Abstract
Members of the conserved RecQ helicase family are important for the preservation of genomic stability. Multiple RecQ homologs within one organism raise the question of functional specialization. Whereas five different homologs are present in humans, the model plant Arabidopsis (Arabidopsis thaliana) carries seven RecQ homologs in its genome. We performed biochemical analysis of AtRECQ3, expanded upon a previous analysis of AtRECQ2, and compared their properties. Both proteins differ in their domain composition. Our analysis demonstrates that they are 3′ to 5′ helicases with similar activities on partial duplex DNA. However, they promote different outcomes with synthetic DNA structures that mimic Holliday junctions or a replication fork. AtRECQ2 catalyzes Holliday junction branch migration and replication fork regression, while AtRECQ3 cannot act on intact Holliday junctions. The observed reaction of AtRECQ3 on the replication fork is in line with unwinding the lagging strand. On nicked Holliday junctions, which have not been intensively studied with RecQ helicases before, AtRECQ3, but not AtRECQ2, shows a clear preference for one unwinding mechanism. In addition, AtRECQ3 is much more efficient at catalyzing DNA strand annealing. Thus, AtRECQ2 and AtRECQ3 are likely to perform different tasks in the cell, and AtRECQ3 differs in its biochemical properties from all other eukaryotic RECQ helicases characterized so far.
RecQ helicases are important players in the maintenance of genomic stability in prokaryotes and eukaryotes (for reviews covering several aspects of RecQ helicases, see Bachrati and Hickson, 2003; Hickson, 2003; Opresko et al., 2004; Sharma et al., 2006; Brosh and Bohr, 2007; Hanada and Hickson, 2007; Bachrati and Hickson, 2008). Interestingly, organisms contain different numbers of RecQ homologs. In humans, mutations in three of the five RecQ helicases have been linked to distinct, cancer-associated genetic diseases. Consequently, the human RecQ helicases have been intensively studied. All RecQ helicases share a characteristic helicase domain, due to which they are classified as RecQ helicases. In addition, a RecQ C-terminal (RQCt) and/or a Helicase and RNAseD C-terminal (HRDC) domain can be identified in some RecQ helicases. The possible functions of the latter in protein stability and DNA and protein binding are summarized, for example, in a recent review (Chu and Hickson, 2009). From the enzymatic point of view, most RecQ helicases both unwind DNA and promote the opposite reaction, called strand annealing. In addition, some RecQ homologs combine these activities to catalyze branch migration.
One of the central questions regarding RecQ helicases is the extent to which the homologs have unique or overlapping functions (Ellis et al., 2008). This question originally raised for the human RecQ helicases is of equal importance for other organisms, such as plants. Analysis of plant RecQ homologs could reveal species- or kingdom-specific tasks, and such comparisons could shed light on evolutionary questions, such as how selection pressures induce multiple homologs within a single species.
In the dicotyledonous model plant Arabidopsis (Arabidopsis thaliana), seven RecQ helicase homologs have been identified (Hartung et al., 2000; Hartung and Puchta, 2006). A biochemical comparative analysis may provide information about the molecular basis of their cellular functions, which are likely to be different. As has been shown, sequence homology does not necessarily imply functional homology (Hartung et al., 2007).
Previously, we were able to characterize basic biochemical properties of the helicase RECQ2 of the model plant Arabidopsis (Kobbe et al., 2008). In this study, we chose to study AtRECQ3 as a RecQ helicase that differs in the domain structure from AtRECQ2, lacking the HRDC domain and the winged helix subdomain of the RQCt domain (Fig. 1A). AtRECQ3 has the same domain structure as human RECQ5β. AtRECQ3 was expressed in Escherichia coli, purified, and biochemically characterized. The specificity of AtRECQ2 and AtRECQ3 was compared with several substrates, extending the analyzed spectrum of substrates for AtRECQ2 and for RecQ helicases in general. We identified several differences in the enzymatic properties of AtRECQ2 and AtRECQ3, suggesting that these two enzymes might play different roles in vivo.
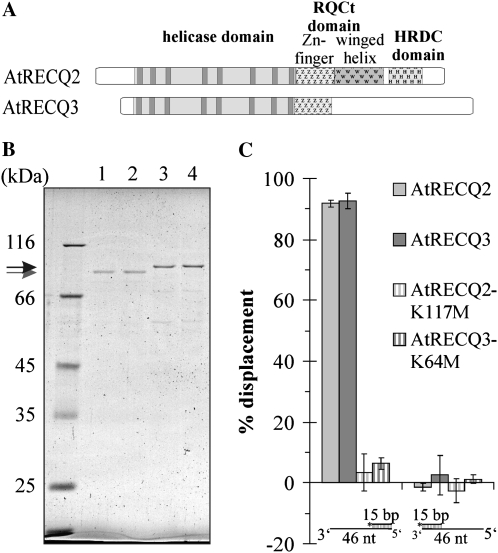
Figure 1.
Domain structure, purification, and directionality of AtRECQ2 and AtRECQ3. A, Schematic drawing of the domain structure of AtRECQ2 and AtRECQ3. In the helicase domain, the different helicase motifs (0, I, Ia, II, III, IV, V, and VI) are depicted in dark gray. B, 10% SDS-PAGE analysis stained with colloidal Coomassie Brilliant Blue of representative purifications of AtRECQ2-K117M (lane 1, 0.5 pmol), AtRECQ2 (lane 2, 0.8 pmol), AtRECQ3-K64M (lane 3, 0.9 pmol), and AtRECQ3 (lane 4, 1.25 pmol). The proteins were overexpressed in E. coli and purified by nickel-immobilized metal ion affinity chromatography and Calmodulin affinity chromatography. The gray arrow indicates AtRECQ2/-K117M with a predicted molecular mass of 85.5 kD, and the black arrow indicates AtRECQ3/-K64M with a predicted molecular mass of 86 kD. C, AtRECQ2 (8 nm), AtRECQ3 (8 nm), AtRECQ3-K64M (8 nm), and AtRECQ2-K117M (5 nm) were incubated with 150 pm of the indicated substrates for 30 min at 37°C. Error bars indicate sd of the mean of three independent experiments.
RESULTS
Purification of the RECQ Helicases
While the Arabidopsis RECQ2 protein contains the conserved helicase, RQCt, and HRDC domains, AtRECQ3 only contains the helicase domain and the zinc finger of the RQCt domain (Fig. 1A). The purification procedure for AtRECQ2 and AtRECQ2-K117M was described previously (Kobbe et al., 2008, 2009). The invariant Lys in the Walker A motif for AtRECQ3 was determined to be Lys-64 (counted without tags). As an amino acid substitution at this position to Met was shown to inactivate the ATPase and therefore the helicase activity (Brosh et al., 1999; Kobbe et al., 2008), we used AtRECQ3-K64M preparations as a negative control. Like the AtRECQ2 proteins, AtRECQ3 and AtRECQ3-K64M (total calculated mass with affinity tags 86.0 kD) were overexpressed in E. coli and purified by consecutive nickel-immobilized metal ion affinity chromatography and calmodulin affinity chromatography (Fig. 1B). Using antibodies directed against the N-terminal FLAG-tag and the C-terminal His-tag, it could be demonstrated that all characterized proteins were full length (data not shown).
Directionality of AtRECQ3
Using partial duplex DNA structures with 15 bp and either a 31-nucleotide 3′ or 5′ overhang, we were able to demonstrate that AtRECQ3 can unwind DNA. More precisely, we could demonstrate that AtRECQ3, like AtRECQ2, is a 3′ to 5′ DNA helicase, as only the substrate providing a 3′ overhang was unwound (Fig. 1C). AtRECQ3-K64M did not significantly unwind the DNA structures. This means that the AtRECQ3 preparation is unlikely to contain contaminant proteins from E. coli and is thus suitable for further biochemical characterization.
Characteristics of the Unwinding of Partial Duplex DNA
The helicases were tested for their ability to unwind partial duplex DNA structures with increasing duplex lengths. In line with previous results for AtRECQ2, AtRECQ3 is also able to disrupt at least 23 bp. This held true at different concentrations of magnesium ion (data not shown).
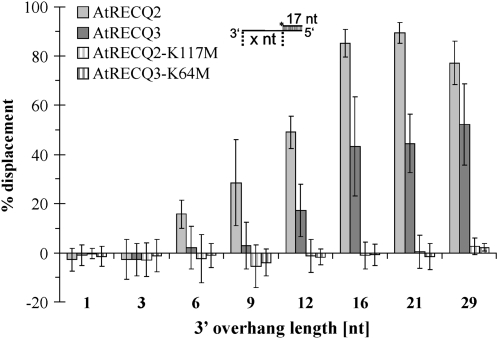
Using a duplex region of 17 bp, the length of the 3′ overhang was varied to characterize the requirements of the enzymes. While AtRECQ2 could catalyze >10% of unwinding with a 6-nucleotide 3′ overhang, AtRECQ3 needed a longer overhang. Under the conditions tested, a 12-nucleotide 3′ overhang was needed for unwinding of >10% of the substrate (Fig. 2).
Figure 2.
Requirements for the unwinding of partial duplex DNA. A total of 8 nm of each respective enzyme was incubated with 150 pm of the indicated substrates for 30 min at 37°C. Error bars indicate sd of the mean of five to eight experiments. nt, Nucleotide.
AtRECQ2 and AtRECQ3 Differ in Their Requirements for (Deoxy)nucleoside Triphosphates for Strand Unwinding
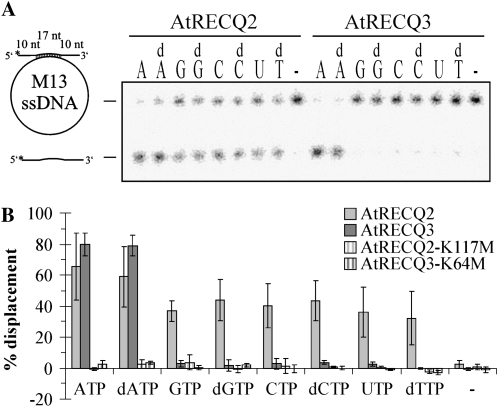
Using an M13-based substrate, the enzymes were compared in their ability to use different (deoxy)nucleoside triphosphates to catalyze unwinding. Both enzymes failed to work in the absence of a nucleotide cofactor, and both catalyzed strand unwinding with ATP and dATP. However, in contrast to AtRECQ2, AtRECQ3 could not use nucleotide cofactors other than ATP and dATP to promote unwinding (Fig. 3).
Figure 3.
Requirements for (deoxy)nucleoside triphosphates for strand unwinding. A, An amount of 5 nm AtRECQ2, AtRECQ2-K117M, AtRECQ3, and AtRECQ3-K64M were incubated with 150 pm of the indicated substrate for 20 min at 37°C. ATP (A) was substituted by dATP (dA), GTP (G), dGTP (dG), CTP (C), dCTP (dC), UTP (U), dTTP (dT), and water (−). Helicase reaction products were resolved by native PAGE. B, Quantification of the results of four experiments. Error bars indicate sd. nt, Nucleotide.
AtRECQ2 and AtRECQ3 Differ in Their Processing of Holliday Junctions
Different synthetic Holliday junctions (HJs; Supplemental Fig. S1) were prepared and incubated with both enzymes. As previously reported, AtRECQ2 was able to branch migrate the X12-HJ, a HJ composed of a homologous core of 12 bp flanked by arms of 19 bp (Kobbe et al., 2008). Comparative analysis of AtRECQ3 and AtRECQ3-K64M with this X12 junction did not reveal significant branch migration or unwinding (data not shown).
Both enzymes were incubated with the static X0-HJ, but no significant conversion was observed. Different enzyme concentrations, different incubation time points, and a wide range of magnesium ion concentrations did not lead to significant conversion of this substrate (data not shown).
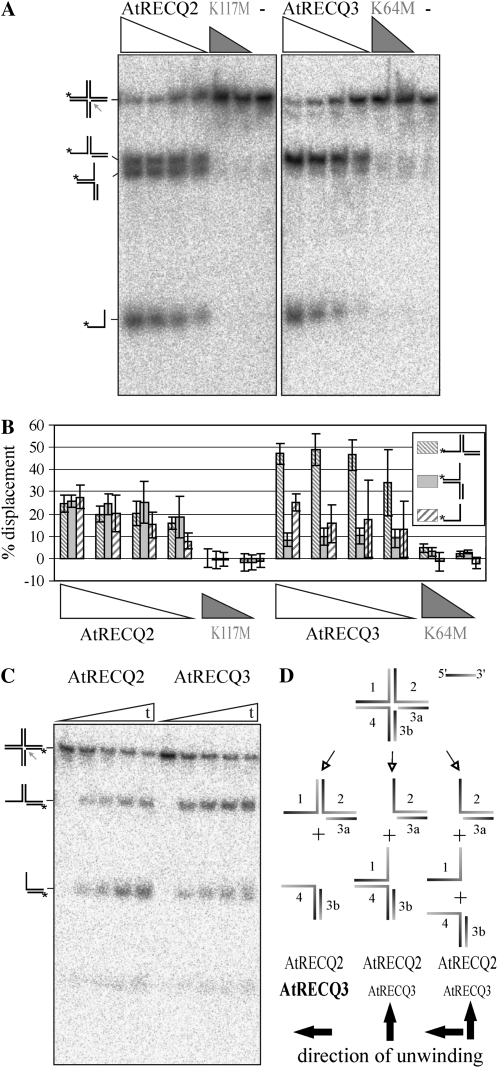
However, when the nicked X0-HJ (nX0), as schematically drawn in Figure 4C, was substituted for the intact X0-HJ, conversion was observed. Interestingly, both enzymes could process this substrate, but they acted on it differently. nX0 junctions were prepared to include labels on the different component oligonucleotides. The reaction of these substrates was analyzed as a function of enzyme concentration (Supplemental Fig. S2), time, and magnesium ion concentration. An enzyme concentration dependency was observed, as exemplified in Figure 4A and quantified in Figure 4B. Figure 4C illustrates a kinetic effect observed with a differently labeled substrate. A synopsis of the results is schematized in Figure 4D. Quantification of each product allowed unwinding outcomes to be grouped into three distinct classes. The observed products conform to a mechanism in which the nick guides the helicases in the center of the HJ, where they subsequently translocate on a sterically favorable strand in a 3′ to 5′ direction. Two of the three classes can be explained by translocation of the helicase on either strand 1 or 2, respectively, as diagrammed in Figure 4D. The third class is explained by helicase translocation on strands 1 and 2. In principle, these two unwinding events could take place simultaneously or sequentially. However, over a time course of 30 min, no significant increase or decrease of one specific intermediate product was observed. This seems to indicate that there is not a specific, favored reaction that terminates prior to a second unwinding step. Interestingly, no significant amount of strand 3b is visible, although the structure composed of strand 4 and 3b is a 3′ overhang partial duplex DNA structure with 23 bp and a 24-nucleotide overhang.
Figure 4.
Processing of the nX0-HJ. A, Decreasing concentrations of AtRECQ2 and AtRECQ3 (8, 5, 4, and 2.5 nm), AtRECQ2-K117M (5 and 4 nm), and AtRECQ3-K64M (8 and 5 nm) were incubated with 150 pm of the nX0-HJ labeled on strand 1 for 30 min in the presence of 1.35 mm MgCl2. Helicase reaction products were analyzed on 12% native TBE polyacrylamide gels. On the left, the substrate and products are symbolized. The asterisk indicates the 32P-label; the arrow highlights the position of the nick; − indicates no enzyme. B, Quantification of the data shown in A. The mean and sd of the results of four experiments are shown. C, AtRECQ2 (8 nm) and AtRECQ3 (8 nm) were incubated with the nX0-HJ (150 pm) labeled on strand 3a in the presence of 1.35 mm MgCl2. After 0, 2, 5, 15, and 30 min, aliquots of the reaction were removed and analyzed. D, Synopsis of all results obtained with the nX0 junction. Three different classes of outcomes from the unwinding reactions and their proposed origins are schematized. A preference of AtRECQ3 or AtRECQ2 for one outcome or the other is symbolized by the size of the lettering.
A clear difference can be observed in the unwinding reaction preferences exhibited by AtRECQ2 and AtRECQ3. AtRECQ3 preferentially produces products compatible with a 3′ to 5′ translocation on strand 1. For AtRECQ2, no such preference can be detected.
These trends can be seen for different time points and different enzyme and magnesium ion concentrations
AtRECQ2 and AtRECQ3 Differ in Their Processing of Replication Forks
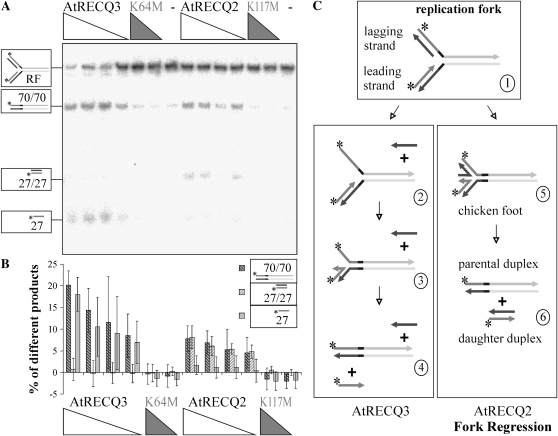
Some standard models of replication forks are composed of three different regions of homology: one double-stranded region corresponding to the parental DNA and two different parental-daughter complexes. In nature, the parental DNA strands are fully complementary and, therefore, also the daughter strands. A replication fork of this kind allows fork regression to occur, in which the daughter strands pair, and a so-called chicken foot (a HJ; see Fig. 5C, structure 5) is formed. If an oligonucleotide-based model replication fork has these characteristics, branch migration after formation of a chicken foot will lead to the appearance of daughter and parental duplexes (Fig. 5C). The model replication fork used in this study is of this type, except for 5 nucleotides of heterology at the junction to prevent spontaneous reannealing of the parental DNA arms (depicted in black in Fig. 5).
Figure 5.
Conversion of a synthetic replication fork (RF). A, Decreasing concentrations of AtRECQ2 and AtRECQ3 (8, 5, 4, and 2.5 nm) and AtRECQ2-K117M and AtRECQ3-K64M (8 and 5 nm) were incubated with 100 pm of the replication fork for 20 min. Helicase reaction products were analyzed on 12% native TBE polyacrylamide gels. On the left, the substrate and products are symbolized. Asterisks mark the 32P-labels. B, Quantification of the main products. The mean and sd of the results of seven experiments are shown. C, Proposed mechanisms of replication fork conversion by AtRECQ2 and AtRECQ3. In the scheme of the replication fork, identical sequences share a common color; complementarities can be deduced from the figure. On the left, the proposed mechanism for AtRECQ3 is shown. Unwinding of the lagging daughter strand enhances the chance of pairing the parental strands in the context of DNA breathing. This will lead to the displacement of the leading daughter strand. On the right, fork regression with the chicken foot intermediate is shown. Further branch migration leads to parental and daughter duplexes.
The main reaction products of AtRECQ2 are daughter and parental duplexes (Fig. 5, A and B). This indicates that AtRECQ2 probably combines strand unwinding and strand annealing to catalyze fork regression.
AtRECQ3 produces different products than AtRECQ2. The main products that can be detected are the parental duplex and leading daughter strands. This means that the unlabeled lagging daughter strand is also a product. It can be explained as follows: unwinding of the lagging daughter strand in 3′ to 5′ direction enhances the chance of pairing the parental strands in the context of DNA breathing. If this pairing takes place, the leading daughter strand will be displaced, too. Reactions with this replication fork were performed with different enzyme concentrations (Fig. 5, A and B) as well as at different magnesium ion concentrations (0.09–14.2 mm free Mg2+). The fact that AtRECQ2 produces mainly daughter duplexes characteristic of fork regression while the main products of AtRECQ3 are daughter leading and lagging strands was confirmed under all conditions tested.
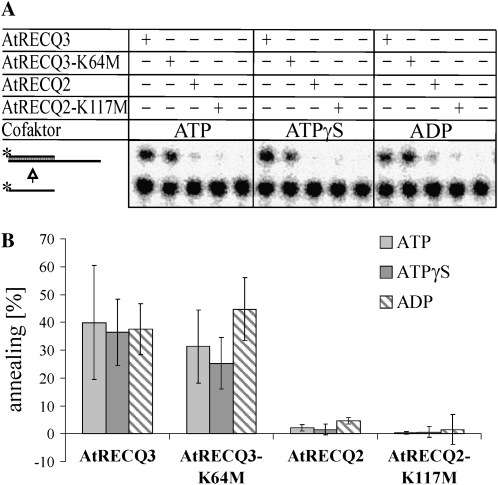
Enhanced DNA Strand Annealing Activity of AtRECQ3
As fork regression is thought to require strand unwinding and strand annealing, we analyzed strand annealing with 40-bp partial duplexes. Interestingly, AtRECQ3 revealed a very strong strand annealing activity under the conditions tested, whereas the activity was hardly detectable for AtRECQ2 (Fig. 6). ATPase activity is not required for the reaction, since AtRECQ3-K64M showed the same annealing propensity as was observed for AtRECQ3. At least for AtRECQ3, the presence of either ATPγS or ADP did not strongly change the effect observed with ATP.
Figure 6.
DNA strand annealing. A, A total of 8 nm of each enzyme was incubated with 40- and 80-nucleotide-long single-stranded oligonucleotides at 37°C for 20 min. Annealing of these two oligonucleotides leads to the appearance of partial duplex DNA with a 5′ overhang as diagrammed. The reaction was performed in the presence of ATP, ATPγS, or ADP. The reaction products were analyzed by native PAGE. Asterisk marks the 32P-label. B, Quantification of the results of seven experiments. The mean and sd are given.
DISCUSSION
Some members of the RecQ helicase family have been shown to be of great importance for the maintenance of genomic stability. Plant genomes appear to be particularly rich in such helicases, as demonstrated by the seven RecQ homologs in the model plant Arabidopsis (Hartung and Puchta, 2006). This diversity naturally raises questions of functional specialization and molecular evolution.
With previous biochemical characterizations of AtRECQ2 and the AtWRNexo protein (Plchova et al., 2003), we were able to substantiate the hypothesis of a functional homology between AtRECQ2 and the RecQ helicase part of HsWRN (Kobbe et al., 2008). In this work, we expanded the analysis of AtRECQ2 and compared its biochemical properties to those of the previously uncharacterized homolog AtRECQ3. This allows on one hand the detection of possibly overlapping activities of those two enzymes and of their biochemical differences. On the other hand, it allows the comparison of the properties of AtRECQ3 with RecQ helicases from other organisms. Under standard conditions, the specific activity of AtRECQ2 and AtRECQ3 appears to be similar, as demonstrated by the M13-based substrate (Fig. 3). Therefore, it was possible to compare the enzymatic efficiency of those two proteins with more sophisticated substrates.
Like all RecQ helicases characterized so far, AtRECQ3 and AtRECQ2 are 3′ to 5′ DNA helicases. AtRECQ3 also exhibits another previously described behavior of AtRECQ2, in that increasing the length of the duplex region leads to a decreased percentage of unwinding. This is consistent with the limited efficiency of other RecQ helicases to displace one strand of a long stretch of duplex DNA in the absence of the cognate single-strand binding protein. Except for RECQ from E. coli (Umezu et al., 1990) and HsRECQ4 (Xu and Liu, 2009), AtRECQ3 and AtRECQ2 share with the other characterized RecQ homologs the necessity of a sufficient 3′ overhang to initiate the unwinding of partial duplex DNA of the kind tested. As the experiments for AtRECQ2 and AtRECQ3 were conducted in parallel under the exact same conditions, it is possible to conclude that for this kind of substrate, the requirements for AtRECQ2 are less demanding. The information that approximately 6 nucleotides of 3′ overhang are sufficient to unwind a 17-bp duplex extends previously published information—that approximately 5 nucleotides are sufficient to promote unwinding of a 15-bp duplex—and is in good accordance with this data.
In contrast to AtRECQ2, but similar to most other characterized RecQ homologs (e.g. ScSGS1 [Bennett et al., 1998], HsRECQ5β [Garcia et al., 2004], and EcRECQ [Umezu et al., 1990]), AtRECQ3 can only use ATP and dATP to catalyze strand unwinding.
We next tested DNA structures that mimic intermediates of DNA repair, recombination, and replication. We started with a structure that can occur in all three mentioned pathways, the HJ. AtRECQ2 and AtRECQ3 both lack the ability to process intact, static HJs. In contrast to the two Arabidopsis homologs, some other RecQ homologs can process static HJ, most prominently HsRECQ1 (Popuri et al., 2008), for which it is a favorite substrate. Both HsBLM (Huber et al., 2002; Popuri et al., 2008) and ScSGS1 (Huber et al., 2002) can also process static HJs but not as favorite substrates. Although static HJs are often used for biochemical characterization of nucleic acids enzymes, they do not occur in vivo.
In contrast to the static HJ, as already published, AtRECQ2 catalyzes branch migration of the X12-HJ (Kobbe et al., 2008). This substrate, with a homologous core, comes much closer to the natural HJs that exist in cells. In the processing of this structure, the two Arabidopsis homologs differ, as AtRECQ3 is not able to significantly process this partially mobile X12-HJ. Considering the other RecQ homologs characterized so far, this property of AtRECQ3 is unique. HsRECQ1 (Sharma et al., 2005; Popuri et al., 2008), HsWRN (Mohaghegh et al., 2001; Cheng et al., 2006), HsBLM (Mohaghegh et al., 2001; Janscak et al., 2003; Bachrati et al., 2006; Popuri et al., 2008), HsRECQ5β (Garcia et al., 2004), EcRECQ (Harmon and Kowalczykowski, 1998), and DmRECQ5β (Özsoy et al., 2003) can process this or another partially mobile oligonucleotide-based HJ. Branch migration of HJs is an important issue for double HJ dissolution catalyzed by some RecQ helicases in complexes with topoisomerases (Wu and Hickson, 2003; Hartung et al., 2007, 2008). Thus, our data suggest that AtRECQ3 does not play such a role. Double HJ dissolution is considered as an alternative pathway to double HJ resolution by nucleases. It leads, in contrast to the latter, to gene conversion events.
Nicked HJs might, for example, occur according to the classical double-strand break repair model prior to the ligation step (Szostak et al., 1983; Puchta, 2005). No extensive characterization of RecQ helicase activity against this kind of substrate has been performed thus far. However, these substrates are important for the biochemistry of DNA repair and recombination enzymes. They are central in the characterization of the Mus81 endonuclease complex, which is assumed to act in parallel to RecQ helicases (Boddy et al., 2000; Mullen et al., 2001; Hartung et al., 2006; Trowbridge et al., 2007; Geuting et al., 2009). They are also central to the analysis of the helicase SRS2, which also acts in parallel to RecQ helicases (Fabre et al., 2002). Interestingly, both Arabidopsis enzymes tested could process a nicked, static HJ. Even more interestingly, the outcome of the reaction differs between them. As visualized in Figure 4D, the main products of AtRECQ3 are in line with a translocation on one specific strand, while no such preference is apparent for AtRECQ2. The behavior of AtRECQ2 toward the nicked HJ might be in accordance with the observed branch migration with the partially mobile X12-HJ: it is assumed that branch migration combines the activities of strand unwinding and strand annealing. Unwinding in the two directions shown in Figure 4D, in combination with annealing, might lead to branch migration, but this mechanism is not possible within a static HJ.
If the unique behavior of AtRECQ3 is examined within the context of DNA double-strand break repair models, its action might lead to gene conversion events. The enzyme might be involved in displacing the invading, elongated strand from its homologous template. This could occur while both 3′ ends of the DSB are elongated but not ligated, so that they could anneal to one another. Repair synthesis and ligation would result in a gene conversion event.
In addition to differences in the processing of HJs, AtRECQ2 and AtRECQ3 also differ in their reactions toward a regressable replication fork. AtRECQ2 catalyzes fork regression, as has been shown with this substrate and HsWRN (Machwe et al., 2006). This is in line with the hypothesis that AtRECQ2 and the HsWRN might be functionally homologous (Kobbe et al., 2008). Additionally, HsBLM catalyzes fork regression with the appearance of single daughter strands at high enzyme concentrations (Machwe et al., 2006). In contrast, EcRECQ does not catalyze fork regression (Machwe et al., 2006). The replication fork data is extended by plasmid-based fork regression catalyzed by BLM (Ralf et al., 2006) but not by HsRECQ1 (Popuri et al., 2008). Fork regression is claimed to be important, as it can provide an alternative DNA synthesis template to prevent replication fork collapse.
In this study, the reaction of AtRECQ3 on the replication fork produced daughter strands and the parental duplex. Since AtRECQ3 is a 3′ to 5′ DNA helicase, the most likely reaction that yields those products is shown in Figure 5C. AtRECQ3 might bind to the junction or to the single-stranded region at the junction on the parental strand that forms the template for the lagging strand and translocate in the 3′ to 5′ direction to displace the lagging strand. This reaction has also be shown to occur when DmRECQ5β acts on a nonregressable replication fork (Özsoy et al., 2003). If the regressable replication fork is processed this way, the corresponding three-way structure is not very stable. Our attempts to purify this proposed intermediate structure by gel extraction to test it as a possible substrate were not successful. However, AtRECQ3 autoradiography samples exhibit a band that might correspond to this structure (Fig. 5A). This three-way structure may then be transformed (either enzymatically or nonenzymatically by breathing of the DNA) into parental duplex DNA with the release of the labeled leading strand. As the daughter duplex itself is not significantly processed by either AtRECQ2 or AtRECQ3, it is improbable that AtRECQ3 catalyzes fork regression and unwinds the daughter duplex DNA later on. In respect to this substrate, AtRECQ3 behaves like the prokaryotic helicase EcUvrD (Machwe et al., 2006). Also, the reaction catalyzed by HsRECQ5β on a slightly different synthetic replication fork is to some extent similar to what we propose for AtRECQ3. HsRECQ5β also unwinds the lagging strand; however, it also promotes the annealing of liberated leading and lagging strands, which we did not observe for AtRECQ3 under our conditions (Kanagaraj et al., 2006).
As detailed below, DNA strand annealing was shown for all human RecQ helicases. Thus, we also analyzed whether AtRECQ2 and AtRECQ3 can catalyze strand annealing. Interestingly, although AtRECQ3 does not catalyze branch migration of a HJ or regression of a replication fork, its annealing activity is enhanced over that of AtRECQ2, which can catalyze the two mentioned reactions.
All human RecQ helicases promote DNA strand annealing, but neither mapping of the annealing activity (Garcia et al., 2004; Cheok et al., 2005; Muftuoglu et al., 2008; Ren et al., 2008) nor the influence of nucleotide cofactors on annealing reveals a uniform picture valid for all RecQ helicases. Concerning the influence of nucleotide cofactors, the strand annealing activity of HsRECQ5β is inhibited by ATPγS but not by ATP or ADP. This inhibition is not seen in HsRECQ5β-K58R (Garcia et al., 2004).
Another study revealed that HsRECQ5α is inhibited by both ATPγS and ATP (Ren et al., 2008). Also for HsRECQ1, strand annealing efficiency is reduced by ATPγS, although the magnitude of this effect depends upon both the substrate and the ATPγS concentration (Sharma et al., 2005). The inhibitory effect of ATPγS on strand annealing in general (Machwe et al., 2005) and, more specifically, by increasing concentrations of ATPγS (but not of ADP) was also shown for HsBLM (Cheok et al., 2005), and an inhibitory effect by ATPγS upon HsWRN was also reported (Machwe et al., 2005). Strand annealing activity was also shown for HsRECQ4 (Macris et al., 2006).
Therefore, it is interesting that in this work, strand annealing by AtRECQ3 and AtRECQ3-K64M did not dramatically change in the presence of ATP, ATPγS, and ADP. The annealing activity of AtRECQ3 is much more enhanced than the annealing activity of AtRECQ2. For AtRECQ2, strand annealing is most prominent, but still much less pronounced, in the presence of ADP.
Both this effect and the observed differences in the substrate preference might be a consequence of domain composition. Bioinformatical analysis indicates that AtRECQ3, similar to HsRECQ5β, lacks both the winged helix subdomain for the RQCt domain and the HRDC domain present in AtRECQ2 (Fig. 1A; Kobbe et al., 2008).
In summary, our data show a different substrate specificity for AtRECQ2 and AtRECQ3, especially toward substrates mimicking intermediates of DNA repair and recombination and of stalled replication forks. Therefore, they are likely to perform different functions in the cell. While AtRECQ2 might have similar functions as HsWRN, AtRECQ3 might have specific functions in the plant, which are not comparable on a one-to-one basis to any of the human RecQ homologs. However, taking the domain structure and the biochemical properties into account, HsRECQ5β seems to be the most similar human RECQ homolog.
As there is no AtRECQ3 mutant available in any of the Arabidopsis T-DNA mutant collections worldwide, it will be interesting to use alternative approaches, such as RNA interference, to test whether knockdown of its expression will reveal its function in vivo.
MATERIALS AND METHODS
Plasmids for the Overexpression of AtRECQ3 and AtRECQ3-K64M
AtRECQ3 and AtRECQ3-K64M were subcloned in several steps. AtRECQ3 was on the one hand cloned into pCAL-n-FLAG (Stratagene) using the ligation-independent cloning strategy. On the other hand, AtRECQ3 was cloned into a derivative of pIEx3 providing a thrombin recognition site and a His-tag. The vector used in this study was built by exchanging the BamHI/XhoI fragment. The final constructs having the vector pCAL-n-FLAG (Stratagene) as its scaffold leads to the addition of an N-terminal calmodulin-binding peptide, a thrombin recognition site, a FLAG epitope, and an enterokinase target (MKRRWKKNFIAVSAANRFKKISSSGALLVPRGSDYKDDDDK) to the N terminus, and a thrombin recognition site and a 6-His-tag (His-tag; LVPRGSHHHHHH) at the C terminus of AtRECQ3 (accession no. CAC14867) and AtRECQ3-K64M. The mutations leading to the K64M amino acid substitution were created by overlap extension PCR and inserted via MfeI and EcoRI into pCAL-n-FLAG-AtRECQ3. For the final construct, the NdeI/EcoRI fragment of pCAL-n-FLAG-AtRECQ3-K64M was exchanged for the corresponding fragment of the pCAL-n-FLAG-AtRECQ3-c-His vector. The open reading frames were confirmed by sequencing. The constructs for AtRECQ2 and AtRECQ2-K117M have been described previously (Kobbe et al., 2008).
Expression and Purification of AtRECQ3
The protein was overexpressed in the Escherichia coli strain BL21-CodonPlus(DE3)-RIPL (Stratagene) by 0.2 mm isopropyl-β-d-thiogalactopyranoside induction for approximately 4 h at 28°C. All purification steps were performed at 4°C. The frozen cells were resuspended (approximately 0.1 g/mL) in buffer A (10 mm Tris-HCl, pH 7.5, 200 mm NaCl, 20 mm imidazole, 5% glycerol, and 10 mm 3-mercapto-1,2-propanediol [thioglycerol]) and disrupted by lysozyme (100 μg/mL) and sonication. After centrifugation at 40,000g for 30 min, the supernatant was filtered and loaded onto a 1 mL Ni2+-charged HiTrap Chelating HP column (GE Healthcare) at a flow rate of 1 mL/min using the low-pressure liquid chromatography system BioLogic LP (Bio-Rad Laboratories). The column was first washed with 45 mL of buffer A plus 0.5% Triton X-100, followed by 15 mL of buffer A. Contaminant proteins were removed by flushing the column for 20 min at 29% of buffer B (buffer A with 400 mm imidazole). AtRECQ3 eluted with 80% buffer B. The buffer of the protein-containing fractions was exchanged to buffer C [50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 2 mm CaCl2, 1 mm Mg(CH3COO)2, 1 mm imidazole, and 10 mm thioglycerol] on a PD-10 column following the instructions of the manufacturer (GE Healthcare). The volume of the eluate was adjusted to 10 mL with buffer C and applied to 0.75 mL of equilibrated Calmodulin affinity resin (Stratagene). The column was washed with 10 mL of buffer C and 15 mL of buffer D (50 mm Tris-HCl, pH 7.5, 2 mm EGTA, and 10 mm thioglycerol). The protein was then eluted with buffer E (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 2 mm EGTA, and 10 mm thioglycerol). The buffer of the protein-containing fractions was exchanged to 50 mm Tris-HCl (pH 7.5), 300 mm NaCl, 10% glycerol, and 10 mm thioglycerol by a PD-10 column. The eluate was concentrated via dialysis against Suc and then mixed with an equal volume of glycerol. Aliquots of the purified protein were stored at −80°C. AtRECQ3-K64M was purified in an identical manner as described for AtRECQ3. The proteins were quantified on colloidal Coomassie-stained SDS-PAGE gels using bovine serum albumin (Bio-Rad) as a standard. The purification procedure for AtRECQ2 and AtRECQ2-K117M has been described previously (Kobbe et al., 2008).
DNA Substrates
Appropriate oligonucleotides were labeled with [γ-32P]ATP (3000 or 6000 Ci/mmol; GE Healthcare) and T4 polynucleotide kinase (NEB). The preparation of the DNA substrates largely followed literature procedures (Bachrati and Hickson 2006; Brosh et al., 2006). The sequence of the oligonucleotides used to prepare the substrate diagrammed in Figures 1B, 2A, and 2B was derived (Shen et al., 1998). The M13mp18ss DNA-based substrate (Kobbe et al., 2008), the X12-HJ (Mohaghegh et al., 2001), the X0- and nX0-HJs (Boddy et al., 2001; Gaillard et al., 2003), and the replication fork (Machwe et al., 2006) were already described. The sequences of the oligonucleotides used for annealing were also previously published (Van Komen et al., 2003; H1 and H4).
For the helicase assay, the reactions were performed at 37°C with 150 pm of DNA substrate (100 pm for the replication fork) in 40 mm Tris acetate (pH 8.0), 50 mm potassium acetate, 6 mm dithiothreitol, 50 μg/mL bovine serum albumin (NEB), 1.8 mm ATP, and 1.8 mm MgCl2 (unless otherwise indicated) and generally started by mixing the enzyme with the other reaction components. They were terminated with one-third of the volume of stop solution (50 mm EDTA, 0.6% SDS, 0.1% xylene cyanol, 0.1% bromphenol blue, and 20% glycerol). For time-course experiments, reaction aliquots were removed at the indicated time points. To determine background values, parallel reactions were performed without any enzyme. After running native TBE-PAGE gels at 4°C, the radioactive DNA was visualized by autoradiography with either the BIO-Imaging-Analyzer BAS-1500 (FUJIFILM) or the Instant Imager (Canberra Packard Company) and quantified with the Packard Imager for Windows (version 2.05) software from Canberra Packard. Except for the replication fork, the fraction of DNA unwound was calculated as described (Mohaghegh et al., 2001). For the replication fork, the amount of each DNA species was standardized with the help of the specific activity of the labeled component oligonucleotides. The product amounts were subsequently expressed as percentage of the original amount of substrate, and background values from reactions without enzyme were subtracted.
For the annealing assay, the annealing reactions were performed essentially as the helicase assays; 3 fmol of oligonucleotide H4 and the enzyme were pipetted as separate drops in a reaction tube. The reaction was started by addition of the labeled oligonucleotide H1 (3 fmol) together with the other reaction components to yield a total volume of 20 μL. To calculate the percentage of annealing, the percentage of annealed substrate in reactions without enzyme was subtracted from the percentage obtained with enzyme. As indicated, reactions were either performed in the presence of ATP, ATPγS, or ADP.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CAC14866 (AtRECQ2) and CAC14867 (AtRECQ3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of the DNA substrates used in this study
Supplemental Figure S2. Concentration dependency of processing of the nX0-HJ.
Supplemental Table S1. Sequences of the oligonucleotides used for the preparation of the DNA substrates.
Supplementary Material
Acknowledgments
We thank H. Plchova for the cDNA constructs used for the initial cloning. Additionally, we thank V. Geuting and S. Thies for providing some labeled DNA structures used in this study, K. Demand for initial experiments with AtRECQ2, and D. Wedlich for providing the BAS-1500 reader. We greatly appreciated skillful technical assistance by C. Moock and K. Bourquin.
This work was supported by the Deutsche Forschungsgemeinschaft grant “Pu 137/10” to H.P. and M.F. and the Concept for the Future of Karlsruhe Institute of Technology within the framework of the German Excellence Initiative “YIG 9-109” to D.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Holger Puchta (holger.puchta@bio.uka.de).
The online version of this article contains Web-only data.
References
- Bachrati CZ, Borts RH, Hickson ID (2006) Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res 34 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J 374 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID (2006) Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol 409 86–100 [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID (2008) RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117 219–233 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Sharp JA, Wang JC (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J Biol Chem 273 9644–9650 [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR III, Russell P (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107 537–548 [DOI] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol 20 8758–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Bohr VA (2007) Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res 35 7527–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Opresko PL, Bohr VA (2006) Enzymatic mechanism of the WRN helicase/nuclease. Methods Enzymol 409 52–85 [DOI] [PubMed] [Google Scholar]
- Brosh RM Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA (1999) Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem 274 18341–18350 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA (2006) Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res 34 2751–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID (2005) The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res 33 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID (2009) RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9 644–654 [DOI] [PubMed] [Google Scholar]
- Ellis NA, Sander M, Harris CC, Bohr VA (2008) Bloom's syndrome workshop focuses on the functional specificities of RecQ helicases. Mech Ageing Dev 129 681–691 [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P (2003) The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell 12 747–759 [DOI] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 23 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuting V, Kobbe D, Hartung F, Durr J, Focke M, Puchta H (2009) Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol 150 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Hickson ID (2007) Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci 64 2306–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon FG, Kowalczykowski SC (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev 12 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Plchova H, Puchta H (2000) Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res 28 4275–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2006) The RecQ gene family in plants. J Plant Physiol 163 287–296 [DOI] [PubMed] [Google Scholar]
- Hartung F, Suer S, Bergmann T, Puchta H (2006) The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res 34 4438–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H (2008) Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet 4 e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Puchta H (2007) Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA 104 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3 169–178 [DOI] [PubMed] [Google Scholar]
- Huber MD, Lee DC, Maizels N (2002) G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res 30 3954–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA (2003) Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J Mol Biol 330 29–42 [DOI] [PubMed] [Google Scholar]
- Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P (2006) Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res 34 5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbe D, Blanck S, Demand K, Focke M, Puchta H (2008) AtRECQ2, a RecQ-helicase homologue from Arabidopsis thaliana, is able to disrupt different recombinogenic DNA-structures in vitro. Plant J 55 397–405 [DOI] [PubMed] [Google Scholar]
- Kobbe D, Focke M, Puchta H (2009) Purification and characterization of RecQ helicases of plants. Methods Mol Biol 587 195–209 [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Matson SW, Orren DK (2005) RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem 280 23397–23407 [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK (2006) The werner and bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45 13939–13946 [DOI] [PubMed] [Google Scholar]
- Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P (2006) Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 5 172–180 [DOI] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh Jr RM Jr, Bohr VA, Hickson ID (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res 29 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA (2008) Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry 47 10247–10254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, Cheng WH, Bohr VA (2004) Junction of RecQ helicase biochemistry and human disease. J Biol Chem 279 18099–18102 [DOI] [PubMed] [Google Scholar]
- Özsoy AZ, Ragonese HM, Matson SW (2003) Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res 31 1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plchova H, Hartung F, Puchta H (2003) Biochemical characterization of an exonuclease from Arabidopsis thaliana reveals similarities to the DNA exonuclease of the human Werner syndrome protein. J Biol Chem 278 44128–44138 [DOI] [PubMed] [Google Scholar]
- Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A (2008) The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 283 17766–17776 [DOI] [PubMed] [Google Scholar]
- Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56 1–14 [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L (2006) The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281 22839–22846 [DOI] [PubMed] [Google Scholar]
- Ren H, Dou SX, Zhang XD, Wang PY, Kanagaraj R, Liu JL, Janscak P, Hu JS, Xi XG (2008) The zinc-binding motif of human RECQ5beta suppresses the intrinsic strand-annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem J 412 425–433 [DOI] [PubMed] [Google Scholar]
- Sharma S, Doherty KM, Brosh RM Jr (2006) Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J 398 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM Jr (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 280 28072–28084 [DOI] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Loeb LA (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res 26 2879–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW (1983) The double-strand-break repair model for recombination. Cell 33 25–35 [DOI] [PubMed] [Google Scholar]
- Trowbridge K, McKim K, Brill SJ, Sekelsky J (2007) Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176 1993–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Nakayama K, Nakayama H (1990) Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA 87 5363–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Reddy MS, Krejci L, Klein H, Sung P (2003) ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J Biol Chem 278 44331–44337 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 870–874 [DOI] [PubMed] [Google Scholar]
- Xu X, Liu Y (2009) Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J 28 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.