Abstract
Hippocampal atrophy rates are useful in both diagnosing and tracking Alzheimer’s disease (AD). However, cohorts and methods used to determine such rates are heterogeneous, leading to differences in reported annualised rates. We performed a meta-analysis of hippocampal atrophy rates in AD patients and matched controls from studies reported in the peer-reviewed literature. Studies reporting longitudinal volume change in hippocampi in AD subjects together with controls were systematically identified and appraised. All authors were contacted either to confirm the results or to provide missing data. Meta-analysis and meta-regression were then performed on this data. Nine studies were included from seven centres, with data from a total of 595 AD and 212 matched controls. Mean (95% CIs) annualised hippocampal atrophy rates were found to be 4.66% (95% CI 3.92, 5.40) for AD subjects and 1.41% (0.52, 2.30) for controls. The difference between AD and control subject in this rate was 3.33% (1.73, 4.94).
Keywords: Meta-analysis, Hippocampus, Atrophy, AD, Alzheimer’s, Rates, Longitudinal
1. Introduction
Alzheimer’s disease (AD) is a large and growing problem with increasing financial and social burdens to the individual, carers and society. AD affects over 5% of the population over 60 years (Dawbarn and Allen, 2001) and its prevalence doubles every 5–10 years above that age (Small et al., 1997). A definitive diagnosis of AD can only be given following pathologic examination of the brain, usually at post-mortem. The disease is pathologically characterised by the presence of microscopic extracellular neuritic plaques and intracellular neurofibrillary tangles. AD tangle pathology progresses from medial temporal lobe structures such as the entorhinal cortex and hippocampus, to encompass the whole cortex, whereas plaque pathology is largely cortical and increases with disease severity (Braak et al., 1993). One of the results of this pathology is cerebral atrophy which can be visualised using structural MRI (Scheltens et al., 2002). The atrophy can be seen even at a single time-point, as brain structures in Alzheimer’s subjects are smaller on average compared with controls. In addition, loss of tissue volume over time can also be detected in large regions such as the whole brain (Jack et al., 2004) and in smaller temporal areas such as the hippocampus (Jack et al., 2004) and entorhinal cortex (Du et al., 2004). This has led to a number of studies that have suggested that hippocampal atrophy rates may be useful both diagnostically, and to track disease progression.
Many studies assessing longitudinal hippocampal change have been reported in the literature (Fox et al., 1996; Jack et al., 2004; Kaye et al., 2005). Different methods have been used to generate these rates of atrophy including automated (Wang et al., 2003; Du et al., 2004) and manual techniques (Jack et al., 2004; Barnes et al., 2005). In addition, different populations of patients have also been included. To the best of our knowledge, no statistical review of the hippocampal atrophy rate literature has been conducted to date. Such a review is required to assess heterogeneity of reported studies, to better understand how age and disease severity affect the calculated atrophy rates, and to pool the results from these studies to more accurately estimate the rate of atrophy of the hippocampus in AD and matched control groups. This type of analysis may also be useful in identifying outlier results where the methodology employed may deserve critical review. In addition measuring the inter-site variance in hippocampal atrophy rates may be useful for anyone planning a multi-site trial, as it may define the level of consistency to expect in the imaging measures across sites.
1.1. Study objective
The objective of this study is to perform a meta-analysis of hippocampal atrophy rates in patients with AD and matched controls from studies reported in the peer-reviewed literature.
2. Methods
The following protocol was employed in the study.
2.1. General study inclusion criteria
We aimed to identify all observational and randomised controlled trials (RCT) where hippocampal volume was measured on MRI at two or more points in time, and annualised hippocampal atrophy rate was reported for patients with AD. We included data from both arms of randomised controlled trials if symptomatic treatment was given, and only the placebo arm if a disease-modifying treatment was given as atrophy rate might have been altered by the treatment. Disease-modifying treatments were classified as those that had been shown to significantly alter whole brain atrophy rates.
We included MRI studies reporting results from patients with AD of any age or gender. There were no restrictions on the method used to diagnose AD. As MCI was not a subject group being formally assessed as part of this meta-analysis, cognitively impaired subject groups with a mean MMSE >26 were excluded. No restriction was placed on scanning protocol, strength of magnet used to image the patients or segmentation protocol used to determine the atrophy rate.
Unpublished studies were not included, unless accepted papers had been published ahead of print online, and studies where the mean annualised rates of atrophy could not be provided by the author were excluded. Since people with Down’s syndrome/Alzheimer’s disease and mutation carriers of the known AD-causing genes may have a different course of disease, studies based on their populations were also excluded.
2.2. Outcome measure
Mean (arithmetic) atrophy rate specified as % loss of baseline volume per year.
2.3. Search methods for identification of studies
Searches for relevant studies were performed using electronic and other sources.
2.3.1. Electronic sources
The published literature was searched in MEDLINE (1950 to February Week 2 2007) through OVID using the following strategy:
(hippocamp$.tw) AND ((EXP magnetic resonance imaging OR((magnetic AND resonance AND imag$).
tw OR mri$.tw)) AND ((EXP Alzheimer’s Disease/)OR Alzheimer$.tw))
Abstracts were read for all papers with relevant titles and full papers obtained and examined for all relevant abstracts. This search was extended by examining reference lists for the papers thus identified. Further papers citing these papers were examined using Web of Science (1985–2007).
2.3.2. Individual searching of journals
In addition the following journals were searched (1996–2006 unless specified) including Neurology, Annals of Neurology, Archives of Neurology, Neurobiology of Aging, American Journal of Neuroradiology, Brain, Cerebral Cortex, Cortex (1997–2006), European Journal of Neurology, Hippocampus, Journal of Neurology, Journal of Neurology, Neurosurgery and Psychiatry, Lancet Neurology (2002–2006), NeuroImage, Neuroradiology, Dementia and Geriatric Cognitive Disorders, Journal of Alzheimer’s Disease, American Journal of Alzheimer’s disease and other dementias, Alzheimer’s Disease and Associated Disorders, Lancet, PNAS, and Nature.
2.3.3. Personal communication with content experts
The list of published studies generated were shown to another researcher in this field from a different research group (LvdP) who checked whether there were any studies that may have not been included. Once the potential list was finalised between the two research groups, all authors of the studies were contacted to ensure that correct information was being used and any information that was lacking (such as MMSEs, or mean rates of atrophy) was requested. In addition, the full list was sent to each author to ensure no other studies had been missed by the searches. Published studies resulting from searches that are not in English were to be included when possible through contact with authors where necessary.
2.4. Quality assessment
After studies were selected for the meta-analysis, each was assessed for quality by tabulating variables which may introduce bias or explain heterogeneity of the results. These include: cohorts used (population, case/control, case series), blinding of raters to patient information, chronology of scan ordering, drop-out rate if RCT, assessment of and exclusion criteria for controls, AD diagnostic criteria and exclusion criteria, white matter damage exclusion criteria, white matter lesion quantification, apolipoprotein E (APOE) genotyping, other medications, magnet strength, number of scanning sites, scan “slice” thickness, hippocampal measurement method, number of raters, registered images or total intracranial volume (TIV) corrected, whole hippocampus measured and reliability of method.
2.5. Statistical analysis
STATA Version 9 (Stata Corporation, College Station, TX, USA) and SAS proc mixed Version 8 (SAS Institute Inc., Cary, NC, USA) were used for the statistical analysis. From each study, the mean atrophy rate and standard deviation were obtained for the AD patients (and separately for controls where available), together with means and standard deviations of the ages, MMSEs and inter-scan intervals in each subject group. Calculation of the standard error required for meta-analysis was based on the standard deviation and the number of patients for each study.
2.5.1. Data synthesis (meta-analysis)
A random-effects meta-analysis model was fitted using restricted maximum likelihood (REML), consistent with the assumption that mean atrophy rates measured in individual studies vary around an overall average atrophy rate. The random-effect for between-study heterogeneity was assumed to be normally distributed, as were the within-study errors. In addition, we tested the hypothesis that the random-effects were needed, by comparing the REML log likelihood ratio statistic to its asymptotic distribution under the null hypothesis that the random-effect variance is 0, which is given by a mixture of two chi-squared distributions (Morrell, 1998).
Since some studies reported results separately for different subgroups of AD patients, we attempted to fit a random-effects model that allowed for both between- and within-study variation. Unfortunately this was not possible, due to the limited number of studies that reported results separately by subgroup. In view of this the non-independence of results from subgroups of the same study was accounted for through sharing the study-specific random-effect. For studies containing both a control and an AD group, the mean difference in atrophy rates between the two groups was calculated. These differences were then entered in a further random-effects meta-analysis to estimate the overall mean within-study difference in rates between AD and control groups.
2.5.2. Assessment of reporting bias
Due to the focus of this meta-analysis, it is unlikely that studies would be denied publication on the basis of the estimated size of mean atrophy in AD patients. However, publication bias was examined with funnel plots. Funnel plots graphically demonstrate publication bias: as the precision of the studies increases, there should be a decrease in the variability of results leading to a funnel shape. Lack of symmetry implies evidence of a publication bias.
2.5.3. Subgroup analysis and investigation of heterogeneity
Heterogeneity in atrophy rates between studies may be explained by the mean age or disease severity (MMSE score) of study participants, or inter-scan interval. This was explored using the data on AD groups through use of meta-regression (Sharp, 1998). This meta-regression analysis can only reveal associations between mean age, MMSE, or scan interval and mean atrophy rates, rather than revealing associations between individual ages, MMSEs, or scan intervals and individual atrophy rates. We added age, MMSE, and scan interval separately as a linear fixed effect in our random-effects model to estimate any such associations. It was recognised that estimates of such effects may be imprecise if only a small number of studies are available and if between-study variability in such factors is small.
2.5.4. Sample size calculation
Of the studies included in the meta-analysis, those with mean scan intervals of 1 year ± 2 months were used to calculated sample sizes. In these studies there was little variation in scan interval between patients. We performed a random-effects meta-analysis (on the variance scale) of these studies’ estimated atrophy rate standard deviations, to give a pooled estimate of the standard deviation. We assumed that a drug would decrease the atrophy of the hippocampus by a certain percentage. Standard techniques were used estimate sample sizes that give 90% statistical power to detect 20 and 50% reduction in hippocampal atrophy rates (using two-sided significance test at the 0.05 level).
3. Results
Our Medline search strategy identified 535 papers, 19 of which satisfied our inclusion criteria. Eleven of these 19 papers were excluded, because: six presented overlapping data with the primary report already included (Jack et al., 1998, 2000; Mori et al., 2002; Silbert et al., 2003; Barnes et al., 2004, 2007a), three did not report annualised atrophy rate (Laakso et al., 2000; Krishnan et al., 2003; Hampel et al., 2005) and two included patients with Vascular Dementia (Cardenas et al., 2003; Mungas et al., 2005). For the eight papers thus identified (Jack et al., 2003; Wang et al., 2003; Du et al., 2004; Jack et al., 2004; Thompson et al., 2004; Fox et al., 2005; Hashimoto et al., 2005; Barnes et al., 2005), we extended the search by examining reference lists and Web of Science, which yielded no further papers. Hand searching and personal communication with content experts yielded an additional paper that does not contain the word ‘hippocampus’ in its title or abstract (Kaye et al., 2005). We were also aware of one study available online ahead of print at time of searching (Barnes et al., 2007b) which replaced (Barnes et al., 2005) owing to overlap of patient populations. The resulting studies included in this meta-analysis are displayed in Table 1. The quality of these studies is reported in Table 2, and those studies that were excluded are detailed in Table 3.
Table 1.
Studies included in the meta-analysis
| Study author, year |
Checked with author |
Research group |
Subject groups |
N | MMSE/30 at baseline mean (S.D.) <number of subjects> |
Age, years at first MRI mean (S.D.) |
Annualised loss of total volume (left plus right hippocampi) mean % year−1 (S.D.) |
Interval (years unless specified) mean (S.D.) |
|---|---|---|---|---|---|---|---|---|
| Barnes et al. (2007b) | Yes | Fox et al. | C | 19 | 29.5 (0.7) | 68.7 (7.0) | 0.32 (0.93) | 365 (5) days |
| AD | 36 | 19.4 (4.1) | 69.6 (7.3) | 4.52 (2.94) | 365 (18) days | |||
| Du et al. (2004) | Yes | Weiner et al. | C | 25 | 29.0 (1.0) | 76.8 (7.8) | 0.8 (1.7) | 2.0 (0.7) |
| AD | 20 | 21.0 (7.2) | 75.3 (7.2) | 5.9 (2.4) | 1.8 (0.7) | |||
| Fox et al. (2005) | Yes | Fox et al. | AD -placebo | 57 | 20.2 (3.5) | 70.7 (8.2) | 3.16 (3.51) | 10.9 (1.1) months |
| Hashimoto et al. (2005) | Yes | Hashimoto et al. | AD no treat | 93 | 21.6 (2.8) | 70.5 (9.1) | 5.04 (2.54) | 392 (35) days |
| AD treat | 54 | 21.8 (3.9) | 69.5 (9.5) | 3.82 (2.84) | 388 (29) days | |||
| Jack et al. (2004) | Yes | Jack et al. | C stab | 40 | 29.0 (0.9) <34> | 78.2 (7.7) | 1.4 (0.7) | 4.3 (0.7) |
| AD slow | 31 | 22.3 (4.4) <30> | 73.9 (6.9) | 3.9 (2.6) | 1.4 (0.4) | |||
| AD Fast | 33 | 19.5 (4.0) <32> | 77.8 (8.4) | 4.8 (3.2) | 1.4 (0.4) | |||
| Jack et al. (2003) | Means and S.D.s given by personal communication |
Jack et al. | AD | 192 | 20.8 (4.1) | 72.8 (7.7) | 5.5 (3.3) | 1.0 (0.1) |
| Kaye et al. (2005) | Yes | Kaye et al. | C | 88 | 28.3 (1.5) | 83.0 (7.0) | 2.2 (6.0) | 2.04 (1.42) |
| AD mild | 27 | 21.7 (4.5) | 76.1 (7.1) | 2.9 (7.8) | ||||
| AD moderate | 17 | 16.8 (6.7) | 75.1 (6.2) | 3.2 (6.8) | ||||
| Thompson et al. (2004) | Erratum in Neuroimage 2007 36,1397–8. Values here are correct |
Thompson et al. | C | 14 | 29.5 (0.9) | 71.4 (3.2) | 2.1 (2.8) | 2.6 (1.0) |
| AD | 17 | 16.8 (6.6) | 68.7 (6.8) | 6.8 (10.2) | 1.3 (0.9) | |||
| Wang et al. (2003) | Yes | Csernansky et al. | C | 26 | 29.0 (1.3) <13> | 73 (7.0) | 2.3 (1.9) | 2.2 (0.53) |
| DAT | 18 | 25.7 (3.9) <12> | 74 (4.4) | 5.1 (2.8) | 2.0 (0.37) |
Key: AD, Alzheimer’s disease; C, control; DAT, dementia of Alzheimer type; <> number of subjects on which information has been obtained; italicized: information from personal communication; C stab, stable controls; AD fast, fast progressing AD; AD slow, slow progressing AD; AD mild, mildly affected AD subjects; AD moderate, moderately affected AD subjects; AD placebo, AD subjects on placebo; AD treat, treated AD (symptomatic treatment); AD non-treat, AD not treated with symptomatic treatment.
Table 2.
Quality assessment of studies included in the meta-analysis
| Barnes et al. (2007b) | Du et al. (2004) | Fox et al. (2005) | Hashimoto et al. (2005) | Jack et al. (2004) | Jack et al. (2003) | Kaye et al. (2005) | Thompson et al. (2004) | Wang et al. (2003) | |
|---|---|---|---|---|---|---|---|---|---|
| Study type | Case series with controls | Case series with controls | Trial | Case series | Case series with controls | Trial | Case series with controls | Case series with controls | Case series with controls |
| AD Diagnostic criteria (imaging used in criteria) |
NINCDS–ADRDA | NINCDS–ADRDA (imaging used to exclude other neuropathol.) |
NINCDS–ADRDA (imaging supporting AD) |
NINCDS–ADRDA (MMSE >15) |
NINCDS–ADRDA/DSM IIIR (general committee consensus) |
Mild–moderate AD (>50 years old) |
NINCDS–ADRDA (mixture of community and clinical cohorts) |
NINCDS–ADRDA/DSM- IV |
DAT = CDR of 0.5 |
| AD exclusion criteria | Change in diagnosis at subsequent follow-up or post-mortem confirmation of a differing disease |
Other neuropathol., head trauma, alcoholism, psychiatric illness, epilepsy, hypertension, diabetes, major heart disease |
Significant other neurological diseases. Medications which affect cognition |
Other neuropathol./mental disorders/congnitive-affecting treatments/focal brain lesions seen on MRI |
Symptoms unrelated to AD, depression |
Non-AD disorders, major and/or unstable conditions such as seizure, PD or tumour |
Substance abuse, depression, Parkinson’s disease, depression blood test used to screen for exclusionary medical conditionsa |
Substance abuse, depression, psychiatric illness/head injury |
Other confounding neuropsychol. Genetic family history of AD |
| Specific white matter damage exclusion criteria |
N/R | N/R | N/R | N/R | Stroke | Substantial cerebro-vascular disease |
N/R | White matter lesions >3mm on T2 images |
N/R |
| White matter lesion quantification |
N/R | Volume of white matter intensities measured at baseline. Distribution similar between subject groups |
N/R | N/R | N/R | N/R | N/R | N/R | N/R |
| APOE genotype (% E4) |
N/R | N/R | AD = 54 | AD-C = 54, AD T = 65 | C = 10, slow = 55, fast = 56 |
N/R | C = 22, mild AD =59, moderate=71 |
N/R | N/R |
| Gender (% male) | C = 47, AD = 39 | C = 44, AD = 40 | AD = 40 | AD-C = 19, AD-T = 22 | C = 42, slow = 39, fast = 42 |
AD-C =40b, AD-T = 41b |
C = 48, mild = 52, moderate = 71 |
C = 50, AD = 41 | C = 46, AD = 61 |
| Treatments | N/R | N/R | Donepezil 60%, galantamine 5%, rivastigmine 21%, HRT 21%, Vit E 32% |
Donepezil (AD-T). AD-C, Both groups allowed Vit E, non-steroidal anti-inflammatory, ginko biloba, lecithin |
N/R | N/R | N/R | N/R | N/R |
| Drop-out if RCT | N/A | N/A | 15 due to no post-baseline scan |
N/A | N/A | N/R | N/A | N/A | N/A |
| Assessment of controls |
N/R | MRI excluding neuropathol. |
N/A | N/A | Normal neurological exam, Community dwelling |
N/A | Yearly assessment of neuropsychol., physical and medical exam |
N/R | CDR 0.0 |
| Exclusion criteria for controls |
N/R | Clinical history of alcoholism, psychiatric illness, epilepsy, hypertension, diabetes, major heart disease or head trauma (Du et al., 2003) |
N/A | N/A | No active neurological disorder, psychoactive medication |
N/A | As for AD, plus diabetes mellitus, hypertension, angina, arrhythmia, seizure disorder, cancer, COPD or renal diseasea |
Same as AD | Other neuropsychiatric disorders, genetic family history of AD |
| Magnet strength (Tesla) |
1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 0.5–1.5 | 1.5 | 2 | 1.5 |
| Sites | N/R | 1 | 17 | 1 | N/R | 38 | 1 | 1 | 1 |
| Protocol (thickness (mm)) |
1.5 | 1.4 | 1.5–1.8 | 1.5 | 1.6 | 1.6 | 4 (detailed in Kaye et al., 1997) | N/R | 1 |
| Measurement method Number of raters |
Manual 1 |
Semi-automated Checked by 1 |
Manual 3a |
Manual 1 |
Manual 1 |
Manual 1 |
Manual 3a |
Manual 1 |
Semi-automated Team of experts for template as in Haller et al. (1997), N/R for landmark placement |
| Registered scans or TIV corrected |
Registered | N/R | Registered | N/R | Registered | Registered as referenced in Jack et al. (1998) |
TIV correcteda | Registered | N/R |
| Whole Hc measured? | No (small part of tail excluded) |
No (small part of tail excluded) as in Hsu et al. (2002) |
No (small part of tail excluded) |
No (white matter excluded), Tail excluded |
No (small part of tail excluded) |
No (small part of tail excluded) |
No (body of hippocampus only as in Kaye et al., 1997) |
No (small part of tail excluded) |
No (exclusion of white matter of alveus and fimbria some of tail excluded) |
| Reliability, inter-/intra-rater, same/different acquisitions, repeated × n, number subjects, volume/rate |
N/R | ICC 0.94, intra-rater different acquisitions. Repeated × 2 (assumed), N = 9, volume |
N/R | ICC 0.98, CV 2.8%c, intra-rater, same acquisition. Repeated × 3, N = 10, volume |
ICC 0.91, IQR of difference in rate: 2.3 × 10−3, intra-rater, same acquisition. Repeated × 2, N = 18, rate |
CV 0.28%c, intra-rater, different acquisitions. Repeated × 2, N= 10, volume |
ICC 0.90, intra-rater, same acquisition. Repeated × 5, N= 5, volume |
ICC 0.99 ICC 0.98, intra- and inter-rater, respectively. Same acquisition. Repeated × 2, N= 10, volume |
ICC 0.93, intra-rater (assumed), different acquisitions. Repeated × 2, N= 8, volume |
| Blinding of raters to diagnosis |
Yes | Visually checked blinded to diagnosis |
Blinded to treatment group |
Blinded to AD-C/AC-T | Yes | Blinded to treatment group |
Yesa | Yes | N/R |
| Blinding of raters to order of scans |
Yes | N/R | Yes | Yes | Yes | Yes | Yesa | Yes | N/R |
Key: AD, Alzheimer’s disease; C, control; DAT, dementia of Alzheimer type, ICC, intra-class correlation coefficient, CV, coefficient of variation; AD-C, control group of AD patients; AD-T, treated AD patients; APC, annualised percentage change; N/A, not applicable; N/R, not reported; CDR, clinical dementia rating; MMSE, mini mental state examination; neuropathol., neuropathologies; neuropsychol., neuropsychologies, COPD, chronic obstructive pulmonary disease; slow, slowly progressing AD; fast, quickly progressing AD; mild, mild AD; moderate, moderate AD.
Conveyed by personal communication.
Based on a wider group.
CV quoted by Jack et al. (2003) is calculated as a median (range 0.02–0.70%). CV quoted by Hashimoto 2005 was quoted for left and right hippocampus separately (right = 2.84% and left = 2.78%). This was calculated as standard deviation/mean, where standard deviation indicated square root value of the arithmetic mean of 10 variance estimates.
Table 3.
Studies excluded from the meta-analysis
| Study | Reason for exclusion | Group | Subject groups | N | Mean (S.D.) MMSE | Mean (S.D.) age in years | Rate of atrophy | Interval (years unless specified) mean (S.D.) |
|---|---|---|---|---|---|---|---|---|
| Barnes et al. (2005) | Overlap with Barnes et al. (2007a) | Fox et al. | C | 50 | 29.4 (0.8) | 59.6 (13.8) | 1.15 (1.86) | 418 (172) days |
| AD | 32 | 19.4 (5.0) | 59.0 (11.3) | 5.49 (3.12) | 456 (311) days | |||
| Cardenas et al. (2003) | VaD included | Weiner et al. | C | 16 | 29.1 (1.0) | 76 (5) | 1.8 (0.8) | 2.6 (1) |
| Dementia | 7 (5 AD), 2 (AD+VaD) | 24.6 (2.6) | 76 (8) | 5.4 (2.8) | 2.6 (1) | |||
| Hampel et al. (2005) | Mean rates not published | Hampel et al. | AD | 22 | 23.1 (4.0) | 67.8 (7.9) | 15.6 (10.5) | 18.4 (9.4) months |
| Jack et al. (1998) | Overlap with Jack et al. (2000) paper | Jack et al. | C | 24 | 28.79 (1.28) | 81.04 (3.78) | 1.55 (1.38) | 1.96 (0.75) |
| AD | 24 | 20.74 (4.60) | 80.42 (4.02) | 3.98 (1.92) | 1.89 (0.68) | |||
| Jack et al. (2000) | Overlap of subjects with Jack et al. (2004) | Jack et al. | C total [StabConv] | 58 [48/10] | [28 (1.6)/28 (1.7)] | [80.4 (6.4)/82.3 (5.8)] | 1.9 (1.1), total [1.7 (0.9)/2.8 (1.7)] | [3.0 (0.5)/3.3 (0.4)] |
| AD | 28 | 22 (4.3) | 73.8 (11.3) | 3.5 (1.8) | 2.9 (0.5) | |||
| Jack et al. (2004) | Subgroup excluded (MMSE >26) |
Jack et al. | C conv | 15 | 28.4 (1.4) <12> | 80.2 (4.6) | 3.1 (1.6) | 3.6 (0.9) |
| Kaye et al. (2005) | Subgroup excluded (MMSE >26) |
Kaye et al. | v.mild | 23 | 26.2 (2.0) | 83.0 (7.8) | 4.3 (7.4) | 2.04 (1.42) |
| Krishnan et al. (2003) | No annualised atrophy rates provided |
Rogers et al. | Placebo AD | 33a | 19.0 (4.6)a | 72.4 (10.1)a | 8.2 (9.9)b | 24 weeks |
| Treated AD | 34a | 19.5 (4.8)a | 74.4 (7.0)a | −0.4 (2.9)b | 24 weeks or endpoint | |||
| Laakso et al. (2000) | Cannot provide original data annualised |
Soininen et al. | C | 8 | 28.4 (1.6) | 69 (8) | 1.2 (5.0)c | 36 (8.9) months |
| AD | 27 | 22.0 (3.6) | 70 (5) | 2.4 (6.7)c | ||||
| Moffat et al. (2000) | Controls only | Resnick et al. | C ε4+ | 13 | 27.9 (2.5) | 68.5 (5.9) | 2.86d | 2.7 (0.6) |
| C ε4 − | 13 | 29.1 (1.1) | 69.7 (6.8) | 0.85d | 2.6 (0.8) | |||
| Mori et al. (2002) | Overlap with Hashimoto et al. (2005) | Hashimoto et al. | AD ε4+ | 38 | 19.2 (5.1) | 72.4 (5.5) | 9.76 (4.27) | 378 (18) days |
| AD ε4 − | 17 | 19.1 (6.3) | 71.1 (6.4) | 6.99 (4.24) | 372 (20) days | |||
| Mungas et al. (2005) | VaD included | Weiner DeCarli et al. | C (CDR 0.0) | 58 | 29.0 (1.4) | 74.1 (6.7) | 1.1 (1.4) | 4.0 (1.1–6.9) |
| Dementia: AD and VaD (CDR ≥ 1.0) | 11 | 23.6 (3.4) | 76.5 (9.7) | 2.9 (2.0) | 2.9 (1.1–5.1) | |||
| Silbert et al. (2003) | Overlap with Kaye et al. (2005) | Kaye et al. | C | 8 | 28.4 (1.4) | 86.6 (8.5) | 2.6 (3.2) | 4.1 (2.2) |
| Demented | 20 | 22.8 (7.4) | 81.4 (8.5) | 3.1 (5.7) |
Key: AD, Alzheimer’s disease; C, control; Stab, stable controls; Conv, converter; VaD, vascular dementia; italicized figures are provided by the author.
Summary statistics given for a wider group than that used to calculate hippocampal changes.
Percentage decline over scanning interval rather than rate.
Calculated rates of atrophy (loss quoted in publication over 3-year period actual rates over 3 years 3.6 (15.1) for controls and 7.2 (20.1)% of hippocampus).
No S.D. given on the % loss. Mean (S.D.) rate initially quoted as 71.2 (19.8) ε4+ and 20.1 (16.5) ε4− mm3 loss per year.
3.1. Data synthesis and heterogeneity
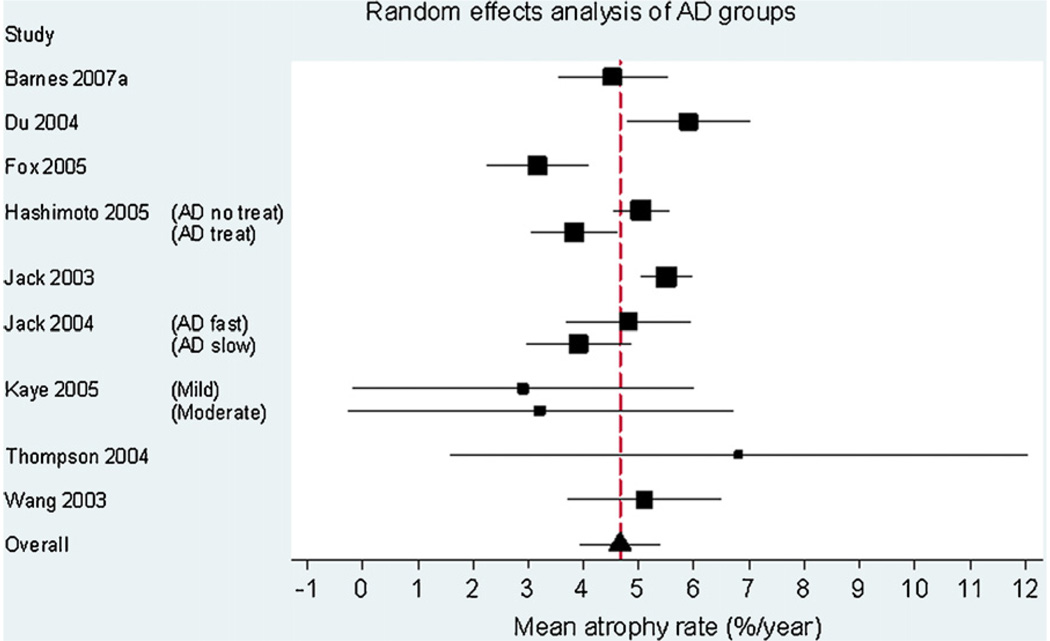
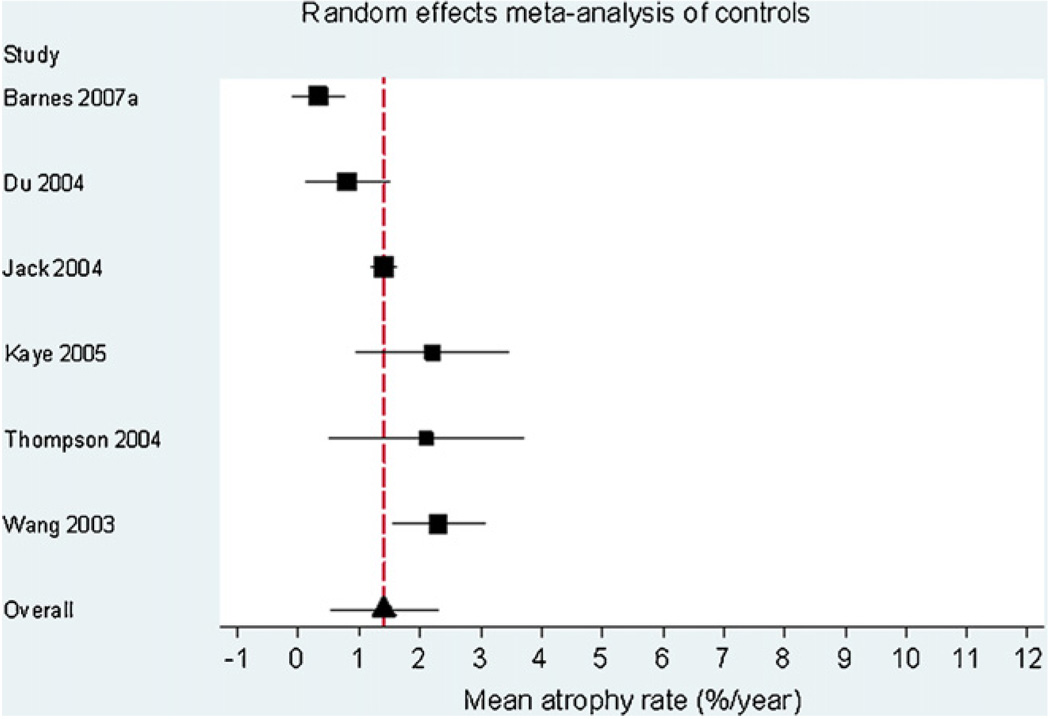
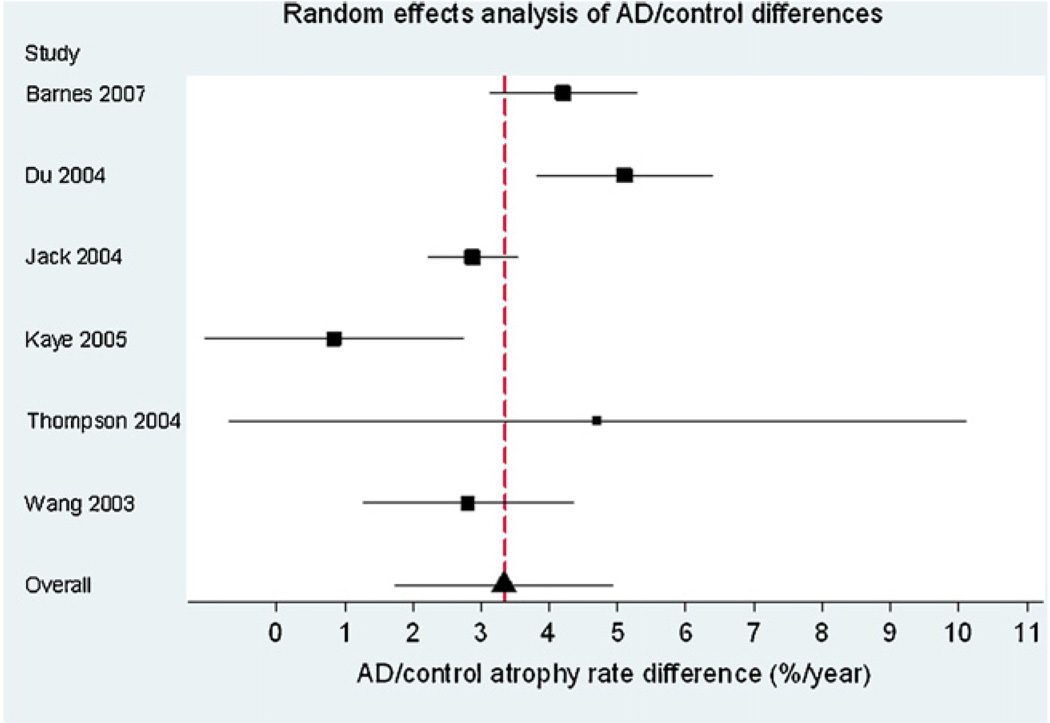
Fig. 1 is a Forest plot demonstrating the results for the AD subjects from the individual studies included in the meta-analysis (n = 595). The estimate of the overall mean atrophy rate in these groups from a random-effects meta-analysis was 4.66% per year (95% CI 3.92, 5.40). The estimate of the between-study standard deviation was 0.77%. The likelihood ratio test for the need for the between-study random-effect was highly statistically significant (p = 0.0001), confirming that there was strong evidence of between-study heterogeneity. Fig. 2 is a Forest plot of the matched control groups (n = 212). The estimated overall mean atrophy rate in these groups from a random-effects meta-analysis was 1.41% per year (95% CI 0.52, 2.30). The estimate of between-study standard deviation was 0.74%. The likelihood ratio test for the presence of between-study heterogeneity was highly statistically significant (p < 0.0001). Fig. 3 shows the differences between the control and AD groups in those studies where both groups were studied (six studies in total). Using data from these studies, and combining AD subgroups within studies, using a random-effects meta-analysis the estimated mean difference in atrophy rates between controls and AD subjects was 3.33% per year (95% CI 1.73, 4.94). The estimate of between-study standard deviation in the AD/control difference was 1.28%, and the likelihood ratio test showed strong evidence for between-study heterogeneity in AD-control rate differences (p = 0.002).
Fig. 1.
Forest plot of rates of atrophy in the hippocampus in AD subjects. The sizes of the squares are proportional to 1/(within-study variance +between-study variance). The solid lines represent 95% CIs.
Fig. 2.
Forest plot of rates of atrophy in the hippocampus in matched control subjects. The sizes of the squares are proportional to 1/(within-study variance + between-study variance estimate). The solid lines represent 95% CIs.
Fig. 3.
Forest plot showing the difference between AD and control subjects in studies where both were reported. The size of the squares are proportional to 1/(within-study variance + between-study variance estimate). The solid lines represent 95% CIs.
3.2. Publication bias
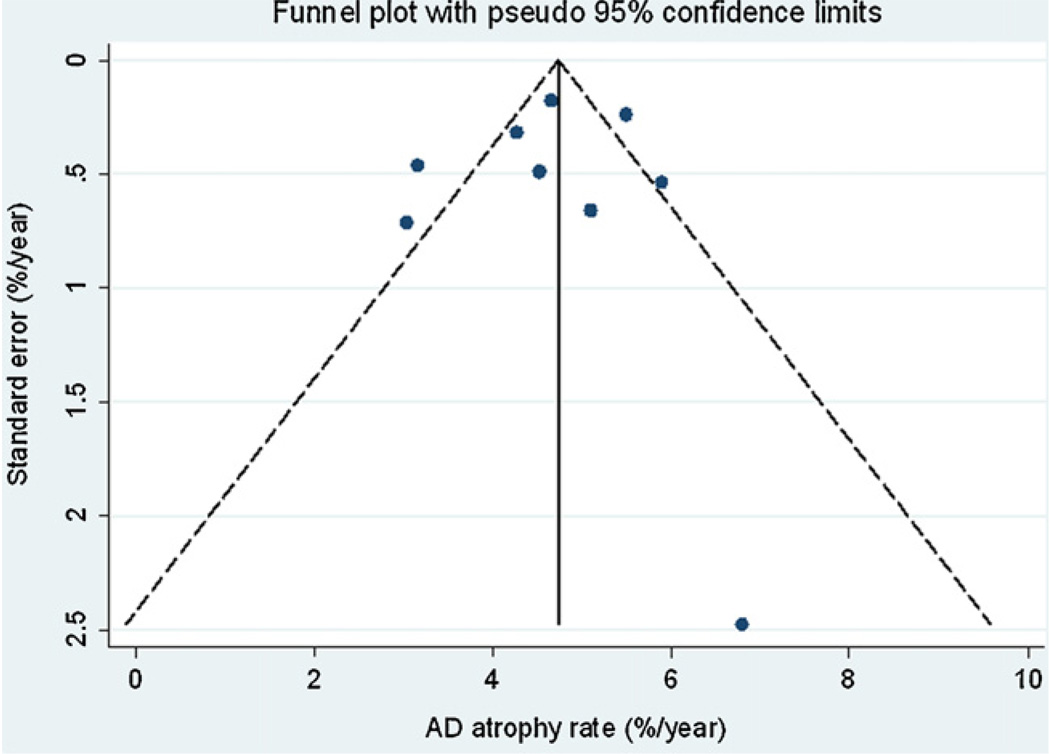
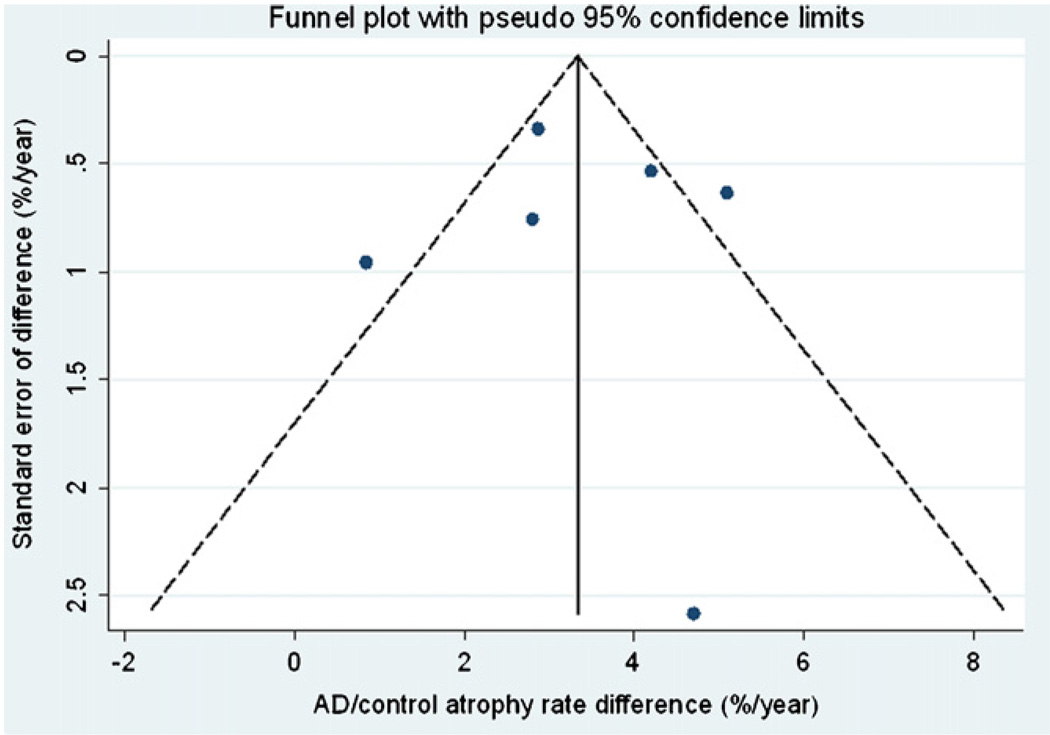
Fig. 4 and Fig. 5 show funnel plots of AD and AD-control differences respectively against the standard error of the estimate. Although few conclusions can be drawn given the small number of studies, there was no suggestion that studies with either large or small atrophy rates were more likely to be published.
Fig. 4.
Funnel plot of AD rates of atrophy with pseudo 95% confidence limits.
Fig. 5.
Funnel plot of AD-control differences with pseudo 95% confidence limits.
3.3. Subgroup analysis and meta-regression
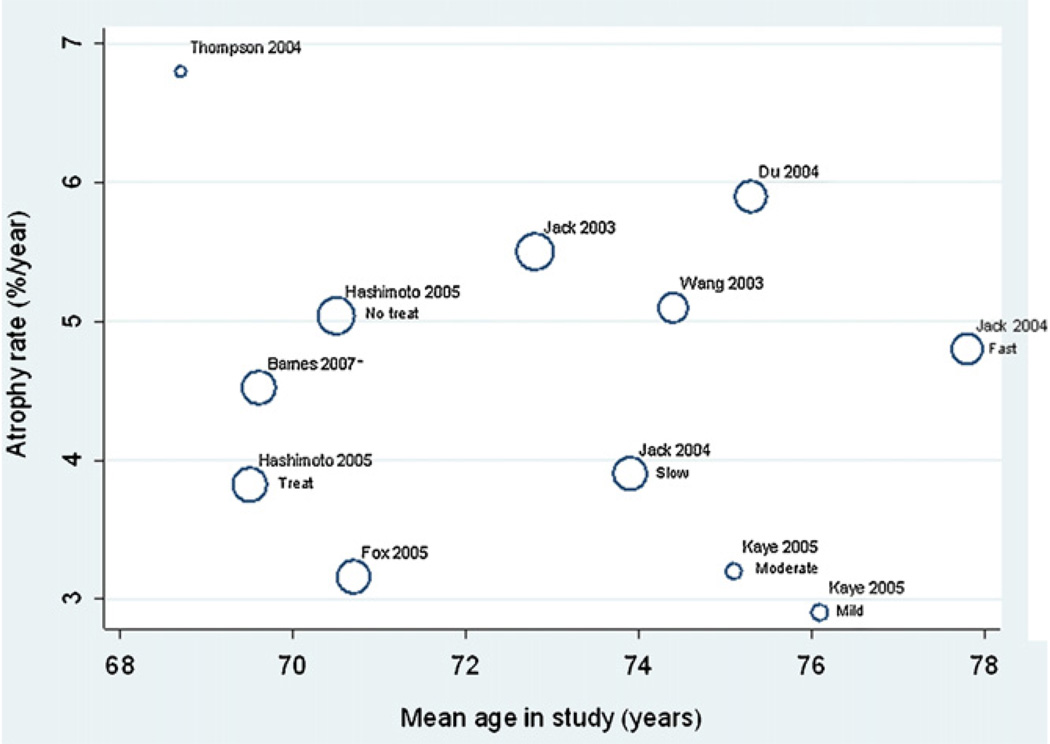
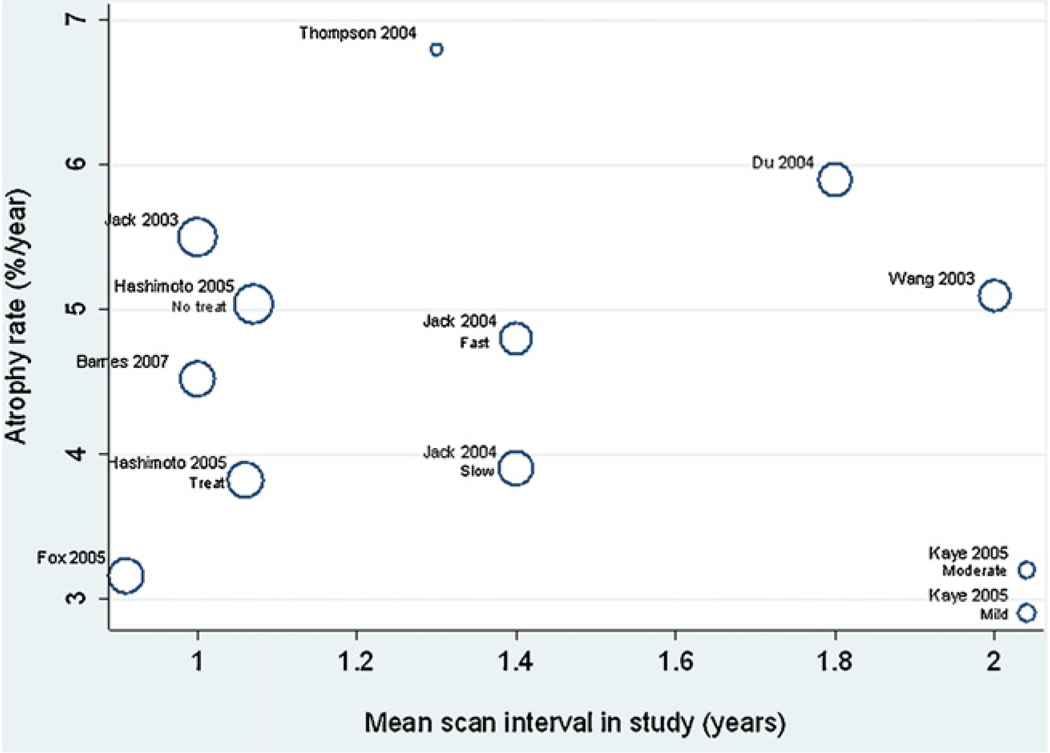
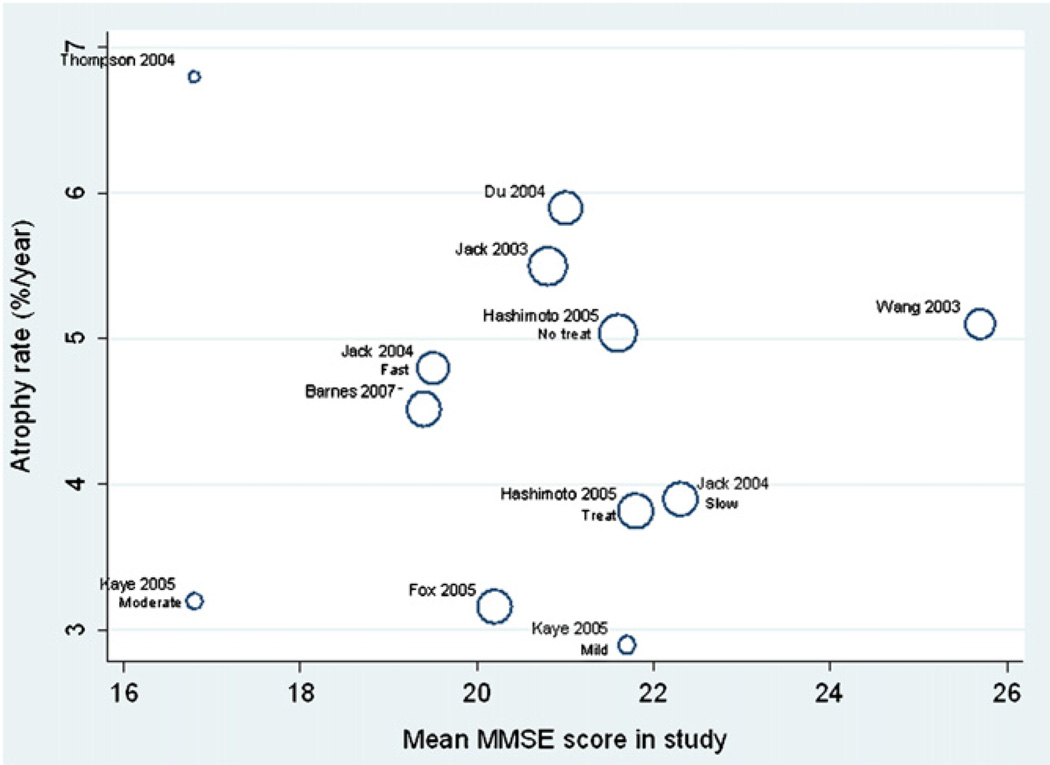
Plots of the AD study/subgroup mean atrophy rates against age, MMSE and inter-scan interval are shown in Fig. 6–Fig. 8, respectively. There were no apparent relationships between these variables and mean atrophy rate. An increase of 1 year in study mean age was associated with a 0.18% (95% CI −0.29, 0.64%) in mean atrophy rate, though this was not statistically significant (p = 0.24). There was no evidence of an association between study mean MMSE score and mean atrophy rate, with a one unit increase in study mean MMSE score being associated with a reduction in mean atrophy rate of 0.07% (−0.56 to 0.70% p = 0.68). Each 1 year increase in mean interscan interval was associated with a 0.45% (95% CI −3.07, 3.97%) increase in mean study atrophy rate, but again this association was not statistically significant (p = 0.64).
Fig. 6.
Meta-regression of atrophy rate and mean age. The size of the circles are proportional to 1/(within-study variance + between-study variance estimate), i.e. larger circles indicate studies with more precise results.
Fig. 8.
Meta-regression of atrophy rate and mean scan interval. The sizes of the circles are proportional to 1/(within-study variance + between-study variance), i.e. larger circles indicate studies with more precise results.
3.4. Sample sizes
Four studies were included in sample size calculations (Barnes et al., 2007b); Fox et al., 2005); (Hashimoto et al., 2005); and (Jack et al., 2003). For these groups the pooled mean atrophy rate was 4.51% per year, while the corresponding pooled estimate of the standard deviation of measured atrophy rates was 3.06%. The estimated numbers of subjects per treatment arm using these estimates were 242 and 39 subjects for a 20 and 50% reduction in hippocampal atrophy rates, respectively.
4. Discussion
This study aimed to estimate the mean rate of hippocampal atrophy across a number of published studies. In addition, this study aimed to formally assess some aspects of the heterogeneity in these studies. Meta-analysis plays an important role in summarising results from studies owing to between-study heterogeneity. Effects of confounders such as disease severity or age may be lessened by pooling results from all studies, making the results more applicable to the wider population.
Hippocampal rates of atrophy were consistently greater in AD subjects than controls in all studies included in this meta-analysis. The pooled mean rates of atrophy were 1.41% in the control subjects and 4.66% in AD subjects, respectively. The AD/control differences were significantly different in the 5/7 studies where both controls and AD subjects had been assessed and reported. The two studies where the differences were not statistically significant had the largest confidence intervals (Thompson et al., 2004; Kaye et al., 2005). Overall, the pooled mean difference between AD and control groups was 3.33% and was significantly greater than 0 (Fig. 3). There was a large amount of between-study variability in both AD rates and control rates. The between-study variability in AD/control rate differences was also relatively large, which cannot be attributed to factors that affect both controls and ADs in a study, such as scanning protocol. Our results therefore suggest that there is genuine variation between studies in AD/control rate difference, possibly as a result of differences in disease severity between studies. It has been shown that rates of atrophy can change as the disease progresses (Kaye et al., 2005; Ridha et al., 2006). Other factors such as concurrent vascular disease may also affect atrophy rates. Some studies had higher variances that others, this may be due to the methods employed by these studies or it may be chance owing to the relatively small numbers within the subject groups.
Some aspects of the heterogeneity of studies are difficult to formally assess because certain variables were not reported (see Table 2). These variables include patient-related information such as other administered medications and APOE genetic status. Treatments may be particularly difficult for case–control or population-based studies to report accurately, since many of these studies will be conducted at a tertiary referral centre whereas it may be the general physician at the primary level who prescribes and monitors the administration of medication. Another example of the differences in reporting in studies is reliability of segmentation technique. Most studies did report reliability, however the results are largely incomparable in most cases since the measures were performed on different numbers of subjects, and with different numbers of repetitions and in some cases with same scanning acquisitions and others with different scanning acquisitions.
Other scan parameters and hippocampal delineation methods may also affect the measured atrophy rates such as scan “slice” thickness or segmentation protocol. Differences in anatomical structures included in the delineation of hippocampi may not have substantial effects on the atrophy rate determined if the same protocol is used for both baseline and follow-up scans. Data from centres using different delineation protocols would only agree if rates of atrophy were similar in separate anatomical locations within the hippocampus (for example, rate of atrophy of the hippocampal tail is similar to the hippocampal head). Meta-analysis on the AD-control differences should be robust to these potential confounders since the hippocampal rates determined in both subject groups should have been determined in the same way.
Formal meta-regression analyses showed no significant associations between mean atrophy rates quoted in the studies included in the meta-analysis, and age, MMSE and interval. However, it may be that these relationships do exist; meta-regression can detect such associations only if there is sufficient variability in the explanatory variable between studies. There was little variability in mean age between studies relative to within studies, perhaps explaining the lack of association with age. A number of studies (not included here) have examined rates of hippocampal atrophy in normal ageing and have shown that age does influence hippocampal atrophy rates in “healthy controls” (Scahill et al., 2003). There was some variability in mean MMSE score between studies however this was not sufficient to show any potential association between this variable and rates of atrophy. Most AD subjects included in the studies were mild to moderately affected, perhaps because of the need to comply with imaging; as a result 11 of the 12 AD groups included had a mean MMSE between 17 and 22 (one group had an MMSE of 25.7); this lack of variability in mean MMSE reduced the chance of finding an association. There was no evidence of an association between mean interval and mean atrophy rate, despite relatively large variation in mean interval between scans and between studies.
Sample sizes were estimated using data from 432 subjects which is an estimate based on a greater number of subjects than previously reported. Compared with other brain regions reported for a similar interval, a greater number of subjects were required for hippocampal atrophy rates than required for whole brain, or ventricular atrophy rates (approximately 150 subjects required per treatment arm for either brain atrophy or ventricular enlargement for 20% reduction in atrophy rates, n = 38) (Schott et al., 2005). However, it may be that disease-specific effects require investigation, and therefore hippocampal atrophy rates, or temporal horn enlargement may be of interest in clinical trials. The 39 subjects estimated for a 50% reduction in atrophy rates were similar to, but lower than, the numbers required for temporal horn enlargement (number required per treatment arm = 65, n = 192) (Jack et al., 2003).
This study has a number of limitations. Although this meta-analysis attempted to include as many studies as possible and there was no evidence of publication bias, only nine studies analysed owing to a number of reasons (see Table 3). Larger numbers of studies would allow a more precise estimate of the mean rates to be calculated and for associated meta-regression analyses to be more robust. Collating the individual patient data from these studies would enable much more precise estimation of relationships between factors such as age and disease severity with hippocampal atrophy rates. Associations using individual patient data within studies would not be confounded by study-level factors, such as scan acquisition protocols and methods to determine hippocampal atrophy rate. Also, variability both within and between studies could be used to estimate these associations more precisely. In addition, standard meta-analysis techniques assume that the precision of individual study estimates are known (assumed to be the estimated value). When some contributing studies are small, as in our meta-analysis, this assumption may not be reasonable. Consequently, our reported confidence intervals may be too narrow.
Because some specific study variables were not reported consistently (see Table 2), and this was an impediment to meta-analysis, we suggest that new studies explicitly report the following information (including presence or lack of information) as part of the subject demographics, scanning, and hippocampal atrophy methods:
4.1. Subject demographics
Cohort: population/case/control/case series
Age
Gender: % male/n male
APOE genotype: % E4/n E4
Neuropsychology: MMSE
AD diagnostic criteria: which used (imaging used in diagnosis?)
AD exclusion criteria
Symptomatic/disease modifying treatments
Drop-out rate if RCT
Details of clinical assessment of controls
Exclusion criteria for control group
Post-mortem confirmation of disease
4.2. Imaging
Magnetic field strength (Tesla)
Number of scanning sites
Acquisition protocol: full details including “slice” thickness (mm)
4.3. Post-processing of scans
Region measurement method: manual/automated
Number of raters
Measurement on registered scans or TIV-corrected scans
Details of anatomy included in the segmentation
Intra-rater segmentation reliability using ICC (preferably quote statistics based on 10 subjects segmented twice by the same rater)
Blinding of raters to diagnosis
Blinding of raters to order of scans
5. Conclusions
The overall hippocampal atrophy rate is 1.4% in normal controls with the range of the quoted mean age being 69–83 years. In AD subjects the overall atrophy rate is 4.6%.
Fig. 7.
Meta-regression of atrophy rate and mean MMSE score. The size of the circles are proportional to 1/(within-study variance + between-study variance), i.e. larger circles indicate studies with more precise results.
Acknowledgements
This work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. The Dementia Research Centre is an Alzheimer’s Research Trust Co-ordinating Centre. JB is funded by the Alzheimer’s Research Trust (UK). NF is funded by the Medical Research Council (UK). PT is supported by the National Institute on Aging (grant AG016570).
The authors would like to thank all those who contributed to this paper including Clifford Jack, An-Tao Du, Mamoru Hashimoto, Lei Wang (supported by NIH R01 MH60883 and P01 AG03991) and Jeffrey Kaye.
Footnotes
Disclosures
None of the authors have any disclosures to make.
References
- Barnes J, Godbolt AK, Frost C, Boyes RG, Jones BF, Scahill RI, Rossor MN, Fox NC. Atrophy rates of the cingulate gyrus and hippocampus in AD and FTLD. Neurobiol. Aging. 2007a;28:20–28. doi: 10.1016/j.neurobiolaging.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Barnes J, Boyes RG, Lewis EB, Schott JM, Frost C, Scahill RI, Fox NC. Automatic calculation of hippocampal atrophy rates using a hippocampal template and the boundary shift integral. Neurobiol. Aging. 2007b;28:1657–1663. doi: 10.1016/j.neurobiolaging.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Boyes RG, Frost C, Lewis EB, Rossor CL, Rossor MN, Fox NC. Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates. Neuroimage. 2004;23:574–581. doi: 10.1016/j.neuroimage.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Barnes J, Scahill RI, Schott JM, Frost C, Rossor MN, Fox NC. Does Alzheimer’s disease affect hippocampal asymmetry? Evidence from a cross-sectional and longitudinal volumetric MRI study. Dement. Geriatr. Cogn. Disord. 2005;19:338–344. doi: 10.1159/000084560. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, Bohl J. Staging of Alzheimer-related cortical destruction. Eur. Neurol. 1993;33:403–408. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- Cardenas VA, Du AT, Hardin D, Ezekiel F, Weber P, Jagust WJ, Chui HC, Schuff N, Weiner MW. Comparison of methods for measuring longitudinal brain change in cognitive impairment and dementia. Neurobiol. Aging. 2003;24:537–544. doi: 10.1016/s0197-4580(02)00130-6. [DOI] [PubMed] [Google Scholar]
- Dawbarn D, Allen SJ. Neurobiology of Alzheimer’s disease. Edition 2. Oxford University Press; 2001. [Google Scholar]
- Du AT, Schuff N, Kramer JH, Ganzer S, Zhu XP, Jagust WJ, Miller BL, Reed BR, Mungas D, Yaffe K, Chui HC, Weiner MW. Higher atrophy rate of entorhinal cortex than hippocampus in AD. Neurology. 2004;62:422–427. doi: 10.1212/01.wnl.0000106462.72282.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Zhu XP, Jagust WJ, Miller BL, Reed BR, Kramer JH, Mungas D, Yaffe K, Chui HC, Weiner MW. Atrophy rates of entorhinal cortex in AD and normal aging. Neurol. 2003;60:481–486. doi: 10.1212/01.wnl.0000044400.11317.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Black RS, Gilman S, Rossor MN, Griffith SG, Jenkins L, Koller M. Effects of A beta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005;64:1563–1572. doi: 10.1212/01.WNL.0000159743.08996.99. [DOI] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. Presymptomatic hippocampal atrophy in Alzheimer’s disease: a longitudinal MRI study. Brain. 1996;119:2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Hampel H, Burger K, Pruessner JC, Zinkowski R, DeBernardis J, Kerkman D, Leinsinger G, Evans AC, Davies P, Moller HJ, Teipel SJ. Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch. Neurol. 2005;62:770–773. doi: 10.1001/archneur.62.5.770. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Kazui H, Matsumoto K, Nakano Y, Yasuda M, Mori E. Does donepezil treatment slow the progression of hippocampal atrophy in patients with Alzheimer’s disease? Am. J. Psychiatry. 2005;162:676–682. doi: 10.1176/appi.ajp.162.4.676. [DOI] [PubMed] [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, Gado M, Joshi SC, Miller MI, Sheline YI, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomical atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, Weiner MW. Comparison of Automated and Manual MRI Volumetry of Hippocampus in Normal Aging and Dementia. J. Magn. Reson. Imaging. 2002;16:305–310. doi: 10.1002/jmri.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Tangalos EG, Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer’s disease. Neurology. 1998;51:993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O’Brien PC, Cha R, Knopman D, Petersen RC. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60:253–260. doi: 10.1212/01.wnl.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CRJ, Shiung MM, Gunter JL, O’Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Moore MM, Dame A, Quinn J, Camicioli R, Howieson D, Corbridge E, Care B, Nesbit G, Sexton G. Asynchronous regional brain volume losses in presymptomatic to moderate AD. J. Alzheimers Dis. 2005;8:51–56. doi: 10.3233/jad-2005-8106. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Swihart T, Howieson D, Dame A, Moore MM, Karnos T, Camicioli R, Ball M, Oken B, Sexton G. Volume loss of the hippocampus and temporal lobe in healthy elderly persons destined to develop dementia. Neurol. 1997;48:1297–1304. doi: 10.1212/wnl.48.5.1297. [DOI] [PubMed] [Google Scholar]
- Krishnan KR, Charles HC, Doraiswamy PM, Mintzer J, Weisler R, Yu X, Perdomo C, Ieni JR, Rogers S. Randomized, placebo-controlled trial of the effects of donepezil on neuronal markers and hippocampal volumes in Alzheimer’s disease. Am. J. Psychiatry. 2003;160:2003–2011. doi: 10.1176/appi.ajp.160.11.2003. [DOI] [PubMed] [Google Scholar]
- Laakso MP, Lehtovirta M, Partanen K, Riekkinen PJ, Soininen H. Hippocampus in Alzheimer’s disease: a 3-year follow-up MRI study. Biol. Psychiatry. 2000;47:557–561. doi: 10.1016/s0006-3223(99)00167-5. [DOI] [PubMed] [Google Scholar]
- Moffat SD, Szekely CA, Zonderman AB, Kabani NJ, Resnick SM. Longitudinal change in hippocampal volume as a function of apolipoprotein E genotype. Neurol. 2000;55:134–136. doi: 10.1212/wnl.55.1.134. [DOI] [PubMed] [Google Scholar]
- Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, Matsui M. Accelerated hippocampal atrophy in Alzheimer’s disease with apolipoprotein E epsilon4 allele. Ann. Neurol. 2002;51:209–214. doi: 10.1002/ana.10093. [DOI] [PubMed] [Google Scholar]
- Morrell CH. Likelihood ratio testing of variance components in the linear mixed-effects model using restricted maximum likelihood. Biometrics. 1998;54:1560–1568. [PubMed] [Google Scholar]
- Mungas D, Harvey D, Reed BR, Jagust WJ, DeCarli C, Beckett L, Mack WJ, Kramer JH, Weiner MW, Schuff N, Chui HC. Longitudinal volumetric MRI change and rate of cognitive decline. Neurology. 2005;65:565–571. doi: 10.1212/01.wnl.0000172913.88973.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, Fox NC. Tracking atrophy progression in familial Alzheimer’s disease: a serial MRI study. Lancet Neurol. 2006;5:828–834. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch. Neurol. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- Scheltens P, Fox NC, Barkhof F, DeCarli CD. Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion. Lancet Neurol. 2002;1:13–21. doi: 10.1016/s1474-4422(02)00002-9. [DOI] [PubMed] [Google Scholar]
- Schott JM, Price SL, Frost C, Whitwell JL, Rossor MN, Fox NC. Measuring atrophy in Alzheimer disease—a serial MRI study over 6 and 12 months. Neurology. 2005;65:119–124. doi: 10.1212/01.wnl.0000167542.89697.0f. [DOI] [PubMed] [Google Scholar]
- Sharp SJ. Meta-analysis regression. Stata Technical Bulletin. 1998;vol. 42:16–22. [Google Scholar]
- Silbert LC, Quinn JF, Moore MM, Corbridge E, Ball MJ, Murdoch G, Sexton G, Kaye JA. Changes in premorbid brain volume predict Alzheimer’s disease pathology. Neurology. 2003;61:487–492. doi: 10.1212/01.wnl.0000079053.77227.14. [DOI] [PubMed] [Google Scholar]
- Small GW, Rabins PV, Barry PP, Buckholtz NS, DeKosky ST, Ferris SH, Finkel SI, Gwyther LP, Khachaturian ZS, Lebowitz BD, McRae TD, Morris JC, Oakley F, Schneider LS, Streim JE, Sunderland T, Teri LA, Tune LE. Diagnosis and treatment of Alzheimer disease and related disorders. Consensus statement of the American Association for Geriatric Psychiatry, the Alzheimer’s Association, and the American Geriatrics Society. JAMA. 1997;278:1363–1371. [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Wang L, Swank JS, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the Alzheimer type from healthy aging. Neuroimage. 2003;20:667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]