Abstract
A workshop sponsored by the National Cancer Institute and the US Food and Drug Administration addressed past lessons learned and ongoing challenges faced in biomarker development and drug and biomarker codevelopment. Participants agreed that critical decision points in the product life cycle depend on the level of understanding of the biology of the target and its interaction with the drug, the preanalytical and analytical factors affecting biomarker assay performance, and the clinical disease process. The more known about the biology and the greater the strength of association between an analytical signal and clinical result, the more efficient and less risky the development process will be. Rapid entry into clinical practice will only be achieved by using a rigorous scientific approach, including careful specimen collection and standardized and quality-controlled data collection. Early interaction with appropriate regulatory bodies will ensure studies are appropriately designed and biomarker test performance is well characterized.
Remarkable advances in the understanding of neoplastic progression at the cellular and molecular levels have spurred interest in molecularly targeted cancer therapeutics. New imaging and bioassay technologies are providing the basis for developing biomarkers that will facilitate development of these molecularly targeted drugs. Biomarkers may be used in early drug development to elucidate the mechanism of action of a drug and provide preliminary evidence of its effect. As the relationship between a drug or class of drugs and a biomarker becomes better understood, there is hope that clinical assays can be developed to identify patients most likely to benefit from the drug. These biomarkers are termed predictive biomarkers. Although prognostic biomarkers that provide information on the natural course of disease after standard treatments are useful, predictive biomarkers are of greater value in clinical decision making and will be essential tools for tailoring treatments.
Drug and assay developers, regulators, and clinical investigators face many dilemmas in the course of developing targeted drugs and associated predictive biomarkers. Difficult choices must be made regarding use of precious resources (eg, biospecimens and funds) in selecting appropriate candidate biomarkers and determining optimal study design. These choices will be influenced by many factors, including the anticipated business model for the biomarker assay (eg, development as a commercial kit or as a service laboratory test) and the inherent tension between rapidly determining whether any patient group benefits from the new drug vs accurately defining individual patients most likely to benefit. Perhaps the most difficult scientific and business decisions in drug and predictive biomarker development involve whether to use biomarkers to determine patient eligibility for inclusion in clinical studies assessing benefit from a new agent. Using a predictive biomarker to select patients can lead to efficient clinical studies if the biomarker is highly sensitive and specific for benefit. But these studies may not produce the information required to demonstrate efficacy of the drug in an unselected patient population or to adequately characterize the performance of the biomarker. Recognition of the fact that single biomarkers may not adequately reflect the biology of cells has led to increasing use of panels of markers or multianalyte markers. Development and evaluation of these multianalyte biomarkers are more complicated than for single biomarkers, but the principles of development are much the same; for this reason, we do not specifically discuss them in this report.
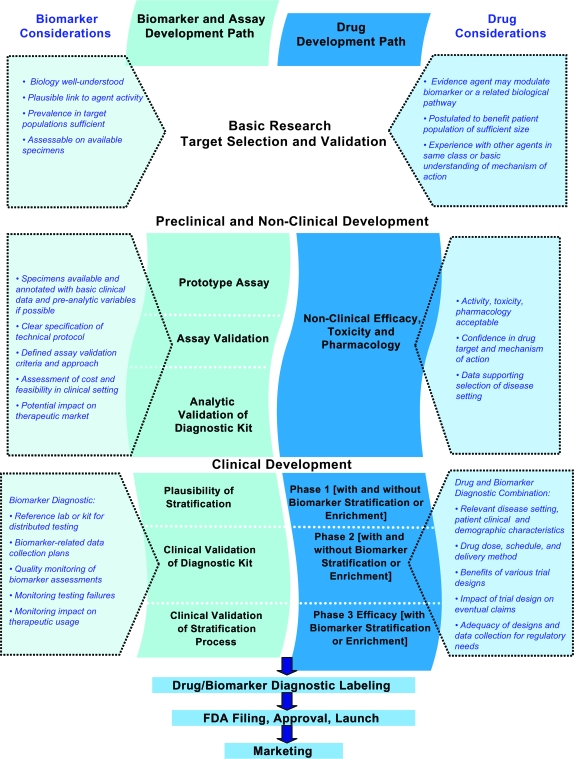
The National Cancer Institute, the US Food and Drug Administration (FDA), and representatives of the drug and biomarker industry convened a workshop on October 29–30, 2007, in Bethesda, Maryland, to address the challenges facing biomarker development and drug and biomarker codevelopment. The goal of this meeting was to consider strategies to assist the research and development community in identifying and addressing issues in predictive biomarker development. This article builds on the discussions that took place at the meeting and presents a set of issues for consideration and proposed development paths. These issues and concepts have been organized graphically in a figure (Figure 1).
Figure 1.
Considerations for drug and biomarker codevelopment. The schematic encompasses the entire life cycle for codevelopment of a drug and biomarker combination from early selection and validation of the biomarker target through preclinical and nonclinical development of the drug and biomarker assay to clinical evaluation of the drug and biomarker assay combination. The center of the diagram lists major steps in the process for the biomarker and assay (left) and the drug (right). The boxes on the left list considerations for the biomarker and biomarker assay at the various phases of development. The boxes on the right list considerations for the drug and drug–biomarker diagnostic combination. The considerations include recommendations for moving development forward and factors that should be taken into account in formulating the development strategy. As an assay and agent progress through the phases of development, continued codevelopment depends on greater confidence in the robustness and performance characteristics of the assay and stronger evidence for correlation of the biomarker with clinical benefit from the agent. At each stage in the development process, there may be different expectations for the marker (its “fitness for purpose”); as development progresses, so do the risks and therefore the expectations and requirements for the marker increase. FDA = Food and Drug Administration.
Design of the Workshop
The workshop began with an overview session that included presentation of two case studies with different clinical study design strategies for evaluating a targeted therapy and its companion predictive biomarker. The presentations reviewed some of the lessons learned from past efforts to develop a targeted drug and companion biomarker and highlighted some critical issues to be addressed. The first case study described development of HER2 as a predictive biomarker for patients who were likely to respond to trastuzumab (Herceptin). The second case study described ongoing research to identify and evaluate predictive biomarkers and biomarker assays for response to epidermal growth factor receptor (EGFR) inhibitors. These case studies are summarized in Boxes 1 (HER2) and 2 (EGFR).
Box 1. HER2 case study
Basic biology
HER2/neu is a member of the human ErbB family of receptors, a group of transmembrane receptors with intracellular tyrosine kinase activity and extracellular binding domain.
HER2 receptor does not appear to have a specific ligand but can signal by forming heterodimers with other members of the ErbB family.
Amplification of the HER2 gene produces overexpression of this cell membrane receptor protein and activation of several downstream signal transduction pathways.
Studies in HER2-transfected cells, as well as transgenic animals, support the hypothesis that amplification and/or overexpression of this proto-oncogene contributes to the pathogenesis and clinical aggressiveness of tumors (1,2).
HER2 is overexpressed in approximately 20%–30% of human breast cancers (3–5). Overexpression rarely occurs in the absence of gene amplification in breast cancer (ie, in approximately 3% of cases).
Clinical observations
HER2 overexpression identifies a subgroup of breast cancer patients with aggressive disease, frequently hormone receptor–negative with poor prognosis (3).
HER2 gene amplification has been associated with resistance to a variety of cytotoxic agents and endocrine therapies (6,7).
Agent description
Trastuzumab (Herceptin) is a humanized monoclonal antibody with high specificity for the HER2 protein.
Trastuzumab has demonstrated antitumor activity when used as a single agent in first- or second-line treatment of HER2-amplified or HER2-overexpressing metastatic breast cancer (8,9).
Development path and clinical trial design(s)
Given strong laboratory and clinical data supporting the importance of the target and antitumor activity noted from target inhibition, clinical development of trastuzumab was initially focused on testing the agent in breast cancer patients with HER2 overexpression and/or HER2 amplification in their tumors.
Trastuzumab received Food and Drug Administration approval in 1998 for the treatment of HER2-overexpressing metastatic breast cancer, as a single agent or in combination with paclitaxel, in patients who have received one or more chemotherapy regimens.
In 2006, trastuzumab was approved for adjuvant treatment of HER2-overexpressing breast cancer, either in combination with doxorubicin, cyclophosphamide, and paclitaxel or as a single agent following chemotherapy based on striking results from pivotal phase III trials (10–14).
Detection of HER2 protein overexpression by immunohistochemistry (IHC) or HER2 gene amplification by fluorescence in situ hybridization (FISH) was advised for selection of patients for trastuzumab therapy.
Assays (trade name of assay, what was measured, method, manufacturer)
HercepTest, HER2 protein (A085 polyclonal antibody), IHC; Dako, Carpinteria, CA.
Pathway, HER2 protein (CB11 monoclonal antibody), IHC; Ventana Medical Systems, Tucson, AZ.
PathVysion, HER2 gene, FISH; Abbott Laboratories, Abbott Park, IL.
INFORM, HER2 gene, FISH; Ventana Medical Systems, Tucson, AZ.
SPoT-Light, HER2 gene, chromogenic in situ hybridization (ISH); Invitrogen, Carlsbad, CA.
EnzMet GenePro, HER2 gene, silver-enhanced ISH; Ventana Medical Systems, Tucson, AZ.
Issues
Box 2. Epidermal growth factor receptor (EGFR) case study
Basic biology
EGFR is a member of the human ErbB family of receptors.
Upon ligand binding, EGFR homodimerizes or heterodimerizes with another member of the ErbB receptor family, activating its protein tyrosine kinase domain and initiating downstream signaling via cellular pathways controlling proliferation, survival, motility, and angiogenesis (20–22).
EGFR is expressed in a variety of malignancies, and experimental evidence suggests that its inhibition can induce tumor stasis or, less commonly, regression (20,21).
Most frequent EGFR abnormality reported in human cancers is receptor overexpression, but unlike HER2, which is in the same family of receptors, high concordance between overexpression and gene amplification has not been well demonstrated.
At the time of initial clinical trials evaluating EGFR inhibitors, gene amplification and/or mutations were known to occur in glioblastoma but not in other tumor types, and markers of sensitivity or resistance were not known.
After antibodies and small molecules were commercially available, additional potential predictive biomarkers emerged, including EGFR gene amplification or increased copy number by fluorescence in situ hybridization (FISH), activating EGFR mutations in lung cancer patients and KRAS mutations that predicted lack of response to EGFR inhibition in colorectal and lung carcinomas.
Clinical observations
EGFR overexpression determined by immunohistochemistry has been associated with poorer clinical outcomes in some settings (20).
High EGFR gene copy number identified by FISH might be a better predictor for survival in tyrosine kinase inhibitor (TKI)–treated non–small cell lung cancer (NSCLC) patients (23,24).
Clinical evidence suggests that EGFR inhibition can induce tumor stasis or regression (20,25–27).
Specific somatic mutations, small deletions, insertions, or point missense mutations in the EGFR tyrosine kinase correlate with better prognosis and increased objective response rate in NSCLC patients treated with small molecule TKIs (25–28) but not with cetuximab (29).
KRAS mutations appear to predict for insensitivity of tumors to both antibodies and small molecules (30–34).
Agent description
Monoclonal antibodies, such as cetuximab and panitumumab, and TKIs, such as erlotinib and gefitinib, targeting EGFR have been developed.
Development path and clinical trial design(s)
Given the paucity of biological information about markers of sensitivity and/or resistance, initial clinical trials focused on cancers that frequently express EGFR, but did not exclude patients with tumors that did not express EGFR.
Monoclonal antibodies were initially evaluated in colorectal and head and neck carcinoma patients, and the initial development of small molecule TKIs focused on non–small cell lung cancer as initial signals of activity were seen in these settings.
Evaluations of monoclonal antibodies in colorectal carcinoma have generally required EGFR protein expression as detected by immunohistochemistry for eligibility; however, EGFR expression has generally not been required for enrollment into trials of inhibitors of EGFR tyrosine kinases.
Assays (trade name of assay, what was measured, method, and manufacturer)
pharmDx, EGFR protein, IHC; Dako, Carpinteria, CA.
HTScan EGFR-phosphorylated protein, IHC; Cell Signaling Technology, Beverly, MA.
CONFIRM EGFR, EGFR protein, IHC; Ventana Medical Systems, Tucson, AZ.
PathVysion, EGFR gene (Locus Specific Identifier for EGFR labeled with Spectrum Orange and Chromosome Enumeration Probe 7 labeled with Spectrum Green), FISH, Abbott Laboratories, Abbott Park, IL.
Issues
Detectable EGFR (high expression not necessarily critical) appears to correlate with clinical benefit from EGFR inhibitors in some cases but fails to provide predictive information in others. It is unclear whether these differences are due to test methodologies, the biology of the disease being evaluated, or a combination of both (35,36).
EGFR mutations that occur more frequently in East Asian patients and never smokers are associated with improved response rate and outcome and rarely occur with KRAS mutations (37–39). Patients with EGFR mutations may develop resistance through emergence of secondary mutations or c-MET amplification (40).
KRAS mutations are associated with lack of activity of EGFR antibodies in colorectal carcinoma and perhaps in NSCLC although data are less definitive because of limited samples analyzed retrospectively from randomized clinical trials (30–34).
A key difference in the development paths followed in these two examples is that a biomarker-enrichment design strategy was used in the case of HER2-trastuzumab (only patients whose tumors tested positive for HER2 were entered into the pivotal phase III trial of trastuzumab and in most of the earlier trials). An enrichment strategy was not uniformly applied in the clinical trials evaluating the therapeutic agents targeting EGFR. Some trials of EGFR antibodies enrolled patients based on EGFR expression detected by immunohistochemical analysis but many trials of EGFR inhibitors enrolled patients without preselection by EGFR status.
Workshop participants divided into three breakout groups to consider different aspects of predictive biomarker development: 1) feasibility studies and choice of assay and biomarker, 2) study design and clinical utility evaluation—for drug, biomarker, drug and biomarker, and 3) decision points and their implications. Summary reports from the three groups were presented to all participants and recommendations were then discussed. These reports form the basis for this article and its recommendations.
Factors to Consider for Biomarker and Drug Codevelopment
A recurring theme in the workshop's discussions was the need to increase understanding of the biology associated with the chosen biomarkers, including their role in tumor behavior and their interplay with the drugs’ mechanisms of action. This biological understanding must be integrated into the clinical context in which the biomarker would be used (Boxes 1 and 2). Critical factors to this understanding are access to well-annotated biospecimens, early attention to characterization of the biomarker and standardization of assay methods, clinical trial designs that allow early evaluation of the effectiveness of the biomarker in predicting responsiveness to the drug, and ability to incorporate advancing technology and emerging data suggesting new or additional biomarkers that better characterize the target populations.
Sound biomarker and drug codevelopment depends on preclinical data that would support use of a biomarker assay in therapeutic decision making and reliable information that would allow development of a robust and reproducible assay. The clinical and laboratory data providing a rationale for codevelopment should show consistent associations between biomarker status and the drug's activity. The method(s) of detecting the biomarker in clinical specimens should show reasonable analytical and clinical performance. Furthermore, the proportion of patients who are likely to benefit from treatment, and the magnitude of the benefit, should be sufficient to warrant use of the biomarker. If only a fraction of the patients with a particular cancer that is diagnosed by standard methods may benefit from treatment but the magnitude of benefit is clinically important, then the codevelopment of a biomarker to identify those patients, if feasible, would be warranted.
Feasibility Studies and Choice of Assay and Biomarker
Biological Rationale
A biological rationale supported by research or observational data is a critical factor in selecting candidate predictive biomarkers for codevelopment with a cancer therapeutic. This would include evidence that the biomarker(s) chosen is meaningfully correlated with the activity of the targeted agent (41). The evidence may derive from existing preclinical and clinical literature; data collected during phase I, expanded phase II, or even early phase III clinical studies of the agent; or modeling and biological inferences using incomplete or partially complete provisional datasets and data mining. All available information on cellular pathways relevant to drug action, mechanisms, or drug interactions should be included in decision making and in generation of study hypotheses.
Technical Feasibility
A robust technology for testing in clinical samples must be available or it must be technically and economically feasible to develop an analytically reliable testing system on a timeline consistent with the development timeline for the drug. Definitive work on establishing even preliminary hypotheses may be at risk if reliable assay performance has not been achieved early in the decision-making process. In pilot work, real-world test performance and cost should all be considered important factors in determining the viability of biomarker testing. The anticipated strength of the association between biomarker status and drug benefit and the impact that use of the biomarker would have on the size of further clinical studies required for the drug development and approval process will likely play a role in the decision regarding whether to pursue codevelopment of a drug and a biomarker.
Standardized Procedures for Specimen and Data Collection and Interpretation
There is increasing appreciation that assay performance can be greatly affected by preanalytic factors, including methods used for sample collection, handling, processing, transport, and storage. Physiological, pharmacological, and pathological features of patients, ranging from underlying patient comorbidities to patient drug treatments, can be the source of spurious biomarker results. Information on these potential confounders should be gathered as comprehensively as possible, and attention should be paid to whether the assay will be robust enough in the real world of sample procurement.
It is critical that standards for test performance be developed and adopted. This need has been addressed by FDA in a concept article on drug and diagnostic codevelopment and in a series of guidance documents (42–47). To ensure success, there must be access to systematically collected, well-annotated, well-preserved patient specimens with linked outcome data. This requires that sufficient sample collection be prospectively built into studies and that standard protocols for collecting the samples and clinical information be implemented. Use of a certification process or reimbursement incentives to promote standardization of sample collection, handling, and preservation might help address these issues. Complete and consistent reporting of the data and analysis of results generated in studies for development of biomarker assays and evaluation of their clinical use are also critical to allow appropriate interpretation and design of subsequent clinical trials. In this regard, the REMARK Guidelines prepared by the National Cancer Institute–European Organization for Research and Treatment of Cancer Working Group on Cancer Diagnostics (48) and the Standards for Reporting of Diagnostic Accuracy initiative (49) may be of considerable value.
There are limitations imposed by informed consent documents and institutional review boards on patient sample collection, storage, and use. An effective, clear, and simplified informed consent process needs to be developed and widely adopted for the purpose of permitting the use of archived biospecimens and associated clinical information in biomarker research.
Timing of Biomarker Development and Drug Development
Because advances in understanding the biology of cancer and in technologies for biomarker assays take place continuously, the decision to develop a companion biomarker may occur at any time during drug development, that is, from preclinical investigations to postmarketing studies conducted after approval of the drug. In ideal circumstances, the biomarker assay should be defined and validated by the end of phase II for definitive evaluation in phase III. However, this may often not be possible. There is also some risk to investing too much on biomarker development early in the drug development process because the biomarker may be irrelevant if the drug does not effectively inhibit the target or if the target is not related to cancer progression in the clinical setting in which it is tested. It is also possible that the putative target of the agent may be shown to be wrong in early clinical testing, which could result in effort and resources being wasted on development of a biomarker that would not be relevant. Alternative approaches for iterative biomarker discovery, design, and validation may need to be considered (see “Study Design and Clinical Utility Evaluation” section below).
The HER2 and EGFR inhibitor case studies highlight two extremes in the spectrum of potential strategies for biomarker and drug codevelopment. As described in Box 1, a wealth of preclinical and clinical biological data were available to provide the rationale for codeveloping trastuzumab and an assay for HER2 in the treatment of breast cancer, including information on use of HER2 as a prognostic marker, data from laboratory experiments linking amplification and/or expression of HER2 to development of breast cancer, and sensitivity of HER2 to blocking antibodies (3). In addition, adequate, although not optimal, assays for HER2 existed, and it was economically advantageous to initiate agent development in the smaller subpopulation of HER2-amplified breast cancer patients. Thus, for trastuzumab, there was a critical mass of information to support the development of the agent in a patient subpopulation identifiable with a biomarker assay.
As suggested in Box 2, the situation for EGFR inhibitors has been less straightforward. Conflicting data exist on the frequency of expression or overexpression of EGFR and its association with clinical outcomes (20,21). Laboratory data do not suggest that EGFR protein overexpression is tightly associated with the anticancer effects of EGFR inhibitors (35,36). For example, some cell lines with low levels of protein expression are sensitive to EGFR inhibition and others with high expression are resistant.
Although cut points and scoring systems based on EGFR protein expression have often been used for patient selection for EGFR inhibitor therapy in clinical trials, this practice is not well supported by clear and consistent clinical or laboratory data. The pivotal positive trial of erlotinib that led to its approval for treatment of patients with locally advanced or metastatic non–small cell lung cancer after failure of at least one prior chemotherapy regimen was conducted in unselected patients (50). Retrospective analyses of results from this study examining a variety of EGFR assays failed to identify subsets of patients that did or did not benefit from treatment with this EGFR inhibitor (23).
Once comprehensive information is gathered on the biology of a biomarker and the analytical performance and practical aspects of using a particular assay (eg, cost, accessibility, stability of analyte under conditions of collection, and use), a decision can be made whether to incorporate the biomarker into further clinical studies.
Study Design and Clinical Utility Evaluation—Drug, Biomarker, and Drug–Biomarker Combination
The intended use of a biomarker must be clearly defined, and the effectiveness and safety data should support the biomarker and its assay for this use. In general, the safety of a biomarker assay is related to the clinical impact of false-positive or false-negative test results (51). Limitations and uncertainties should be described.
There are several strategies for assessing biomarkers and assays for their ability to predict effectiveness of treatment and several choices of trial designs for demonstrating that the biomarker is informative in making treatment decisions. To most efficiently achieve success in meeting regulatory requirements for safety and effectiveness of the biomarker and drug combination, dialog with FDA early in the development of the biomarker and coordination of regulatory work across multiple FDA centers may be helpful (42).
Enrichment Designs Used to Evaluate Treatment Effect
Clinical trial strategies for biomarker and therapeutic development may be broadly divided into enrichment and all-comers designs. Enrichment studies (eg, studies performed solely in biomarker-positive patients or only in biomarker-negative patients for toxicity or resistance signals) may expedite collection of data to demonstrate safety and efficacy of the diagnostic–drug combination compared with existing treatments or placebo. This approach can facilitate rapid and cost-effective market entry. Enrichment studies may be preferred when the biomarker of interest is well understood biologically and analytically, and the supporting data suggest little benefit of the drug in biomarker-negative populations or suggest substantial drug toxicity or resistance in biomarker-positive patients. When efficacy is tested only in biomarker-positive patients, postmarketing studies of biomarker-negative patients to understand the full spectrum of drug utility are valuable but may be challenging. There will always be limitations on how far preclinical and early clinical trial results can be extrapolated to justify later steps in the drug and biomarker development paths.
Clinical trials in enriched populations must be designed with appropriate control groups to identify both the prognostic and predictive value of the biomarker to determine the potential clinical utility of the test for selecting a specific treatment (52). If the biomarker is prognostic for clinical outcomes independent of treatment, then results from single-arm studies may not have a historical reference for comparison. In some cases, imperfect understanding of the biomarker could lead to enrichment studies creating misleading information on the utility of the drug. If only biomarker-positive patients are studied, the sole performance characteristic of the biomarker that can be evaluated is whether or not the drug benefits biomarker-positive patients, and the behavior of the drug in biomarker-negative patients would be undefined. This leaves open the possibility that a new drug could be introduced into the market with restrictions. For example, although cetuximab was initially evaluated in enriched populations and approved for use only in patients with EGFR-positive tumors, it is becoming apparent that some patients with EGFR-negative tumors, as assessed by immunohistochemistry, may benefit (29). Thus, the biological background on the biomarker suggesting that treatment benefit will be limited to those positive for the biomarker must be very strong if one is to consider an enrichment strategy.
All-Comers Designs
An “all-comers” study design does not restrict entry into the clinical trial on the basis of biomarker status. Although the accrual of patients is prospective, the biomarker evaluation may be conducted either prospectively or retrospectively. The biomarker can be measured at trial entry and used for stratified randomization of patients to the treatment groups, or patients can be randomly assigned to treatment groups without stratification by marker status and biomarker assays can be conducted later on the banked biospecimens. For the latter situation, it is even possible that the biomarker assays could be conducted many years later. If biomarker evaluation is not required for study entry and will be conducted later, it is important that the proportion of patients from whom specimens are collected for assay is high. Otherwise, there may be questions about whether the group of patients for whom specimens and assay results are available is representative of the full trial cohort. An advantage of requiring biomarker status for stratified randomization is that it ensures that biomarker results will be available for all patients who enter the trial, and it ensures balanced marker distribution between treatment groups. However, trials requiring up-front biomarker assay results can be logistically more difficult to conduct. For larger studies, major imbalances in biomarker distributions between treatment arms are unlikely to occur, and stratification becomes less important.
Investigators have explored the possibility of a two-step procedure in which drug effect is evaluated first in the total population without knowledge of biomarker results followed by evaluation of drug effect in a biomarker-defined subset, depending on the outcome in the full group (53,54). If a two-step procedure is to be used, it should be specified prospectively in the protocol. The plan should include a precise definition of the subset, specification of the rule that will determine whether testing terminates with the full group or continues to the subset, and an indication of the statistical operating characteristics of the procedure. If a positive drug effect is found when analyzing the full patient cohort, this would be interpreted as indicating benefit of the drug in the entire unselected population. A negative study outcome in the full group would be followed by another statistical test for treatment benefit according to biomarker status. If a statistically significant treatment benefit was observed in biomarker-positive patients, the results would indicate benefit of the drug in this biomarker-selected population. To account for the multiple tests and control the overall probability of reaching incorrect conclusions, a standard approach would be to distribute the typical testing type I error of .05 over the multiple tests. For example, an α-level of .04 could be allocated to the test of treatment effect in the overall patient cohort, and if biomarker exploration is subsequently needed, the remainder of the α-level (.01) can be applied to the test of treatment effect in the biomarker-selected subgroup. This means that the size of the study will have to be larger than it would have been if an enrichment design had been used up front with the full α-level of .05 allocated to the treatment comparison in the biomarker-selected subgroup. However, the two-step design offers valuable flexibility if there is some uncertainty about how to best define the biomarker-positive subgroup or if there is a possibility that the treatment has broad effects resulting in some benefit to all patients, regardless of biomarker status. A potential danger noted in this two-step approach is that a drug benefit observed in the entire population might be a consequence of the strength of response in the biomarker-positive subset rather than truly being a benefit in the entire population. Nonetheless, an advantage of this design is that data will have been collected on biomarker-negative patients, and one could perform exploratory analyses to examine for possible treatment effects in the biomarker-negative subgroup.
To date, no studies using this two-step approach have been reported. Most reports of biomarker and therapeutic studies in the literature and most FDA-approved drug and biomarker assay combinations have been based on enrichment designs. These have been pragmatic and facile but have not occurred without surprises. For example, as noted above, cetuximab appears to benefit both EGFR-positive and EGFR-negative patients when EGFR is assessed by immunohistochemistry (29). Given the specificity of the antibody for target, it is possible that the apparent benefit to the immunohistochemically defined EGFR-negative group might reflect the limitations of the assay or biomarker in defining the patients most likely to benefit, although other biological explanations continue to be explored.
Alternative Approaches
Other approaches using adaptive biomarker enrichment have been suggested (55,56). Adaptive clinical trial designs permit midtrial modifications, which are based on incorporating new interim information either from inside or outside the trial. Adaptive designs may be Bayesian (using previous information gathered outside of the trial updated with information collected in the trial), but do not have to be. The flexibility of adaptive designs permits changes during the conduct of the trial in features such as sample size, treatment allocation ratios, and the number of interim analyses, as well as the selection of treatments and doses. For example, using adaptive designs, patients enrolled in a drug intervention clinical trial could be classified initially on the basis of the presence or absence of selected sets of biomarkers. As new drug efficacy and biomarker data become available, those data could be used to change the probability that newly accrued patients receive certain treatments depending on their biomarker profiles. Adaptive designs, like any other design, must adhere to regulatory requirements for maintaining study integrity and patient safety. They need to be designed with specific operating characteristics in mind. For most study designs, this requires specification of type I and type II error rates; for Bayesian designs, more extensive evaluations, often through a series of computer simulation studies, are required to fully evaluate their operating characteristics.
Other approaches, including biomarker-based strategies (57,58) and the two-step procedures described above (53,54) have recently been published. Further work in these areas should be encouraged.
Decision Points and Their Implications: Go–No Go Decisions for the Study of a Drug–Biomarker Combination
Pharmaceutical and device manufacturers working to market codeveloped products are urged to bring all the needed expertise to the development process, including FDA regulatory staff and third-party payers, to make the appropriate scientific and business decisions. These experts should be convened near the beginning of the process to ensure that challenges and obstacles are identified and addressed early in the product life cycle with minimum surprises and to consider novel business and marketing arrangements to share both risks and rewards for development of these products.
The Timing of Decisions About Studying a Drug–Biomarker Combination
Consideration of the integration of biomarker development into the drug development process should occur as early as possible because of the implications for biospecimen and data collection. Early implementation of standardized procedures for biospecimen and clinical data collection and efforts devoted to characterizing biomarker targets can maximize the utility of the specimens and provide information on the targets useful in addressing future questions regarding variations in biomarker expression or modulation. Decisions on how to study a biomarker in a particular clinical trial depend on the context of the study. Key considerations include factors such as the need to collect safety data, availability of reliable assays for evaluating the biomarker and drug activity, and relationship of results to clinical benefit (see the “Preclinical and Nonclinical Development” and “Clinical Development” sections of Figure 1).
Timing of Decisions Regarding Codevelopment of Biomarker Assays
A long lead time may be needed to develop well-characterized biomarker assays. The goal is to be prepared for phase III by ensuring that appropriate biomarker-based selection strategies are in place. For example, better-developed selection strategies might have more efficiently identified the positive effects of EGFR inhibitors in early phase III trials in patients with lung (23,50) and colorectal carcinomas (31,33,34).
In some cases, practical business considerations may outweigh benefits of a science-based strategy because the incentives for producing biomarker assays may be substantially misaligned for the diagnostics and pharmaceutical companies during different stages of development. The long time to market is a financial risk and critical issue for both the pharmaceutical and diagnostics industries, but it is a higher risk for the diagnostics industry during early biomarker and assay development. At this stage, not much is known about the drug or the biomarker, and the chosen biomarker might not ultimately be suitable or the drug might not prove sufficiently effective to warrant full development. Well-annotated biospecimens collected using standard protocols and appropriate consents are critical to allow biomarker assay development to take place when there is sufficient knowledge available about the drug and its relationship to the biomarker. The development of a shared repository of specimens from patients receiving the treatment of interest could help to address this need.
The lack of a rigorously evaluated biomarker at the end of phase II and the large sample size required to validate a biomarker assay increase the risk to the pharmaceutical company in later development stages. Secondary approvals (cross-labeling studies)—for example, taking a drug and biomarker combination approved in a narrowly defined population into a second population or evaluating a second drug from the same chemical or mechanistic class in combination with a biomarker—are less risky for both drug and biomarker assay development.
The perceived market worth for drugs and biomarkers may also be mismatched. The drug developed with the biomarker is a value generator, whereas the diagnostic may be perceived as a commodity (sometimes offered at no cost by the pharmaceutical company). Higher reimbursement rates for diagnostic assays used in patient management would likely provide needed incentives to the diagnostics industry.
The FDA usually recommends validating biomarkers prospectively to ensure study of appropriate populations, to minimize sampling bias, and to maximize the likelihood of having adequate power to evaluate the hypothesis being tested. This ideal study approach is most appealing if it can be performed in a time- and cost-efficient manner. If timelines for studies are long, and if study size requirements are large, companies may be reluctant to involve themselves in the uncertain process of companion product development, particularly in a competitive market with rapid changes in drugs, diagnostics, and clinical decision making. If companies were able to access well-constructed biological specimen collections and clinical data in early decision making, more robust biomarker-drug candidates and more focused phase III clinical studies might result.
If companies were able to develop well-planned incremental evaluations of drug-companion diagnostic pairs across phase I, II, and III studies that provide for better product decision making or in terms of trial design and size, the cost–benefit of these incremental evaluations could potentially revolutionize the development pipeline for new drugs and diagnostics. Unfortunately, it is too early to fully understand the financial costs and risks of this approach.
Well-planned incremental evaluations of drug-companion diagnostic pairs across phase I, II, and III studies that provide for better decision making in terms of trial design would ideally increase the efficiency of the drug development process. However, limits to biological understanding and resources can make it difficult to identify or carry out an optimal sequence of evaluations in a prospective fashion. Retrospective biomarker analyses using specimens previously collected within treatment trials may also be helpful, either in the discovery process, in fine-tuning developmental plans, or in some cases in supporting submissions to the regulatory agencies. The potential biases, effects of multiplicity, missing data, and other deficiencies of such analyses must be addressed. In addition, the statistical power for detecting biomarker effects in such retrospective studies will often be low. However, the results might identify promising new or improved biomarker assays that could be introduced into phase III studies. The FDA continues to seek input from the research and development community on these issues, but there is still no clear strategy for the approval of a predictive diagnostic with therapeutic agent using the retrospective approach (59).
There are situations in which biomarker assay approvals may be based on retrospective analysis alone. For example, a safety biomarker assay might be approved based on retrospective analysis of biomarker-positive and biomarker-negative specimens from patients who have been treated with approved drugs and in whom the safety outcomes are known. Early consultation with FDA scientists from both drug and device offices is encouraged when considering these strategies.
Recommendations
The workshop participants addressed scientific, regulatory, and business-related issues that affect the success or lack thereof in developing biomarkers for clinical decision making. Discussions of lessons learned in the development processes for trastuzumab and the EGFR inhibitors made it clear that the more known about the biology of the biomarker and its relationship to the therapeutic agent, the easier it is to determine the clinical trial design most appropriate for evaluating the utility of the biomarker assay for predicting the clinical effects of the agent. Major challenges for biomarker assay and therapeutic agent codevelopment relate to the costs of standardizing and evaluating the utility of an assay when neither the clinical activity of the agent nor the relationship of the biomarker to the mechanism(s) of action of the agent being developed is clear.
The following recommendations were made to address the challenges identified:
Consider the potential need for a predictive biomarker assay early in development of the therapeutic agent to facilitate coordinated development of both biomarker assay and therapeutic agent.
Build evidence from literature and preclinical, observational, and early clinical studies supporting a strong biological rationale for the biomarker and therapeutic agent interaction being predictive of therapeutic benefit.
Convene all the relevant diagnostic, pharmaceutical, and regulatory representatives early in the process to establish performance requirements, the developmental path and timeline, and go–no go decision points for the therapeutic agent and biomarker assay combination. Well-defined development goals and milestones should be established for effective go–no go decisions for both the therapeutic agent and the biomarker assay.
Early discussions with scientists from the regulatory agencies are critical. The regulatory preference for data from prospective studies is often problematic for device developers. Alternative trial designs should be considered.
Consider practical aspects of the proposed assay. Will the technology be robust, economical, and otherwise feasible in the proposed clinical setting?
Consider specimen needs early to ensure that appropriate biospecimens are collected with consent for future research use and that they are handled and stored properly. Informed consents that allow for flexibility in the future use of collected samples may be required.
Efforts should be made to collect specimens in a standardized way throughout the biomarker assay and therapeutic agent development process so that valid retrospective analyses can be carried out to facilitate assay performance and clinical utility evaluations.
Careful plans for pathological and demographic data collection as well as clinical data are necessary for the biomarker evaluations. The documentation regarding the collected specimens will be critical because clinical trials often extend for years and new technologies for marker measurements may be developed during this time. Well-planned specimen collection and documentation throughout the therapeutic agent development process (preclinical and clinical phases I, II, and III) could facilitate evaluation of biomarker assays retrospectively.
Finally, the misalignment of incentives for production of biomarker assays vs therapeutic agents continues to hamper codevelopment of agents and biomarkers. The resolution of this problem is not straightforward but some suggestions were made. Higher reimbursement for diagnostic tests, particularly those that are required for choice of therapeutic, would probably provide additional incentive to the assay developer. The many challenges identified during the discussions at the workshop and catalogued in this commentary and in Figure 1 will take time and community effort to address. We focused on discussion of alternative trial designs, the need for early comprehensive planning, and consideration of factors affecting business decisions for assay and drug developers. We hope that this will serve as the beginning of a productive dialog in the community.
Funding
National Cancer Institute. C.S. also received National Cancer Institute support for scientific and editorial assistance.
Footnotes
This article was developed from the presentations and discussions at the workshop entitled, NCI/FDA/Industry Workshop on Development of Markers for Clinical Decision-Making, held at the DoubleTree Hotel Bethesda, Bethesda, MD, on October 29–30, 2007.
During the planning and implementation of the workshop and preparation of the manuscript, Dr Taube was Associate Division Director of the Division of Cancer Treatment and Diagnostics, Director of the Cancer Diagnosis Program, National Cancer Institute; Dr Dancey was in the Cancer Therapy Evaluation Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute; and Dr Gutman was Director of the Office of In Vitro Diagnostic Device Safety and Evaluation at the US Food and Drug Administration.
The authors wish to thank the members of the Workshop Planning Committee (in addition to the authors) for their valuable contributions to the content and the agenda: Deborah Jaffe (NCI, Office of the Director), Francis Kalush (FDA, Center for Devices and Radiological Health, Office of the Center Director), Patricia Keegan (FDA, Center for Drug Evaluation and Review, Office of New Drugs, Office of Oncology Drug Products, Division of Biologic Oncology Products), Soon Myung Paik (NSABP), Raj Puri (FDA, Center for Biologics Evaluation and Research, Office of Cellular Tissue and Gene Therapies, Division of Cellular and Gene Therapies), Richard Schilsky (University of Chicago, Division of Biological Sciences, Department of Hematology/Oncology). The authors also thank the workshop participants for their thoughtful practical presentations and discussions and the several participants who reviewed and provided commentary on the manuscript.
Dr Clark holds stock in OSI Pharmaceuticals, Inc, the maker of erlotinib, an epidermal growth factor receptor inhibitor.
References
- 1.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 2.Pegram MD, Pauletti G, Slamon DJ. HER-2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res Treat. 1998;52(1–3):65–77. doi: 10.1023/a:1006111117877. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 4.Bange J, Zwick E, Ullrich A. Molecular targets for breast cancer therapy and prevention. Nat Med. 2001;7(5):548–552. doi: 10.1038/87872. [DOI] [PubMed] [Google Scholar]
- 5.Ross JS, Fletcher JA, Linette GP, et al. The Her-2/neu gene and protein in breast cancer 2003: biomarker and target of therapy. Oncologist. 2003;8(4):307–325. doi: 10.1634/theoncologist.8-4-307. [DOI] [PubMed] [Google Scholar]
- 6.Pegram MD, Finn RS, Arzoo K, Beryt M, Pietras RJ, Slamon DJ. The effect of HER-2/neu overexpression on chemotherapeutic drug sensitivity in human breast and ovarian cancer cells. Oncogene. 1997;15(5):537–547. doi: 10.1038/sj.onc.1201222. [DOI] [PubMed] [Google Scholar]
- 7.Carlomagno C, Perrone F, Gallo C, et al. c-erb B2 overexpression decreases the benefit of adjuvant tamoxifen in early-stage breast cancer without axillary lymph node metastases. J Clin Oncol. 1996;14(10):2702–2708. doi: 10.1200/JCO.1996.14.10.2702. [DOI] [PubMed] [Google Scholar]
- 8.Vogel CL, Cobleigh MA, Tripathy D, et al. First-line Herceptin monotherapy in metastatic breast cancer. Oncology. 2001;61(suppl 2):37–42. doi: 10.1159/000055400. [DOI] [PubMed] [Google Scholar]
- 9.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17(9):2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 10.Slamon D, Eiermann W, Robert N, et al. BCIRG 006: 2nd interim analysis phase III randomized trial comparing doxorubicin and cyclophosphamide followed by docetaxel (ACT) with doxorubicin and cyclophosphamide followed by docetaxel and trastuzumab (ACTH) with docetaxel, carboplatin and trastuzumab (TCH) in Her2neu positive early breast cancer patients. In: 29th Annual San Antonio Breast Cancer Symposium; December 14–17, 2006; San Antonio, TX. [Google Scholar]
- 11.Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med. 2006;354(8):809–820. doi: 10.1056/NEJMoa053028. [DOI] [PubMed] [Google Scholar]
- 12.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 13.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 14.Smith I, Procter M, Gelber RD, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369(9555):29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 15.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [Also: Arch Pathol Lab Med. 2007;131(1):18] [DOI] [PubMed] [Google Scholar]
- 16.Carlson RW, Moench SJ, Hammond ME, et al. HER2 testing in breast cancer: NCCN Task Force report and recommendations. J Natl Compr Canc Netw. 2006;4(suppl 3):S1–S22. quiz S23–S24. [PubMed] [Google Scholar]
- 17.Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358(13):1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 18.Paik S, Kim C, Jeong J, et al. Benefit from adjuvant trastuzumab may not be confined to patients with IHC 3+ and/or FISH-positive tumors: central testing results from NSABP B-31 [abstract 511] J Clin Oncol. 2007;25(18S):5s. [Google Scholar]
- 19.Ross JS, Fletcher JA, Bloom KJ, et al. HER-2/neu testing in breast cancer. Am J Clin Pathol. 2003;120(suppl):S53–S71. doi: 10.1309/949FPQ1AQ3P0RLC0. [DOI] [PubMed] [Google Scholar]
- 20.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12(18):5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 21.Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33(4):369–385. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23(11):2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 23.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24(31):5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 25.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 27.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 28.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23(31):8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 29.Chung KY, Shia J, Kemeny NE, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23(9):1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 30.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13(10):2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 31.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 32.Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 33.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 34.Raponi M, Winkler H, Dracopoli NC. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol. 2008;8(4):413–418. doi: 10.1016/j.coph.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Rusch V, Klimstra D, Venkatraman E, Pisters PW, Langenfeld J, Dmitrovsky E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3(4):515–522. [PubMed] [Google Scholar]
- 36.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 37.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 38.Tsao AS, Tang XM, Sabloff B, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1(3):231–239. doi: 10.1016/s1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 39.Bell DW, Brannigan BW, Matsuo K, et al. Increased prevalence of EGFR-mutant lung cancer in women and in East Asian populations: analysis of estrogen-related polymorphisms. Clin Cancer Res. 2008;14(13):4079–4084. doi: 10.1158/1078-0432.CCR-07-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park JW, Kerbel RS, Kelloff GJ, et al. Rationale for biomarkers and surrogate end points in mechanism-driven oncology drug development. Clin Cancer Res. 2004;10(11):3885–3896. doi: 10.1158/1078-0432.CCR-03-0785. [DOI] [PubMed] [Google Scholar]
- 42.Drug-diagnostic co-development concept paper. http://www.fda.gov/downloads/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/UCM116689.pdf. Accessed September 22, 2009. [Google Scholar]
- 43.Guidance for industry and FDA staff: class II special controls guidance document: drug metabolizing enzyme genotyping system. http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm077933.htm. Accessed September 22, 2009. [Google Scholar]
- 44.Guidance on informed consent for in vitro diagnostic device studies using leftover human specimens that are not individually identifiable. Guidance for sponsors, institutional review boards, clinical investigators and FDA staff. http://www.fda.gov/RegulatoryInformation/Guidances/ucm127022.htm. Accessed September 22, 2009. [Google Scholar]
- 45.Pharmacogenetic tests and genetic tests for heritable markers. Guidance for industry and FDA staff. http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071075.pdf. Accessed September 22, 2009. [Google Scholar]
- 46.Bracey A. In vitro diagnostic devices: guidance for the preparation of 510(k) submissions. http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm094533.htm. Accessed September 22, 2009. [Google Scholar]
- 47.Guidance for industry and FDA staff: interactive review for medical device submissions: 510(k)s, original PMSa, PMA supplements, original BLAs, and BLA supplements. http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm089402.htm. Accessed September 22, 2009. [Google Scholar]
- 48.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. [Also: J Clin Oncol. 2005;23(36):9067–9072; Br J Cancer. 2005;93(4):387–391; Breast Cancer Res Treat. 2006;100(2):229–235; Nat Clin Pract Oncol. 2005;2(8):416–422; Eur J Cancer. 2005;41(12):1690–1696; Exp Oncol. 2006;28(2):99–105] [DOI] [PubMed] [Google Scholar]
- 49.Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clin Chem Lab Med. 2003;41(1):68–73. doi: 10.1515/CCLM.2003.012. [Also: Am J Clin Pathol. 2003;119(1):18–22; Ann Clin Biochem. 2003;40(pt 4):357–363; Clin Radiol. 2003;58(8):575–580; Clin Chem. 2003;49(1):1–6; Acad Radiol. 2003;10(6):664–669; Am J Roentgenol. 2003;181(1):51–55; Clin Biochem. 2003;36(1):2–7; Ann Intern Med. 2003;138(1):40–44; Radiology. 2003;226(1):24–28; BMJ. 2003;326(7379):41–44] [DOI] [PubMed] [Google Scholar]
- 50.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 51.Gutman S, Richter K, Alpert S. Update on FDA regulation of in vitro diagnostic devices. JAMA. 1998;280(2):190–192. doi: 10.1001/jama.280.2.190. [DOI] [PubMed] [Google Scholar]
- 52.Clark GM. Prognostic and predictive factors. In: Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Diseases of the Breast. Philadelphia, PA: Lippincott Williams & Wilkins; 2000. pp. 489–514. [Google Scholar]
- 53.Freidlin B, Simon R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin Cancer Res. 2005;11(21):7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 54.Jiang W, Freidlin B, Simon R. Biomarker-adaptive threshold design: a procedure for evaluating treatment with possible biomarker-defined subset effect. J Natl Cancer Inst. 2007;99(13):1036–1043. doi: 10.1093/jnci/djm022. [DOI] [PubMed] [Google Scholar]
- 55.Jones CL, Holmgren E. An adaptive Simon two-stage design for phase 2 studies of targeted therapies. Contemp Clin Trials. 2007;28(5):654–661. doi: 10.1016/j.cct.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Chow SC, Chang M. Adaptive design methods in clinical trials—a review. Orphanet J Rare Dis. 2008;3((May 2)):11. doi: 10.1186/1750-1172-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sargent D, Allegra C. Issues in clinical trial design for tumor marker studies. Semin Oncol. 2002;29(3):222–230. doi: 10.1053/sonc.2002.32898. [DOI] [PubMed] [Google Scholar]
- 58.Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23(9):2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 59.Oncology Drugs Advisory Committee Meeting, December 16, 2008. FDA Briefing Document. http://www.fda.gov/ohrms/dockets/ac/08/briefing/2008-4409b1-01-FDA.pdf. Accessed September 22, 2009. [Google Scholar]