Abstract
Many strains of Arabidopsis (Arabidopsis thaliana) require exposure to prolonged cold for rapid flowering, a process known as vernalization. Vernalization in Arabidopsis results in the suppression of FLOWERING LOCUS C (FLC), a repressor of flowering. In a screen for mutants that no longer require vernalization for rapid flowering, we identified a dominant allele of the Enhancer of Zeste E(z) ortholog CURLY LEAF (CLF), clf-59. CLF is a Polycomb Group gene, and the clf-59 mutant protein contains a proline-to-serine transition in a cysteine-rich region that precedes the SET domain. Mutant plants are early flowering and have reduced FLC expression, but, unlike clf loss-of-function mutants, clf-59 mutants do not display additional pleiotropic phenotypes. clf-59 mutants have elevated levels of trimethylation on lysine 27 of histone H3 (H3K27me3) at FLC. Thus, clf-59 appears to be a gain-of-function allele, and this allele represses FLC without some of the components required for vernalization-mediated repression. In the course of this work, we also identified a marked difference in H3K27me3 levels at FLC between plants that contain and those that lack the FRIGIDA (FRI) gene. Furthermore, FRI appears to affect CLF occupancy at FLC; thus, our work provides insight into the molecular role that FRI plays in delaying the onset of flowering.
The switch from vegetative to reproductive growth is an important developmental transition in the life history of flowering plants. The proper timing of this switch is critical for reproductive success, especially in temperate climates where flower and seed production often needs to align with favorable weather conditions and/or the presence of pollinators. To ensure that flowering occurs at an optimal time of the year, plants have evolved mechanisms to sense and respond to seasonal environmental cues. One such cue is the prolonged cold of winter. The promotion of flowering by cold occurs through a process known as vernalization (for review, see Sung and Amasino, 2005; Dennis and Peacock, 2007).
Among accessions of Arabidopsis (Arabidopsis thaliana), there is variation in the requirement for vernalization. In winter-annual accessions, flowering is delayed unless plants undergo vernalization; summer-annual accessions do not require prolonged cold for rapid flowering. Natural variation at two loci in Arabidopsis, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), has a major influence on the requirement for vernalization (Napp-Zinn, 1979; Koornneef et al., 1994; Lee et al., 1994a). Many summer-annual accessions, including Columbia (Col), Wassilewskija (Ws), and Landsberg erecta, flower rapidly without a prolonged cold treatment because they carry mutations in FRI (Johanson et al., 2000). FRI acts to delay flowering through the up-regulation of FLC, a MADS box transcription factor that represses flowering. Vernalization leads to FLC repression in the presence of FRI and is thus able to supersede the ability of FRI to activate FLC (Michaels and Amasino, 1999; Sheldon et al., 1999). Thus, under natural conditions, the role of the FRI/FLC system is likely to repress flowering in autumn. The prolonged cold of winter then results in vernalization, which is manifest in the repression of FLC and rapid flowering in the spring.
Arabidopsis also contains a group of genes known collectively as the autonomous pathway that, in contrast to FRI, act as repressors of FLC. LUMINIDEPENDENS (LD), FCA, FLOWERING LOCUS D (FLD), and FVE are among this group of genes (Lee et al., 1994b; MacKnight et al., 1997; He et al., 2003; Ausin et al., 2004). FCA functions in RNA-mediated gene silencing (Baurle et al., 2007). FLD and FVE function in complexes that modify chromatin (He et al., 2003; Ausin et al., 2004). Mutations in autonomous pathway genes result in elevated FLC expression and delayed flowering (Michaels and Amasino, 2001). Similar to winter-annual accessions with an active FRI allele, delayed flowering in autonomous pathway mutants can be suppressed through vernalization.
Chromatin modification appears to play a large role in setting the expression level of FLC. In genetic backgrounds that favor elevated FLC expression, active chromatin modifications accumulate at FLC, and several proteins required for the deposition of these modifications have been identified (for review, see Schmitz and Amasino, 2007). During the course of cold exposure, FLC expression is suppressed (Michaels and Amasino, 1999; Sheldon et al., 1999), and several signatures of silenced chromatin accumulate at FLC (Bastow et al., 2004; Sung and Amasino, 2004; Mylne et al., 2006; Schubert et al., 2006; Sung et al., 2006b; Finnegan and Dennis, 2007; Schmitz et al., 2008). One such signature is trimethylation on Lys-27 of histone H3 (H3K27me3). Changes in the amount and distribution of H3K27me3 at FLC both during and after cold treatment have been described (Schubert et al., 2006; Sung et al., 2006a; Finnegan and Dennis, 2007). H3K27me3 is carried out by Polycomb-Group (PcG) complexes that contain orthologs of Drosophila melanogaster Polycomb Repressive Complex 2 (PRC2) components and other plant-specific proteins, such as VERNALIZATION INSENSITIVE3 (VIN3; Wood et al., 2006; De Lucia et al., 2008). PcG proteins have also been shown to repress FLC in accessions that do not require vernalization for rapid flowering (Jiang et al., 2008).
The Arabidopsis protein CURLY LEAF (CLF) is an ortholog of the Drosophila PRC2 component Enhancer of Zeste [E(z)], a methyltransferase with specificity for H3K27 (Goodrich et al., 1997; Czermin et al., 2002; Muller et al., 2002). CLF was initially characterized as a suppressor of floral homeotic genes, including AGAMOUS (AG; Goodrich et al., 1997). In addition, recent work has indicated a role for CLF in the suppression of FLC and the floral promoter FT in a nonvernalized, rapid-flowering accession (Jiang et al., 2008). CLF has also been shown to play a role in the repression of FLC by vernalization (Wood et al., 2006).
Here, we describe a gain-of-function allele of CLF that was isolated in a screen for suppressors of delayed flowering in an autonomous pathway mutant background. This allele leads to elevated H3K27me3 at FLC chromatin, FLC mRNA repression, and rapid flowering in backgrounds that would otherwise require vernalization. In addition, we also demonstrate that the presence of FRI has a substantial effect on the degree of H3K27me3 accumulation and CLF occupancy at FLC.
RESULTS
A Mutation in CLF Suppresses FLC-Mediated Delayed Flowering via Reduced FLC Expression
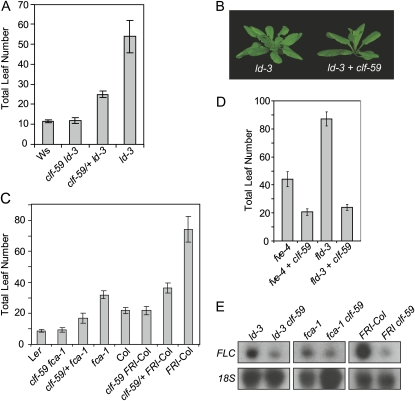
Mutants with lesions in the autonomous pathway gene LD are delayed in flowering due to elevated expression of FLC (Michaels and Amasino, 2001). An ethyl methanesulfonate mutagenesis carried out in an ld-3 mutant background (Ws accession) resulted in the isolation of several mutants with rapid flowering phenotypes. One mutation suppressed delayed flowering in a semidominant manner (Fig. 1A; the mutation is designated clf-59) and mapped to a region that included CLF (Goodrich et al., 1997). Given the dominant nature of this mutation, it was not possible to confirm gene identity by transgenic complementation with the wild-type gene product. Instead, the CLF genomic region was cloned from the mutant and introduced into an ld-3 background. Plants carrying the mutant transgene flowered more rapidly than nontransformed plants (an example of a transgenic versus parental line is shown in Fig. 1B). An introduced copy of wild-type CLF did not alter flowering time (data not shown). Thus, this semidominant mutation is an allele of CLF, which we designate clf-59.
Figure 1.
clf-59 suppresses FLC-mediated late flowering. A, The average total leaf number at flowering, a measure of flowering time in the wild type, ld-3, and ld-3 combined with homozygous and heterozygous clf-59 mutants. B, Images of the ld-3 mutant (left) and ld-3 carrying a transgenic copy of clf-59 (left). C and D, The average total leaf number at flowering of plants containing clf-59 combined with other FLC-mediated late-flowering backgrounds, including fca-1 and FRI-Col (C) and fve-4 and fld-3 (D). Data in D represent the average of at least eight individual T1 plants. E, Northern blot showing the suppression of FLC expression in clf-59 mutants. 18S ribosomal RNA is shown as a loading control. In A to D, all leaf counts include both rosette and cauline leaves and represent the average of at least eight plants. Error bars represent se. In A and C, clf-59/+ indicates a heterozygote. In B and D, a “+” indicates that clf-59 was added transgenically. [See online article for color version of this figure.]
Mutations in autonomous pathway genes lead to delayed flowering via elevated expression of FLC (Michaels and Amasino, 2001). Similarly, plants carrying a functional copy of FRI also display FLC-dependent late flowering (Koornneef et al., 1994; Lee et al., 1994a; Michaels and Amasino, 1999; Sheldon et al., 1999). To evaluate whether the effect on flowering was specific to ld, clf-59 was combined genetically with fca, another autonomous pathway mutant (MacKnight et al., 1997), and a Col line that contains a functional copy of FRI (FRI-Col; Lee et al., 1994a); in both cases, clf-59 suppressed late flowering in a semidominant manner (Fig. 1C). Rapid flowering was also observed when clf-59 was introduced transgenically into the autonomous pathway mutants fld (He et al., 2003) and fve (Ausin et al., 2004; Fig. 1D). Because autonomous pathway mutants and FRI-containing plants are delayed in flowering due to elevated FLC levels (Michaels and Amasino, 2001), rapid flowering in a clf-59 background could result from FLC repression. Indeed, ld-3, fca-1, and FRI-Col all displayed reduced levels of FLC mRNA when combined with clf-59 (Fig. 1E). Thus, rapid flowering in clf-59 results, at least in part, from decreased expression of FLC.
clf-59 Contains a Single Amino Acid Change in a Cys-Rich Motif
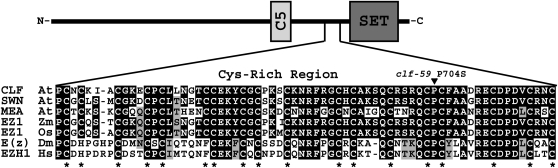
The CLF protein contains several conserved motifs (Fig. 2). The C5 motif functions in protein-protein interactions (Chanvivattana et al., 2004; Ketel et al., 2005), and the SET domain contains the histone methyltransferase catalytic site (Rea et al., 2000). In CLF and other E(z)-like proteins, a conserved Cys-rich region precedes the SET domain. clf-59 contains a single amino acid substitution within this region that converts a conserved Pro to a Ser (P704S; Fig. 2).
Figure 2.
Amino acid alignment of the Cys-rich domain of CLF with other E(z)-like proteins. Sequence alignments were done using ClustalW. An asterisk indicates the location of a conserved Cys. Abbreviations are as follows: At, Arabidopsis; Zm, Zea mays; Os, Oryza sativa; Dm, D. melanogaster; Hs, Homo sapiens.
In addition to CLF, Arabidopsis contains two additional E(z)-like genes, SWINGER (SWN) and MEDEA (MEA; Grossniklaus et al., 1998; Chanvivattana et al., 2004). MEA is essential for endosperm development (Grossniklaus et al., 1998). SWN is partially redundant in function with both CLF and MEA (Chanvivattana et al., 2004; Wang et al., 2006). For example, CLF and SWN act redundantly in the silencing of FLC as a result of vernalization. Both clf and swn single mutants can become vernalized, but down-regulation of both genes simultaneously results in vernalization insensitivity (Wood et al., 2006). Given the pronounced effect of clf-59, it was of interest to determine whether the P-to-S substitution in SWN and MEA would also cause rapid flowering or possibly other developmental aberrations. Accordingly, this substitution was introduced into genomic clones of both SWN and MEA. The mutant genes were subsequently transformed into Col and FRI-Col backgrounds. All transgenic plants flowered synchronously with the nontransformed parents (data not shown). Thus, the P-to-S transition in MEA and SWN does not cause enhanced FLC repression. In addition, no visual phenotypic differences were observed in plants containing modified SWN or MEA at any stage of development. Possible reasons for lack of a phenotype in plants with modified SWN or MEA include, but are not limited to, wild-type CLF masking an effect of these modified transgenes or simply the lack of an effect of this substitution on SWN or MEA activity. As noted below, the only effect of the P-to-S transition in clf-59 that we observe is on flowering and FLC expression.
clf-59 Mutants Do Not Display Phenotypes Seen in Either clf or ag Loss-of-Function Mutants
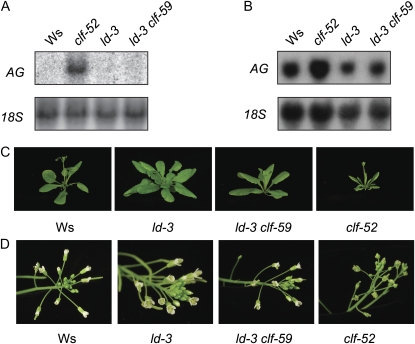
Loss-of-function mutations in clf cause a number of phenotypes, including upward curling of leaves and abnormal flower development (Goodrich et al., 1997). These phenotypes are largely due to ectopic expression of the floral homeotic gene AG (Mizukami and Ma, 1992; Goodrich et al., 1997). On the other hand, plants that do not express AG fail to produce stamens and carpels (Bowman et al., 1989; Yanofsky et al., 1990). Given that CLF is a regulator of AG, it was of interest to look for altered AG expression in a clf-59 background. The presence of clf-59 does not result in hypersuppression AG similar to the effects of this allele on FLC, at either the mRNA or phenotypic level. Specifically, in a clf-59 background, expression of AG in either seedling (Fig. 3A) or inflorescence tissue (Fig. 3B) is indistinguishable from the wild type nor are there any ag reduction-of-function phenotypes in clf-59 mutants (Fig. 3, C and D). In addition, no phenotypes characteristic of clf loss of function (and the associated ectopic expression of AG) are observed in clf-59 mutants. Thus, the clf-59 protein does not appear to affect all targets of wild-type CLF.
Figure 3.
clf-59 mutants do not display phenotypes seen in clf loss-of-function mutants. A and B, Northern blot showing the expression of AG in seedlings (A) and inflorescences (B). 18S ribosomal RNA is shown as a loading control. C and D, Images of rosettes (C) and flowers (D) in Ws, ld-3, ld-3 clf-59, and clf-52. [See online article for color version of this figure.]
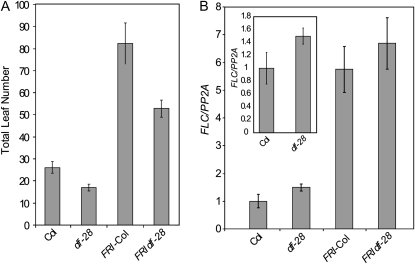
The effect of a clf loss-of-function allele, clf-28, on flowering time and FLC expression was also examined. clf-28 hastened flowering in FRI-Col but to a much lesser degree than clf-59 (Fig. 4A) and, unlike clf-59, did not have a significant effect on FLC expression (Fig. 4B). This distinction indicates that clf-59 is a gain-of-function allele with respect to FLC expression as opposed to a dominant negative.
Figure 4.
Flowering time and FLC expression in clf loss-of-function mutants. A, Average total leaf number of clf-28 in Col and FRI-Col backgrounds. Data represent the average of at least eight plants. Error bars represent se. B, Quantitative reverse transcription PCR showing FLC expression. The inset is shown to highlight the difference between Col and clf-28. All data represent the average of at least three experiments, and error bars represent se.
In the absence of FRI, clf-28 also hastened flowering despite slightly elevated FLC levels (Fig. 4, A and B). Such FLC derepression in a loss-of-function clf mutant has been reported previously (Jiang et al., 2008). Early flowering in clf loss-of-function mutants likely results from derepression of FT (Jiang et al., 2008), a strong promoter of flowering that when overexpressed bypasses the repressive effects of FLC (Michaels et al., 2005).
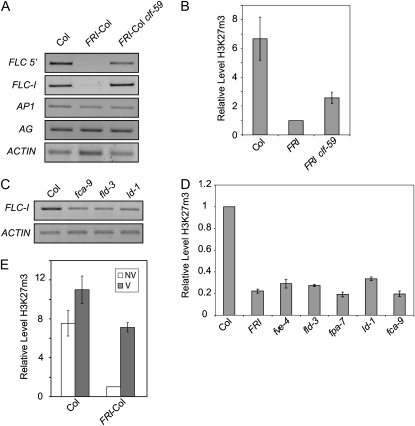
Enrichment of H3K27me3 at FLC Chromatin Is Altered in clf-59 and Is Dependent on Genetic Background
CLF catalyzes the methylation of H3K27. Previous studies have extensively examined the distribution of H3K27me3 across FLC and surrounding chromatin (Bastow et al., 2004; Sung and Amasino, 2004; Schubert et al., 2006; Finnegan and Dennis, 2007). The accumulation H3K27me3 at FLC that occurs as a result of vernalization initiates near the transcriptional start site and then spreads throughout the gene (Finnegan and Dennis, 2007); thus, we have focused our study on regions of FLC near the transcriptional start site. Levels of H3K27me3 were analyzed in nonvernalized FRI clf-59, Col, and FRI-Col using chromatin immunoprecipitation (ChIP). The level of H3K27me3 in FRI clf-59 was greater than that in FRI-Col, indicating that, in a FRI background, clf-59 is capable of elevating H3K27me3 levels in the absence of vernalization (Fig. 5, A and B).
Figure 5.
H3K27me3 accumulation at FLC chromatin in different genetic backgrounds. A and C, Real-time PCR on ChIP samples. FLC 5′ is near the transcriptional start site. FLC-I is in the first intron. APETALA1 (AP1), AG, and Actin were used as controls. B, D, and E, Real-time PCR on ChIP samples showing H3K27me3 levels near the transcriptional start site of FLC. E, NV and V indicate nonvernalized and vernalized samples, respectively. Graphs represent the average of at least three experiments. Error bars represent se. Primer sequences can be found in Supplemental Table S1. Real-time data shown are relative to an AG control.
When assaying H3K27me3 in clf-59 plants, a marked difference in H3K27me3 levels was observed between Col and FRI-Col (Fig. 5, A and B). To our knowledge, this is the first report of the effect of FRI on H3K27 methylation. Reduced H3K27me3 levels in the presence of FRI indicate that, in a genetic sense, FRI is a negative regulator of H3K27me3 at FLC. Moreover, autonomous pathway mutants also display reduced levels of H3K27me3 at FLC, which are comparable to levels seen in FRI-Col (Fig. 5, C and D). Thus, in backgrounds that lack FRI, the autonomous pathway is required for the elevated H3K27me3 levels found at FLC.
Given the elevated levels of H3K27me3 in Col relative to FRI-Col in nonvernalized plants, it was of interest to determine whether vernalization simply elevates H3K27me3 levels to those found in Col or if additional H3K27me3 accumulates in Col upon exposure to prolonged cold. H3K27me3 levels are in fact elevated in Col following cold treatment; however, the fold change is not as great as that seen in a FRI-Col background (Fig. 5E). In both vernalized and nonvernalized plants, FLC had higher levels of H3K27me3 in Col than in FRI-Col. However, it should be noted that the 30-d cold treatment given in these experiments is not saturating with respect to the acceleration of flowering (Lee and Amasino, 1995). Thus, longer cold treatments may equalize the amount of H3K27me3 at FLC in vernalized Col and vernalized FRI-Col.
clf-59 Causes Rapid Flowering in the Absence of Genes Required for Vernalization
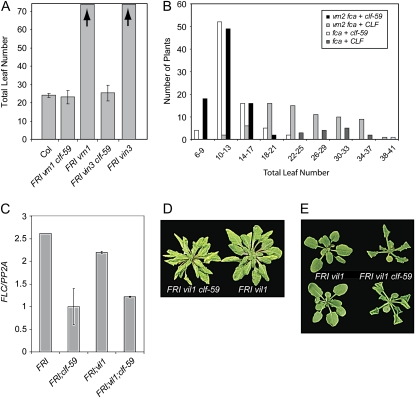
In addition to CLF, several other genes have been shown to play a role in the vernalization-mediated deposition and maintenance of H3K27me3 at FLC. These include the PcG gene VERNALIZATION2 (VRN2; Gendall et al., 2001; Bastow et al., 2004; Sung and Amasino, 2004) as well as VRN1 and the PHD domain proteins VIN3 and VIN3-LIKE1 (VIL1; also known as VRN5; Levy et al., 2002; Bastow et al., 2004; Sung and Amasino, 2004; Sung et al., 2006a; Wood et al., 2006; Greb et al., 2007; De Lucia et al., 2008). clf-59 was introduced into lines carrying mutations in these genes in order to evaluate whether any of these genes are required for the rapid flowering, gain-of-function phenotype in FRI clf-59. However, rapid flowering was observed in all tested lines: FRI vrn1-1, FRI vin3-1, FRI vil1, and fca vrn2-1 (Fig. 6, A and B), indicating that these genes, which are required for vernalization, are not required for the clf-59 phenotype.
Figure 6.
The interaction of clf-59 with vernalization-insensitive mutants. A, Flowering-time data showing the effect of clf-59 on vrn1 and vin3 mutants. Arrows indicate that >70 leaves were produced prior to flowering. B, Flowering time of fca-1 and fca-1 vrn2-1 T1 plants transformed with either wild-type CLF or clf-59. Data represent all T1 plants generated and are shown in part to demonstrate the high transformation efficiency of the clf-59 transgene. Similar efficiency was observed in other transformations such as those represented in Figure 1, B and D. C, Quantitative reverse transcription PCR showing FLC expression. Data represent the average of at least three experiments. Error bars represent se. D and E, Images of pleiotropic phenotypes seen in FRI vil1 clf-59 plants. E, Top and bottom pictures show adaxial and abaxial angles of the same plants, respectively. [See online article for color version of this figure.]
Although the loss of vil1 did not affect the ability of clf-59 to repress FLC and cause early flowering (Fig. 6C), FRI vil1 clf-59 plants did display several pleiotropic phenotypes, including pale green leaves and downward leaf curling (Fig. 6, D and E), the opposite of that seen in clf loss-of-function mutants. Thus, VIL1 appears to be required for wild-type leaf morphology in the presence of clf-59.
Genetic Background Affects Wild-Type CLF Localization to FLC
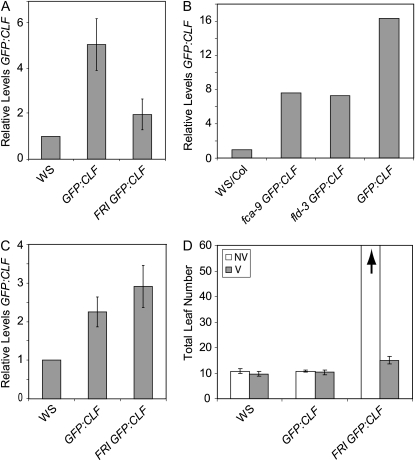
As discussed above, FRI-Col and autonomous pathway mutants have relatively low levels of H3K27me3 at FLC chromatin when compared to Col (Fig. 5). In addition, it has been shown that clf loss-of-function mutants have reduced H3K27me3 levels at FLC chromatin in a Col background (Jiang et al., 2008). Because the presence of FRI or a clf mutation has a similar effect on H3K27me3 levels at FLC, the effect of FRI on CLF localization was investigated. A constitutively expressed, GFP-tagged version of CLF (GFP:CLF; Schubert et al., 2006) was introduced by crossing into a line containing FRI (FRI-Ws: all lines in the FRI/CLF localization experiments are in a Ws background). ChIP analysis using an anti-GFP antibody revealed that levels of CLF were enriched at FLC chromatin in Ws when compared to FRI-Ws (Fig. 7A). Such a reduction in CLF occupancy likely contributes to the lower levels of H3K27me3 at FLC seen in FRI-containing backgrounds (Fig. 5B).
Figure 7.
CLF localization at FLC. A to C, Real-time PCR on ChIP samples showing the enrichment of CLF near the transcriptional start site of FLC under nonvernalized (A and B) and vernalized (C) conditions. Real-time data shown is relative to an AG control. Graphs in A and C represent the average of three experiments. Data in B represent a single experiment. Error bars represent se. Samples were standardized to a Ws line that does not contain GFP. D, Flowering time of lines used in A and C both with (V) and without (NV) vernalization. The arrow indicates that >60 leaves were produced prior to flowering. Leaf counts represent the average of at least 10 plants. Error bars represent se.
GFP:CLF was also introduced into fca-9 and fld-3 backgrounds. In both cases, the autonomous pathway mutant displayed lower levels of CLF occupancy at FLC chromatin relative to the wild type (Fig. 7B). Thus, as with FRI, the fca and fld genetic backgrounds create a condition that leads to reduced occupancy of CLF protein at FLC chromatin.
We also examined the effect of prolonged cold on the enrichment of CLF at FLC. After a 35-d cold exposure, a difference in CLF occupancy at FLC could no longer be detected between plants that contain and those that lack FRI (Fig. 7C). This 35-d cold exposure was sufficient to substantially hasten flowering time in FRI-Ws (Fig. 7D). Thus, with vernalization, the presence of FRI no longer affects the amount of CLF that occupies FLC chromatin, and increased CLF occupancy at FLC after prolonged cold correlates with the vernalization-mediated increase in H3K27me3 at FLC.
DISCUSSION
We have identified a gain-of-function allele of CLF, clf-59, that reduces the level of FLC expression and thus eliminates the requirement for vernalization in winter-annual types of Arabidopsis. CLF is a PcG protein that functions in a complex, PRC2, which has conserved features in plants and animals. Plant genomes typically contain multiple copies of conserved PRC2 components [for example, Arabidopsis has three E(z) homologs: CLF, SWN, and MEA], and these components interact with members of the plant-specific, VIN3-like family of proteins (Wood et al., 2006; De Lucia et al., 2008). Thus, there is potential for much variability in the specific PcG complexes that assemble in plants. The clf-59 protein may either enhance the activity of a particular PcG complex or allow for the formation of a novel PcG complex that possesses high affinity for FLC chromatin.
clf-59 harbors a Pro-to-Ser amino acid transition in a Cys-rich region. Although the specific biochemical role of this region is not known, it contains two contiguous domains with a unique spacing of Cys residues called CXC domains as first described in ENX-1, the human homolog of E(z) (Hobert et al., 1996). Subsequent work has shown this domain to be important for the function of E(z) and E(z)-like proteins (Carrington and Jones, 1996; Kuzmichev et al., 2002; Ketel et al., 2005). Several otherwise unrelated proteins from plants and animals have been described that contain CXC domains separated by sequences of varying length, and these domains have been shown to bind zinc in vitro (Andersen et al., 2007). TSO1 is a CXC-containing protein in Arabidopsis that functions in the regulation of cell division, and both the tso1-1 and tso1-2 loss-of-function alleles contain amino acid substitutions within the CXC domain (Hauser et al., 2000; Song et al., 2000).
Both clf-59 and clf loss-of-function alleles hasten the onset of flowering; however, the two types of alleles do so via different mechanisms. clf-59 causes hyperrepression of FLC expression with no noticeable effect on AG. In contrast, clf loss-of function mutants cause derepression of several genes, including AG, FLC, and FT (Goodrich et al., 1997; Jiang et al., 2008), and it is likely that FT derepression leads to rapid flowering in clf loss-of-function mutants. This occurs despite elevated FLC expression, as ectopic FT expression can bypass the repressive effects of FLC (Michaels et al., 2005).
It is interesting to compare the degree of FLC derepression that we observe in a clf loss-of-function mutant to that reported previously (Jiang et al., 2008). In a Col background, Jiang et al. (2008) report a higher level of FLC derepression than we observe. In a delayed-flowering background, the difference is even greater: Jiang et al. (2008) report extensive derepression in an fca clf double mutant, whereas we do not observe derepression in FRI clf. Therefore, with respect to FLC derepression, clf loss-of-function mutants may have different effects in FRI compared to autonomous pathway mutant backgrounds.
The wild-type function of the autonomous pathway is to repress FLC expression (Michaels and Amasino, 2001). We find reduced levels of both H3K27me3 and CLF at FLC chromatin in several autonomous pathway mutants. Thus, one aspect of autonomous pathway-mediated FLC repression involves a direct or indirect effect on the occupancy of CLF at FLC chromatin. However, given the large degree of FLC derepression previously reported in fca clf mutants (Jiang et al., 2008), it is possible that a sufficient amount of CLF still occupies FLC chromatin in autonomous pathway mutant backgrounds as this would allow for some degree of derepression when CLF is genetically removed.
In this work, we present data that are consistent with FRI playing an antagonistic role with respect to PcG occupancy at FLC. We observe that H3K27me3 levels, a mark deposited by PcG protein complexes, were less abundant at FLC chromatin in a FRI-Col background relative to Col. We provide a possible explanation for the difference in H3K27me3 levels in the presence or absence of FRI by demonstrating that CLF, a PcG protein that catalyzes H3K27me3 deposition, is less abundant at FLC in FRI-Col relative to Col plants. Because CLF occupancy at FLC is reduced in a FRI background, one might expect a clf loss-of-function mutant to have less of an effect on FLC expression in a FRI-Col background; indeed, this is consistent with the lack of FLC derepression we observe in FRI clf plants. A molecular signature present in vernalized Arabidopsis plants is the accumulation of H3K27me3 at FLC (Schubert et al., 2006; Sung et al., 2006a; Finnegan and Dennis, 2007). Consistent with our results, the antagonism between FRI and CLF is relieved upon vernalization; CLF occupancy and H3K27me3 increases at FLC following exposure to prolonged cold. Genes required for vernalization, such as VIN3, may play a role in overcoming the antagonistic effects of FRI by helping to recruit CLF to FLC chromatin.
Chromatin modification has been shown in many systems to reinforce and/or maintain states of gene expression. Much of the data presented here and in other articles on FLC chromatin and flowering show correlations between the transcriptional level of FLC and particular chromatin modifications. We show, for example, that levels of H3K27me3, a repressive chromatin mark, and occupancy of CLF, a PcG protein, are decreased at FLC in two genetic situations in which FLC expression is elevated: the presence of FRI and the lack of autonomous pathway genes. The differential accumulation of marks such as H3K27me3 at FLC may very well be an indirect consequence of the effect that FRI and autonomous pathway genes have on FLC transcription. Indeed, just as we show that FRI leads to a reduction in CLF occupancy at FLC, a recent report has shown that FRI also correlates with the accumulation of a COMPASS complex component at FLC that modifies chromatin to an active state (Jiang et al., 2009). The specific biochemical role of FRI and the autonomous pathway components in FLC regulation remain to be determined.
The unique gain-of-function clf-59 allele results in an increase in the abundance of H3K27me3 at FLC in the presence of FRI. Thus, the clf-59 protein might be immune to the ability of FRI or loss of autonomous pathway components to reduce CLF occupancy at FLC chromatin in the absence of vernalization; thus, clf-59 may be able to accumulate to higher levels than wild-type CLF at FLC in these genetic backgrounds. Alternatively, clf-59 may produce a hyperactive protein that can elevate H3K27me3 levels without increased occupancy at FLC. It is intriguing that clf-59 can suppress FLC expression in nonvernalized plants to a level comparable to that found in the wild type after vernalization in the absence of proteins known to mediate the vernalization-induced silencing of FLC, including VRN2, VIN3, and VIL1. Future studies addressing the unique biochemical properties of the clf-59 protein may further our knowledge of FLC regulation and PcG targeting in general by providing a better understanding of the molecular events that either recruit or exclude PcG genes to FLC chromatin.
MATERIALS AND METHODS
Plant Growth Conditions and Mutant Stocks
Plants were grown under cool-white fluorescent lights at 22°C in long-day photoperiods (16 h light: 8 h dark). Cold treatments were carried out at 4°C under cool-white fluorescent lights. The clf-59 mapping population was created by crossing ld-1 to ld-3 clf-59. The ld-1 and ld-3 mutants are from the Col and Ws-2 backgrounds, respectively (Lee et al., 1994b; Michaels and Amasino, 2001). F2 plants segregated 1:2:1 for the fully suppressed, partially suppressed, and the parental delayed flowering phenotypes, respectively. The latest flowering F2 plants were used to map the locus, as these plants were known to be homozygous Col at the locus of interest. A PCR-based derived cleaved amplified polymorphic sequence marker (Michaels and Amasino, 1998) was designed to detect the clf-59 mutation in both genetic studies and in transgenic plants. The marker was designed such that the wild-type CLF sequence contained an NcoI restriction site at the lesion that was absent in the clf-59. Primer sequences are listed in Supplemental Table S1. Other lines used in this work have been previously described. The fca-1 allele is from the Landsberg erecta but contains a Col allele of FLC (Sanda and Amasino, 1996). Alleles of other autonomous pathway mutants used in this work are in the Col background: fpa-7 (Michaels and Amasino, 2001), fca-9 (Bezerra et al., 2004), ld-1 (Redei, 1962), fld-3 (He et al., 2003), and fve-4 (Michaels and Amasino, 2001). clf-28 is from the Salk T-DNA collection (SALK_139371), and clf-52 is a T-DNA allele from the Ws-2 background (Noh and Amasino, 2003). The Ws-2 line with an introgressed FRI and the CLF:GFP line have been described previously (Noh and Amasino, 2003; Schubert et al., 2006). CLF:GFP is under the control of the cauliflower mosaic virus 35S promoter as described. CLF:GFP segregates as a single locus, and all lines containing this construct carried homozygous null mutations in the endogenous CLF gene. The CLF genomic region was amplified out of both ld-3 and ld-3 clf-59 using PCR. DNA fragments were then cloned into the binary vector pPZP221B using BamHI sites engineered into the primers. These primers and primers used to clone and make site-directed mutants in MEA and SWN are listed in Supplemental Table S1.
Analysis of RNA Abundance
Tissue was harvested from 7-d-old seedlings. RNA was isolated using TRI reagent (Sigma-Aldrich). For northern analysis, 12 μg of total RNA was run on a 1% agarose denaturing formaldehyde gel and transferred to a nylon filter (Hybond). A DNA probe was used that was complementary to the 3′ untranslated region of FLC. Real-time PCR was performed with the 7000 Real-Time PCR System (Applied Biosystems) using the DyNAmo Flash SYBR Green qPCR Kit (Finnzymes). The PCR parameters were as follows: one cycle of 15 min at 95°C, 40 cycles of 15 s at 95°C, 20 s at 58°C, and 45 s at 72°C. The constitutively expressed gene encoding the A3 subunit of protein phosphatase 2A was used as a control (Czechowski et al., 2005). PCR primer sequences are listed in Supplemental Table S1. All results presented are an average of at least three biological replicates.
ChIP
Tissue was harvested from 7-d-old seedlings. Chromatin samples were prepared as described (Gendrel et al., 2005). Antibodies used in this work were as follows: anti-GFP (Invitrogen catalog no. A-11122) and anti-H3K27me3 (Upstate catalog no. 07-449). Real-time PCR was done as described above. Changes at FLC chromatin are shown relative to an AG control. Because AG has abundant H3K27me3, it is possible that this mark at AG may behave similarly to that at FLC. However, observed patterns of H3K27me3 and CLF localization at FLC were also evident relative to an Actin control. Actin does not contain high levels of H3K27me3 (data not shown). PCR primer sequences are listed in Supplemental Table S1. All ChIP results presented are an average of at least two biological replicates unless noted otherwise.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CLF, At2g23380; FLC, At5g10140; FRI, At4g00650; LD, At4g02560; FCA, At4g16280; FLD, At3g10390; FPA, At2g43410; FVE, At2g19520; AG, At4g18960; VIL1, At3g24440; VRN2, At4g16845; VIN3, At5g57380; VRN1, At3g18990; MEA, At1g02580; and SWN, At4g02020.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Sequences of oligos used in cloning and gene expression analysis.
Supplementary Material
Acknowledgments
We thank Dr. Scott Michaels for identifying the clf-59 mutant and for helpful discussions. We also thank Drs. Justin Goodrich and Daniel Schubert for supplying constructs and seed used in this work and the Salk Institute Genomic Analysis Laboratory for providing the Arabidopsis T-DNA insertion lines.
This work was supported by the University of Wisconsin, the National Institutes of Health (grant no. 1R01GM079525), and the National Science Foundation (grant no. 0446440).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard M. Amasino (amasino@biochem.wisc.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andersen SU, Algreen-Petersen RG, Hoedl M, Jurkiewicz A, Cvitanich C, Braunschweig U, Schauser L, Oh SA, Twell D, Jensen EO (2007) The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J Exp Bot 58 3657–3670 [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36 162–166 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 [DOI] [PubMed] [Google Scholar]
- Baurle I, Smith L, Baulcombe DC, Dean C (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318: 109–112 [DOI] [PubMed]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM (2004) Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40 112–119 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington EA, Jones RS (1996) The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development 122 4073–4083 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 5263–5276 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185–196 [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed]
- Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10 520–527 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES (2007) Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17 1978–1983 [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107 525–535 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 213–218 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–51 [DOI] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17 73–78 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450 [DOI] [PubMed] [Google Scholar]
- Hauser BA, He JQ, Park SO, Gasser CS (2000) TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 127 2219–2226 [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed]
- Hobert O, Jallal B, Ullrich A (1996) Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol 16 3066–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed]
- Jiang D, Wang Y, Wang Y, He Y (2008) Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3 e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in arabidopsis flowering time. Science 290 344–347 [DOI] [PubMed] [Google Scholar]
- Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA (2005) Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol 25 6857–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6 911–919 [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM (1995) Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol 108 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994. b) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM (1994. a) The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6 903–909 [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297 243–246 [DOI] [PubMed]
- MacKnight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745 [DOI] [PubMed] [Google Scholar]
- Michaels S, Amasino R (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous-pathway mutations, but not responsiveness to vernalization. Plant Cell 13 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino R (1999) Flowering Locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1998) A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J 14 381–385 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71 119–131 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 197–208 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K (1979) On the genetical basis of vernalization requirement in Arabidopsis thaliana (L.) Heynh. In P Champagnat, R Jaques, eds, La Physiologie de la Floraison. Coll. Int. CNRS, Paris, pp 217–220
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 593–599 [DOI] [PubMed] [Google Scholar]
- Redei GP (1962) Supervital mutants in Arabidopsis. Genetics 47 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda SL, Amasino RM (1996) Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol Gen Genet 251 69–74 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Amasino RM (2007) Vernalization: a model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim Biophys Acta 1769 269–275 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Sung S, Amasino RM (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA 105 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Leung T, Ehler LK, Wang C, Liu Z (2000) Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development 127 2207–2217 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2005) Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56 491–508 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006. b) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38 706–710 [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM (2006. a) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tyson MD, Jackson SS, Yadegari R (2006) Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA 103 13244–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.