Abstract
The stamen, which consists of an anther and a filament, is the male reproductive organ in a flower. The specification of stamen identity in Arabidopsis (Arabidopsis thaliana) is controlled by a combination of the B genes APETALA3 (AP3) and PISTILLATA, the C gene AGAMOUS (AG), and the E genes SEPALLATA1 (SEP1) to SEP4. The “floral organ-building” gene SPOROCYTELESS/NOZZLE (SPL/NZZ) plays a central role in regulating anther cell differentiation. However, much less is known about how “floral organ identity” and floral organ-building genes interact to control floral organ development. In this study, we report that ectopic expression of SPL/NZZ not only affects flower development in the wild-type background but also leads to the transformation of petal-like organs into stamen-like organs in flowers of ap2-1, a weak ap2 mutant allele. Moreover, our loss-of-function analysis indicates that the spl/nzz mutant enhances the phenotype of the ag weak allele ag-4. Furthermore, ectopic expression and overexpression of SPL/NZZ altered expression of AG, SEP3, and AP2 in rosette leaves and flowers, while ectopic expression of SPL/NZZ resulted in ectopic expression of AG and SEP3 in the outer whorls of flowers. Our results indicate that the SPL/NZZ gene is engaged in controlling stamen identity via interacting with genes required for stamen identity in Arabidopsis.
The Arabidopsis (Arabidopsis thaliana) flower contains four types of organs that are arranged in four concentric whorls. Four sepals are found in the outermost whorl, four petals in the second whorl, six stamens in whorl 3, and two carpels in the innermost whorl. As in most angiosperms, flower development in Arabidopsis occurs in four steps: transition from vegetative growth to reproductive growth, establishment of floral meristem identity, specification of floral organ identity, and floral organ morphogenesis (Zhao et al., 2001a; Jack, 2004). Extensive molecular genetic studies have not only uncovered a large number of genes that are required for flower development but also elucidated regulatory relationships among key genes.
The ABC model describes three classes of homeotic genes, termed A, B, and C, which specify floral organ identity in four whorls (Haughn and Somerville, 1988; Bowman et al., 1991b; Coen and Meyerowitz, 1991; Meyerowitz et al., 1991; Ma, 1994; Weigel and Meyerowitz, 1994). In Arabidopsis, the class A genes APETALA1 (AP1) and AP2 are active in whorls one and two, the class B genes AP3 and PISTILATA are active in whorls two and three, and the class C gene AGAMOUS (AG) is active in whorls three and four. Class A genes alone are required for sepal identity. Class A and B genes together direct petal identity. Class B and C genes control stamen identity, while the class C gene solely specifies carpels. Moreover, class A and C genes function antagonistically (Drews et al., 1991; Mizukami and Ma, 1992). The activity of class A genes in whorls one and two prevents the function of the class C gene in the same whorls and vice versa in whorls three and four. A new addition to the ABC model is that the E genes (SEPALLATA1 [SEP1]–SEP4) are involved in specifying organ identity in all four whorls via interacting with ABC regulators (Pelaz et al., 2000; Honma and Goto, 2001; Ditta et al., 2004; Melzer et al., 2009). The ABC model is applicable to diverse plant species in terms of regulation of floral organ identity. However, much less is known about genes that are necessary for building floral organs.
Stamens are male reproductive organs in a flower. Each stamen consists of an anther, where the male gametophyte develops, and a filament, which provides water and nutrients to the anther and anchors the anther to the flower. The anther is a four-lobed structure in Arabidopsis. Each mature anther lobe is composed of five types of well-organized cell layers: epidermis, endothecium, middle layer, tapetum, and microsporocyte (pollen mother cell; Goldberg et al., 1993; Sanders et al., 1999). Microsporocytes are reproductive cells that undergo meiosis and eventually develop into pollen grains. The remaining nonreproductive (somatic) cells are required for the normal development and release of pollen. In particular, the tapetum is essential for pollen development. After the specification of stamen identity, anther development involves a series of cell division, cell differentiation, and cell death, resulting in the formation of reproductive microsporocytes and somatic cell layers. Although microarray experiments uncovered many genes that are important for anther development (Zik and Irish, 2003; Hennig et al., 2004; Wellmer et al., 2004; Lu et al., 2006; Alves-Ferreira et al., 2007; Ma et al., 2007, 2008; Wijeratne et al., 2007), so far only a few genes have been identified to play direct roles in controlling differentiation of various anther cells.
Besides genes that encode signaling proteins (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003; Albrecht et al., 2005; Colcombet et al., 2005; Hord et al., 2006; Mizuno et al., 2007; Jia et al., 2008; Zhao, 2009), the SPOROCYTELESS/NOZZLE (SPL/NZZ) gene plays a central role in controlling early anther cell differentiation (Schiefthaler et al., 1999; Yang et al., 1999). Anthers in the spl/nzz mutant do not produce microsporocytes or anther walls, suggesting that the SPL/NZZ gene is required for the differentiation of microsporocytes and anther walls, including the tapetum. The SPL/NZZ gene encodes a novel protein related to MADS box transcription factors. The direct activation of SPL/NZZ by AG is required for early anther development (Ito et al., 2004). Detected as early as the stage when stamen primordia are generated, the expression of SPL/NZZ coincides with the expression of AG during early anther development (Yanofsky et al., 1990; Bowman et al., 1991a; Drews et al., 1991; Ito et al., 2004). Therefore, we hypothesize that SPL/NZZ might be involved in controlling stamen identity.
Here, we report our characterization of the SPL/NZZ function in specifying stamen identity. We show that ectopic expression of SPL/NZZ affects both leaf and flower development. Furthermore, the ectopic expression of SPL/NZZ leads to the transformation of petal-like organs into stamen-like organs in flowers of ap2-1, a weak ap2 mutant allele. The spl/nzz mutant also enhances the phenotype of the ag weak allele ag-4. In addition, ectopic expression and overexpression of SPL/NZZ alter the expression of AG, SEP3, and AP2 in rosette leaves and flowers. Our results indicate that the “floral organ-building” gene SPL/NZZ is not only essential for early anther cell differentiation but also is engaged in controlling stamen identity in Arabidopsis.
RESULTS
Ectopic Expression and Overexpression of SPL/NZZ Cause Abnormal Plant Development
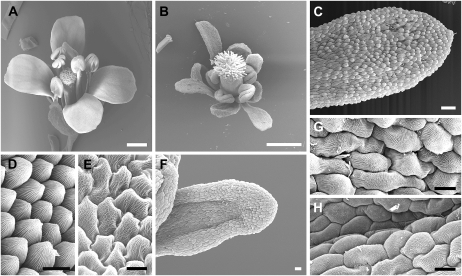
The SPL/NZZ gene is critical for anther morphogenesis, since spl/nzz mutant plants fail to produce the main structures of the anther, including microsporocytes and anther walls (Schiefthaler et al., 1999; Yang et al., 1999). To further test the function of SPL/NZZ in anther development, we ectopically expressed SPL/NZZ under the control of the constitutively active cauliflower mosaic virus 35S promoter (35S). Eighty-seven percent of the 237 Pro35S:SPL/NZZ transgenic plants examined were defective in both vegetative and reproductive growth. Pro35S:SPL/NZZ plants were shorter than wild-type plants (Fig. 1A). In addition, fertility of most Pro35S:SPL/NZZ plants was reduced. Interestingly, Pro35S:SPL/NZZ plants produced curled rosette leaves, although the curling degree and leaf size were variable (Fig. 1, B–E). In the Pro35S:SPL/NZZ-GLUCOCORTICOID RECEPTOR (GR) transgenic line, the SPL/NZZ activity can be induced by continuous treatment of the steroid hormone dexamethasome (DEX; Ito et al., 2004). The formation of curled rosette leaves in Pro35S:SPL/NZZ-GR plants after DEX treatment indicated that curled rosette leaves were caused by the ectopic expression of SPL/NZZ (Fig. 1F). Additionally, an earlier study showed that the spl-D mutant, in which SPL/NZZ was overexpressed, exhibited similar phenotypes to Pro35S:SPL/NZZ transgenic plants, including curled rosette leaves (Supplemental Fig. S1, A and C; Li et al., 2008). Furthermore, most curled rosette leaves in both Pro35S:SPL/NZZ and spl-D plants were up-curled (Fig. 1, B–E; Supplemental Fig. S1, A–C) and resembled the rosette leaf phenotype of Pro35S:AG plants (Fig. 1G). Our results indicate that ectopic expression and overexpression of SPL/NZZ caused abnormal vegetative and reproductive growth.
Figure 1.
Pro35S:SPL/NZZ plants are abnormal in development. A, Compared with the wild type (left), the Pro35S:SPL/NZZ transgenic plant (right) had shorter stature and reduced fertility. B, Wild-type seedling. C to E, Pro35S:SPL/NZZ seedlings showing variabilities in their up-curled rosette leaves. F, Pro35S:SPL/NZZ-GR seedling exhibiting wrinkled and up-curled rosette leaves after treatment with DEX. G, Pro35S:AG seedling displaying up-curled rosette leaves. Bars = 1 cm (A) and 0.5 cm (B–G).
Ectopic Expression of SPL/NZZ Affects Flower Development
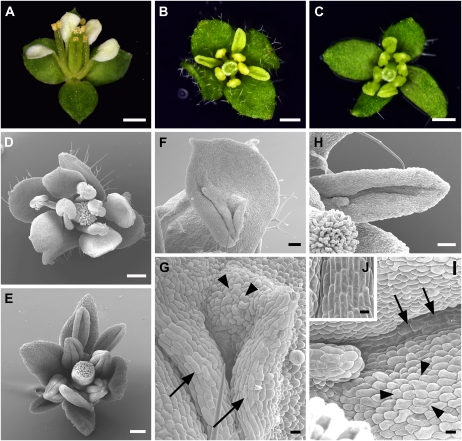
Pro35S:SPL/NZZ plants were abnormal in flower development. Pro35S:SPL/NZZ flowers were smaller than those of the wild type (Fig. 2, A and B). In addition, Pro35S:SPL/NZZ plants frequently produced buds with unfurled sepals (Fig. 3A). The development of petals in Pro35S:SPL/NZZ flowers was delayed, resulting in the late emergence of petals. More interestingly, Pro35S:SPL/NZZ flowers sometimes produced small and narrow petals in whorl 2 (Figs. 2, B and C, and 3B). Occasionally, narrow petals were up-curled (Fig. 2F; Table I). Although no morphological defects of stamens and carpels were observed, Pro35S:SPL/NZZ plants had reduced fertility. Anthers in many mature flowers could not reach the stigma due to elongation defects in filaments (Fig. 2, A and B). To further test the phenotype of Pro35S:SPL/NZZ flowers, we analyzed the surface features of petals using scanning electron microscopy (SEM). Wild-type floral organs have distinctive epidermal cell morphologies. In the wild type, petal epidermal cells are uniform in size and conical in shape (Fig. 2D), while stamen epidermal cells exhibit irregular cell edges and have wavy ridges (Fig. 2E). In Pro35S:SPL/NZZ flowers, epidermal cells of narrow petals were long and uneven in size, although some cells still had dome shapes (Fig. 2, C and G). Cells of some Pro35S:SPL/NZZ petals were similar to those of wild-type stamens, indicated by irregular cell edges and wavy ridges (Fig. 2, F and H). Our results indicate that the ectopic expression of SPL/NZZ affects floral organ development. The stamen feature in some Pro35S:SPL/NZZ petals suggests that the SPL/NZZ gene might be involved in specifying stamen identity by promoting C function in whorl 2.
Figure 2.
Scanning electron micrographs showing defects in flower development in Pro35S:SPL/NZZ plants. A, A wild-type flower showing normal petals. B, A Pro35S:SPL/NZZ flower showing narrow petals and shortened stamens. C, A narrow petal in whorl 2 of a Pro35S:SPL/NZZ flower. D, Wild-type petal epidermal cells showing uniform size and conical shape. E, Wild-type anther epidermal cells exhibiting irregular cell edges and wavy ridges. F, A narrow and up-curled petal in whorl 2 of a Pro35S:SPL/NZZ flower. G, High-magnification view of epidermal cells of the organ shown in C displaying elongated shape and uneven sizes. H, High-magnification view of epidermal cells of the organ shown in F exhibiting the resemblance to wild-type anther epidermal cells in E. Bars = 0.5 mm (A and B), 50 μm (C and F), and 10 μm (D, E, G, and H).
Figure 3.
The petal phenotype caused by Pro35S:SPL/NZZ may require a functional AG. A, A Pro35S:SPL/NZZ young flower showing unclosed sepals and small petals. B, A Pro35S:SPL/NZZ flower showing four narrow petals. C, An ag-1 Pro35S:SPL/NZZ young flower. D, An ag-1 Pro35S:SPL/NZZ flower showing normal petals in the second whorl. E, An ag-1 flower. Bars = 0.5 mm.
Table I.
Comparison of floral organs
Leaf like in whorl 1, sepals with leaf or stigma structures; stamen like in whorl 2, narrow, up-curled, or with stamen structures; stamen like in whorl 3, stamen with petal structures. The first 10 flowers from 10 plants were examined for each genotype.
| Phenotype | Genotype |
||||
|---|---|---|---|---|---|
| Wild Type | spl | Pro35S:SPL/NZZ | ap2-1 | ap2-1Pro35S:SPL/NZZ | |
| Whorl 1 sepal | 4.00 ± 0.00 | 4.00 ± 0.00 | 4.00 ± 0.00 | 0 | 0 |
| Leaf like | 4.00 ± 0.00 | 4.00 ± 0.00 | |||
| Whorl 2 petal | 4.00 ± 0.00 | 4.00 ± 0.00 | 3.47 ± 0.10 | 2.37 ± 0.11 | 0.65 ± 0.09 |
| Stamen like | 0 | 0 | 0.24 ± 0.06 | 0.62 ± 0.11 | 2.43 ± 0.14 |
| Whorl 3 stamen | 6.00 ± 0.00 | 5.44 ± 0.06 | 4.90 ± 0.12 | 5.25 ± 0.07 | 4.68 ± 0.08 |
| Stamen like | 0 | 0 | 0 | 0.21 ± 0.05 | 0.03 ± 0.01 |
| Carpel |
2.00 ± 0.00 |
2.00 ± 0.00 |
2.00 ± 0.00 |
2.00 ± 0.00 |
2.00 ± 0.00 |
Ectopic Expression of SPL/NZZ Enhances the ap2-1 Flower Phenotype in Whorl 2
The ectopic expression of SPL/NZZ led to the subtle phenotype of floral organ transformation from petal to stamen in the second whorl, while ap2-1 mutant flowers displayed a mild but similar phenotype in whorl 2. To test whether SPL/NZZ promotes the stamen identity, we transformed ap2-1 mutant plants using the Pro35S:SPL/NZZ construct. In ap2-1 flowers, the whorl-one organs are transformed into leaf-like organs from sepals and sometimes have carpel-like structures (Fig. 4, A and D; Table I). In whorl 2, most petals are close to normal, although stamen-like structures were observed in some petals (Table I). In ap2-1 Pro35S:SPL/NZZ flowers, a majority of petals in whorl 2 were narrow and up-curled (Fig. 4, B and E; Table I). In some ap2-1 Pro35S:SPL/NZZ flowers, all of the petals were converted into stamens (Fig. 4C). Further SEM analyses revealed that stamen-like organs in ap2-1 Pro35S:SPL/NZZ flowers produced cells that resembled anther and filament epidermal cells (Fig. 4, F–J). Moreover, we did not detect narrow petals in the second whorl of ag-1 Pro35S:SPL/NZZ flowers (Fig. 3). Our results indicate that the ectopic expression of SPL/NZZ enhances the flower phenotype of ap2-1 in whorl 2, which further supports the idea that SPL/NZZ is involved in promoting C function in whorl 2.
Figure 4.
Ectopic expression of SPL/NZZ promoted stamen identity in the second floral whorl of the ap2-1 mutant. A, An ap2-1 flower showing leaf-like organs in whorl 1 and petals in whorl 2. B, An ap2-1 Pro35S:SPL/NZZ flower exhibiting narrow and up-curled petal-like organs in whorl 2. C, An ap2-1 Pro35S:SPL/NZZ flower showing all stamens in whorls two and three. D, SEM image showing a Pro35S:SPL/NZZ flower. E, SEM image exhibiting an ap2-1 Pro35S:SPL/NZZ flower with up-curled stamen-like organs in whorl 2. F and G, SEM images showing a stamen-like organ (F) as well as filament-like (arrows) and anther-like (arrowheads) epidermal cells (G; high-magnification view of F) in an ap2-1 Pro35S:SPL/NZZ flower. H and I, SEM images displaying an up-curled stamen-like organ (H) as well as filament-like (arrows) and anther-like (arrowheads) epidermal cells (I; high-magnification view of H) in an ap2-1 Pro35S:SPL/NZZ flower. J, SEM image exhibiting wild-type filament cells. Bars = 0.5 mm (A–D), 100 μm (E), 50 μm (F and H), and 10 μm (G, I, and J).
Loss-of-Function Analysis Indicates That SPL/NZZ Is Involved in Controlling Stamen Identity
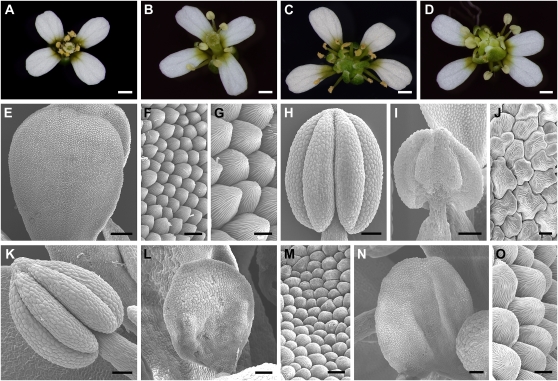
Our gain-of-function analyses indicate that the SPL/NZZ gene promotes the specification of stamen identity. To further test whether SPL/NZZ controls stamen identity, we created the spl ag-4 double mutant. Although SPL/NZZ (At4g27330) and AG (At4g18960) genes are located 3.3 Mb apart on chromosome 4, we found three spl ag-4 double mutants from 287 F2 plants. Previous studies have shown that spl/nzz petals are normal, while mutant anthers do not produce microsporocytes and anther walls (Schiefthaler et al., 1999; Yang et al., 1999). Compared with the wild type (Fig. 5, A and H), mature spl anthers were somewhat flattened and did not produce pollen grains (Fig. 5, B and I). ag-4, a weak ag allele, produced normal petals in whorl 2 and stamens in whorl 3 (Fig. 5, C and K). However, although anthers in spl ag-4 double mutant flowers were similar to those of spl (Fig. 5, B and D), SEM analyses revealed stamen-to-petal transformation of organs in the third whorl of spl ag-4 flowers. Epidermal cells from spl stamens (Fig. 5J) and ag-4 stamens in the third whorl (Sieburth et al., 1995; Chen and Meyerowitz, 1999) are similar to those of wild-type stamens (Fig. 2E). Conversely, in spl ag-4 double mutant flowers, the third whorl organs (petaloid organs) exhibited cone-shaped cells (Fig. 5, L–O), which resembled wild-type petal epidermal cells (Fig. 5, E–G). Our results provided strong evidence to support the idea that the SPL/NZZ gene plays a role in promoting stamen identity by interacting with AG.
Figure 5.
Loss-of-function analysis suggested that SPL/NZZ was involved in regulating stamen identity. A, A wild-type flower showing petals in whorl 2 and stamens in whorl 3. B, An spl flower exhibiting normal petals in whorl 2 and sterile stamens (no pollen released) in whorl 3. C, An ag-4 flower showing normal petals in whorl 2 and normal stamens in whorl 3. D, An spl ag-4 double mutant flower displaying normal petals in whorl 2 and stamens resembling those of spl inward whorl 2. E, SEM image exhibiting a wild-type petal. F and G, Low-magnification (F) and high-magnification (G) views of wild-type petal epidermal cells showing uniform size and cone shape. H, A wild-type anther. I, An spl anther showing flattened shape and undeveloped lobes. J, High-magnification view of epidermal cells of the spl anther, which are the same as wild-type anther epidermal cells (Fig. 2D). K, An ag-4 anther is similar to the wild-type anther. L, An spl ag-4 double mutant petal-like (petaloid) organ from whorl 3 showing no developed anther lobes. M, High-magnification view of epidermal cells of the organ shown in L showing the resemblance to wild-type petal epidermal cells in F. N, An spl ag-4 double mutant petal-like organ from whorl 3. O, High-magnification view of epidermal cells of the organ shown in N, which is similar to wild-type petal epidermal cells in G. Bars = 0.5 mm (A–D), 200 μm (E), 50 μm (L and N), 20 μm (H, I, and K), 10 μm (F and M), and 5 μm (G, J, and O).
Ectopic Expression and Overexpression of SPL/NZZ Alter the Expression of Genes Required for Stamen Identity
Previous studies showed that class B, C, and E genes cooperate to control stamen identity. The expansion of C gene function to whorls one and two leads to the formation of carpel and stamen-like structures in two outer whorls (Drews et al., 1991; Mizukami and Ma, 1992). In addition, ectopic expression of AG and SEP3 resulted in the formation of curled rosette leaves (Fig. 1G; Mizukami and Ma, 1992; Goodrich et al., 1997; Castillejo et al., 2005). Plants impaired in function of the CURLY LEAF (CLF) gene, which is required for maintaining the repression of AG epigenetically, also produced curled rosette leaves (Goodrich et al., 1997). Our results demonstrated that the ectopic expression of SPL/NZZ promotes stamen identity in the second whorl and causes the formation of curled rosette leaves. Therefore, to test how SPL/NZZ controls stamen identity, we carried out regular semiquantitative and quantitative real-time reverse transcription (RT)-PCR experiments to examine the expression of AP1, AP2, AP3, AG, SEP3, and CLF in Pro35S:SPL/NZZ and spl-D plants.
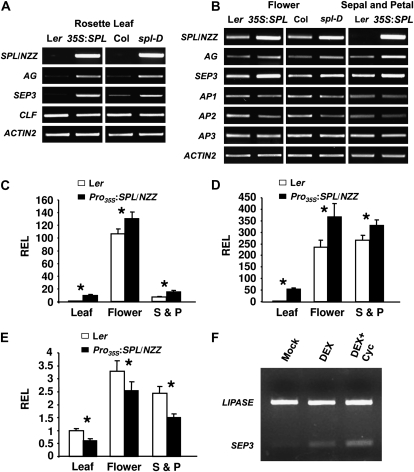
As expected, the expression of SPL/NZZ was greatly increased in both Pro35S:SPL/NZZ and spl-D rosette leaves (Fig. 6A). The expression of AG and SEP3 was also significantly increased, while there was no detectable change in the expression of CLF. We further examined gene expression in flowers. Compared with wild-type flowers, RT-PCR and semiquantitative RT-PCR results demonstrated that expression of SPL/NZZ, AG, and SEP3 was increased in both Pro35S:SPL/NZZ and spl-D flowers (Fig. 6B; Supplemental Fig. S2). Conversely, the expression of AP2 was decreased. The expression of AP1 was decreased only in Pro35S:SPL/NZZ flowers, while the expression of AP3 was unaffected (Fig. 6B; Supplemental Fig. S2). Our quantitative real-time RT-PCR results confirmed that expression of AG and SEP3 was significantly increased, while the expression of AP2 was decreased, in rosette leaves and flowers of both Pro35S:SPL/NZZ and spl-D plants (Fig. 6, C–E).
Figure 6.
Ectopic expression and overexpression of SPL/NZZ altered the expression of genes required for stamen identity in rosette leaves and flowers. A, RT-PCR results showing greatly increased expression of SPL/NZZ, AG, and SEP3 in both Pro35S:SPL/NZZ and spl-D rosette leaves. There was no detectable change of CLF expression. B, RT-PCR results showing that expression of SPL/NZZ, AG, and SEP3 was increased in Pro35S:SPL/NZZ and spl-D flowers as well as Pro35S:SPL/NZZ sepals and petals. The expression of AP2 was slightly decreased, while the expression of AP1 and AP3 seemed unchanged. C, Quantitative real-time RT-PCR results showing the increased expression of AG in rosette leaves, flowers, sepals, and petals of Pro35S:SPL/NZZ plants. D, Quantitative real-time RT-PCR results showing the increased expression of SEP3 in rosette leaves, flowers, sepals, and petals of Pro35S:SPL/NZZ plants. E, Quantitative real-time RT-PCR results illustrating the decreased expression of AP2 in rosette leaves, flowers, sepals, and petals of Pro35S:SPL/NZZ plants. F, Semiquantitative RT-PCR results showing the increased expression of SEP3 in Pro35S:SPL/NZZ-GR ag-1 inflorescences after both DEX and DEX + Cyc treatments for 4 h. The LIPASE gene was used as a control. The transcripts in wild-type rosette leaves in C to E were used as a standard for normalization. Asterisks indicate that the difference is significant (P < 0.01 or P < 0.05). REL, Relative expression level; S & P, sepal and petal; 35S:SPL, Pro35S:SPL/NZZ.
To examine whether ectopic expression SPL/NZZ affects the expression of AG, SEP3, and AP2 in sepals and petals, young buds from both wild-type and Pro35S:SPL/NZZ plants were dissected and then sepals and petals were collected with the dissection microscope. Both RT-PCR and quantitative real-time RT-PCR results showed that AG and SEP3 were ectopically expressed in sepals and petals of Pro35S:SPL/NZZ flowers (Fig. 6, B–D). Additionally, the expression of AP2 was decreased in sepals and petals (Fig. 6, B and E). In summary, our results indicate that both ectopic expression and overexpression of SPL/NZZ increase the expression of AG and SEP3 in rosette leaves and flowers. Furthermore, the ectopic expression of SPL/NZZ causes ectopic expression of AG and SEP3 in flower whorls one and two, which may affect the A gene function.
To test whether SPL/NZZ directly activates the expression of AG or SEP3, the expression of AG and SEP3 was examined in nzz-2 Pro35S:SPL/NZZ-GR plants after DEX as well as both DEX and cycloheximide (Cyc) treatments. The expression of AG was induced by DEX alone, while when Cyc was present, the expression of AG did not change (data not shown). The expression of SEP3 was rapidly induced in nzz-2 Pro35S:SPL/NZZ-GR inflorescences in the presence of DEX as well as both DEX and Cyc after 4 h (Fig. 6F).
To further examine whether AG is required for the formation of curled rosette leaves, the Pro35S:SPL/NZZ construct was transformed into ag-1 heterozygous plants and the spl-D mutant was crossed to ag-1 heterozygous plants. The rosette leaves in both ag-1 Pro35S:SPL/NZZ and ag-1 spl-D plants also exhibited the curled-leaf phenotype (Supplemental Fig. S1). However, plants with the most severe curled-leaf phenotype were not observed. Therefore, the formation of curled leaves was not fully dependent on a functional AG. Our results suggest that the induction of AG expression might be caused by the direct activation of SEP3 by SPL/NZZ in Pro35S:SPL/NZZ and spl-D plants.
DISCUSSION
Anther development entails a series of processes, including specification of stamen identity, establishment of anther adaxial and abaxial polarity, and differentiation of reproductive microsporocytes and somatic cell layers. To date, interactions between “stamen identity” and “stamen-building” genes have not been extensively studied in the context of development, although excellent advances have been made in understanding stamen identity, early anther cell differentiation, male meiosis, pollen development, and anther dehiscence (Goldberg et al., 1993; Walbot and Evans, 2003; Jack, 2004; McCormick, 2004; Scott et al., 2004; Ma, 2005; Feng and Dickinson, 2007; Borg et al., 2009; Wilson and Zhang, 2009; Zhao, 2009). Stamen identity is controlled by B, C, and E genes. The SPL/NZZ transcription factor plays a central role in regulating early anther cell differentiation, since the spl/nzz mutant anther is defective in producing the majority of anther cells, including endothecium, middle layer, tapetum, and microsporocyte (Schiefthaler et al., 1999; Yang et al., 1999). Different from SPL/NZZ, genes encoding Leu-rich repeat receptor-like protein kinases directly regulate the differentiation of fewer anther cell types in Arabidopsis (Zhao, 2009). EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1/2 determine the tapetum (Canales et al., 2002; Zhao et al., 2002; Albrecht et al., 2005; Colcombet et al., 2005). BARELY ANY MERISTEM1/2 is required for the differentiation of anther somatic cell layers (Hord et al., 2006). In addition, RECEPTOR-LIKE PROTEIN KINASE2 specifies middle layer cells (Mizuno et al., 2007). TAPETUM DETERMINANT1 defines the tapetum by acting as a potential ligand of EMS1/EXS (Yang et al., 2003; Jia et al., 2008). Therefore, SPL/NZZ may control anther cell differentiation by regulating a subset of downstream genes necessary for anther establishment. On the other hand, in this study, our gain-of-function and loss-of-function analyses demonstrated that SPL/NZZ is involved in controlling stamen identity. Furthermore, we showed that the ectopic expression and overexpression of SPL/NZZ altered the expression of several key genes that are required for stamen identity. Besides the role played in early anther cell differentiation, our results suggest that the floral organ-building gene SPL/NZZ might regulate stamen identity by interacting with floral organ identity genes such as AG, SEP3, and AP2.
SPL/NZZ may be involved in controlling stamen identity through interacting with AG. The AG gene, which encodes a MADS box protein, is essential for specifying stamens and carpels as well as repressing stem cell proliferation in the center of the flower (Yanofsky et al., 1990; Lenhard et al., 2001; Lohmann et al., 2001). The AG transcription factor controls early stamen development by directly activating the expression of SPL/NZZ (Ito et al., 2004). In addition, AG is necessary for regulating anther dehiscence, a late event in stamen development, by directly activating the expression of DEFECTIVE IN ANTHER DEHISCENCE1, which encodes a catalytic enzyme involved in the biosynthesis of jasmonic acid (Ito et al., 2007). Therefore, AG not only controls early stamen development, including stamen identity and early anther cell differentiation, but also regulates late stamen development, such as anther dehiscence. Previous studies have shown that AG is ectopically expressed in two outer whorls in ap2 mutant flowers, while plants ectopically expressing AG form ap2-like flowers, which have stamen-like structures in the second whorl (Drews et al., 1991; Mizukami and Ma, 1992). Our results showed that both ectopic expression and overexpression of SPL/NZZ result in the increased expression of AG in rosette leaves and flowers. In particular, the ectopic expression of SPL/NZZ led to ectopic expression of AG in sepals and petals. Therefore, the ectopic expression of AG may cause the flower phenotypes in Pro35S:SPL/NZZ plants and enhance the stamen phenotype in the second whorl of ap2-1 Pro35S:SPL/NZZ flowers. Although Pro35S:SPL/NZZ flowers produced abnormal sepals (Fig. 3A), we have not detected carpel-like structures in whorl 1 organs of Pro35S:SPL/NZZ and ap2-1 Pro35S:SPL/NZZ flowers. Hence, SPL/NZZ seemed to have no carpel identity function. It is possible that the expression level of AG might not reach the threshold for promoting the formation of carpel-like structures in sepal-like organs of both Pro35S:SPL/NZZ and ap2-1 Pro35S:SPL/NZZ flowers. The other reason could be that AG was not ectopically expressed in whorl 1. In ag-1 flowers, ectopic expression of SPL/NZZ is able to induce microsporogenesis in whorl 3 petal-like structures, which, however, lack normal anther structure and filament morphology (Ito et al., 2004). In this study, we did not detect narrow petals in the second whorl of ag-1 Pro35S:SPL/NZZ flowers. Although the third whorl stamens in spl ag-4 double mutant flowers showed petal identity, the null SPL/NZZ mutants appeared normal in stamen identity. Thus, SPL/NZZ should not play an equal role to AG in specifying stamen identity. The expression of SPL/NZZ coincides with the expression of AG during early anther development, when anther cell division and differentiation actively occur (Yanofsky et al., 1990; Bowman et al., 1991a; Drews et al., 1991; Ito et al., 2004; Xing and Zachgo, 2008). After the anther structure is established, AG is expressed in anther connective tissues and anther walls (Bowman et al., 1991a; Ito et al., 2004). Therefore, SPL/NZZ may contribute to maintaining the expression of AG during anther development.
The spatial and temporal specificity of AG expression is achieved by both positive and negative regulators. The transcription factor LEAFY (LFY) is a key positive regulator of AG, which binds to cis-elements in an unusually large intron of AG to activate its expression (Schultz and Haughn, 1991; Weigel et al., 1992; Parcy et al., 1998; Busch et al., 1999). In addition, the homeodomain transcription factor WUSCHEL activates the expression of AG in the center of developing flowers via binding to the same regulatory region as LFY (Mayer et al., 1998; Lenhard et al., 2001; Lohmann et al., 2001). Also, several other positive regulators of AG have been identified with enhancer screenings. The ag-4 enhancers HUA1 and HUA2, as well as HUA ENHANCER4, are involved in facilitating the processing of AG pre-mRNA, particularly splicing the large intron (Chen and Meyerowitz, 1999; Li et al., 2001; Cheng et al., 2003). The expression of AG is decreased after stage 6 in hen2-1 hua1-1 hua2-1 flowers, suggesting that HEN2 is important for maintaining the expression of AG (Western et al., 2002). Recent studies found that the bZIP transcription factor PERIANTHIA directly activates the expression of AG (Das et al., 2009; Maier et al., 2009).
AG expression is also negatively regulated by repressors. AP2 represses the expression of AG in outer whorls, possibly by binding to the AG large intron (Drews et al., 1991; Mizukami and Ma, 1992; Jofuku et al., 1994; Sieburth and Meyerowitz, 1997; Bomblies et al., 1999). Moreover, microRNA172 acts as a negative regulator of AP2 (Aukerman and Sakai, 2003; Chen, 2004; Zhao et al., 2007). LUG and SEU serve as corepressors to form a transcriptional repressor complex of AG together with DNA-binding transcription factors, including BELLRINGER, AP1, SEP3, and AGAMOUS-LIKE 24 (Liu and Meyerowitz, 1995; Conner and Liu, 2000; Bao et al., 2004; Sridhar et al., 2004, 2006; Franks et al., 2006; Gregis et al., 2006). Furthermore, polycomb group proteins CLF and EMBRYONIC FLOWER2, as well as the plant-specific protein EMF1, repress the expression of AG epigenetically (Goodrich et al., 1997; Sieburth and Meyerowitz, 1997; Aubert et al., 2001; Yoshida et al., 2001; Calonje et al., 2008).
Our results suggest that SPL/NZZ induces the expression of AG via SEP3. Both ectopic expression and overexpression of SPL/NZZ caused the increased expression of AG. Furthermore, ectopic expression of SPL/NZZ could directly induce the expression of SEP3. Previous studies showed that the ectopic expression of SEP3 is sufficient to ectopically activate AG (Castillejo et al., 2005). Depending on regulatory conditions, SEP3 may play a positive or negative role in regulating the expression of AG (Sridhar et al., 2006). Very recently, studies using the ChIP-Seq technique revealed that the SEP3 binding site is located in the second intron of AG (Kaufmann et al., 2009). AG controls early stamen development by directly activating the expression of SPL/NZZ (Ito et al., 2004). Our results suggest that the positive feed-forward regulation between AG and SPL/NZZ plays an important role in stamen identity specification and stamen morphogenesis.
It is worthwhile to point out that ectopic expression and overexpression of SPL/NZZ led to the decreased expression of AP2. AG prevents the expression of AP1 in whorls three and four of wild-type flowers (Gustafson-Brown et al., 1994). However, no report has shown that AG represses AP2 (Jofuku et al., 1994; Chen, 2004). In spl/nzz anthers, the expression of AP2 is increased 5-fold, suggesting that SPL/NZZ may repress the expression of AP2 in the third whorl (Wijeratne et al., 2007). Therefore, it might be possible that SPL/NZZ plays a direct role in negatively regulating the expression of AP2. Further studies of interactions among SPL/NZZ, AG, SEP3, and AP2 as well as identifying the target genes of SPL/NZZ should lead to a better understanding of anther development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants are in the Landsberg erecta (Ler) background, except for the spl-D mutant, which is in the Columbia (Col-0) ecotype. To construct the spl ag-4 double mutant, pollen from the ag-4 mutant was used to pollinate spl heterozygous plants. In the F2 generation, plants exhibiting the ag-4 flower phenotype were genotyped for the spl mutant by PCR (Supplemental Table S1; Yang et al., 1999; Zhao et al., 2002). The Pro35S:SPL/NZZ-GR line was generated in the Meyerowitz Laboratory (Ito et al., 2004). Plants were grown on Metro-Mix 360 soil at 22°C under a 16-h-light/8-h-dark cycle.
Vector Construction and Plant Transformation
The cDNA of SPL/NZZ was amplified by Phusion High-fidelity DNA polymerase (Supplemental Table S1; New England Biolabs) and then was cloned into the pENTR TOPO vector (Invitrogen). After verification by sequencing, the cDNA fragment was introduced into the Gateway binary vector (a gift from Dr. T. Nakagawa, Shimane University) by an LR recombination reaction using Gateway LR Clonase II enzyme mix (Invitrogen). The resulting construct was introduced into Agrobacterium tumefaciens strain GV3101. Wild-type, ap2-1 mutant, and ag-1 heterozygous plants were transformed. T0 seeds were screened for transformants on half-strength Murashige and Skoog agar plates containing 50 mg L−1 kanamycin and 50 mg L−1 hygromycin (Clough and Bent, 1998). The Pro35S:AG construct was kindly provided by Dr. Hong Ma (Penn State University).
DEX Treatment
The Pro35S:SPL/NZZ-GR T3 seedlings were screened on half-strength Murashige and Skoog plates containing 10 μg mL−1 phosphinothricin. The phosphinothricin-resistant seedlings were transferred to soil. Seedlings were then treated with a DEX solution containing 10 μm DEX and 0.015% Silwet L-77 and a mock solution (0.015% Silwet L-77 and the same concentration of ethanol used for dissolving DEX; Wagner et al., 1999; Ito et al., 2004). Rosette leaves were collected for total RNA extraction after 6 h, 12 h, 3 d, and 6 d. Inflorescences of nzz-2 Pro35S:SPL/NZZ-GR and ag-1 Pro35S:SPL/NZZ-GR plants were treated similarly for 4 h.
Phenotype Analyses and Microscopy
Micrographs were taken with an Olympus DP70 digital camera through a stereomicroscope (Olympus SZX-RFL). Samples for SEM were fixed, dried, dissected, and coated as described previously (Bowman et al., 1989; Zhao et al., 2001b). Specimens were then examined using a Hitachi S-570 scanning electron microscope.
RT-PCR and Quantitative Real-Time RT-PCR
Two-week-old rosette leaves and young buds from Ler, Col-0, Pro35S:SPL/NZZ, spl-D, and ag-1 Pro35S:SPL/NZZ-GR plants were collected. To examine gene expression in sepals and petals, about 500 Ler and Pro35S:SPL/NZZ young buds were dissected, and then sepals and petals were collected using a dissection microscope (Olympus SZ51). Total RNAs were extracted from different plant tissues using the RNeasy Plant Mini Kit (Qiagen). RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer. RT reactions were carried out using the QuantiTect Reverse Transcription Kit (Qiagen).
Primers for regular RT-PCR are listed in Supplemental Table S1. PCR cycles varied with expression levels for examined genes: for ACTIN2, 21 cycles in rosette leaves, flowers, sepals, and petals; for SPL/NZZ, 28 cycles in rosette leaves, flowers, sepals, and petals; for AG, 33 cycles in leaves, sepals, and petals and 29 cycles in flowers; for CLF, 33 cycles in leaves; for AP1, 28 cycles in flowers and 26 cycles in sepals and petals; for AP2, 28 cycles in flowers, sepals, and petals; for AP3, 26 cycles in flowers and 30 cycles in sepals and petals; for SEP3, 34 cycles in leaves and 28 cycles in flowers, sepals, and petals.
Quantitative real-time PCR was performed with a DNA Engine Opticon 2 system (Bio-Rad) using Fast SYBR Green PCR Master Mix (Applied Biosystems). The ACTIN2 gene was used as a control. The quantitative RT-PCR results were analyzed as described previously (Pfaffl et al., 2002). Three independent experiments were repeated. Each value indicates the average and se.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf phenotypes.
Supplemental Figure S2. Semiquantitative RT-PCR.
Supplemental Table S1. Primer list.
Supplementary Material
Acknowledgments
We thank S. Forst, D. Heathcote, and C. Starrett for technical assistance and critical comments on the manuscript, Y. Wang for helping with SEM, T. Schuck for plant care, and T. Nakagawa for providing Gateway binary vectors. We also thank E.M. Meyerowitz for providing the Pro35S:SPL/NZZ-GR line, H. Ma for providing the Pro35S:AG construct, and L. Qu for providing the spl-D mutant.
This work was supported by the National Science Foundation (grant no. IOS–0721192 to D.Z.), the Research Growth Initiative Program at the University of Wisconsin-Milwaukee (to D.Z.), the Shaw Scientist Award from the Greater Milwaukee Foundation (to D.Z.), and the American Society of Plant Biologists Summer Undergraduate Research Fellowship (to A.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dazhong Zhao (dzhao@uwm.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM (2007) Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol 145 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D, Chen L, Moon YH, Martin D, Castle LA, Yang CH, Sung ZR (2001) EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z (2004) Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Dagenais N, Weigel D (1999) Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev Biol 216 260–264 [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D (2009) Male gametophyte development: a molecular perspective. J Exp Bot 60 1465–1478 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM (1991. a) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991. b) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112 1–20 [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285 585–587 [DOI] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen L, Sung ZR (2008) EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12 1718–1727 [DOI] [PubMed] [Google Scholar]
- Castillejo C, Romera-Branchat M, Pelaz S (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J 43 586–596 [DOI] [PubMed] [Google Scholar]
- Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Meyerowitz EM (1999) HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell 3 349–360 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Kato N, Wang W, Li J, Chen X (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Liu Z (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 97 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Ito T, Wellmer F, Vernoux T, Dedieu A, Traas J, Meyerowitz EM (2009) Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136 1605–1611 [DOI] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14 1935–1940 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002 [DOI] [PubMed] [Google Scholar]
- Feng X, Dickinson HG (2007) Packaging the male germline in plants. Trends Genet 23 503–510 [DOI] [PubMed] [Google Scholar]
- Franks RG, Liu Z, Fischer RL (2006) SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224 801–811 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–51 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF (1994) Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76 131–143 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1988) Genetic control of morphogenesis in Arabidopsis. Dev Genet 9 73–89 [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Kohler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529 [DOI] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM (2007) The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19 3516–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 356–360 [DOI] [PubMed] [Google Scholar]
- Jack T (2004) Molecular and genetic mechanisms of floral control. Plant Cell (Suppl) 16 S1–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Liu X, Owen HA, Zhao D (2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acad Sci USA 105 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7 e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814 [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13 2269–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Qin GJ, Tsuge T, Hou XH, Ding MY, Aoyama T, Oka A, Chen Z, Gu H, Zhao Y, et al (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179 751–764 [DOI] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121 975–991 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803 [DOI] [PubMed] [Google Scholar]
- Lu XC, Gong HQ, Huang ML, Bai SL, He YB, Mao X, Geng Z, Li SG, Wei L, Yuwen JS, et al (2006) Molecular analysis of early rice stamen development using organ-specific gene expression profiling. Plant Mol Biol 61 845–861 [DOI] [PubMed] [Google Scholar]
- Ma H (1994) The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev 8 745–756 [DOI] [PubMed] [Google Scholar]
- Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56 393–434 [DOI] [PubMed] [Google Scholar]
- Ma J, Duncan D, Morrow DJ, Fernandes J, Walbot V (2007) Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell types. Plant J 50 637–648 [DOI] [PubMed] [Google Scholar]
- Ma J, Skibbe DS, Fernandes J, Walbot V (2008) Male reproductive development: gene expression profiling of maize anther and pollen ontogeny. Genome Biol 9 R181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AT, Stehling-Sun S, Wollmann H, Demar M, Hong RL, Haubeiss S, Weigel D, Lohmann JU (2009) Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136 1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815 [DOI] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell (Suppl) 16 S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R, Verelst W, Theissen G (2009) The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res 37 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM, Bowman JL, Brockman LL, Drews GN, Jack T, Sieburth LE, Weigel D (1991) A genetic and molecular model for flower development in Arabidopsis thaliana. Dev Suppl 1 157–167 [PubMed] [Google Scholar]
- Mizukami Y, Ma H (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71 119–131 [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50 751–766 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395 561–566 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11 297–322 [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW (1991) LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16 S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM (1995) Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133 3159–3166 [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584 [DOI] [PubMed] [Google Scholar]
- Walbot V, Evans MM (2003) Unique features of the plant life cycle and their consequences. Nat Rev Genet 4 369–379 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78 203–209 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Cheng Y, Liu J, Chen X (2002) HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis. Development 129 1569–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H (2007) Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J 52 14–29 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60 1479–1492 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53 790–801 [DOI] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39 [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D (2009) Control of anther cell differentiation: a teamwork of receptor-like kinases. Sex Plant Reprod (in press) [DOI] [PubMed]
- Zhao D, Wang GF, Speal B, Ma H (2002) The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Yu Q, Chen C, Ma H (2001. a) Genetic control of reproductive meristems. In MT McManus, B Veit, eds, Annual Plant Reviews: Meristematic Tissues in Plant Growth and Development. Sheffield Academic Press, Sheffield, UK, pp 89–142
- Zhao D, Yu Q, Chen M, Ma H (2001. b) The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128 2735–2746 [DOI] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X (2007) miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J 51 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003) Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.