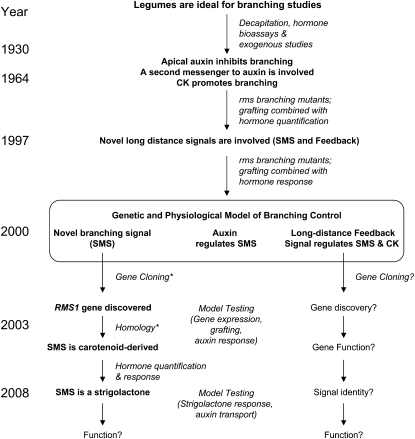
Shoot branching was one of the first developmental processes found to be controlled by plant hormones including auxin and cytokinin (Dun et al., 2009). Later, a novel branching hormone was proposed (Fig. 1), and recently strigolactones were discovered as this hormone (Gomez-Roldan et al., 2008; Umehara et al., 2008). In this Update, we focus on the important contributions to this discovery made by legumes, particularly garden pea (Pisum sativum; Fig. 1). Legumes are useful for shoot-branching research because of several features that facilitate studies of axillary buds and long-distance signaling. They have long internodes separating axillary buds and the shoot tip, are easy to graft, are amenable to root xylem-sap extraction, and their axillary buds are accessible for hormone applications, growth measurements, and other related analyses. Additionally, for many pea varieties, most axillary buds are dormant but have the potential for release throughout development. Some of these traits, in addition to the availability of mutants, made working with pea and other legumes attractive to the early plant physiologists, and remain relevant today.
Figure 1.
Research date line for the discovery of strigolactones as the new branching hormone, its regulation by auxin, and the involvement of long-distance feedback signaling. Approximate date line is shown on left, major hypotheses in bold, and experimental approaches in italics. Question mark (?) highlights knowledge gaps; asterisk (*) denotes discoveries that relied on research in Arabidopsis. CK, Cytokinin; SMS, novel branching signal.
Early studies of shoot branching focused on decapitation-induced bud outgrowth, comparing branched decapitated plants with nonbranched control plants (Fig. 1). The term apical dominance was coined because decapitation and auxin application studies provided evidence that auxin, produced in the shoot tip, was involved in the inhibition of axillary bud outgrowth at nodes below (Cline, 1991). Studies of axillary bud release on different shoots of plants with two shoots (e.g. Snow, 1929; Fig. 2A) suggested the existence of an inhibitory signal that moves upward in the shoot and is controlled by auxin supplied from the shoot tip.
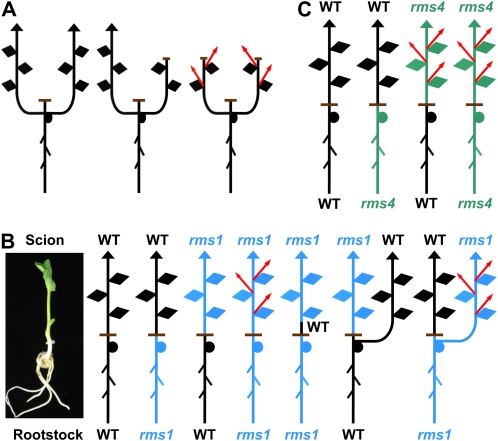
Figure 2.
Classic experiments supporting the idea of a novel, upward-moving bud outgrowth inhibitor. Two-shoot experiments, where the main shoot was decapitated to allow for the outgrowth of two cotyledonary branches, conducted by Snow (1929) helped show that a signal that was not auxin moves upward to inhibit axillary bud outgrowth (A). Grafting experiments using the pea rms mutants, combined with auxin and cytokinin measurements, showed that a signal that is made in the rootstock and scion, and moves upward only, inhibits axillary bud outgrowth (B and C). The rms1 mutant is defective in the synthesis of this signal (B), while the rms4 mutant is defective in response to the signal (C). Arrows represent growing shoots including the vertical main stem and axillary branches (shown in red). Horizontal lines correspond to the site of decapitation (A) or grafting (B and C). WT, Wild type.
Like any area of modern plant biology, the investigation of mutants has been an essential component of shoot-branching studies. In pea, several increased branching mutants are named ramosus (rms), meaning with many branches. Generated through various mutagenesis programs, these mutants began to be characterized in the 1990s (Fig. 1; for review, see Beveridge, 2000).
GRAFTING COMBINED WITH HORMONE QUANTIFICATION
As long-distance signaling was clearly an important process in the control of shoot branching (Fig. 2A), grafting studies, easily performed in legumes, were used to investigate the rms mutants (e.g. Fig. 2, B and C; for review, see Beveridge, 2000, 2006).
There are five different rms mutants that have axillary branches at basal and aerial nodes of the plant (rms1–rms5; for review, see Beveridge, 2006). Others show branching only at basal nodes and are less well characterized (Beveridge et al., 2003). The mutants rms1, rms2, and rms5 show inhibition of bud outgrowth when grafted to wild-type rootstocks (Fig. 2B; Beveridge et al., 1997a; Morris et al., 2001), demonstrating the presence of a long-distance signal moving from the root to the shoot. It was clear that this signal was also produced in shoots, because wild-type shoots did not branch when grafted to mutant rootstocks (Fig. 2B; e.g. Beveridge et al., 1997a; Morris et al., 2001). Such grafting studies allowed these mutants (rms1, rms2, and rms5) to be characterized as long-distance signaling mutants controlling the level or transport of a long-distance signal(s). In contrast, bud release in the shoots of the other mutants, rms3 and rms4, was not reduced by grafting to wild-type rootstocks (Fig. 2C), and hence these mutants could be considered as response mutants. As described below, rms4 does indeed lack response to the novel hormone strigolactone (Gomez-Roldan et al., 2008).
The long-distance signaling mutants provided an opportunity to isolate and investigate branching regulatory hormones, both known and unknown. Consequently, the major hormones previously thought to account for shoot branching, root-derived cytokinin, and shoot-derived auxin were quantified in these mutants (Beveridge et al., 1997a, 1997b; Beveridge, 2000). In two of the mutants affecting a long-distance branch-regulation signal (rms1 and rms5), auxin content and transport were not reduced and xylem-sap cytokinin content was not elevated (Beveridge et al., 1997a, 2000; Morris et al., 2001). This led to the hypothesis that a novel long-distance signal, other than auxin or cytokinin, must explain the branching phenotype of these plants (Fig. 1).
Further grafting studies, where plants had two shoots (Fig. 2B) or two rootstocks, led to the hypothesis that the novel long-distance signal was an inhibitor of branching that could only move upward in shoots, not down and then up (Fig. 3; Foo et al., 2001). Similarly, grafting studies showed that RMS1 and RMS5 were required in the same cell or tissue to produce this inhibitor (Fig. 3; Morris et al., 2001). Moreover, this signal is produced in potent quantities in the stem as a small piece of wild-type epicotyl was sufficient to repress branching in an rms1 mutant shoot (Fig. 2B; Foo et al., 2001). Similar grafting studies have been reported in Arabidopsis (Arabidopsis thaliana) and petunia (Petunia hybrida; Beveridge, 2006).
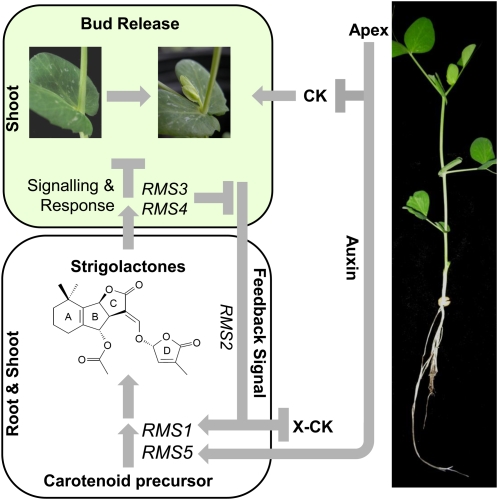
Figure 3.
Strigolactone branching pathway. RMS1 and RMS5 are together required for the synthesis of the strigolactone bud outgrowth inhibitor that is carotenoid derived. Strigolactones (orobanchyl acetate, a strigolactone found in pea, is shown) are synthesized in the root and shoot, and move upward. RMS3 and RMS4 are both required local to the bud (in the shoot) to respond to the strigolactone branching inhibitor; response results in inhibition of bud release (as seen in the wild-type plant on the right) and down-regulation of the RMS2-mediated feedback signal. The feedback signal, in addition to auxin from the shoot apex, moves downward and up-regulates strigolactone biosynthesis via transcriptional regulation of RMS1 and RMS5 expression. The feedback signal also represses xylem-sap cytokinin (X-CK) export from the roots, and auxin represses local cytokinin (CK) production in the shoot. While mutations in any of the genes shown result in an increased branching phenotype, rms1 and rms5 are referred to as synthesis mutants, and rms3 and rms4 as response mutants. Flat-ended lines represent inhibition; arrows represent promotion or flow.
Hormone measurements in one of the long-distance signaling mutants of pea, rms2, revealed both elevated shoot auxin content and elevated xylem-sap cytokinin content (Beveridge et al., 1997a). In the same year, Faiss et al. (1997) showed that root-derived cytokinins have little effect on axillary bud release in shoots, an observation that remains unchallenged to date (Shimizu-Sato et al., 2009). As such, an alternative hypothesis for the long-distance signaling role of RMS2 was suggested (Fig. 3; discussed below; Beveridge, 2000).
Cytokinin quantification showed that rather than enhanced xylem-sap cytokinin content, as in an rms2 mutant, the other increased branching mutants had greatly depleted xylem-sap cytokinin levels (Beveridge et al., 1997a, 1997b). This depletion in the xylem sap does not cause depleted cytokinin content in shoot tissue (Foo et al., 2007). Grafting studies indicated that a second long-distance signal may be involved in the branching regulatory system because the reduced xylem-sap cytokinin content in rootstocks was caused by the scion (Beveridge et al., 1997b; Beveridge, 2000). This signal was named the feedback signal (Fig. 3; Beveridge, 2000). Since rms2 plants did not induce this feedback, and branching in rms2 shoots could not be inhibited by grafting to rms1 long-distance signaling mutant rootstocks, yet could be inhibited by grafting to wild-type rootstocks, it was concluded that RMS2 may be required to feedback regulate xylem-sap cytokinin content and the RMS1 long-distance signal. This was later supported by RMS1 gene expression studies; RMS1 gene expression is feedback up-regulated in all rms mutants except rms2 (Foo et al., 2005). The genes required for long-distance signaling could therefore be separately classified as synthesis (RMS1 and RMS5) and feedback (RMS2) genes (Beveridge, 2006).
GENE DISCOVERY
Unfortunately, until recently cloning genes responsible for pea mutant phenotypes has largely been via gene waiting, a process where candidate genes are identified by waiting for the genes to be characterized in other plant species, usually Arabidopsis (Fig. 1). This approach has been quite successful in pea, as mutants are generally well characterized, including their genetic map location, which helps to guide identification of the best candidate genes. Given that pea is difficult to transform and has a genome 30 times larger than that of Arabidopsis, it is not an ideal species for molecular genetics studies. The recent direct gene cloning of a leaf morphology gene in pea demonstrates the potential for future discoveries in pea (Hofer et al., 2009). The availability of pea bacterial artificial chromosome libraries, a TILLING platform, EST data sets, and genome sequences from pea and other model legumes, together with strong macrosyntenic relationships among pea, Medicago truncatula, and Lotus japonicus, will greatly enhance the rate of gene discoveries in legumes including pea (for review, see Cannon et al., 2009).
In the case of shoot branching, research in Arabidopsis has yielded four branching genes (for review, see Ongaro and Leyser, 2008). Three of these were found to be orthologs of branching genes in pea and rice (Oryza sativa; Dun et al., 2009). The response gene in Arabidopsis, MORE AXILLARY GROWTH2 (MAX2), which was identified as encoding an F-box protein, is orthologous to the response gene RMS4 in pea (for review, see Dun et al., 2009). F-box proteins and the ubiquitin-proteasome pathway play an important role in hormone signaling; in particular for auxin, it has been demonstrated that the F-box TRANSPORT INHIBITOR RESPONSE1 (TIR1) functions as an auxin receptor. This provides a framework to test the hypothesis that the response genes MAX2 and RMS4 are involved in hormone signal transduction and may encode a receptor for the branching inhibitor.
Two of the synthesis genes, identified first in Arabidopsis (MAX3 and MAX4) and then in pea (RMS5 and RMS1), petunia (DECREASED APICAL DOMINANCE1), and rice (DWARF10 [D10] and D17/HIGH TILLERING DWARF1), encode carotenoid cleavage dioxygenases (CCD7 and CCD8, respectively; Dun et al., 2009). These proteins are targeted to the plastid and are likely involved in sequential cleavage of a carotenoid substrate. This supported the likely enzymatic function of RMS1 and RMS5, explained the requirement of RMS1 and RMS5 to function in the same cell, and led to the hypothesis that the novel signal may be carotenoid derived. Unfortunately, RMS2 and RMS3 have not yet been identified at the molecular level.
STRIGOLACTONE DISCOVERY
The putative carotenoid cleavage function of the synthesis genes led researchers to consider that the novel branching inhibitor may be a strigolactone (Gomez-Roldan et al., 2008; Umehara et al., 2008; Fig. 3). Strigolactones, which are likely derived from carotenoids, are known to function in the rhizosphere in the symbiosis of plants with arbuscular mycorrhizae and in the seed germination of parasitic weeds.
In a simultaneous breakthrough in pea (Gomez-Roldan et al., 2008) and rice (Umehara et al., 2008), the increased branching synthesis mutants (rms1 and rms5 in pea) were shown to be deficient in strigolactones. The synthetic strigolactone, GR24, was shown to inhibit bud outgrowth to the level seen in wild type and was active down to 10 nm when supplied directly to the xylem stream in the internode (Gomez-Roldan et al., 2008; Umehara et al., 2008). In contrast, the response mutants were not deficient and did not respond to strigolactone treatments. Strigolactones are therefore likely to be the novel shoot-branching inhibitor (Figs. 1 and 3).
Importantly, mutants that do not respond to strigolactones may not necessarily be directly affected in the strigolactone signaling pathway. For example, it is unclear whether TEOSINTE BRANCHED1 (TB1), another gene implicated in shoot branching control but not yet reported in pea, acts in a strigolactone-dependent or -independent pathway (for review, see Dun et al., 2009), even though the Arabidopsis mutant lacks strigolactone response (Brewer et al., 2009). An additional argument that RMS4/MAX2 is likely part of the strigolactone signaling pathway is that the corresponding mutants in all species show feedback regulation of the strigolactone biosynthesis genes (Foo et al., 2005; Johnson et al., 2006; Arite et al., 2007; Hayward et al., 2009), whereas the tb1-branching mutant, examined only in rice so far, does not show the feedback regulation (Arite et al., 2007). This observation does not preclude TB1 from being a target of the strigolactone pathway, but simply highlights the likelihood that RMS4/MAX2 is directly involved.
AUXIN RESPONSE: GRAFTING, DECAPITATION, AND APPLICATION STUDIES
Many legume species are excellent for decapitation studies because buds of intact plants are completely inhibited, yet they are fully released after decapitation. After decapitation these buds can be substantially, but not completely (e.g. Morris et al., 2005), inhibited by exogenous auxin applications. As also suggested by early studies (Snow, 1929), this implies that auxin alone may not be sufficient to inhibit branching.
While there are generally two or three axillary buds present at most nodes of pea plants, one feature of the rms mutants is that only one of these grows out to produce a branch under standard conditions (e.g. Beveridge et al., 2000). If the growing branch is removed, a previously inhibited mutant bud is left behind at the node. Subsequent decapitation leads to bud outgrowth at this node. Such observations indicate that rms mutant buds are sensitive to correlative inhibition by growing shoot tips (Ferguson and Beveridge, 2009). Consequently, comparisons can be made between the outgrowth of previously inhibited buds on mutant and wild-type plants. Interestingly, using this system, rms branching mutants show a reduced response to auxin after decapitation compared with wild-type plants (Beveridge et al., 2000). This could have led to the classification of these mutants as auxin-response or auxin-resistant mutants; however, such a classification can be misleading. The rms mutants are all defective in auxin response (Beveridge et al., 2000), but some are also deficient in strigolactones and it is this hormone rather than auxin that accounts for their branching phenotype (Gomez-Roldan et al., 2008).
The question then arises: What is the relationship between strigolactones and auxin? Using grafting, bud outgrowth is inhibited in intact rms synthesis mutant shoots, by wild-type rootstocks, presumably by restoring strigolactone content to the shoot (Fig. 2B). Importantly, this restoration of strigolactones to synthesis mutant shoots of grafted plants also fully restores auxin response to decapitated shoots (Beveridge et al., 2000). This early work gave rise to the current hypothesis that branching induced after decapitation is due to a reduction in strigolactone levels caused by auxin depletion.
Once the synthesis genes (RMS1 and RMS5) were identified at the molecular level in pea, their transcript levels were found to be auxin responsive (Fig. 3; Foo et al., 2005; Johnson et al., 2006), consistent with the proposition that auxin regulates strigolactone levels (Beveridge, 2000; Dun et al., 2009). Indeed, bud outgrowth in decapitated plants was inhibited by strigolactone applications (Brewer et al., 2009). Moreover, branching in auxin-response mutants of Arabidopsis, such as auxin resistant1 (axr1) and the tir1 auxin signaling f-box1 (afb1) afb2 afb3 quadruple mutant, was also inhibited by strigolactone treatments. Although this does not rule out an additional function of auxin downstream of strigolactones, it provides strong support for auxin depletion promoting bud outgrowth via strigolactone depletion. Auxin regulation of strigolactone synthesis genes has also been reported in rice and Arabidopsis (Arite et al., 2007; Hayward et al., 2009). In Arabidopsis, this may occur via a TIR1/AXR1-mediated pathway involving degradation of an Aux/IAA protein (Hayward et al., 2009).
Auxin regulation of strigolactone synthesis genes contributes to the feedback up-regulation of these genes in shoots of mutant plants of pea and Arabidopsis (Dun et al., 2009; Hayward et al., 2009). The long-distance component of feedback regulation is substantial in pea and involves auxin and a nonauxin signal (Dun et al., 2009). In Arabidopsis, where the feedback is mostly shoot controlled, the feedback is mostly regulated by auxin.
AUXIN-INDEPENDENT DECAPITATION RESPONSE
By using pea plants with long internodes, and hence a considerable distance between the shoot tip and axillary buds, it became evident that auxin dynamics were inadequate to account for the early bud outgrowth observed after decapitation (Morris et al., 2005). For example, measurable bud outgrowth at the base of a 20-cm-tall pea plant can occur as early as 6 h after decapitation. Meanwhile, polar auxin transport and the rate of endogenous auxin depletion in stem segments after decapitation, as measured in Morris et al. (2005), occurs at about 1 cm per hour. This suggests that a signal faster than the depletion of auxin caused the observed early bud outgrowth of these decapitated plants. Consistent with this, auxin addition did not prevent early bud outgrowth, but did prevent sustained outgrowth, from about 20 h after treatment (Morris et al., 2005). Moreover, treatments causing an auxin depletion in intact shoots, including treatment with the auxin transport inhibitor naphthylphthalamic acid, or stem girdling (described below), though similar to decapitated plants in the magnitude of auxin depletion, were considerably less effective than decapitation at inducing outgrowth (Morris et al., 2005; Ferguson and Beveridge, 2009). Combined, these experiments suggest that factors other than auxin, perhaps even including turgor or electrochemical signaling, prime particular axillary buds to be responsive to auxin depletion.
LOCAL CYTOKININ
The role of locally synthesized cytokinin, regulated by auxin, has recently been reviewed by Shimizu-Sato et al. (2009). Additional studies in pea using stem girdling indicate that low local cytokinin levels (at or below wild-type levels) may limit bud outgrowth even in auxin- and strigolactone-depleted plants (Ferguson and Beveridge, 2009). Girdling via placement of hot wax in a reservoir encircling the stem destroys living stem tissue and is facilitated by the long internodes in pea. Like decapitation, stem girdling reduces expression of auxin response and strigolactone synthesis genes and yet allows ongoing growth of the shoot tip. Furthermore, as expected from auxin-cytokinin interactions (Shimizu-Sato et al., 2009), stem girdling usually enhances cytokinin biosynthesis gene expression and bud outgrowth at nodes below (Ferguson and Beveridge, 2009). An interesting finding from this study is that bud outgrowth does not occur at basal nodes of older plants when the girdle is positioned at the internode immediately above those nodes. In this case, cytokinin biosynthesis gene expression is not elevated. Importantly, bud outgrowth will occur under these conditions if the bud is supplied with exogenous cytokinin. This indicates that low local cytokinin levels might limit bud outgrowth even when auxin and strigolactone levels are reduced. It should be noted that this local cytokinin is different to xylem-sap cytokinin, and that while some strigolactone mutants have depleted xylem-sap cytokinin, shoot cytokinin content (nodes and shoot tip) is not different from wild type (Foo et al., 2007).
AUXIN TRANSPORT
A major direction in the study of strigolactone function in shoot branching is the effect on auxin transport. Initial auxin transport experiments in strigolactone mutants indicated the mutants transport more auxin than wild type and raised the notion that differences in auxin transport capacity might be important (Bennett et al., 2006). Later experiments in pea and Arabidopsis showed that wild-type stems do not have a limited auxin transport capacity (Brewer et al., 2009). A recent computational model of auxin transport and shoot branching proposes a role for an auxin transport switch that is established by competing auxin sources (Prusinkiewicz et al., 2009). This model focuses in part on interpreting studies such as that of branching in plants with two shoots (Fig. 2), and accounts for strigolactone effects on auxin transport and shoot branching. Experimental verification of this model will need to account for the observation that direct strigolactone application to growing axillary buds reduces outgrowth but does not have a rapid effect on their polar auxin transport (Brewer et al., 2009). In contrast, the auxin transport inhibitor naphthylphthalamic acid immediately blocks auxin transport from buds but does not affect their initial outgrowth (Brewer et al., 2009).
STAGES OF BUD OUTGROWTH: STRIGOLACTONES DO NOT COMPLETE THE STORY
In pea, the rms mutants do not branch at every node, and the outgrowth of a bud at a particular node depends on environmental factors such as photoperiod (Beveridge et al., 2003). Photoperiod is unlikely to exert its influence simply via strigolactones, as the rms mutants already lack strigolactones or strigolactone signaling. The variable branching patterns of the rms mutants, the evidence described above for a fast-moving decapitation signal, and the inability of auxin depletion to consistently induce outgrowth in wild-type plants made us consider the concept of stages of bud outgrowth. Dividing bud outgrowth into separate stages (for review, see Dun et al., 2006) provides a conceptual framework to consider the mélange of factors controlling shoot branching. Whereas some interactions may be via cross talk of signaling pathways, such as auxin regulation of strigolactone synthesis gene expression (Foo et al., 2005; Arite et al., 2007; Hayward et al., 2009), others may operate in a developmental context, where a particular hurdle must be removed or passed before another hormone or signaling response is effective. As such, strigolactones are proposed to inhibit the release of competent or responsive buds (Fig. 3); once a bud is released, additional factors are required for its outgrowth.
RECENT FINDINGS AND FUTURE DIRECTIONS
The physiological function and target genes and pathways involved in strigolactone response are yet to be revealed (Fig. 1). Two genes recently identified in rice, D14, a response gene (Arite et al., 2009), and D27, a synthesis gene (Lin et al., 2009), provide new opportunities for analysis. In particular, D14 is an exciting candidate for RMS3. The possibility that shoot branching control is not simply an on-off switch with multiple interacting triggers and inhibitors will need further attention in future studies. The roles of various signals and the cross talk among them need to be elucidated.
The discovery of strigolactones as a hormone controlling shoot branching could have substantial implications in horticulture and other plant industries. For example, the unwanted flush of growth after pruning could be suppressed by strigolactone application. In this case, inexpensive yet highly active and perhaps more stable strigolactone-type molecules need to be found. Conversely, to promote branching, inhibitors of strigolactone synthesis or response are needed.
Acknowledgments
We thank Drs. Julie Hofer, Philip Brewer, and Michael Mason for helpful comments on the manuscript. We apologize for literature not referred to due to the limit on the number of references.
This work was supported by the Australian Research Council Centre of Excellence for Integrative Legume Research and the Australian Research Council Discovery Grants Scheme (C.A.B., E.A.D.), and the Agence Nationale de la Recherche and the European Union FP6 Grain Legumes Integrated Project (C.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christine A. Beveridge (c.beveridge@uq.edu.au).
References
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50 1416–1424 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16 553–563 [DOI] [PubMed] [Google Scholar]
- Beveridge CA (2000) Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul 32 193–203 [Google Scholar]
- Beveridge CA (2006) Axillary bud outgrowth: sending a message. Curr Opin Plant Biol 9 35–40 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Murfet IC, Kerhoas L, Sotta B, Miginiac E, Rameau C (1997. b) The shoot controls zeatin riboside export from pea roots: evidence from the branching mutant rms4. Plant J 11 339–345 [Google Scholar]
- Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C (1997. a) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol 115 1251–1258 [Google Scholar]
- Beveridge CA, Symons GM, Turnbull CGN (2000) Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulates by genes Rms1 and Rms2. Plant Physiol 123 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Weller JL, Singer SR, Hofer JMI (2003) Axillary meristem development: budding relationships between networks controlling flowering, branching and photoperiod responsiveness. Plant Physiol 131 927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, May GD, Jackson SA (2009) Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant Physiol 151 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MG (1991) Apical dominance. Bot Rev 57 318–358 [Google Scholar]
- Dun EA, Brewer PB, Beveridge CA (2009) Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci 14 364–372 [DOI] [PubMed] [Google Scholar]
- Dun EA, Ferguson BJ, Beveridge CA (2006) Apical dominance and shoot branching: divergent opinions or divergent mechanisms? Plant Physiol 142 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiss M, Zalubìlová J, Strnad M, Schmülling T (1997) Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J 12 401–415 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA (2009) Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol 149 1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Morris SE, Parmenter K, Young N, Wang H, Jones A, Rameau C, Turnbull CGN, Beveridge CA (2007) Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiol 143 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CGN, Beveridge CA (2001) Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol 12 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais JC, et al (2008) Strigolactone inhibition of shoot branching. Nature 455 189–194 [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Turner L, Moreau C, Ambrose M, Isaac P, Butcher S, Weller J, Dupin A, Dalmais M, Le Signor C, et al (2009) Tendril-less regulates tendril formation in pea leaves. Plant Cell 21 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogné K, Beveridge CA, Rameau C (2006) Branching genes are conserved across species: genes controlling a novel signal in pea are co-regulated by other long-distance signals. Plant Physiol 142 1014–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, et al (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Cox MCH, Ross JJ, Kristantini S, Beveridge CA (2005) Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol 138 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SE, Turnbull CGN, Murfet IC, Beveridge CA (2001) Mutational analysis of branching in pea: evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol 126 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Leyser O (2008) Hormonal control of shoot branching. J Exp Bot 59 67–74 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed]
- Shimizu-Sato S, Tanaka M, Mori H (2009) Auxin-cytokinin interactions in the control of shoot branching. Plant Mol Biol 69 429–435 [DOI] [PubMed] [Google Scholar]
- Snow R (1929) The transmission of inhibition through dead stretches of stem. Ann Bot (Lond) 43 261–267 [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200 [DOI] [PubMed] [Google Scholar]