Abstract
We describe an adaptation of the rolling circle amplification (RCA) reporter system for the detection of protein Ags, termed “immunoRCA.” In immunoRCA, an oligonucleotide primer is covalently attached to an Ab; thus, in the presence of circular DNA, DNA polymerase, and nucleotides, amplification results in a long DNA molecule containing hundreds of copies of the circular DNA sequence that remain attached to the Ab and that can be detected in a variety of ways. Using immunoRCA, analytes were detected at sensitivities exceeding those of conventional enzyme immunoassays in ELISA and microparticle formats. The signal amplification afforded by immunoRCA also enabled immunoassays to be carried out in microspot and microarray formats with exquisite sensitivity. When Ags are present at concentrations down to fM levels, specifically bound Abs can be scored by counting discrete fluorescent signals arising from individual Ag–Ab complexes. Multiplex immunoRCA also was demonstrated by accurately quantifying Ags mixed in different ratios in a two-color, single-molecule-counting assay on a glass slide. ImmunoRCA thus combines high sensitivity and a very wide dynamic range with an unprecedented capability for single molecule detection. This Ag-detection method is of general applicability and is extendable to multiplexed immunoassays that employ a battery of different Abs, each labeled with a unique oligonucleotide primer, that can be discriminated by a color-coded visualization system. ImmunoRCA-profiling based on the simultaneous quantitation of multiple Ags should expand the power of immunoassays by exploiting the increased information content of ratio-based expression analysis.
Keywords: prostate-specific antigen, human IgE, IgG, ELISA, immuno-microarrays
Antibody-based detection systems for specific Ags are versatile and powerful tools for various molecular and cellular analyses, as well as clinical diagnostics (1). The power of such systems originates from the considerable specificity of Abs for particular antigenic epitopes. There are, however, numerous examples where important biological markers for cancer, infectious disease, or biochemical processes are present at too low a concentration in body fluids or tissues to be detected by using conventional immunoassays. Recent advances in the field of low-level Ag detection include the development of stronger fluorochromes and chemiluminescent substrates for use in ELISAs, immunofluorescence-based staining and immunoblotting (2), and the application of signal amplification methods such as tyramide deposition (3). Although these techniques can be quite powerful, greater sensitivity and specificity are often required, particularly when working with limited amounts of sample material or when Ag density is extremely low. With these needs in mind, we have adapted the recently described rolling circle amplification (RCA) reporter system (4) for the detection of protein Ags.
The use of DNA amplification for the detection of Abs bound to Ag has been documented previously (5–10). In immuno-PCR, a unique DNA sequence tag is associated with a specific Ab using streptavidin-biotin interactions, alternative bridging moieties, or covalent linkage. Abs bound to Ag are then detected by PCR amplification of the associated DNA tag. Multiple Abs and multiple DNA tags have been used (10) to analyze several Ags simultaneously. Although immuno-PCR was shown to be significantly more sensitive than ELISA, gel electrophoresis was required after DNA amplification in solution to separate and/or quantitate the different amplified DNA tags. The requirements for thermal cycling and product separation by gel electrophoresis have restricted the widespread adoption of immuno-PCR as an alternative to ELISA and have precluded its utility in immunohistochemical or array formats.
RCA driven by DNA polymerase can replicate circularized oligonucleotide probes with either linear or geometric kinetics under isothermal conditions (4). Using a single primer, RCA generates hundreds of tandemly linked copies of the circular template within a few minutes. In ImmunoRCA, the 5′ end of this primer is attached to an Ab. In the presence of circular DNA, DNA polymerase, and nucleotides, the rolling circle reaction results in a DNA molecule consisting of multiple copies of the circle DNA sequence that remains attached to the Ab (Fig. 1). The amplified DNA can be detected in a variety of ways, including direct incorporation of hapten-labeled or fluorescently labeled nucleotides, or by hybridization of fluor-labeled or enzymatically labeled complementary oligonucleotide probes. ImmunoRCA, therefore, represents a novel approach for signal amplification of Ab–Ag recognition events. Although RCA reactions can be carried out with either linear or geometric kinetics (4), the signal-generation paradigm we have used in this study is based exclusively on the linear RCA model. Here, we describe the construction of Ab–DNA conjugates and demonstrate the utility of using these conjugates to detect Ags in several different immunoRCA formats.
Figure 1.
Schematic of immunoRCA assay. (Top Left) A reporter Ab conjugated to an oligonucleotide binds to a test analyte that is captured on a solid surface by covalent attachment or by a capture Ab. (Top Right) A DNA circle hybridizes to a complementary sequence in the oligonucleotide. (Bottom Left) The resulting complex is washed to remove excess reagents, and the DNA tag is amplified by RCA. (Bottom Right) The amplified product is labeled in situ by hybridization with fluor-labeled oligonucleotides.
Methods
Oligonucleotide Synthesis.
All oligonucleotides used in this study were synthesized on a PerSeptive Biosystems (Framingham, MA) Expedite DNA Synthesizer and purified by reverse-phase HPLC. Thiol- and fluor-modified phosphoramidites were purchased from Glen Research (Sterling, VA) and Amersham Pharmacia, respectively. Circle DNAs were constructed as previously described (4). The sequences used for immunoRCA detection of (i) prostate-specific Ag (PSA) and avidin were as follows: conjugate primer 1, 5′-thiol-AAA AAA AAA AAA AAA CAC AGC TGA GGA TAG GAC AT-3′; circle 1, 5′-CTC AGC TGT GTA ACA ACA TGA AGA TTG TAG GTC AGA ACT CAC CTG TTA GAA ACT GTG AAG ATC GCT TAT TAT GTC CTA TC-3′; detectors, 5′-FITC-AAC AAC ATG AAG ATT GTA-DNP-3′, 5′-FITC-TCA GAA CTC ACC TGT TAG-DNP-3′, 5′-FITC-ACT GTG AAG ATC GCT TAT-DNP-3′; (ii) for IgE and sheep IgG: conjugate primer 2, 5′-thiol-GTA CCA TCA TAT ATG TCC GTG CTA GAA GGA AAC AGT TAC A-3′; circle 2, 5′-TAG CAC GGA CAT ATA TGA TGG TAC CGC AGT ATG AGT ATC TCC TAT CAC TAC TAA GTG GAA GAA ATG TAA CTG TTT CCT TC-3′; detectors, 5′-Cy3-TAT ATG ATG GTA CCG CAG-Cy3–3′, 5′-Cy3-TGA GTA TCT CCT ATG ACT-Cy3–3′, 5′-Cy3-TAA GTG GAA GAA ATG TAA-Cy3–3′ [for CACHET (condensation of amplified circles after hybridization of encoding tags) experiments, detectors for circle 2 had DNP instead of Cy3 on the 3′ ends].
Ab–DNA Conjugates.
The following Abs were used to construct DNA conjugates in the course of this study: rabbit anti-mouse IgG (generously provided by DAKO), mouse monoclonal anti-human IgE, mouse monoclonal anti-biotin, rabbit anti-sheep IgG, and rabbit anti-avidin. The conjugation of one of the Abs, the mouse mAb against human IgE, is described in the supplemental data on the PNAS web site (www.pnas.org) as an example for the other conjugations.
ELISA.
Ninety-six-well plates (Nunc Maxisorb) were incubated with 100 μl of 2 μg/ml goat polyclonal anti-human IgE per well for 2 h at 37°C and then overnight at 4°C. Plates were washed three times with 100 μl of Tris-buffered saline (TBS)/0.05% Tween 20, and then blocked with 5% nonfat dry milk for 2 h at 37°C. Plates were washed again, followed by addition of the IgE analyte at variable concentrations in a 100-μl volume. After a 37°C incubation for 30 min, plates were washed three times. In the conventional ELISA experiments, anti-human IgE–alkaline phosphatase conjugate was added to each well and incubated at 37°C for 30 min. After plates were washed, the alkaline phosphatase substrate 4-methylumbelliferyl phosphate (MUP) was added, and fluorescence levels were read after 20 min on a BioTek (Winooski, VT) FL600 plate reader at an excitation wavelength of 360 nm and an emission wavelength of 460 nm. In the immunoRCA experiments, anti-human IgE–DNA conjugate (5 ng/μl) was added to each well in a 60-μl volume and incubated at 37°C for 30 min. After plates were washed three times, circle 2 DNA (170 nM) in 60 μl of ϕ29 buffer (250 mM Tris⋅HCl (pH 7.5)/50 mM MgCl2/1 mg/ml BSA/1 mM dATP/1 mM dCTP/1 mM dGTP/0.75 mM dTTP/0.25 mM FITC-12-dUTP) was added to each well, and incubated at 37°C for 30 min. RCA reactions were initiated by addition of 1.5 μl of ϕ29 DNA polymerase (0.4 units/μl, generously provided by Amersham Pharmacia) and continued for 30 min at 37°C. RCA products were detected by addition of an anti-FITC–alkaline phosphatase conjugate. After a 37°C incubation for 30 min, plates were washed three times, 4-methylumbelliferyl phosphate (MUP) substrate was added, and fluorescence levels were read after 20 min.
Magnetic Bead Immunoassay.
Streptavidin-coated magnetic beads (Bangs Laboratories, Carmel, IN) were coated with a solution of 16 μg/ml biotinylated polyclonal anti-human IgE (PharMingen) in TBS, washed three times in TBS/0.05% Tween 20, and blocked overnight with 2 mg/ml BSA. Beads were incubated with human IgE (25 ng/ml) in TBS for 20 min at room temperature and washed three times. Detection of IgE by using a conventional anti-IgE-alkaline phosphatase conjugate or an immunoRCA conjugate was carried out as described above for the ELISA.
Detection of PSA on Microspots.
Preparation of microspots.
Clean glass slides were chemically functionalized by immersing in a solution of mercaptopropyltrimethoxysilane [1% vol/vol in 95% ethanol (pH 5.5)] for 1 h. Slides were rinsed in 95% ethanol for 2 min, dried under nitrogen, and heated at 120°C for 4 h. Thiol-derivatized slides were activated by immersing in a 0.5 mg/ml solution of the heterobifunctional crosslinker N-[γ-maleimidobutyryloxy]succinimide ester (GMBS; Pierce) in 1% dimethylformamide, 99% ethanol for 1 h at room temperature. Slides were rinsed with ethanol, dried under nitrogen, and stored in a vacuum dessicator until use. Spotting of goat anti-PSA polyclonal Ab (BioSpecifics, Lynbrook, NY) onto the slides was accomplished by pipetting 0.2 μl of 0.5 mg/ml solution in a grid pattern. Hand-spotted arrays were blocked with 2% BSA (protease free), air-dried, and stored under nitrogen at 4°C until use.
Ag capture.
Purified human PSA (BiosPacifics, Emeryville, CA) was diluted in PBS to the desired concentration. Ten μl of the PSA dilution was spotted onto a coverslip (Hybrislip, Grace Biolabs Inc., Bend, OR) that was then inverted onto the slide over the area of the array. The slide was incubated at 37°C for 30 min in a humidified chamber, washed twice for 2 min in PBS/0.05% Tween 20, and tapped dry. Ten microliters of a 1:5,000 dilution in PBS of monoclonal anti-PSA Ab was added to the array. The slide was incubated at 37°C for 30 min in a humidified chamber, washed twice, and tapped dry. Finally, rabbit anti-mouse IgG-DNA conjugate (10 ng/μl in PBS) was added to the array and incubated at 37°C for 30 min in a humidified chamber. Slides were washed twice and tapped dry.
RCA reaction.
Circle 1 DNA (200 nM) in 10 μl ϕ29 buffer [250 mM Tris⋅HCl (pH 7.5)/50 mM MgCl2/1 mg/ml BSA] was added to the array and incubated at 45°C for 30 min. Ten microliters of RCA reaction mixture (2 mM dATP/2 mM dCTP/2 mM dGTP/1.5 mM dTTP/0.5 mM FITC-dUTP/ϕ29 buffer/0.4 units/μl ϕ29 polymerase) was added to the array and incubated at 37°C for 30 min. The slide was washed in 2× SSC/0.05% Tween 20 for 5 min with agitation at room temperature, followed by a wash in 2× SSC. Slides were dried under nitrogen, and the array portion of the slide was covered with Prolong Antifade (Molecular Probes). Fluorescent imaging was done on a Zeiss epifluorescence microscope equipped with a charge-coupled device (CCD) imaging system and a ×100 objective. Fluorescence quantitation was performed by using iplab software (Scanalytics, Inc., Fairfax, VA).
Detection of IgE on Microarrays.
Preparation of microarrays.
Glass slides were functionalized with thiol-silane and activated with GMBS as described above. A solution of polyclonal goat anti-human IgE (BiosPacifics; 0.5 mg/ml) was spotted onto the slides by using a pin-tool type microarrayer (GeneMachines, San Carlos, CA).
Antigen capture.
Each microarray was blocked by adding a 50-μl volume of a 2 mg/ml BSA solution in 50 mM glycine (pH 9.0) and incubating for 1 h at 37°C in a humidity chamber. After blocking, slides were twice washed by immersion of the slides into a coplin jar containing 1× PBS/0.05% Tween 20 and allowing the slides to stand in wash for 2 min followed by a 1-min 1× PBS wash. A 10-μl volume of human IgE at various concentrations was immediately added to each microarray and incubated for 30 min at 37°C in a humidity chamber, and then washed as described above.
ImmunoRCA.
Goat anti-human IgE (BioSpecifics) was labeled with biotin using the BiotinTag Micro Biotinylation Kit (Sigma). Ten microliters of the Ab at 2.5 ng/μl in PBS/0.05% Tween 20/1 mM EDTA was applied to each array and incubated at 37°C for 30 min in a humid chamber. Slides were washed twice for two minutes in PBS/0.05% Tween 20. A mouse monoclonal anti-biotin Ab conjugated to primer 1 was annealed with 50 nM circle 1 in PBS/0.05% Tween 20/1 mM EDTA at 37°C for 30 min. Ten microliters was applied to each array and incubated at 37°C for 30 min in a humid chamber, and then slides were washed twice. A 20-μl volume of reaction solution containing T7 native DNA polymerase (0.01 units/μl)/1 mM dNTPs/0.04 mg/ml ssDNA-binding protein/20 mM Tris⋅HCl (pH 7.4)/10 mM MgCl2/25 mM NaCl was then added to each microarray. The slides were incubated at 37°C for 45 min, and the reaction was stopped by washing the slides in a 2× SSC/0.05% Tween 20 solution at room temperature. A 20-μl solution of 0.5 μM DNA decorators was added to each array and allowed to hybridize to the RCA product for 30 min at 37°C. Slides were washed in 2× SSC at room temperature and spin-dried. Slides were scanned in a General Scanning (Watertown, MA) Luminomics 5000 microarray scanner, and fluorescence was quantitated by using quantarray software.
Detection of Single Ab-Ag Complexes.
Preparation of capture slides.
Slides were coated with 4-aminobutyl-dimethylmethoxysilane and derivatized with 1,4-phenylene-diisothiocyanate as previously described (11). A mixture of an avidin capture reagent, the oligonucleotide 5′-NH2-GG18G-biotin-3′, and an anti-digoxigenin IgG capture reagent, digoxigenin-succinyl-ɛ-aminocaproic acid hydride, each at a concentration of 5 μM, were spotted onto the activated slide surface in an array format (8–10 spots/array). Chemical coupling was allowed to proceed for 2 h before blocking unreacted functional groups as previously described (11).
Binding of Ags to capture slides.
Various concentrations of avidin and sheep antidigoxigenin IgG, either singly or in defined molar ratios, were spiked into normal human serum. The spiked serum (1.0 μl) was applied to the capture arrays, and the slides were incubated in a moist chamber at 37°C for 30 min. The slides then were washed twice in PBS/Tween 20 and air-dried. Five microliters of a mixture of 7.5 nM rabbit anti-avidin-primer-1 conjugate and 7.5 nM rabbit anti-sheep IgG Ab-primer-2 conjugate was applied to each array spot and incubated at 37°C for up to 2.5 h. The slides were washed eight times (1 min each) and air-dried. Five microliters of 0.2 μM solution of the two circular probes (circle 1 and circle 2) in 2× SSC/0.1% Tween 20/3% BSA/0.1% sonicated herring sperm DNA was applied to each spot. After hybridization at 37°C for 20 min, the slides were washed with 2× SSC/0.1% Tween 20 at 37oC for 5 min and air-dried.
RCA-CACHET Reactions.
Five microliters of RCA reaction mixture (50 mM NaCl/50 mM Tris⋅HCl (pH 7.2)/5 mM MgCl2/0.5 mM each dNTP/1 mM DTT/0.04 mg/ml ssDNA-binding protein/0.5 units/μl Sequenase) was applied to each spot, and the slides were incubated at 37°C for 15 min. After the slides were washed, they were air-dried, and 5 μl of a solution containing 0.2 μM double labeled (fluor + 2,4-DNP) detector probes were applied to each array. Slides were incubated in the moist chamber for 30 min at 37°C, then washed five times and air-dried. To collapse the RCA products into point sources of fluorescence so that single Ag–Ab complexes could be enumerated, 5 μl of 33 nM sheep anti-DNP IgM in PBS was added to each array spot, and slides were incubated at 37 oC for 45 min, washed for 3 min in 2× SSC at room temperature, and then air-dried. Prolong Antifade solution (Molecular Probes) was applied to the slide, and the slide was covered with a 20× 20-mm coverslip. Separate FITC and Cy3 fluorophore microscope images were captured by using a ×63 objective lens, and individual RCA products in each field were counted manually. The FITC and Cy3 image were merged electronically, and RCA signals were pseudocolored green and red.
Results
Preparation and Characterization of Ab–DNA Conjugates.
The covalent coupling strategy described by Hendrickson et al. (10) was used with several modifications to construct Ab–DNA conjugates for immunoRCA applications. Each batch of conjugate synthesized was subjected to several quality control checks, including agarose gel (Fig. 6, see the supplemental data) and SDS/PAGE gel analyses (not shown). In addition, competitive ELISA experiments were carried out to assess the ability of the conjugate to bind cognate Ag. In these assays, the matching unconjugated and DNA-conjugated Abs were assessed in parallel for their ability to compete with a reporter Ab for binding to Ag. The conjugated Abs, each coupled to ≈3 oligonucleotides per mole of protein, exhibited nearly equivalent avidity for Ag as the unconjugated forms (Fig. 7, see the supplemental data). Finally, the ability of the Ab–DNA conjugates to serve as primers for RCA reactions was examined. Ab–primer conjugate gave more RCA reaction product than an equimolar amount of unconjugated primer in the presence of a complementary circle DNA (Fig. 8, see the supplemental data), consistent with the observation that each Ab is conjugated to more than one primer. Neither form of primer gave an appreciable product in the absence of complementary circle or in the presence of a noncomplementary circle (data not shown).
Comparison of Conventional and RCA-Based ELISAs.
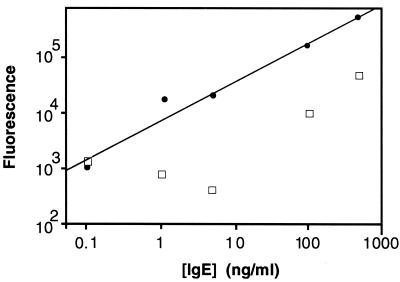
ImmunoRCA assays were first investigated in a single analyte ELISA format to demonstrate the feasibility of the sandwich configuration and to confirm the utility of the Ab–DNA conjugates. Human IgE was selected as the first test analyte because of its clinical importance in the assessment of allergic disorders (12). The immunoRCA sandwich assay for human IgE was performed by using microtiter plates coated with a polyclonal anti-human IgE capture Ab. Two ELISAs were carried out. In one assay, the reporter Ab was a monoclonal anti-human IgE Ab conjugated to a 40-mer oligonucleotide (primer-2) consisting of a primer sequence that is complementary to a portion of a DNA circle designated circle 2; this conjugate was used in an immunoRCA reaction as described in Methods. The lowest level of IgE that could be detected over background (no IgE) was 0.1 ng/ml (Fig. 2). This result indicated that the Ab–DNA conjugate formed an effective sandwich complex in response to IgE; furthermore, it established that a covalently coupled oligonucleotide can function effectively as an RCA primer both in solution (see above) and as a surface-immobilized protein adduct. In the conventional ELISA, the reporter Ab was the monoclonal anti-human IgE Ab conjugated to alkaline phosphatase. As shown in Fig. 2, the immunoRCA assay gave a dose-response over a greater range of IgE concentration than the conventional assay. In addition, the immunoRCA assay, even in its less amplified linear mode, could detect IgE levels approximately two orders of magnitude lower than those detected by the conventional ELISA assay.
Figure 2.
Comparison of immunoRCA and conventional immunoassay in an ELISA format. Filled circles, ELISA of human IgE with immunoRCA, using an anti-human IgE-DNA conjugate; open squares, ELISA of human IgE with an anti-human IgE-alkaline phosphatase conjugate.
Comparison of Conventional and RCA-Based Microparticle Immunoassays.
The utility of immunoRCA in a microparticle format was also investigated to examine the flexibility of the method. Microparticle assays are often the format of choice for clinical immunoanalyzers because of favorable performance characteristics for high-throughput automated platforms (13). Human IgE was again selected as the test analyte, but this sandwich assay was performed by using avidin-coated magnetic microparticles and biotinylated polyclonal anti-human IgE capture Abs. ImmunoRCA with the anti-IgE-DNA conjugate gave approximately 75-fold more signal than detection with an anti-IgE-alkaline phosphatase conjugate with the same amount of input IgE (25 ng IgE/ml).
Ultrasensitive Detection of PSA on Microspots.
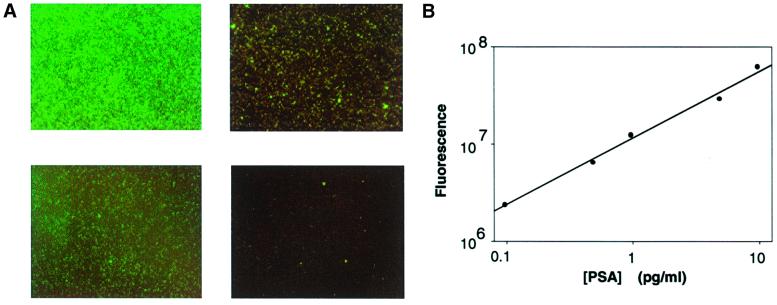
Several features of immunoRCA that make it suitable for solid-phase detection were demonstrable by using detection of PSA as a model system. For this application, immunoRCA was configured in an indirect sandwich assay format. Briefly, goat anti-PSA polyclonal Ab was immobilized on thiol-silane-coated microscope slides that had been activated with the heterobifunctional crosslinker GMBS. Purified human PSA in various concentrations was added, and the slide was incubated at 37°C and then washed to remove unbound protein. A mouse monoclonal anti-PSA Ab was used to form the second part of the immuno-sandwich complex. This complex was detected with a polyclonal rabbit anti-mouse IgG Ab that had been conjugated to an oligonucleotide containing a sequence for priming an RCA reaction. RCA was carried out by the addition of reaction mixture containing circle DNA, DNA polymerase, and dNTPs including FITC-dUTP. As shown in Fig. 3A, in this direct labeling protocol, fluorescence was clearly detectable over background (no PSA) at 0.1 pg/ml PSA. Quantitation of the fluorescence with a CCD-camera-equipped microscope indicated that the signal was linear over at least 2 logs of PSA concentration (Fig. 3B).
Figure 3.
Detection of PSA by immunoRCA in a microspot assay. (A) Fluorescent microscope images of immunoRCA signals obtained with different concentrations of human PSA captured on microspots of anti-human PSA Ab. Concentrations: (Upper Left) 5 pg/ml PSA; (Lower Left) 0.5 pg/ml PSA; (Upper Right) 0.1 pg/ml PSA; (Lower Right) 0 pg/ml PSA. (B) Fluorescence in microscope images was quantitated as described in Methods and plotted versus PSA concentrations incubated on microspots.
Detection of IgE on Glass Microarrays.
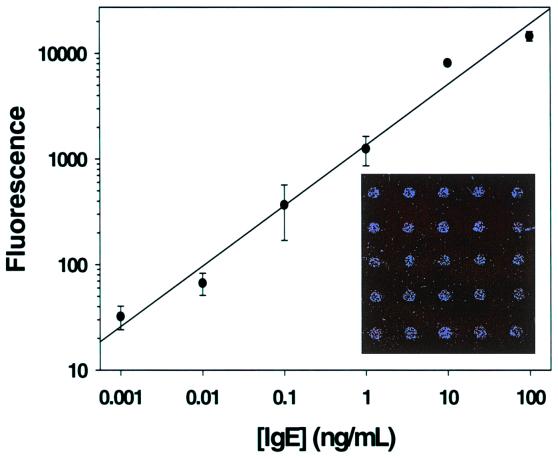
The results with PSA demonstrated that immunoRCA could be used for ultrasensitive Ag detection on a glass surface. These experiments, however, were carried out by using hand-spotted arrays and a CCD-camera for detection, limiting the density of the arrays and the dynamic range of the detection, respectively. Therefore, similar immunoRCA experiments were performed in a sandwich format on microarrays of polyclonal goat anti-human IgE Ab spotted onto glass slides by using a pin-tool type microarraying robot. In these microarrays, approximately 0.5 nl of Ab solution was deposited in each spot, spots had a diameter of approximately 200 μm, and the spot-to-spot spacing was 250 μm. The anti-IgE microarrays were incubated with human IgE, and bound Ag was detected with a biotinylated anti-human IgE Ab and an anti-biotin mAb that had been conjugated to an oligonucleotide containing an RCA-priming sequence. RCA was carried out as described in Methods, and the RCA product was detected by hybridization with a complementary oligonucleotide labeled with the fluorophore Cy3. Fluorescence was measured with a microarray scanner, with a dynamic range of approximately 5 logs. As shown in Fig. 4 Inset, microarray spots were clearly seen at low (1 ng/ml) levels of IgE. In other experiments, as little as 1 pg/ml IgE was detected by immunoRCA, and with a dynamic range of approximately 5 logs (Fig. 4).
Figure 4.
ImmunoRCA anti-human IgE microarray dose-response for purified IgE. Signals from six microarray spots were averaged for each point, and the background (no IgE) signal was subtracted. (Inset) Microarray scanner image of anti-human IgE array incubated with 1 ng/ml IgE.
Direct Quantitation of Individual Ag–Ab Complexes.
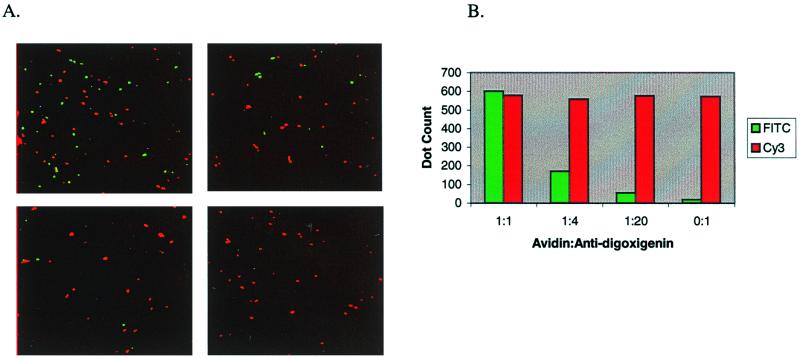
RCA is a generic reporter system for the detection of immobilized nucleic acid analytes and has already been shown to permit the visualization and quantitation of individual nucleic acid hybridization events, including discrimination of point mutations by imaging of fluorescent dots by using a two-color labeling system (4). To test the feasibility of detecting two (or more) proteins simultaneously in a single-molecule-counting mode, avidin and sheep anti-digoxigenin IgG were chosen as test analytes because of their wide-spread use as detector reagents in molecular studies. Biotin and digoxigenin, in a 1:1 ratio, were covalently coupled to glass slides to act as analyte capture reagents and were spotted in an array format. Avidin and anti-digoxigenin IgG were mixed in different ratios, diluted in serum to simulate complex biological samples, and then spotted onto the array. The two Ags then were detected with anti-avidin-primer-1 and anti-sheep IgG-primer-2 conjugates. The immunoRCA products were condensed to 0.3- to 0.7-μm spots by crosslinking with an anti-DNP IgM (the CACHET reaction), and single fluorescence spots were counted during microscopic examination. The discrete fluorescence signals had either a pure fluorescein or pure Cy3 spectra (Fig. 5A). Whereas the possibility exists that some of these signals arose from aggregates of analyte molecules, the absence of signals with mixed spectra (yellow) indicated that each dot was generated by a single Ab–Ag complex. Quantitation of the avidin (green) and sheep anti-digoxigenin IgG (red) signals demonstrated that the ratio of green/red signals closely corresponded to the known input ratios of the two protein Ags (Fig. 5B), further suggesting a 1-to-1 correspondence between Ab–Ag complexes and signals.
Figure 5.
Dual Ag detection using immunoRCA-CACHET. (A) The RCA products from the anti-avidin and the anti-sheep Ab conjugates were decorated with fluorescein- and Cy3-labeled detector oligonucleotides, respectively. Each image shown is the superimposition of two separate images, with fluorescein and Cy3 signals pseudocolored in green and red, respectively. (Upper Left) Avidin:anti-digoxigenin (anti-dig) = 1:1; (Upper Right) avidin:anti-dig = 1:4; (Lower Left) avidin:anti-dig = 1:20; (Lower Right) avidin:anti-dig = 0:1. (B) Quantitation of avidin (FITC) and anti-digoxigenin (Cy3) signals.
We have examined the efficiency of the single molecule detection process as illustrated in the two-color experiment described above by titrating a known amount of a single Ag (avidin) and counting the number of RCA signals. A comparison of the number of input molecules with RCA signals produced indicated that the overall efficiency of molecular detection ranged from 0.5% to 5% in different experiments. These studies provided further confirmation that the sensitivity of immunoRCA was in the subattomole range (data not shown). It remains to be seen whether implementation of the geometric mode of RCA will provide further improvements in sensitivity.
Discussion
DNA tags are ideal molecular labels for multiple analyte detection because different specific sequences can be arbitrarily associated with each individual analyte. In most published examples of immunoPCR, a specific DNA tag is associated with an Ab by means of biotin–streptavidin bridges. Direct covalent coupling of DNA to Ab, as described by Hendrickson et al. (10), has advantages for the implementation of simple assay formats, because fewer reagent mixing and washing steps are required; furthermore, variability in the stoichiometry of the assembled components can be avoided. We have adopted the covalent coupling strategy used by Hendrickson et al., for use in a signal amplification strategy termed ImmunoRCA. By employing several modifications and improvements in the synthetic and purification strategies, these conjugates can be produced in high yields with a high degree of purity (supplemental Fig. 6). Characterization of these conjugates indicated that the Abs had not lost avidity for Ags (supplemental Fig. 7); in addition, the oligonucleotides retained efficiency for priming an RCA reaction (supplemental Fig. 8).
The detection of IgE by using an anti-IgE Ab-DNA conjugate in an ELISA format was used to establish the feasibility of immunoRCA-based assays (Fig. 2). Covalent linkage of the RCA primer to the reporter Ab guaranteed that the surrogate relationship between the analyte and the amplified RCA product was maintained throughout the assay. One tangible benefit of immunoRCA over conventional methods of signal amplification was a significantly enhanced immunoassay sensitivity (Fig. 2): the immunoRCA IgE sandwich assay exhibited a two to three order of magnitude increase in sensitivity over the conventional ELISA. This level of increased sensitivity was similar to that observed for immunoPCR in an ELISA format (10). In the immunoPCR assay, however, the amplified DNA products were first generated by thermocycling and then quantified separately by gel electrophoresis. Because the amplification products of the RCA reaction remain associated with the Ab in the immunoRCA assay, the signal amplification and read-out can be carried out in the same multiwell plate. Finally, immunoRCA is compatible with a number of DNA polymerases; three different polymerases (ϕ29, T7 native, and T7 Sequenase) were used in various experiments reported in the current study.
The features that make immunoRCA a powerful technology for signal amplification in ELISAs also give it an advantage for other immunoassay formats. ImmunoRCA was compatible with a microparticle-based immunoassay format; in fact, the signal observed in the immunoRCA experiment was significantly greater than that obtained with conventional signal amplification for the same amount of analyte. This result suggests that immunoRCA may be adaptable for use in automated clinical immunoanalyzers, which often use microparticles as their assay format (13). It should also be noted that no effort was made in this assay (or in any other assay described in this study) to optimize Ab choice, Ab matching, etc. for the immunoRCA process. Such optimization measures are anticipated to improve sensitivity of immunoRCA reactions further.
ImmunoRCA is ideally suited for microarray applications. During the entire isothermal RCA reaction, the resulting amplified DNA molecule remains covalently attached to the Ab–Ag complex. On microarrays, this process results in an approximately 3 log increase in detectable fluorescent signal over nonamplified signal detection approaches (data not shown). A distinctive feature of RCA is the ability to precisely localize signals arising from a single DNA reporter molecule, thus enabling the visualization of individual recognition events on a solid surface. When the long ssDNA product of RCA is decorated by hybridization to many complementary oligonucleotides labeled with both a reporter fluorophore and a hapten, such as 2,4-dinitrophenol, all of the fluorophores can be collapsed into a point source of light. When the number of molecular signals is extremely high, the signal from a spot on a microarray can be read by using aggregate fluorescence. When the number of surface-bound Ags is smaller, however, the signals can be scored as discrete single molecule counts (Figs. 4 and 5), and subattomoles of analyte can be visualized. ImmunoRCA carried out on microarrays thus provides assays with an extremely wide dynamic range; combining single molecule counting and total fluorescence output indicates a dynamic range between 6 and 7 logs. Incorporation of molecular beacon type detector probes (14) into the immunoRCA assay could also eliminate post hybridization washing steps because unhybridized beacons are nonfluorescent.
Proof-of-principle for the use of immunoRCA in a microspot or microarray format was obtained by using PSA, IgE, avidin, and anti-digoxigenin IgG as the test analytes. PSA is the current “gold-standard” serum tumor marker for the screening of men for prostate cancer and for persistence or recurrence of disease after therapy (15). Although elevation of PSA levels in peripheral blood in advanced disease can usually be monitored by standard immunodiagnostic procedures such as ELISAs, which have a sensitivity limit of approximately 100 pg/ml, this level of sensitivity may be insufficient for detecting residual disease after radical prostatectomy (16). As shown in Fig. 3, immunoRCA can detect as little as 0.1 pg/ml PSA (300 zeptomoles) in a microspot assay, which is three orders of magnitude more sensitive than standard immunoassays for PSA. The Ab used as the conjugate in this experiment was a rabbit anti-mouse IgG polyclonal Ab; this reagent can serve as a “universal” conjugate to detect any mouse mAb with high sensitivity. The ultimate sensitivity of any immuno-RCA assay will undoubtedly be limited by the specificity and avidity of the Ab–Ag binding recognition and the impact of large RCA products bound to the Ab on these parameters. Additional studies will be required to more precisely define these issues.
The utility of immunoRCA in microarray-based immunoassays was examined further by using high-density arrays of anti-IgE on a glass slide. The results from these experiments (Fig. 4) indicate that immunoRCA has high sensitivity, a wide dynamic range, and excellent spot-to-spot reproducibility. Recently, a microarray-based immunoassay was reported that used alkaline phosphatase conjugates and the fluorescent substrate ELF for detection (17). The ELF-based assay required a specially constructed CCD-based camera to read the signal, and a sensitivity of approximately 10 ng/ml was achieved. In another report, Abs labeled with a near-infrared dye were used to detect IgG subclasses on a microarray (18); this system also required a specially designed imaging system and achieved a detection limit of approximately 15 ng/ml. In contrast, signal amplification by immunoRCA gives sensitivity in the pg/ml range and allows the assay results to be read with commonly available microarray scanners. In other work not presented here, we have demonstrated that immunoRCA can be used to detect allergen-specific IgEs on microarrays with excellent clinical sensitivity and specificity (S.W., S.O., J.L., K.K., S.F.K., and B.S., unpublished data). The Ab used as the conjugate in this experiment was a monoclonal anti-biotin Ab; this reagent can be used to detect any biotinylated polyclonal or monoclonal Ab, as well as any other protein, nucleic acid, or small molecule that can be biotinylated.
A logical extension of immunoRCA is the detection of Ags in tissues and cytological specimens. Indeed, initial experiments indicate that immunoRCA can readily detect low abundance membrane receptor molecules and intranuclear Ags that are very difficult to visualize by using conventional immunohistochemical techniques (Maltzman, W., Visconti, R., Wheeler, V., and Kingsmore, S., unpublished data). Future applications of immunoRCA include array-based protein-expression-profiling (where multiple Ags can be detected and quantitated simultaneously, yielding precise information on their relative abundance), the measurement of protein–protein interactions, and drug discovery or toxicology. Finally, it is conceivable that DNA or RNA aptamers may be substituted for Abs as the recognition moieties in immunoRCA. For example, combinatorial selection has permitted the generation of DNA aptamers of relatively high affinity that bind the anion-binding exosite of thrombin (19), or the prion protein (20). If aptamer affinities could be improved to the point that they approached the binding constants typical of Ag–Ab interactions, then it may become possible to implement the use of aptamer-RCA reporter systems of considerable simplicity and power. The detection reagent in such an assay would be a synthetic oligonucleotide, combining in a single DNA molecule the dual functions of molecular recognition and RCA priming.
Supplementary Material
Acknowledgments
We thank Dr. Linhua Fang for characterizing the performance of the conjugates in solution.
Abbreviations
- CACHET
condensation of amplified circles after hybridization of encoding tags
- RCA
rolling circle amplification
- GMBS
N-[γ-maleimidobutyryloxy]succinimide ester
- PSA
prostate-specific Ag
- TBS
Tris-buffered saline
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170237197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170237197
References
- 1.Gosling J P. Clin Chem. 1990;36:1408–1427. [PubMed] [Google Scholar]
- 2.Diamandis E P, Christopoulos T K, editors. Immunoassay. San Diego: Academic; 1996. [Google Scholar]
- 3.van Gijlswijk R P, Zijlmans H J, Wiegant J, Bobrow M N, Erickson T J, Adler K E, Tanke H J, Raap A K. J Histochem Cytochem. 1997;45:375–382. doi: 10.1177/002215549704500305. [DOI] [PubMed] [Google Scholar]
- 4.Lizardi P M, Huang X, Zhu Z, Bray-Ward P, Thomas D C, Ward D C. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 5.Sano T, Smith C L, Cantor C R. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 6.Ruzicka V, Marz W, Russ A, Gross W. Science. 1993;260:698–699. doi: 10.1126/science.8480182. [DOI] [PubMed] [Google Scholar]
- 7.Zhou H R, Fisher J, Papas T S. Nucleic Acids Res. 1993;21:6038–6039. doi: 10.1093/nar/21.25.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano T, Smith C, Cantor C R. Science. 1993;260:699. doi: 10.1126/science.260.5108.699. [DOI] [PubMed] [Google Scholar]
- 9.Sano T, Cantor C R. BioTechnology. 1991;9:1378. doi: 10.1038/nbt1291-1378. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson E R, Hatfield T M, Joerger R D, Majarian W R, Ebersole R C. Nucleic Acids Res. 1995;23:522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Z, Guifoyle R A, Thiel A J, Wang R, Smith L M. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knauer K A, Adkinson N F. In: Allergy: Principles and Practice. Middleton P, Reed D, Ellis M, editors. St. Louis: Mosby; 1983. pp. 673–688. [Google Scholar]
- 13.Chan D W, editor. Immunoassay Automation: An Updated Guide to Systems. San Diego: Academic; 1996. [Google Scholar]
- 14.Tyagi S, Kramer F R. Nat Biotech. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Catalona W J, Smith D S, Wolfert R L, Wang T J, Rittenhouse H G, Ratliff T L, Nadler R B. J Am Med Assoc. 1995;274:1214–1220. [PubMed] [Google Scholar]
- 16.Lu-Yao G L, Mclerran D, Wasson J, Wennberg J E. J Am Med Assoc. 1993;269:2633–2655. doi: 10.1001/jama.269.20.2633. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza L G, McQuary P, Mongan A, R, Gangadharan R, Brignac S, Eggers M. Biotechniques. 1999;27:778–788. doi: 10.2144/99274rr01. [DOI] [PubMed] [Google Scholar]
- 18.Silzel J W, Cercek B, Dodson C, Tsay T, Obremski R J. Clin Chem. 1998;44:2036–2043. [PubMed] [Google Scholar]
- 19.Bock L C, Griffin L C, Latham J A, Vermaas E H, Toole J J. Nature (London) 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 20.Weiss S, Proske D, Neumann M, Groschup M H, Kretzschmar H A, Famulok M, Winnacker E L. J Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.