All eukaryotic organisms have a diversity of transcription factor (TF) gene families, encoding key proteins regulating gene expression. TF families are strongly conserved across eukaryotic organisms, especially plants. The specific function of each of these TF genes is of interest due to their role in controlling plant developmental processes and responses to environmental conditions, including functions of key importance to agronomic performance. In this review, we focus on the role of TF genes in legume species. The review also provides an update on the identification and categorization of TF genes in several eukaryotes, including three partially or completely sequenced legume genomes (soybean [Glycine max], Medicago truncatula, and Lotus japonicus). The role of TF genes in legumes is discussed in an evolutionary context based upon a comprehensive comparison of TF gene distribution and direct experimental data obtained for a significant number of legume TF genes.
TF genes are present in all eukaryotic phyla. They encode regulatory proteins that interact with genomic DNA promoter and enhancer sequences. These interactions facilitate the transcriptional activation or repression of proximal genes and enable cells to respond to changes in their environment (e.g. biotic and abiotic stresses), to regulate the cell cycle, and, in the case of the most complex organisms, to control cell fate. As mentioned by Carroll (2001), the expansion of regulatory protein numbers and interactions, as well as changes to their spatial and temporal expression, is part of the evolutionary process leading to increasingly complex organisms. Therefore, determining the repertoire of TF genes in genomes, the regulation of their expression, and their biochemical properties (i.e. DNA- and protein-binding affinities) is important to the understanding of TF regulatory networks and organism evolution.
The immobile nature of plants represents a major disadvantage compared with animals, which can flee many environmental assaults. The ability to face environmental challenges implies that plants must possess complex regulatory systems to respond appropriately. This sometimes involves changing developmental programs, which is facilitated by the fact that plants maintain active stem cells, called meristems, which can differentiate and develop into various organs depending on environmental and endogenous cues. As regulators of transcription, TFs play important roles in helping plants meet and master environmental challenges. Therefore, it is not surprising that plants have more TF genes than animals (Riechmann et al., 2000; this study).
Most of the extant knowledge of plant TF genes was obtained from studies of the major genetic model in plant biology, Arabidopsis (Arabidopsis thaliana). However, while Arabidopsis is a useful model for many developmental and other processes common to all higher plants, it lacks certain traits that are of immense value to agriculture, such as the ability to form nitrogen-fixing symbioses with rhizobia and soil nutrient-scavenging symbioses with mycorrhizal fungi. Legumes, on the other hand, are able to establish such beneficial symbioses and, as a result, have been mainstays for sustainable agriculture for thousands of years. The legume family includes important food plants such as common bean (Phaseolus vulgaris), soybean, and pea (Pisum sativum) and important forage species such as alfalfa (Medicago sativa) and clover (Trifolium spp.). There is also growing interest in the use of legumes as a source of biomass for biofuel production. Although mycorrhizal symbioses are widespread among plant families, occurring in approximately 90% of all species, symbiotic nitrogen fixation (SNF) is restricted to legumes and a few nonlegume families. This makes legumes special, but just how SNF evolved in legumes remains largely unknown. Answers to this question may emerge from comparative analysis of the genomes of legumes and nonlegumes.
Genome sequencing of three legume species, L. japonicus (http://www.kazusa.or.jp/lotus), soybean (http://www.phytozome.net/soybean), and M. truncatula (http://www.medicago.org/genome), is nearing completion, and the genome sequences of several nonlegumes, including Arabidopsis (Arabidopsis Genome Initiative, 2000), Vitis vinifera (Jaillon et al., 2007), Sorghum bicolor (Paterson et al., 2009), Physcomitrella patens (Rensing et al., 2008), Chlamydomonas reinhardtii (Merchant et al., 2007), Oryza sativa (Yu et al., 2002), and Populus trichocarpa (Tuskan et al., 2006), are known (Table I).
Table I.
Classification of main plant species based on their class, order, family, and genus
The genome size, ploidy level of each genome, number of predicted protein-coding genes, and number of TF genes identified are also indicated.
| Plant Type | Organism | Organism Genome Size | Chromosome No. and Ploidy | Predicted Total Gene No. | Predicted TF Gene No. | Predicted TF Gene Percentage |
|---|---|---|---|---|---|---|
| % | ||||||
| Alga | Chlamydomonas reinhardtii | 120 Mb | n = 17 (diploid) | 16,709 | 349 | 2.09 |
| Moss | Physcomitrella patens | 511 Mb | n = 27 (diploid) | 35,938 | 1,316 | 3.66 |
| Monocotyledon | Oryza sativa | 430 Mb | n = 12 (diploid) | 67,393 | 4,432 | 6.58 |
| Monocotyledon | Zea mays | 2,400 Mb | n = 10 (diploid) | 125,435 | 5,383 | 4.29 |
| Monocotyledon | Sorghum bicolor | 730 Mb | n = 10 (tetraploid) | 36,338 | 2,464 | 6.78 |
| Dicotyledon, rosid, Malvidae | Arabidopsis thaliana | 115 Mb | n = 5 (diploid) | 32,825 | 2,269 | 6.91 |
| Dicotyledon, rosid, Fabidae, Fabales | Medicago truncatula | 500–550 Mb | n = 8 (diploid) | 38,835 | 1,473 | 3.79 |
| Dicotyledon, rosid, Fabidae, Fabales | Lotus japonicus | 450 Mb | n = 6 (diploid) | 42,395 | 1,637 | 3.86 |
| Dicotyledon, rosid, Fabidae, Fabales | Glycine max | 1,115 Mb | n = 20 (tetraploid) | 66,153 | 5,557 | 8.40 |
| Dicotyledon, rosid, Fabidae, Malpighiales | Ricinus communis | 425 Mb | n = 10 (diploid) | 31,221 | 1,543 | 4.94 |
| Dicotyledon, rosid, Fabidae, Malpighiales | Populus trichocarpa | 550 Mb | n = 19 (diploid) | 45,555 | 2,758 | 6.05 |
| Dicotyledon, rosid, Vitaceae |
Vitis vinifera |
504.6 Mb |
n = 19 (diploid) |
30,434 |
1,675 |
5.50 |
Given the central role of TFs in regulating plant gene expression and consequently development, differentiation, and responses to the environment, as well as their key roles in evolution (Ramalingam et al., 2003; Miller et al., 2006; Francia et al., 2007), the set of genes encoding TFs is an obvious place to start looking for evolutionary innovations that define key legume traits, such as SNF. More generally, the study of legume TFs will shed light on their roles in plant processes common to all plant species.
When comparing legumes with other plant phyla, a key question is, “What traits define a legume” This question was raised several years ago by Doyle and Luckow (2003) and by Zhu et al. (2005). Morphogenesis and life styles of different plant species and more generally of eukaryotes depend heavily on the control of gene expression. Therefore, it can be assumed that the repertoire of TF genes within a plant species, their expression pattern, and their function largely determine the unique aspects of the species. Unfortunately, as discussed below, the functions of only a few legume TF genes are known. Therefore, a tremendous effort is required to characterize the role of TF genes in legumes. The emerging high-throughput genome-based technologies that build upon the sequencing of several legume genomes open new fields of investigation, including a complete identification and classification of legume TF genes.
In this review, we summarize knowledge of legume TFs while updating the classification of legume, nonlegume, and nonplant eukaryotic TF genes. Legume TF function and TF gene expression are also discussed in the light of recently published studies. Altogether, these data will be discussed in an evolutionary context to better understand the impact of TF genes on legume development.
LEGUME TF GENES: WHAT IS KNOWN?
The last comprehensive review of legume TFs predated the completion of any of the legume genome sequencing projects but noted that more than 99% of predicted legume TFs remained to be characterized functionally (Udvardi et al., 2007). In the interim, the genome sequences of three legumes have been completed, or nearly so, enabling both comparative and functional genomics studies, and the roles of new legume TFs have been determined.
Recent studies have elucidated the roles of several TFs in legume development. Initially, several legume TF genes were identified based upon similarity of mutant phenotypes in legumes and Arabidopsis and sequence homology between legume genes and the Arabidopsis genes known to confer specific mutant phenotypes. Legume TF genes discovered in this way include those involved in the control of floral meristems, such as LjFLO, PsFLO/LFY, PsPEAM4, and MtPIM (Hofer et al., 1997; Berbel et al., 2001; Taylor et al., 2002; Dong et al., 2005; Benlloch et al., 2006). In the same way, three L. japonicus MYB TFs with homology to Arabidopsis TRANSPARENT TESTA2 (AtTT2) were shown to regulate proanthocyanidin biosynthesis after their transient expression in Arabidopsis leaves (Yoshida et al., 2008). More recently, the role of legume TF genes was investigated using reverse-genetic approaches. For example, expression of the soybean GmWRKY13 gene in Arabidopsis positively affected lateral root formation (Zhou et al., 2008). RNA interference knockdown of MtSERF1, which encodes an AP2-EREBP TF, altered the production of somatic embryos (Mantiri et al., 2008). Legume TF genes were also recently implicated in responses to environmental challenges. For instance, after identifying TF genes regulated in response to phosphate deprivation in common bean (Hernández et al., 2007), Valdés-López et al. (2008) characterized the role of one MYB TF, PvPHR1, as a key protein in phosphorus uptake. Similarly, after quantifying the expression levels of soybean Dof genes in various organs, including flowers and pods, the overexpression of GmDof4 and GmDof11 TF genes in Arabidopsis was shown to increase seed lipid content (Wang et al., 2007).
A large number of studies also highlighted the role of legume TF genes in plant responses to abiotic stresses. For example, a common method for examining gene function is to overexpress the gene of interest from a strong, constitutive promoter (e.g. cauliflower mosaic virus 35S) and then to gauge the response of the resulting transgenic plants to a variety of treatments (e.g. abiotic stress). For example, the overexpression of two TFIIIA-related TF genes (MtZPT2-1 and MtZPT2-2) in M. truncatula led to an increase in the size of the plant root system under salt stress (de Lorenzo et al., 2007). GmWRKY54 and GmDREB2 increased tolerance to both salt and drought when overexpressed in Arabidopsis (Chen et al., 2007; Zhou et al., 2008). In the same study, Zhou et al. (2008) also implicated two other WRKY TF genes, GmWRKY21 and GmWRKY13, in tolerance to cold, salt, and mannitol stresses by overexpressing these genes in Arabidopsis plants. A similar approach established a role for GmDREB3 in Arabidopsis tolerance to cold, drought, and high-salt stresses (Chen et al., 2009). Zhu et al. (2006) observed a higher tolerance of soybean plants to high temperature following overexpression of the GmHSFA1 TF gene. Interestingly, Zhang et al. (2008) identified two GmAP2-EREBP TF genes involved not only in abiotic stress response but also in plant-pathogen defense mechanisms. Overexpression of GmAP2-EREBPs in tobacco (Nicotiana tabacum) enhanced plant resistance to drought, salt stress, and pathogen infection. This study suggested that some TF genes are central to the general plant stress response. With regard to pathogen response, Park et al. (2007) reported that GmZF-HD1 and GmZF-HD2 were induced in response to Pseudomonas syringae infection.
LEGUME TF GENES AND NODULATION
In addition to the activation of plant defense systems through the activation of TF genes, legume TFs are also involved in the control of mutualistic interactions between plant root and soil microorganisms. For example, nodulation involves the interaction between root and soil bacteria leading to SNF. This complex interaction is mainly restricted to legumes and, for this reason, makes legumes special. The infection of plant roots by symbiotic bacteria begins by the invasion of root hair cells by the symbiont through the newly formed infection thread. This infection is dependent on several genes. Genes that are specifically expressed during nodulation are termed nodulins. Several years ago, protein factors that bound to the AT-rich promoter sequences of nodulin genes were identified (Jensen et al., 1988; Metz et al., 1988; Forde et al., 1990; Laursen et al., 1994; Hansen et al., 1999). More recently, TF genes specifically involved in the rhizobial infection process have been identified. Among them, the M. truncatula NSP1 and NSP2 genes, which encode two GRAS TFs, are essential to root hair infection. The root hairs of Mtnsp1 and Mtnsp2 mutants are not infected by the symbiont (Catoira et al., 2000; Oldroyd and Long, 2003). More recently, based on a mutant screen for plants lacking infection threads upon inoculation with rhizobia, the L. japonicus NSP1 and NSP2 genes were identified (Heckmann et al., 2006).
The first TF shown to have an essential role in nodulation was the L. japonicus NIN gene (Schauser et al., 1999). Mutations in this gene abolish both infection and the induction of nodule primordia. However, NIN appears to act downstream of the initial steps in symbiont recognition by the plant (Oldroyd and Downie, 2008). By screening fast-neutron and Tnt1 transposon-tagged mutagenized populations, Marsh et al. (2007) characterized the M. truncatula ortholog of LjNIN (Catoira et al., 2001). The PsSym35 locus defines the pea NIN ortholog (Borisov et al., 2003). The likely soybean ortholog of NIN is clearly evident in the soybean genomic sequence, and this genomic region shows significant microsynteny to the NIN-encoding regions of M. truncatula and L. japonicus (M. Libault, X.C. Zhang, and G. Stacey, unpublished data).
Additional TF genes critical to the nodulation process were also identified by direct screening for nodulation-defective mutants. For example, this approach led to the identification of the L. japonicus ASTRAY gene encoding a bZIP TF (Nishimura et al., 2002). L. japonicus astray mutants exhibit a hypernodulation phenotype (i.e. increased nodule numbers). Similarly, Middleton et al. (2007) identified the MtERN gene, encoding an AP2-EREBP TF, as necessary for invasion of plant cells by symbiotic bacteria. Recently, an additional AP2-EREBP TF gene was identified as a positive regulator of nodulation in L. japonicus (Asamizu et al., 2008). Other strategies also allowed the identification of important TF genes involved in nodulation. For example, based on its interaction with LjSymRK (a receptor kinase critical for rhizobial infection; Stracke et al., 2002), Zhu et al. (2008) identified LjSIP1, a protein previously described for its role during nodulation that encodes an ARID TF. The authors proposed that LjSIP1 controlled the expression of the Nod factor-induced NIN gene. El Yahyaoui et al. (2004) utilized DNA microarray analysis to identify several genes regulated during M. truncatula nodulation, including an AP2-EREBP TF gene named MtEFD. Based on the phenotype of an edf1 null mutant, as well as plants either silenced for EFD expression or overexpressing the gene, Vernié et al. (2008) defined MtEFD as an important regulator controlling Sinorhizobium meliloti infection of M. truncatula.
In summary, recent analyses highlight the roles of a diverse group of TF genes in a wide variety of legume biological processes. Overexpression of legume TF genes in nonlegume plants often produces phenotypes consistent with strong conservation of TF function among higher plant species. Despite the redundancy of TF gene families across the plant kingdom, it is interesting that legumes nodulate while most other plant species do not. Although specific TF genes have been implicated in nodule development, these belong to families common to nonlegumes. This suggests that neofunctionalization of TF genes was important in the evolution of SNF, rather than the invention of novel TF genes/families.
EVOLUTION OF TF GENES AND FAMILIES IN THE EUKARYOTIC KINGDOM
To better understand the evolution of TFs, comparative studies of TF gene families of several eukaryote organisms, including Arabidopsis, have been performed (Riechmann et al., 2000). However, limited genome sequences from plant species and underrepresentation of TFs in cDNA and EST databases have hampered such studies until recently. Now, however, genome sequences of a large diversity of organisms, including several plant species, have been released or will be soon (Table I). The release of genomic sequences combined with the use of improved gene prediction software (e.g. FGENESH) and protein function prediction software (e.g. Pfam [Finn et al., 2008], InterProScan [Hunter et al., 2009], and UniProt [UniProt Consortium, 2009]) allows a more complete identification of the gene content in each genome, including TF genes. Furthermore, prediction tools are now well supported by the detection of gene transcripts via ultra-high-throughput sequencing methods. Based on such characteristics, it is now possible to characterize the entire TF gene population of a few plant species, at least as far as known TF families are concerned. Highlighting similarities and differences in TF gene populations among eukaryotes, and more specifically among plants, may help to answer the question, “What makes a legume a legume” (Doyle and Luckow, 2003).
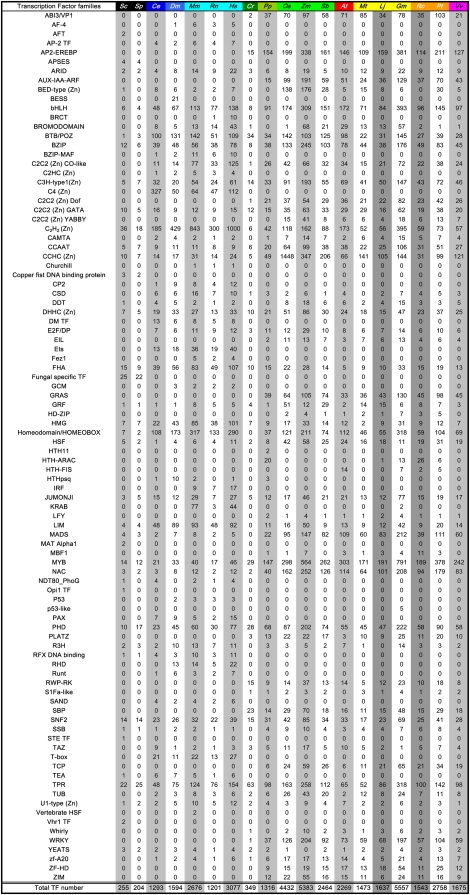
We investigated TF gene evolution in eukaryotic phyla based on their family membership. By mining protein sequence databases of 19 major eukaryotes, we identified signature Pfam domains conserved in the different TF families (e value e−3; Table II) and categorized them according to their family membership (Fig. 1). The percentage of identified TF genes compared with the total number of protein-encoding genes analyzed fluctuates between 2% and 9% among the 19 organisms studied. Not surprisingly, the smallest populations of TF genes were identified in the most primitive organisms (e.g. Saccharomyces cerevisiae, Schizosaccharomyces pombe, and C. reinhardtii, where TF genes represent 2%–4% of the genes annotated). In higher eukaryotes, the higher complexity of form and function likely dictates the need for an increased number of TF genes (7.36% in Drosophila melanogaster, 9.12% in Rattus norvegicus, 7.65% in Mus musculus, and 8.15% in Homo sapiens; an average of 5.7% of plant genes are TF genes; Table II).
Table II.
Number of TF genes in 19 of the main eukaryotic organisms
TF proteins were identified for 19 model eukaryotes: Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Caenorabditis elegans (Ce), Drosophila melanogaster (Dm), Mus musculus (Mm), Rattus norvegicus (Rn), Homo sapiens (Hs), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Oryza sativa (Os), Zea mays (Zm), Sorghum bicolor (Sb), Arabidopsis thaliana (At), Ricinus communis (Rc), Populus trichocarpa (Pt), Medicago truncatula (Mt), Lotus japonicus (Lj), Glycine max (Gm), and Vitis vinifera (Vv). The TF genes were distributed across 94 families based on the identification of the TF family signature protein domain (Kakar et al. [2008] and the DBD database [http://dbd.mrc-lmb.cam.ac.uk/DBD/index.cgi?Home; Wilson et al., 2008] were used as references for this classification). The color code used is based on phylogenetic relationships: yeast (black), animal (blue; nematode, arthropod, and mammal in dark blue, blue, and light blue, respectively), algae (dark green), moss (green), monocotyledon (light green), dicotyledon (warm colors; Malvidae, Fabidae, and Vitaceae in red, yellow/orange (Fabales/Malpighiales), and pink, respectively.
Figure 1.
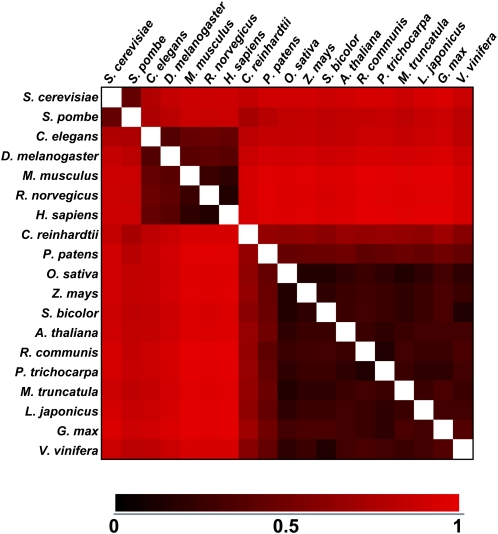
Comparison of TF gene distribution based on family membership across 19 different eukaryotic species. For each species, the representation of each TF family in the total pool of TF genes identified was expressed as a percentage. The color scale indicates the degree of correlation (red, low correlation; black, strong correlation). The heat map was generated using the Euclidean distances between each organism using the MultiExperiment Viewer (http://www.tm4.org/mev.html).
Considering the distribution of genes among the 94 TF families identified, 22 families are specific to plants and 20 families are specific to animals (Table II). Eight families are found exclusively in yeast, while one TF family is represented in D. melanogaster only. Forty-three families were common to all phyla represented in this study. No legume-specific TF gene family was found, which may reflect the paucity of knowledge about legume TFs but more likely indicates conservation of TF families among plants. Consistent with this notion, the distribution of legume genes among the various TF families is similar to that of other plant species. Overall, based on this census of genes in the various TF families, there does not appear to be any enrichment of known TF families in legumes.
CAN THE KNOWLEDGE ACQUIRED ON TF GENES IN ONE PLANT BE USED IN LEGUMES?
The absence of significant differences in TF gene distribution across TF families between legume and nonlegume plants suggests that legume-specific traits are likely dependent on TF gene expression patterns and TF protein function. So far, most of the plant TFs characterized belong to Arabidopsis. Therefore, an attractive strategy is to apply the knowledge established in Arabidopsis to legume TFs (Hofer et al., 1997; Berbel et al., 2001; Taylor et al., 2002; Dong et al., 2005; Benlloch et al., 2006). The expression of legume TF genes in Arabidopsis or tobacco plants also supports a conservation of functions for TF homologs across plant species (e.g. in plant development [GmWRKY13; Zhou et al., 2008], metabolite biosynthesis [LjMYB TFs homologous to AtTT2; Yoshida et al., 2008], plant resistance to pathogens [GmAP2-EREBP TFs; Zhang et al. 2008], and plant resistance to abiotic stresses [GmWRKY TFs; Zhou et al. 2008; GmDREB3; Chen et al., 2009]). However, complete functional redundancy of TF proteins between plants is not always found. For example, the AtPAP1 gene encoding a MYB TF gene controls anthocyanin synthesis in Arabidopsis but it does not activate this pathway when expressed in M. truncatula (Peel et al., 2009).
Based upon these few examples, TF function appears highly but not absolutely conserved across plant species. This conclusion is also supported by the strong conservation of TF signature protein domains and the tertiary structure of TFs (Dr. Jianlin Cheng, personal communication). However, one difficulty in applying this strategy is the accurate identification of true orthologs between plant species. As described below, due to the evolutionary distance existing between plants, syntenic relationships are difficult to define. For example, in the rosid clade, Arabidopsis and legumes fall in the Malvidae (Eurosidae II) and the Fabidae (Eurosidae I) subclades, respectively (Table I). These two subclades diverged approximately 115 to 93 millions years ago (Mya). According to Wang et al. (2009), a diversification of rosids occurred suddenly (in less than 15 millions years) shortly after the divergence between Fabidae and Malvidae. Such diversification might explain the vast heterogeneity in rosid habitats, morphology, and development. This evolutionary distance between crop plants and Arabidopsis is also highlighted by the strong changes in the size and organization of their genomes (Table I), such as the ancient genome duplication in legumes occurring around 45 to 55 Mya (Pfeil et al., 2005; Cannon et al., 2006) followed by a more recent duplication in soybean around 10 to 15 Mya (Schlueter et al., 2004, 2007). Despite the evolutionary distances existing between legumes and Arabidopsis, microsynteny is still found in some regions (Grant et al., 2000; Yan et al., 2003). For example, 14% of analyzed contig groups showed microsynteny between Arabidopsis and soybean (Zhu et al., 2003; Yan et al., 2004; Mudge et al., 2005). Likewise, a comparison between soybean, Medicago, and Arabidopsis identified two blocks with strong microsynteny (Shultz et al., 2007). Conversely, macrosynteny between Arabidopsis and legumes is difficult to identify due to genome duplication, recombination, and gene loss (Kevei et al., 2005; Schlueter et al., 2008).
Extensive macrosyntenic relationships exist between legume species (Choi et al., 2004; Cannon et al., 2006; for review, see Young and Udvardi, 2009). The establishment of genome-wide colinearity between legumes will be an important advantage for transferring information to soybean, an economically important plant, from the knowledge established in the legume models M. truncatula and L. japonicus. The fact that all four major legume genetic models (L. japonicus, M. truncatula, soybean, and common bean) fall in the Papilionoideae group, one of the three groups of legumes (i.e. legumes are divided into three groups named Caesalpinioideae, Mimosoideae, and Papilionoideae), supports the strong macrosynteny found in the available legume genome sequences. The Papilionoideae group diverged from the two other legume clades approximately 50 Mya, while the Lotus and Medicago lineages diverged from one another around 40 Mya (Wojciechowski, 2003). This recent divergence among major legumes makes it easier to identify orthologs by direct genome comparison. For example, this approach allowed the identification of key TF gene orthologs involved in the nodulation process and in floral meristem development (e.g. LjNIN and PsSym35; LjNSP1 and MtNSP1; LjNSP2 and MtNSP2; and LjFLO and PsFLO [Hofer et al., 1997; Schauser et al., 1999; Catoira et al., 2000; Borisov et al., 2003; Oldroyd and Long, 2003; Dong et al., 2005; Kalo et al., 2005; Smit et al., 2005; Heckmann et al., 2006]).
SOME LEGUME TF GENES ARE INDUCED SPECIFICALLY DURING NODULATION
As described above, legume TF distribution across families and their basic functions appear to be conserved compared with other plant families. Consequently, special legume traits may derive from unique TF gene expression patterns. During the last decade, the availability of cDNA and oligonucleotide arrays allowed the quantification of gene expression patterns in a large number of organisms in different tissues and under differing environmental conditions. However, several studies clearly demonstrated the limit of this technology to accurately quantify the expression of low-abundance transcripts, such as TF genes (Czechowski et al., 2004; Libault et al., 2007). To better characterize the expression of TF genes, large-scale quantitative reverse transcription-PCR platforms were developed to quantify TF gene expression in Arabidopsis, rice, M. truncatula, soybean, and L. japonicus (Czechowski et al., 2004; Caldana et al., 2007; Kakar et al., 2008; Libault et al., 2009; O. Montanari and M.K. Udvardi, unpublished data). Several studies used these resources to quantify TF gene expression in different tissues and in response to different treatments (McGrath et al., 2005; Libault et al., 2007; Gruber et al., 2009; Libault et al., 2009). Complementary to the use of large-scale quantitative reverse transcription-PCR platforms, ultra-high-throughput sequencing technologies, such as the 454 Life Sciences (Margulies et al., 2005) and Illumina Solexa (Bennett et al., 2005) platforms, allow an accurate quantification of low-abundance transcripts. These resources allow the characterization of the TF gene transcriptome. However, the use of such technology is fully informative only if the genome of the organism of interest is sequenced and accurately annotated.
To assess the potential of divergent gene expression patterns as a potential reason for legume-specific attributes, we investigated the expression of members of the NIN-like gene family. This family was selected based upon the involvement of some of its members in root hair infection and nodule development (Schauser et al., 1999; Catoira et al., 2001; Borisov et al., 2003, Marsh et al., 2007). Two recent studies place NIN gene function just downstream to NSP1 and NSP2 GRAS in the signaling cascade regulating legume nodulation. For example, LjNIN gene expression is under the control of LjNSP2 (Murakami et al., 2006). Moreover, MtNSP1 and MtNSP2 interact with the MtNIN gene promoter to regulate its expression (Hirsch et al., 2009). In the same study, Hirsch et al. (2009) showed that the molecular interaction of MtNSP1 and MtNSP2 was also required to activate MtENOD11 gene expression during nodulation. In addition, Gonzalez-Rizzo et al. (2006) showed that MtNIN gene expression was under the control of cytokinin, a hormone previously described to influence legume nodulation (Murray et al., 2007; Frugier et al., 2008).
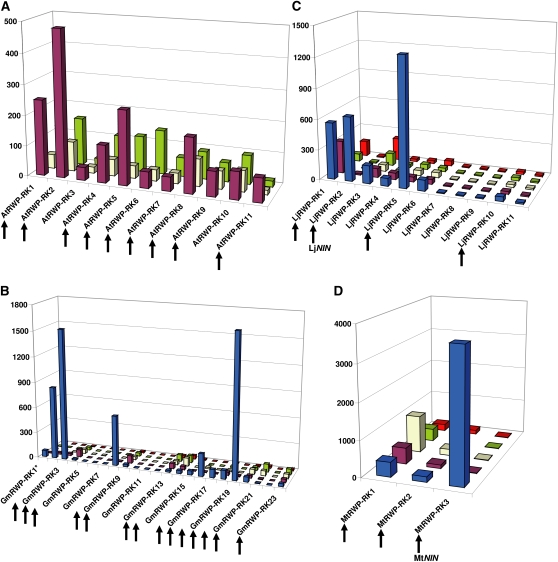
Based on a bioinformatic analysis and similarly to Schauser et al. (2005), we identified two distinctive domains in the plant NIN-like amino acid sequences: an RWP-RK domain and a Phox and Bem1p (PB1) domain (the PB1 domain is found in a large number of eukaryotic cytoplasmic signaling proteins; Sumimoto et al., 2007; Fig. 2). In addition, TF proteins carrying only the RWP-RK domain were also identified in different plant genomes. The RWP-RK gene TF family is a small one, with 14 members in Arabidopsis (0.62% of all identified TF genes), five in M. truncatula (0.34%), 12 in L. japonicus (0.73%), and 23 in soybean (0.41%; Fig. 2). Among them, we identified nine, three, four, and 14 NIN-like genes in Arabidopsis, M. truncatula, L. japonicus, and soybean, respectively. The expression of 11 RWP-RK genes Arabidopsis, three in M. truncatula, 12 in L. japonicus, and 23 in soybean was quantified in a large variety of tissues using Affymetrix arrays or ultra-high-throughput sequencing (Schmid et al., 2005; Benedito et al., 2008; Høgslund et al., 2009; M. Libault, A. Farmer, G.D. May, and G. Stacey, unpublished data; Fig. 2). RWP-RK genes were expressed at least 10-fold higher in nodules than in other organs analyzed in the case of one gene in M. truncatula, two in L. japonicus, and seven in soybean (Fig. 2). All of these legume RWP-RK genes expressed specifically in nodules share the PB1 domain and, consequently, are NIN-like genes. Interestingly, M. truncatula and soybean nodule-specific NIN-like genes are not expressed (or expressed at very low levels) in other tissues, indicating specialization of these genes for nodulation and SNF. Because the proportion of NIN-like genes in the Arabidopsis and soybean genomes is similar, this analysis supports the hypothesis that legumes coopted a subset of preexisting NIN-like genes for use in symbiotic establishment by modifying their expression patterns.
Figure 2.
Arabidopsis, soybean, L. japonicus, and M. truncatula RWP-RK gene expression in various plant tissues. Relative expression levels of Arabidopsis (A), soybean (B), L. japonicus (C), and M. truncatula (D) RWP-RK genes (y axis) are reported for five different tissues (nodule [blue], root [purple], leaf [yellow], flower [green], and pod [red]). For each plant species, the identity of the RWP-RK genes is reported on the x axis. Details are available in Supplemental Table S1. Genes encoding proteins carrying a PB1 domain (NIN-like genes) are highlighted by arrows. LjNIN (Schauser et al., 1999) and MtNIN (Marsh et al., 2007) involved in nodulation are highlighted. *, GmRWP-RK1 was also strongly expressed in soybean apical meristem and root tip; consequently, GmRWP-RK1 is not considered a nodule-specific RWP-RK.
FUTURE DIRECTIONS IN LEGUME TF RESEARCH
An analysis of TF genes among the sequenced plant genomes does not reveal a legume-specific family, nor does it identify a specific TF family that appears to have been preferentially expanded in legumes. However, legumes have clearly succeeded in developing specific traits by diverting some TF genes for a more specialized function. In the case of NIN-like TFs, this likely involved changes in the sequences of promoters that allowed activation of gene expression during nodule development.
What is now needed is a clearer picture of the legume TF gene transcriptome, interactome (protein-protein interaction), and elucidation of the regulon controlled by each TF. The legume TF transcriptome should be elucidated soon with the emergence of ultra-high-throughput sequencing approaches, making the establishment of a TF interactome the next important challenge. Such an interactome will encompass TF-TF interactions as well as TF-DNA interactions. Identifying TF-TF interactions will highlight the complexity of legume gene regulation. Studies focusing on bacteria and yeast cells have already established TF-TF interaction networks, giving the first insights into the complexity of these systems (Babu et al., 2004; Luscombe et al., 2004; Ye et al., 2009).
The in vivo identification of cis-regulatory regions of TF genes is now possible by combining chromatin immunoprecipitation (ChIP) methods with the use of tiling arrays developed after sequencing of the whole genome (Gregory et al., 2008). The most recent approach utilizes ChIP coupled with ultra-high-throughput sequencing technologies (ChIP-Seq; Barski and Zhao, 2009). These approaches allow the identification of accessible promoter elements that interact with TF proteins with respect to chromatin remodeling. In fact, chromatin rearrangements are an important tool for regulation of gene expression at the epigenetic level by controlling access to TF-binding sites (Barrera and Ren, 2006). Analyzing TF interactome (TF-TF and TF-DNA) data in the context of chromatin structure will provide a clearer picture of legume gene regulation.
CONCLUSION
Legumes are a fascinating family of plants due, in part, to their ability to develop unique organs, called nodules, which harbor nitrogen-fixing rhizobia. Among the genes involved in the nodulation process, several TFs have been characterized. The similar distribution of TF genes in the various known TF families in legumes and nonlegumes, as well as the conserved nature of their basic biochemical functions, cannot readily explain how legume-specific traits such as SNF evolved. Based upon analysis of the NIN-like TF family in soybean, L. japonicus, M. truncatula, and Arabidopsis, we assume that legumes coopted existing TF genes for use in legume-specific processes by modifying their expression patterns. Analysis of synteny between legume and nonlegume plants and subsequent dissection of the expression patterns of orthologous TF genes will help to test this hypothesis. Furthermore, identification of DNA-binding sequences of TF proteins and detailed analysis of TF gene expression will provide a means to understand the impact of TF activity on the legume transcriptome.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Expression levels of Arabidopsis, soybean, L. japonicus, and M. truncatula RWP-RK genes used to create Figure 2.
Supplementary Material
This work was supported by the National Science Foundation Plant Genome Program (grant no. DBI–0421620).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gary Stacey (staceyg@missouri.edu).
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Shimoda Y, Kouchi H, Tabata S, Sato S (2008) A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol 147 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14 283–291 [DOI] [PubMed] [Google Scholar]
- Barrera LO, Ren B (2006) The transcriptional regulatory code of eukaryotic cells: insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol 18 291–298 [DOI] [PubMed] [Google Scholar]
- Barski A, Zhao K (2009) Genomic location analysis by ChIP-Seq. J Cell Biochem 107 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55 504–513 [DOI] [PubMed] [Google Scholar]
- Benlloch R, d'Erfurth I, Ferrandiz C, Cosson V, Beltran JP, Canas LA, Kondorosi A, Madueno F, Ratet P (2006) Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol 142 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ST, Barnes C, Cox A, Davies L, Brown C (2005) Toward the 1,000 dollars human genome. Pharmacogenomics 6 373–382 [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25 441–451 [DOI] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, et al (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA 103 14959–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB (2001) Chance and necessity: the evolution of morphological complexity and diversity. Nature 409 1102–1109 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128 1507–1518 [DOI] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ (2007) GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem Biophys Res Commun 353 299–305 [DOI] [PubMed] [Google Scholar]
- Chen M, Xu Z, Xia L, Li L, Cheng X, Dong J, Wang Q, Ma Y (2009) Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J Exp Bot 60 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- de Lorenzo L, Merchan F, Blanchet S, Megias M, Frugier F, Crespi M, Sousa C (2007) Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol 145 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D (2005) Floral patterning in Lotus japonicus. Plant Physiol 137 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA (2003) The rest of the iceberg: legume diversity and evolution in a phylogenetic context. Plant Physiol 131 900–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al (2008) The Pfam protein families database. Nucleic Acids Res 36 D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Freeman J, Oliver JE, Pineda M (1990) Nuclear factors interact with conserved A/T-rich elements upstream of a nodule-enhanced glutamine synthetase gene from French bean. Plant Cell 2 925–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia E, Barabaschi D, Tondelli A, Laido G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115 1083–1091 [DOI] [PubMed] [Google Scholar]
- Frugier F, Kosuta S, Murray JD, Crespi M, Szczyglowski K (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci 13 115–120 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Cregan P, Shoemaker RC (2000) Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc Natl Acad Sci USA 97 4168–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Yazaki J, Ecker JR (2008) Utilizing tiling microarrays for whole-genome analysis in plants. Plant J 53 636–644 [DOI] [PubMed] [Google Scholar]
- Gruber V, Blanchet S, Diet A, Zahaf O, Boualem A, Kakar K, Alunni B, Udvardi M, Frugier F, Crespi M (2009) Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol Genet Genomics 281 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AC, Busk H, Marcker A, Marcker KA, Jensen EO (1999) VsENBP1 regulates the expression of the early nodulin PsENOD12B. Plant Mol Biol 40 495–506 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Ramirez M, Valdes-Lopez O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Munoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7 581–587 [DOI] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS One 4 e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37 D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467 [DOI] [PubMed] [Google Scholar]
- Jensen EO, Marcker KA, Schell J, Bruijn FJ (1988) Interaction of a nodule specific, trans-acting factor with distinct DNA elements in the soybean leghaemoglobin Ibc(3) 5# upstream region. EMBO J 7 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC, et al (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kevei Z, Seres A, Kereszt A, Kalo P, Kiss P, Toth G, Endre G, Kiss GB (2005) Significant microsynteny with new evolutionary highlights is detected between Arabidopsis and legume model plants despite the lack of macrosynteny. Mol Genet Genomics 274 644–657 [DOI] [PubMed] [Google Scholar]
- Laursen NB, Larsen K, Knudsen JY, Hoffmann HJ, Poulsen C, Marcker KA, Jensen EO (1994) A protein binding AT-rich sequence in the soybean leghemoglobin c3 promoter is a general cis element that requires proximal DNA elements to stimulate transcription. Plant Cell 6 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Joshi T, Takahashi K, Hurley-Sommer A, Puricelli K, Blake S, Xu D, Nguyen HT, Stacey G (2009) Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol 151 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact 20 900–911 [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431 308–312 [DOI] [PubMed] [Google Scholar]
- Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang XD, Vandenbosch KA, Rose RJ (2008) The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol 146 1622–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz BA, Welters P, Hoffmann HJ, Jensen EO, Schell J, de Bruijn FJ (1988) Primary structure and promoter analysis of leghemoglobin genes of the stem-nodulated tropical legume Sesbania rostrata: conserved coding sequences, cis-elements and trans-acting factors. Mol Gen Genet 214 181–191 [DOI] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, Galiba G, Dubcovsky J (2006) A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol Genet Genomics 275 193–203 [DOI] [PubMed] [Google Scholar]
- Mudge J, Cannon SB, Kalo P, Oldroyd GE, Roe BA, Town CD, Young ND (2005) Highly syntenic regions in the genomes of soybean, Medicago truncatula, and Arabidopsis thaliana. BMC Plant Biol 5 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S (2006) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13 255–265 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Kawaguchi M (2002) The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol 43 853–859 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim ML, Lee SM, Bahk JD, Yun DJ, Lim CO, Hong JC, Lee SY, Cho MJ, Chung WS (2007) Pathogen-induced binding of the soybean zinc finger homeodomain proteins GmZF-HD1 and GmZF-HD2 to two repeats of ATTA homeodomain binding site in the calmodulin isoform 4 (GmCaM4) promoter. Nucleic Acids Res 35 3612–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556 [DOI] [PubMed] [Google Scholar]
- Peel GJ, Pang Y, Modolo LV, Dixon RA (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59 136–149 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54 441–454 [DOI] [PubMed] [Google Scholar]
- Ramalingam J, Vera Cruz CM, Kukreja K, Chittoor JM, Wu JL, Lee SW, Baraoidan M, George ML, Cohen MB, Hulbert SH, et al (2003) Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact 16 14–24 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Schauser L, Wieloch W, Stougaard J (2005) Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol 60 229–237 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47 868–876 [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Lin JY, Schlueter SD, Vasylenko-Sanders IF, Deshpande S, Yi J, O'Bleness M, Roe BA, Nelson RT, Scheffler BE, et al (2007) Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Scheffler BE, Jackson S, Shoemaker RC (2008) Fractionation of synteny in a genomic region containing tandemly duplicated genes across Glycine max, Medicago truncatula, and Arabidopsis thaliana. J Hered 99 390–395 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Shultz JL, Ray JD, Lightfoot DA (2007) A sequence based synteny map between soybean and Arabidopsis thaliana. BMC Genomics 8 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Kamakura S, Ito T (2007) Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE 2007 re6. [DOI] [PubMed] [Google Scholar]
- Taylor SA, Hofer JM, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis TH (2002) PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol 129 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang JY, Benedito V, Hofer JM, Chueng F, et al (2007) Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol 144 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res 37 D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Reyes JL, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vernié T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P (2008) EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE (2009) Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA 106 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52 716–729 [DOI] [PubMed] [Google Scholar]
- Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA (2008) DBD: taxonomically broad transcription factor predictions. New content and functionality. Nucleic Acids Res 36 D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF (2003) Reconstructing the phylogeny of legumes (Fabaceae): an early 21st century perspective. In BB Klitgaard, A Bruneau, eds, Advances in Legume Systematics. Part 10. Higher Level Systematics. Royal Botanic Garden, Kew, UK, pp 5–35
- Yan HH, Mudge J, Kim DJ, Larsen D, Shoemaker RC, Cook DR, Young ND (2003) Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor Appl Genet 106 1256–1265 [DOI] [PubMed] [Google Scholar]
- Yan HH, Mudge J, Kim DJ, Shoemaker RC, Cook DR, Young ND (2004) Comparative physical mapping reveals features of microsynteny between Glycine max, Medicago truncatula, and Arabidopsis thaliana. Genome 47 141–155 [DOI] [PubMed] [Google Scholar]
- Ye C, Galbraith SJ, Liao JC, Eskin E (2009) Using network component analysis to dissect regulatory networks mediated by transcription factors in yeast. PLOS Comput Biol 5 e1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49 157–169 [DOI] [PubMed] [Google Scholar]
- Young ND, Udvardi M (2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12 193–201 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92 [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Guan S, Li LC, Li A, Guo J, Mao L, Ma Y (2008) Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot 59 4095–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6 486–503 [DOI] [PubMed] [Google Scholar]
- Zhu B, Ye C, Lu H, Chen X, Chai G, Chen J, Wang C (2006) Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J Plant Res 119 247–256 [DOI] [PubMed] [Google Scholar]
- Zhu H, Chen T, Zhu M, Fang Q, Kang H, Hong Z, Zhang Z (2008) A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Choi HK, Cook DR, Shoemaker RC (2005) Bridging model and crop legumes through comparative genomics. Plant Physiol 137 1189–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kim DJ, Baek JM, Choi HK, Ellis LC, Kuester H, McCombie WR, Peng HM, Cook DR (2003) Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol 131 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.