it is still unclear what role ABA plays in plant response to nutrient deprivation. It is clear, however, that the NPX1-ox plants contained higher levels of ABA than other lines and that the overexpression of NPX1 resulted in a moderate increase in tolerance to potassium deprivation (Supplemental Fig. S3). It is well known that root growth is stimulated by ABA when applied at a low concentration, but it inhibits growth at higher concentrations (Pilet and Saugy, 1987). It is possible that ABA is needed for growth under normal conditions and that higher concentrations may be required for maintaining root growth under potassium-deprived conditions. This finding links ABA to the potassium deprivation response circuits, which include auxin (Shin et al., 2007), ethylene (Jung et al., 2009), and reactive oxygen species (Shin and Schachtman, 2004). Further work will be needed to fully understand the role of ABA plant response to low potassium.
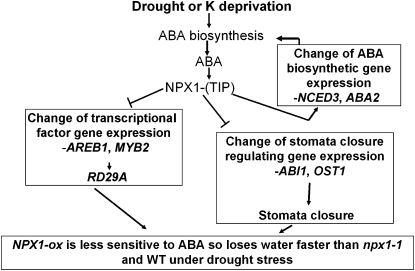
In conclusion, the data presented here provide evidence that a novel nuclear factor modulates ABA signaling and plant sensitivity to abiotic stress, as summarized in Figure 8. The isolation and characterization of NPX1 provides new insight into the control mechanisms for modulating the ABA responsiveness of plants and may provide new approaches for the genetic engineering of drought tolerance in plants.
Figure 8.
A model for NPX1 modulation of ABA signaling. NPX1 does not contain a classical DNA BD and does not exhibit transcriptional activity. NPX1 functions as a transcriptional regulator by interacting with factors such as TIP. This NPX1-TIP complex negatively regulates the expression of genes coding for ABA-dependent stress-responsive signaling and ABA-induced stomatal closing. The reduced expression of these genes may contribute to the reduced ABA sensitivity and faster water loss in the NPX1-ox plants. In addition, drought and low-potassium stresses induce ABA biosynthesis, and the increased ABA further stimulates the expression of some of the biosynthesis genes through positive feedback regulation. NPX1 may also regulate this feedback loop, thereby leading to increased ABA accumulation in NPX1-ox plants. WT, Wild type.
MATERIALS AND METHODS
DNA Cloning
The NPX1 (At5g63320) open reading frame was amplified using Pfu-polymerase (Stratagene) and the following primers: 5′-CCTGCAGATGCAAGTTGGAGTGCTCGG-3′ and 5′-TCCCGGGTCAATCAATTTCTACATCTATATC-3′. The YFP-NPX1 and GAL4 DNA BD fusion proteins were constructed by amplifying the NPX1 open reading frame using Pfu-polymerase (Stratagene) and the following primers: 5′-CACCATGGATGCAGTAGTTCTCGTTCCAG-3′ and 5′-TCAATCAATTTCTACATCTATATCTTCCT-3′. The PCR product was introduced into pENTR D/Topo (Invitrogen) following the manufacturer's instructions. The insert was sequenced to ensure that no changes were introduced by PCR. The resulting entry clone was introduced into the destination plasmid pEARLYGATE N-YFP (Earley et al., 2006) to yield pEARLYGATE YFP-NPX1, which allows the expression of fusion proteins with YFP at the N-terminal position under control of the cauliflower mosaic virus 35S promoter.
Quantitative Real-Time PCR Analysis
To analyze the expression of NPX1 by real-time PCR, the gene-specific primers (5′-AAAAGCAACCAGGAGTTGGTATAGACAAAA-3′ and 5′-ATCCATGGTGGTTGACTTTTCCCCAA-3′) were used. The gene-specific primers for quantifying the transcripts of ABA signaling genes were described previously (Chen et al., 2006; Shen et al., 2006). β-Tubulin gene (At5g62690) was amplified with the primers 5′-GCCAATCCGGTGCTGGTAACA-3′ and 5′-CATACCAGATCCAGTTCCTCCTCCC-3′ and was used to normalize the reactions. Real-time PCR was performed according to the instructions provided for the Bio-Rad iCycler iQ system (Bio-Rad Laboratories) with platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). The fold change of transcripts was calculated based on an efficiency-calibrated model (Yuan et al., 2006) and compared with the transcript level under normal conditions. Statistical differences between samples were evaluated by Student's t test using delta Ct values (Yuan et al., 2006). In each experiment, the mean of three biological replicates is used to generate means and statistical significance. Two experiments on independently grown plant material were performed to confirm that the results are reproducible.
Plant Material and Growth Conditions
For constitutive expression of NPX1 in planta, we used pCambia2300 with the figwort mosaic virus promoter (Sanger et al., 1990) and nopaline synthase terminator. Arabidopsis (Arabidopsis thaliana) plants were transformed using Agrobacterium tumefaciens by the floral dip method (Clough and Bent, 1998). The insertion mutant in the NPX1 locus (SAIL_123) or TIP (SALK_150356C) was obtained from the Arabidopsis Biological Resource Center. Homozygous plants were identified by PCR as described (http://signal.salk.edu/tdnaprimers.2.html). The absence of NPX1 transcript or TIP was confirmed by reverse transcription-PCR. Prior to growth assays, both mutant alleles and the corresponding wild type (ecotype Columbia) were grown together under the same conditions to generate wild-type, overexpression, and knockout seeds for further experiments. For the nutrient-deficiency assays, plants were grown on plates containing a nutrient solution described previously (Shin et al., 2005) with the addition of 2% Suc and 0.6% SeaKem agarose (Cambrex). After stratification of the seeds at 4°C for 2 to 3 d, the plates were transferred to the growth chamber at 22°C with a 16-h daylength at 200 μmol m–2 s–1. For K+-deficiency growth assays, 10 μm KCl was used instead of 1.75 mm KCl. Four-day-old seedlings were transferred to nutrient-deficient medium, and then the length of primary roots and number of lateral roots were measured after 7 d.
Germination and Root Growth Assays
Germination and root growth assays with exogenous ABA were performed as described (Kuhn et al., 2006). Sterilized seeds were plated on minimal medium (0.25× Murashige and Skoog medium, no Suc) supplemented with increasing ABA concentrations. To score seed germination, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined in three independent experiments (36 seeds per genotype and experiment).
Root growth assays to assess ABA sensitivity were carried out by transferring 6-d-old seedlings onto minimal medium supplemented with the indicated ABA concentrations on 0.8% agar (Phytagel; Sigma) plates. Root growth was measured 6 d after the transfer in three independent experiments with 70 individuals per genotype and experiment.
Measurements of the Stomatal Aperture
For stomatal closing experiments, fully expanded leaves from 3- to 4-week-old plants were excised, and epidermal pieces were peeled from the abaxial surface. The epidermal peels were floated for 2.5 h in stomatal opening solution (Pei et al., 1997) containing 50 mm KCl, 50 μM CaCl2, and 10 mm MES (pH 6.15). After incubation in ABA for 2 h, the stomatal aperture was measured. Control experiments were performed in parallel with no ABA added. Double-blind stomatal movement assays were performed such that the genotype and applied ABA concentrations were unknown. Three independent experiments were performed, and the same results were obtained.
Drought Tolerance Experiment
For the drought tolerance experiment, the soil in pots containing 3-week-old plants was covered with plastic film and exposed to a period of 14 d without added water. Stomatal conductance (gs) was obtained using the AP3 porometer (Delta-T Devices) at 3 and 7 d after watering was stopped. Two independent experiments were performed, and similar results were obtained.
Visualization of YFP-NPX1 Localization
For transient expression of YFP-NPX1, biolistic bombardment of onion (Allium cepa) epidermal cells was performed as described previously (Marc et al., 1998) using 5 μg of plasmid DNA precipitated onto gold particles. Transformed Arabidopsis plants were generated expressing YFP-NPX1 as a fusion protein under the control of the cauliflower mosaic virus 35S promoter. Seedlings were grown on plates containing 0.25× MS + 0.8% agar and were imaged using a Nikon Eclipse E800 microscope. To stain the nucleus of Arabidopsis cells, 1 μg mL−1 Hoescht dye (Sigma) was used for 30 min. Successive images were collected either with transmitted light or with appropriate YFP and Hoescht filters.
Yeast Two-Hybrid Screening
Full-length NPX1 cDNA was amplified by PCR and subcloned into the vector pDEST32 (Invitrogen). An Arabidopsis root cDNA library was constructed using the activation domain expression vector pDEST22 (Invitrogen). Plasmids were cotransformed into Saccharomyces cerevisiae strain MaV203 according to the manufacturer's instructions (Invitrogen).
Yeast Repression Assay
Full-length TIP (At5g24590) cDNA was generated by PCR with the primers 5′-AGAATTCATGAAAGAAGACATGGAAGTACT-3′ and 5′-ACTCGAGGAATTGATCGGAACAAACATCAC-3′ and subcloned into the EcoRI/SalI sites in pGBKT7 (Clontech). As a result, TIP was fused with GAL4 DNA BD. BD-NPX1 and BD-YFP were expressed from pDEST22 (Invitrogen). The plasmids were transformed into S. cerevisiae Y187, and β-galactosidase assays were performed as described (Sridhar et al., 2006). BD-TIP, BD-NPX1, or BD-YFP was selected by –Trp and –Leu. A liquid culture assay using o-nitrophenyl β-d-galactopyranoside as a substrate was performed to determine transcriptional activation. Data shown for these assays are averages of triplicates, and the experiment was repeated twice.
Measurement of ABA Concentration
The ABA concentrations in leaves or roots were measured with a Phytodetek ABA enzyme immunoassay test kit (Agdia) Mature leaves or roots of 3-week-old plants grown in medium were harvested, freeze dried, frozen in liquid nitrogen, and ground into powder. ABA was extracted by suspending 200 mg of lyophilized tissue in 1 mL of water and agitating overnight at 4°C. The suspension was centrifuged at 4,000 rpm for 20 min, and the supernatant was transferred to a clean tube and dried under vacuum. The dry residue was dissolved with 60 μL of water, and a 1:100 dilution with Tris-buffered saline (45 mm Tris-HCl, pH 7.8, 90 μm MgCl2, 0.135 m NaCl, and 3 mm sodium azide) was made. The ABA concentration was then determined with a Phytodetek ABA enzyme immunoassay test kit (Agdia) The color absorbances were detected with the Spectramax 250 microplate reader (Molecular Devices) at 405 nm. Two independent experiments were performed, and the same results were obtained.
Sequence data from this article can be found in the GenBank/EMBL data libraries as described in “Materials and Methods” and Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR analysis of NPX1 expression in Arabidopsis plants.
Supplemental Figure S2. Analysis of additional NPX1 overexpression and knockout lines.
Supplemental Figure S3. Overexpression of NPX1 enhances primary root growth.
Supplemental Figure S4. The expression of TIP was up-regulated by nutrient deprivation and abiotic stresses.
Supplemental Figure S5. Disruption of TIP alters the expression of ABA signaling genes.
Supplemental Table S1. Proteins that interact with NPX1 as determined by yeast two-hybrid screening.
Supplemental Table S2. Information on genes used for real-time PCR analysis.
Supplementary Material
Acknowledgments
We thank Ji-Yul Jung and Sung Chul Bahn for measurements of the stomatal aperture and Howard Berg for microscopy assistance. We also thank Sara Lee Schupf for her support through the Lubin Foundation.
This work was supported by the Korea Research Foundation funded by the Korean Government (grant no. KRF–2006–352–C00069) and by a grant from the Lubin Foundation to D.P.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel P. Schachtman (dschachtman@danforthcenter.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM (2003) OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci 8 151–153 [DOI] [PubMed] [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J (2006) Overexpression of the regulator of G-protein signalling protein enhances ABA-mediated inhibition of root elongation and drought tolerance in Arabidopsis. J Exp Bot 57 2101–2110 [DOI] [PubMed] [Google Scholar]
- Christmann A, Weiler EW, Steudle E, Grill E (2007) A hydraulic signal in root-to-shoot signalling of water shortage. Plant J 52 167–174 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46 521–529 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39 863–876 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9 436–442 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M, Harper J, Cushman J, Mittler T, Girke T, Zhu JK, Bailey-Serres J, Mittler R (2006) What makes species unique? The contribution of proteins with obscure features. Genome Biol 7 R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M, Harper J, Cushman J, Mittler T, Mittler R (2007) POFs: what we don't know can hurt us. Trends Plant Sci 12 492–496 [DOI] [PubMed] [Google Scholar]
- Haeder HE, Beringer H (1981) Influence of potassium nutrition and water stress on the content of abscisic acid in grains and flag leaves of wheat during grain development. J Sci Food Agric 32 522–526 [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53 383–397 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Jeschke WD, Peuke AD, Pate JS, Hartung W (1997) Transport, synthesis and catabolism of abscisic acid (ABA) in intact plants of castor bean (Ricinus communis L.) under phosphate deficiency and moderate salinity. J Exp Bot 48 1737–1747 [Google Scholar]
- Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49 199–222 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ (2008) Gene networks in Arabidopsis thaliana for metabolic and environmental functions. Mol Biosyst 4 199–204 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ (1998) A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H, editor (1995) Mineral Nutrition of Higher Plants. Academic Press, London
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM (2007) The control of transpiration: insights from Arabidopsis. Plant Physiol 143 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T (2005) Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44 972–984 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuke AD, Jaschke W, Hartung W (1994) The uptake and flow of C, N and ions between roots and shoots in Ricinus communis L. II. Long distance transport of abscisic acid depending on nitrogen nutrition and salt stress. J Exp Bot 45 741–747 [Google Scholar]
- Peuke AD, Jeschke WD, Hartung W (2002) Flows of elements, ions and abscisic acid in Ricinus communis and site of nitrate reduction under potassium limitation. J Exp Bot 53 241–250 [DOI] [PubMed] [Google Scholar]
- Pilet PE, Saugy M (1987) Effect on root growth of endogenous and applied IAA and ABA: a critical reexamination. Plant Physiol 83 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ (2000) HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12 1917–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M, Valon C, Fenzi F, Giraudat J, Leung J (2005) The genetics of adaptive responses to drought stress: abscisic acid-dependent and abscisic acid-independent signalling components. Physiol Plant 123 111–119 [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23 319–327 [DOI] [PubMed] [Google Scholar]
- Sanger M, Daubert S, Goodman RM (1990) Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14 433–443 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQ (2008) Chemical root to shoot signaling under drought. Trends Plant Sci 13 281–287 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D (2001. a) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52 627–658 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ (2001. b) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330 [DOI] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, et al (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443 823–826 [DOI] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46 1350–1357 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58 221–227 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133 3159–3166 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1 771–781 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K (2006) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.