Abstract
Zebrafish teeth develop on pharyngeal jaws in the 5th branchial arch, but early tooth development is remarkably similar to mammals (Borday-Birraux et al., Evol Dev 8:130, 2006). Recently, eve1 has been shown to be associated with the primary tooth (4V1) and early ameloblast development, the enamel organ precursor (Laurenti et al., Dev Dyn 230:727, 2004). dax1 is initially expressed in the 5th branchial arch in zebrafish at approximately 26 h postfertilization (hpf) and colocalizes with eve1 expression at ~48 hpf. Embryos injected with dax1 morpholino show downregulation of eve1 expression. Based on the zebrafish observations, we demonstrated novel DAX1 expression in normal human dental, benign ameloblastoma, and malignant ameloblastoma tissues. The association of NR0B1 and its protein product DAX1 with primary tooth development and ameloblastoma tumorigenesis is an association not previously described.
Keywords: dax1, Tooth, Ameloblasts, Ameloblastoma
Introduction
NR0B1 and its protein product DAX1 are expressed in the hypothalamic–pituitary–adrenal/gonadal axis, and mutations in the gene encoding this transcription factor result in adrenal hypoplasia congenital and hypogonadotropic hypogonadism (Guo et al. 1995; Ikeda et al. 1996). Zhao et al. (2000) recently identified and described the spatial and temporal expression patterns of the zebrafish ortholog dax1. These in situ hybridization (ISH) studies on zebrafish embryos revealed dax1 expression in a bilateral structure cephalad and medial to the pectoral fins in the region of the 5th branchial arch, appearing around 26 h postfertilization (hpf), peaking about 32 hpf, and nearly lost by 72 hpf. The 5th branchial arch and its association with tooth development in the zebrafish model have been well characterized in numerous other studies (Jackman et al. 2004; Van der Heyden et al. 2000). ISH with dax1 and previously identified tooth markers colocalized these mRNAs to the area of pharyngeal tooth development. The association of dax1 and the 5th branchial arch prompted us to search for a possible link between zebrafish tooth development and this orphan nuclear receptor.
Pitx2, a DNA- and RNA-binding homeodomain protein, is responsible for normal heart morphogenesis, mandibular and maxillary development, and normal pituitary and tooth development (Cox et al. 2002). There are three major isoforms found in human, mouse, chicken, frog, and zebrafish. pitx2a, until this report, was the earliest known marker of tooth development in zebrafish (Jackman et al. 2004). The gene begins expression at 36 hpf and is found in all developing teeth. No previously known tooth marker has been shown to be expressed in zebrafish before pitx2a. lhx6 is expressed in the mesenchyme of the developing tooth germ as early as 28 hpf; however, the role of this gene in tooth development is unclear (Jackman et al. 2004). Previous double ISH showed pitx2a and dax1 to have adjacent expression (Zhao et al. 2000), suggesting a possible role for dax1 in tooth development.
The role of the Dlx/dlx family of genes is well described in mouse and zebrafish (Borday-Birraux et al. 2006; Zhao et al. 2006), and mutations in this gene family are known to cause severe disorders in human dentition (Green et al. 2001). Dlx2 is the earliest known tooth marker in mice (Thomas et al. 2000). Double ISH of dlx2a and dlx2b with dax1 shows adjacent expression in the region of the 5th branchial arch in zebrafish (Zhao et al. 2006, doctoral thesis). Pitx2 and Dlx2 are expressed in the same tissues in early development. Green et al. (2001) recently described the function of Pitx2 as an upstream regulator of Dlx2 expression. Since Pitx2 is known to be an upstream regulator of Dlx2 activity and dax1 and pitx2a do not appear to be closely related developmentally, it is unlikely that dax1 affects expression of dlx2a or dlx2b.
eve1 is the closest paralog to the even-skipped-related (evx) class of genes, which are homeodomain-containing transcription factors with varying expression patterns in different species (Laurenti et al. 2004). The zebrafish genome contains three evx genes, evx1, evx2, and eve1. In zebrafish, evx1 and evx2 are responsible for anterior–posterior axis formation (Avaron et al. 2002). The human genome contains paralogs for only two of these genes, EVX1 and EVX2. There is no known ortholog to eve1 in mammals. Laurenti et al. (2004) recently discovered that the gene eve1 is expressed in the placode of the first primary tooth, 4V1, in zebrafish, suggesting it may be involved in early tooth initiation (Laurenti et al. 2004). eve1 is not expressed in early odontogenesis, like pitx2 and dlx2 isoforms, but expressed at 48 hpf and is restricted only to initiation of the first tooth and early differentiation of ameloblasts, the enamel organ precursor. The EVX gene family has not been previously shown to be involved in mammalian tooth development. Single ISH of eve1 in zebrafish embryos showed a paired structure near the 5th branchial arch, similar to the expression pattern seen with dax1.
The proximity of the expression of dax1 to 5th branchial arch and numerous known tooth markers led us to hypothesize that dax1 is involved in pharyngeal tooth development in the zebrafish.
Materials and methods
Wild-type zebrafish (AB, Tübingen, or SS) were maintained at 28.5°C. Embryos were obtained by natural spawning and cultured following standard described procedures (Westerfield 2000). Of phenylthiourea (Sigma), 0.03% was added to the culture medium at approximately 6 hpf to inhibit pigment formation.
cDNA transformation
Constructs used
cDNA construct for dax1 was isolated by Dr. Yan Zhao from the sequence from the Sanger Centre zebrafish genome database (http://www.sanger.ac.uk/Projects/D_rerio/; Zhao et al. 2000).
The following cDNA/expressed sequence tag (EST) constructs were purchased from Open Biosystems (http://www.openbiosystems.com/) based on their accession numbers. The gene sequence was obtained from The Zebrafish Information Network (www.zfin.org) and a Basic Local Alignment Search Tool alignment of the gene of interest and available constructs was performed to confirm the identity:
dlx2b: Catalog # MDR1738-8985435; Vector name: pBluescript SK+
pitx2a: Catalog # EDR1052-7472648; Vector name: pExpress-1
The following ESTs were purchased from the Zebrafish International Resource Center (http://zebrafish.org/zirc/home/guide.php):
dlx2a: Catalog # ZDB-EST-051103-34; Vector name: pSportI
eve1: Catalog # ZBD-EST-cb872; Vector name: pBSII-SK+
eve1 and dlx2a were transformed according to the protocol as supplied by Stratagene. DNA was isolated using the Mini-Prep Kit and protocol as supplied by Qiagen. The constructs were linearized by restriction enzyme digestion and the products were purified using a Gel Extraction Kit (Bioland).
Probe synthesis for whole-mount in situ hybridization
rNPT mix consists of 20 μl 10 mM ATP, 20 μl 10 mM GTP, 20 μl 10 mM CTP, 13 µl 10 mM UTP, and 7 μl 10 mM digoxigenin-11-UTP (Boehringer Mannheim). The reaction components for probe synthesis for dax1, eve1, pitx2a, dlx2a, and dlx2b were set up as follows: 500-ng linearized DNA template, 5 µl 5× transcription buffer (Promega), 5 μl rNPT mix, 2.5 µl 100 mM dithiothreitol (Promega), 1.0 µl RNasin (Promega), 1.0 µl RNA polymerase (Promega), and nuclease-free water (Ambion) to 25 µl total volume. Probe synthesis was carried out according to standard described procedure (Darby and Hewitson 2006).
Whole-mount in situ hybridization
Solutions:
Hyb−: 50% formamide, 5× standard sodium citrate (SSC), 0.1% Tween-20
hyb+: hyb−, 500 μg/ml torula yeast RNA, 500 μg/ml heparin
Maleic acid buffer: 100 mM maleic acid (Sigma M0375), 150 mM NaCl, pH 7.5
Re-equilibration buffer: 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, Tris-HCl, pH 9.5
Embryos were collected at various stages of development, dechorionated by pronase treatment (2 mg/ml, 3–10 min at room temperature), fixed with 4% paraformaldehyde diluted in 1× phosphate buffered saline (PBS), and allowed to incubate for at least 2 h at room temperature or overnight at 4°C. The embryos were dehydrated in sequential steps for 5 min at room temperature as follows: 25% methanol/75% PBS plus Tween-20 (PBST), 50% methanol/50% PBST, 75% methanol/25% PBST, and 100% methanol. The embryos were placed at −20°C at least overnight prior to use. When ready for ISH, the embryos were sequentially rehydrated for 5 min at room temperature in each of the following: 75% methanol/25% PBST, 50% methanol/50% PBST, 25% methanol/75% PBST, and 100% PBST. Single and double ISH studies were performed according to the protocol outlined by Darby and Hewitson (2006).
Morpholino injection
Danieau’s solution consists of 58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, and 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.6)
Sequences:
dax1 Morpholino (MO) 2: 5′-CAGAGCTGCTTATGTTTCGTTGCTG-3′
dax1 Morpholino 3: 5′-CTGAGCTGCACGCTTGCACATGATC-3′
dax1 mismatch control (mMO): 5′-CAcAGgTGCTTATcTTTCcTTGgTG-3′
Morpholinos were synthesized and purchased from Gene Tools (http://www.gene-tools.com/). A stock solution of 10 μg/μl was prepared by dissolving the lyophilized powder in doubly distilled water. The stock solution was diluted to a working concentration of 2 μg/μl in 1× Danieau’s solution. Freshly laid embryos were collected and placed into embryo culture medium (brackish water) and transferred onto an agarose platform. Ten nanograms of MO2, MO3, or mMO solution was injected in the yolk of one- to two-cell embryos, or four- to eight-cell embryos, from the vegetal pole. Microinjection was performed using a Nanoject injector.
Deparaffinization and ISH on human tissue samples
Solutions:
Denhardt’s solution 100× stock: distilled H2O 500 ml total volume, Ficoll 10 g, polyvinylpyrrolidone 10 g, bovine serum albumin 10 g
Hybridization buffer: distilled H2O 200 ml total volume, formamide 100 ml, dextran sulfate 20 g, SSC (20 stock) 40 ml, Denhardt’s solution (100×) 5 ml, salmon sperm DNA 0.05 g, yeast transfer RNA 0.12 g, SDDS 0.05 g, Dig-block reagent 0.2 g
After obtaining institutional review board exemption, we obtained 5-μm-thick formalin-fixed, paraffin-embedded human tissue samples of normal adrenal gland, normal dental tissue, benign ameloblastoma tumor, and malignant ameloblastoma tumor cut from previously excised tissue using steel microtome blade from the UCLA Tissue Core and Pathology Laboratory. The tissue sections were deparaffinized according to standard described protocol (Abcam). ISH was then performed according to the protocol provided by Roche Applied Science.
Results
ISH using digoxigenin-labeled antisense riboprobes for pitx2a showed that its expression pattern remained identical in dax1 MO2- and dax1 MO3-injected embryos compared to control embryos at all studied stages of development (Fig. 1). These data suggest that dax1 is not an upstream regulator of pitx2a expression.
Fig. 1.
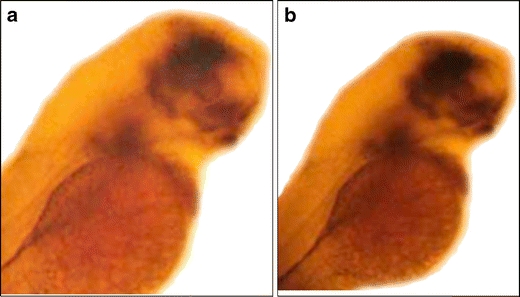
Single ISH of pitx2a at 48 hpf. a Wild-type control embryo showing expression of pitx2a in the developing brain and 5th branchial arch where pharyngeal tooth development occurs. b dax1 MO2-injected embryo showing unchanged expression of pitx2a. dax1 MO3-injected embryos showed a similar unchanged pitx2a expression pattern
Since Pitx2a is a known upstream regulator of the Dlx isoforms and since our results showed that dax1 is not an upstream regulator of pitx2a, we hypothesized that dax1 MO should have no effect on the expression patterns of dlx2a and dlx2b. ISH confirmed our hypothesis by showing no difference in expression of dlx2a or dlx2b on embryos injected with dax1 MO2 or dax1 MO3 compared to the uninjected control embryos (results not shown).
Double ISH studies of eve1 and dax1 showed colocalization of expression in the region of the 5th branchial arch starting at 48 hpf (Fig. 2). Double ISH of eve1 and dax1 at 60 and 72 hpf showed similar results (results not shown). ISH studies showed complete lack of expression of eve1 in the majority of dax1 MO2- and dax1 MO3-injected embryos compared to the control mismatch MO-injected embryos at 48 hpf (Fig. 3). Again, similar results were also seen at 60 and 72 hpf, respectively (results not shown). When the embryos were injected before or during the two-cell stage, nearly all the MO-injected embryos showed complete lack of eve1 expression. However, when we injected embryos in the four- to eight-cell stage, when the MO does not disperse evenly between developing cells to inhibit expression, there was no statistically significant difference in the expression between the MO-injected and mismatch MO-injected embryos (Table 1).
Fig. 2.
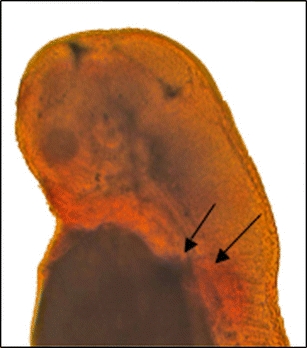
Double ISH of dax1 and eve1. The upper arrow shows purple-staining eve1 expression while the bottom arrow shows red-staining dax1
Fig. 3.
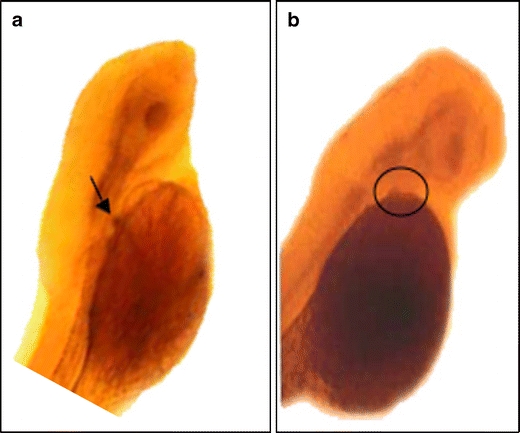
ISH of eve1 on 48-hpf embryos. a Arrow shows eve1 in the 5th branchial arch in control wild-type embryo. b Circle shows absence of eve1 expression in a dax1 MO-injected wild-type embryo
Table 1.
A statistically significant difference in the number of embryos exhibiting expression of eve1 in mismatch MO-injected embryos as compared to dax1 MO2- and dax1 MO3-injected embryos
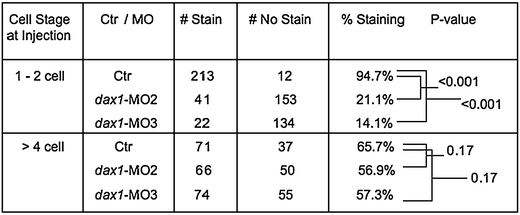
When embryos are injected early in embryogenesis, e.g., one- to two-cell stage, the MO disperses evenly between the cells inhibiting translation of the target mRNA. Injection later in embryogenesis, e.g., more than four-cell stage, does not allow for even dispersion of MO resulting in incomplete inhibition of translation
Ctr mismatch MO
Although no ortholog for eve1 exists in humans, we hypothesized that DAX1 may be involved in primary tooth development and amelogenesis. ISH studies with DAX1 on normal human dental tissue and benign and malignant ameloblastoma tumors were performed. Normal dental tissue showed weak expression of DAX1 in the dental epithelium (Fig. 4a), while benign and malignant ameloblastoma tumors showed strong expression of DAX1 in the clusters of ameloblasts, with higher expression in malignant tumor tissue (Fig. 4b, c) .
Fig. 4.
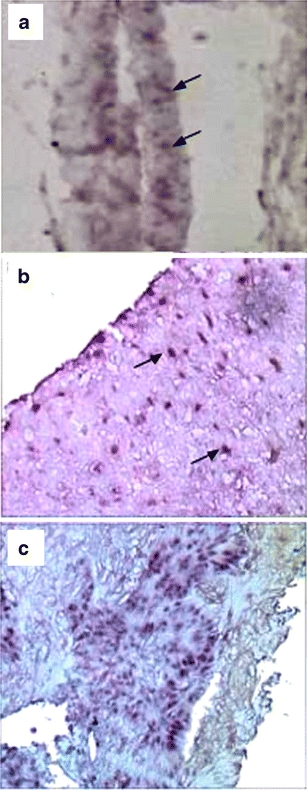
ISH of DAX1 on human dental tissue specimens. a Normal dental epithelium with the developing enamel organ. Arrows indicate ameloblasts. b Benign ameloblastoma tumor. Arrows indicate purple-stained ameloblasts expressing DAX1. c Malignant ameloblastoma tumor with numerous clusters of purple-stained ameloblasts infiltrating normal dental tissue
Discussion
Here, we illustrate another possible novel function of nr0b1 as the earliest marker of zebrafish tooth development. Its expression in the 5th branchial arch 10 h earlier in embryogenesis compared to the previously described pitx2a, appearing around 36 hpf, and 22 h earlier than eve1 suggests that it may be involved in the regulation of tooth development and amelogenesis. This difference is remarkable, considering that zebrafish embryogenesis is complete by 72 h. It is yet to be determined why dax1 expression occurs so early in this region as the primary tooth in zebrafish does not start to form until 36 hpf or appear until 48 hpf. This may speak to the complexity of gene product interactions required to initiate and to continue tooth development in this polyphyodont model.
It remains to be shown whether tooth development occurs in dax1 MO-injected embryos. It also remains to be shown if dax1 and eve1 expression occurs in the same versus adjacent cells. Laurenti et al. (2004) showed eve1 expression in the placode of the first developing tooth at 48 hpf followed by limited expression in differentiating ameloblasts in zebrafish at 72 hpf. Human tissue sections show DAX1 expression in ameloblasts in normal dental tissue and ameloblastoma tumor cells.
The expression pattern of dax1 in zebrafish, including the expression of dax1 in the 5th branchial arch, has been well described by numerous previous authors (Bertrand et al. 2007; Thisse and Thisse 2008; Zhao et al. 2000). None of the previous studies identified a role of dax1 expression in developing cartilage. Lack of evidence from previous studies of dax1 involvement in cartilage formation or differentiation supports our inference that dax1 is likely involve in tooth formation, the only know function of the 5th branchial arch in zebrafish, as opposed to cartilage involvement and that eve1 and dax1 expression occurs in the same cells.
Numerous previous studies have identified functions of NR0B1 outside the HPAG axis. Niakan et al. (2006) showed in the mouse model that Dax1 plays a role in maintaining early embryonic stem cells in a less-differentiated state. Mendiola et al. (2006) showed that DAX1 is upregulated by the EWS/FLI1 oncoprotein in Ewing sarcoma tumors. DAX1 expression in normal human teeth as well as ameloblastoma tumors was a previously unidentified association of this dynamic gene product. The use of the zebrafish model vertebrate has allowed us to elucidate not only a possible gene function that had gone unnoticed but also a marker for the most common odontogenic tumor, the ameloblastoma.
Acknowledgments
We would like to thank the Translational Pathology Core Laboratory, UCLA Department of Pathology, David Geffen School of Medicine at UCLA for providing us with the tissue specimens.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Avaron F, Thaeron-Antono C, Beck CW, Borday-Barraux V, Geraudie J, Casane D, Laurenti P. Comparison of even-skipped related gene expression pattern in vertebrates shows an association between expression domain loss and modification of selective constraints on sequences. Evol Dev. 2002;5:145. doi: 10.1046/j.1525-142X.2003.03021.x. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escrivà H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3(11):e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borday-Birraux V, Van der Heyden C, Debiais-Thibaud M, Verreijdt L, Stock DW, Huysseune A, Sire JY. Expression of Dlx genes during the development of the zebrafish pharyngeal dentition: evolutionary implications. Evol Dev. 2006;8:130. doi: 10.1111/j.1525-142X.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Cox C, Espinoza H, McWilliams B, Chappell K, Morton L, Hjalt T, Semina E, Amendt B. Differential regulation of gene expression by PITX2 isoforms. J Biol Chem. 2002;277:28. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- Darby I, Hewitson T. In situ hybridization protocols. New Jersey: Humana; 2006. [Google Scholar]
- Green PD, Hjalt TA, Kirk DE, Sutherland LB, Thomas BL, Sharpe PT, Snead ML, Murray JC, Russo AF, Amendt BA. Antagonistic regulation of Dlx2 expression by PITX2 and Msx2: implications for tooth development. Gene Expr. 2001;9:265. doi: 10.3727/000000001783992515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Burris TP, McCabe ER. Expression of DAX-1, the gene responsible for X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism, in the hypothalamic-pituitary-adrenal/gonadal axis. Biochem Mol Med. 1995;56:8. doi: 10.1006/bmme.1995.1049. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Swain A, Weber TJ, Hentges KE, Zanaria E, Lalli E, Tamai KT, Sassone-Corsi P, Lovell-Badge R, Camerino G, Parker KL. Steroidogenic factor 1 and Dax-1 colocalize in multiple cell lineages: potential links in endocrine development. Mol Endocrinol. 1996;10:1261. doi: 10.1210/me.10.10.1261. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Laurenti P, Thaeron C, Allizard F, Huysseune A, Sire JY. Cellular expression of eve1 suggests its requirement for the differentiation of the ameloblasts and for the initiation and morphogenesis of the first tooth in the zebrafish (Danio rerio) Dev Dyn. 2004;230:727. doi: 10.1002/dvdy.20080. [DOI] [PubMed] [Google Scholar]
- Mendiola M, Carillo J, Garcia E, Lalli E, Hernandez T, de Alava E, Tirode F, Delattre O, Garcia-Miguel P, Lopez-Barea F, Pestana A, Alonso J. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int J Cancer. 2006;118:1381. doi: 10.1002/ijc.21578. [DOI] [PubMed] [Google Scholar]
- Niakan K, Davis E, Clipsham R, Jiang M, Dehart D, Sulik K, McCabe ERB. Novel role for the orphan nuclear receptor Dax1 in embryogenesis, different from steroidogenesis. Mol Genet Metab. 2006;88:261. doi: 10.1016/j.ymgme.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B (2008) Expression from: unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. ZFIN direct data submission [DOI] [PMC free article] [PubMed]
- Thomas BL, Liu JK, Rubenstein JL, Sharpe PT. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- Van der Heyden C, Huysseune A, Sire JY. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae) Dev Dyn. 2000;219:486. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: guide for the laboratory use of zebrafish (Danio rerio) 4. Eugene: University of OR Press; 2000. [Google Scholar]
- Zhao Z, Stock D, Buchanan A, Weiss K. Expression of Dlx genes during the development of the murine dentition. Dev Genes Evol. 2000;210:270. doi: 10.1007/s004270050314. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang Z, Phelan JK, Wheeler DA, Lin S, McCabe ER. Zebrafish dax1 is required for development of the interrenal organ, the adrenal cortex equivalent. Mol Endocrinol. 2006;20:2630. doi: 10.1210/me.2005-0445. [DOI] [PubMed] [Google Scholar]


