Summary
In two-component regulatory systems, covalent phosphorylation typically activates the response regulator signaling protein and hydrolysis of the phosphoryl group reestablishes the inactive state. Despite highly conserved three-dimensional structures and active site features, the rates of catalytic autodephosphorylation for different response regulators vary by a factor of almost 106. Previous studies identified two variable active site residues, corresponding to Escherichia coli CheY residues 59 and 89, that modulate response regulator autodephosphorylation rates about 100-fold. Here, a set of five CheY mutants, which match other “model” response regulators (ArcA, CusR, DctD, FixJ, PhoB, or Spo0F) at variable active site positions corresponding to CheY residues 14, 59 and 89, were characterized functionally and structurally in an attempt to identify mechanisms that modulate autodephosphorylation rate. As expected, the autodephosphorylation rates of the CheY mutants were reduced six- to 40-fold relative to wild type CheY, but all still autodephosphorylated 12- to 80-fold faster than their respective model response regulators. Comparison of X-ray crystal structures of the five CheY mutants (complexed with the phosphoryl group analogue BeF3−) to wild type CheY or corresponding model response regulator structures gave strong evidence for steric obstruction of the phosphoryl group from the attacking water molecule as one mechanism to enhance phosphoryl group stability. Structural data also suggested that impeding the change of a response regulator from the active to inactive conformation might retard the autodephosphorylation reaction if the two processes are coupled, and that the residue at position '58' may contribute to rate modulation. A given combination of amino acids at positions '14', '59', and '89' adopted similar conformations regardless of protein context (CheY or model response regulator), suggesting that knowledge of residue identity may be sufficient to predict autodephosphorylation rate, and hence, the kinetics of the signaling response, in the response regulator family of proteins.
Keywords: autodephosphorylation, CheY, response regulator, signal transduction, two-component regulatory system
Introduction
Cells utilize signal transduction mechanisms to mediate appropriate responses to environmental stimuli. Diverse signaling strategies have evolved that employ different mechanisms while sharing some functional characteristics1–3. Typically, detection of an extracellular stimulus by a sensory receptor triggers a sequence of reversible protein modification events (e.g. phosphorylation and dephosphorylation) and culminates in the modification and activation of a key signaling molecule that executes the intracellular response. The signaling molecule thus has active (modified) and inactive (unmodified) states and the distribution of these states across the population of signaling molecules determines the output response at any time. Therefore, a successful signal transduction strategy must control the rates of formation and destruction of the active signaling molecule.
Two-component regulatory systems are a widespread type of signal transduction system, present in microorganisms and plants [reviewed in4–6], that regulate diverse cellular processes including metabolism, virulence, sporulation, morphological development, and antibiotic resistance7,8. In their simplest form, two-component systems contain a transmembrane sensor kinase protein and a cytoplasmic signaling protein called a response regulator. Detection of an extracellular stimulus by the sensor kinase dictates the rate at which the kinase autophosphorylates a cytoplasmic histidine residue. The phosphoryl group is then transferred to an aspartate residue within the receiver domain of the partner response regulator protein. Phosphorylation of the receiver domain results in a conformational change that alters a surface necessary for output function9–11. For example, phosphorylation of the chemotaxis response regulator CheY changes the surface that binds to the flagellar motor, whereas phosphorylation of some response regulators that enhance transcription induces dimerization and subsequent DNA-binding of an attached output domain. Although the rate of phosphorylation (activation) of the response regulator is primarily controlled by sensor kinase activity, the rate of dephosphorylation (deactivation) is controlled by an intrinsic autodephosphorylation activity. In some cases, the autodephosphorylation rate is augmented by an independent phosphatase, which appears to function by enhancing the baseline rate set by the intrinsic autodephosphorylation process12–14.
All receiver domains whose structures are known exhibit a (βα)5 topology and very similar three-dimensional configurations5 (Fig. 1a). The active site that mediates both phosphorylation and dephosphorylation of receiver domains is primarily comprised of a quintet of conserved residues located largely on loops that connect secondary structural elements. Atomic resolution structures of multiple receiver domains complexed with the phosphoryl analogue BeF3− serve as models of both the activated state and the initial ground state for the autodephosphorylation reaction. The geometry of the active appears to be well conserved with the BeF3− group positioned such that the three fluorine atoms (representing the three oxygen atoms of the phosphoryl group) form interactions with conserved threonine and lysine side chains and a Mg2+ ion, which in turn is coordinated by three conserved aspartate residues (including the phosphorylated aspartate)11 (Fig. 1a).
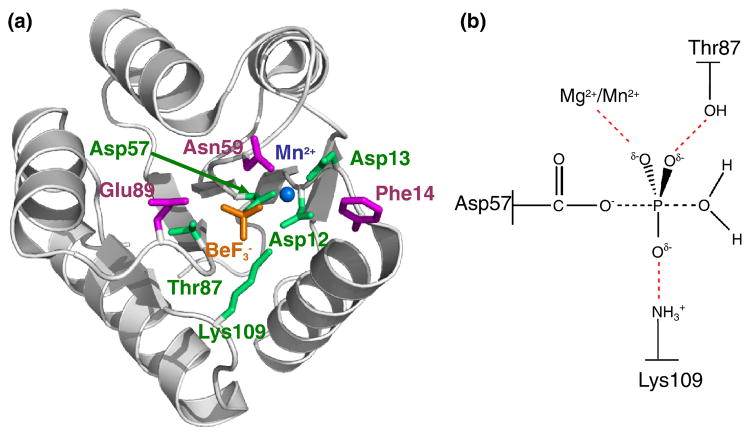
Fig. 1.
Overall structure of activated CheY and proposed transition state for autodephosphorylation. (a) Ribbon representation (gray) of wild type CheY•BeF3−•Mn2+ (pdb 1FQW) with active site features shown in color. Mn2+ is represented by a blue sphere. Conserved active site residues Asp57, Asp12, Asp13, Lys109, and Thr87 (green), BeF3− (orange), and variable active site residues altered in the study Phe14, Asn59, and Glu89 (magenta) are shown as stick models. (b) Schematic of proposed transition state in CheY autodephosphorylation reaction. Black dashed lines, partially formed covalent bonds; red dashed lines, stabilizing interactions between CheY and the phosphoryl group.
Receiver domain autodephosphorylation and subsequent regeneration of the inactive state is believed to involve in-line nucleophilic attack of a water molecule on the phosphorous atom and proceed through a trigonal bipyramidal transition state, a hallmark of phosphorous substitution reactions (Fig. 1b)15. The Mg2+ ion is an absolute requirement for catalysis of autodephosphorylation16, and substitution of the conserved lysine17 or threonine18 residues in CheY (Lys109 and Thr87, respectively) results in reduced autodephosphorylation rates, consistent with catalytic roles such as transition state stabilization (Fig. 1b). Further support for this mechanism comes from structures of several enzymes in the related haloacid dehalogenase family19,20, which show direct interactions between fluorine atoms of the planar transition state inhibitor AlF3 and the conserved threonine, lysine, and Mg2+ (Fig. 1b).
Despite the strongly conserved active sites geometries for the activated states of response regulators, measured rates of autodephosphorylation span almost six orders of magnitude21–23, with faster autodephosphorylation rates correlating with faster biological responses (such as chemotaxis) and slower autodephosphorylation rates correlating with slower biological processes (such as development). How do response regulators, with highly conserved active site geometries and catalytic mechanisms, tailor the kinetics of signaling biochemistry (e.g. autodephosphorylation) to be commensurate with the timescale of the biological events under their control? We previously identified two surface residues that are important determinants in modulating autodephosphorylation rates22. The two residues vary between response regulators and are in close physical proximity to the phosphorylation site. Changing the particular amino acids at variable positions '59' and '89' (numbers in single quotation marks indicates the residue that corresponds to the indicated position number in Escherichia coli CheY) (Fig. 1a) results in at least a 90- or 130-fold range of autodephosphorylation rates when varied in tandem for E. coli CheY or Bacillus subtilis Spo0F, respectively. In many cases, however, the rate of a mutant CheY or Spo0F protein does not equal the rate of a model response regulator with matched residues at positions '59' and '89'22. Therefore, there must be additional determinants that influence response regulator autodephosphorylation rate. Rate modulation by Asn59 and/or Glu89 in CheY is not due to direct participation in catalysis (e.g. orientation of the attacking water molecule) because CheY mutants bearing alanine substitutions at either position have autodephosphorylation rates indistinguishable from wild type CheY24,25.
Our previous work raises two questions potentially amenable to be answered by structural analysis. First, how do particular amino acids at positions '59' and '89' exert their influence on response regulator autodephosphorylation rate? Second, what structural determinants other than amino acids '59' and '89' contribute to autodephosphorylation rate control? Here we report structural and functional characterization of five CheY mutants that mimic the active sites of other response regulators by sharing eight residues (variable positions '14', '59', and '89' and conserved positions Asp'12', Asp'13’, Asp'57', Thr'87', and Lys'109'). We compare the structures of the mutant CheYs to both wild type CheY, with which they share all residues except the three variable active site positions, and to the modeled response regulator, with which they share the eight active site residues. Several such pairwise comparisons provided strong evidence that side chains of residues at positions '59' or '89' can assume conformations that limit steric access of water to the phosphoryl group as a mechanism to increase phosphoryl group stability. However, occlusion of the attacking water molecule does not appear to account for all observed variations in autodephosphorylation rate. The structural analyses also suggested that perturbations to conformational change may influence response regulator autodephosphorylation rate if conformational change and the autodephosphorylation reaction are coupled, and that the residue at position '58' may influence reaction rate.
Results and Discussion
Design of CheY active site mutants that mimic other response regulators
As a first step in defining the location of structural features that influence response regulator autodephosphorylation, we conceptually subdivided the receiver domain into the active site and the remainder of the domain, which we term the “core”. Our general strategy was to create CheY mutants that could be compared to either previously characterized response regulators with the same active site but a different protein core, or to wild type CheY, which has the same protein core but a different active site. To clearly indicate the intended comparisons, we designate the CheY mutants by a subscript with the name of the modeled response regulator that it is designed to mimic. For example, wild type CheY contains variable active site residues Phe14, Asn59, and Glu89 (Fig. 1a). CheYPhoB represents a CheY triple mutant bearing the three substitutions F14E, N59M, and E89R to match the active site of the model response regulator PhoB, which has Glu11, Met55, and Arg85 at the analogous positions. Thus CheYPhoB can be considered to be a chimera in which the active site of PhoB is spliced onto the core of CheY.
Five model response regulators (E. coli PhoB, Sinorhizobium meliloti FixJ, S. meliloti DctD, B. subtilis Spo0F, and E. coli ArcA) were chosen based on the availability of both a high resolution X-ray crystal structure of the activated state of the receiver domain and published in vitro autodephosphorylation rate measurements (Table 1). Four mutants of E. coli CheY were designed such that the identities of three variable active site residues were altered to match the analogous residues of the model response regulators (Table 1). FixJ and DctD are similar to one another in that both contain Arg'59' and His'89'. CheYFixJ DctD, containing Glu14, Arg59, and His89, was considered to be a mimic of both FixJ and DctD, even though DctD has an aspartate (not a glutamate) at position '14', because our previous study22 demonstrated that the residue at position '14' had little (less than two-fold) influence on autodephosphorylation rate. The CheY mutants were constructed before the minor role of position '14' was evident, but the additional extent of identity should not affect our comparisons. The fifth CheY mutant, CheYCusR (Glu14/Met59/Leu89), was designed for direct comparison to CheYPhoB (Glu14/Met59/Arg89) in order to assess the importance of the Arg89 positive charge. Although the Met/Leu combination at positions '59' and '89' is common in response regulator amino acid sequences (Kristin Wuichet and Igor Zhulin, personal communication), we are not aware of any published autodephosphorylation rates or structures for CusR or any other response regulator with the Met'59'/Leu'89' combination of residues.
Table 1.
Autodephosphorylation rate constants of CheY, model response regulators, and CheY mutants.
| Amino acid at positiona |
Wild type CheY & model response regulators |
CheY mutants |
||||
|---|---|---|---|---|---|---|
| '14' | '59' | '89' | Nameb | kdephosph (min−1)c | Name | kdephosph (min−1)c |
| F | N | E | E. coli CheY | 2.5 ± 0.322 | ||
| E | M | R | E. coli PhoB | 0.00944 | CheYPhoB | 0.11 ± 0.0022 |
| E | M | L | E. coli CusR | NAd | CheYCusR | 0.060 ± 0.006 |
| E | R | H | S. mel. FixJ | 0.00545 | CheYFixJ DctD | 0.060 ± 0.008 |
| D | R | H | S. mel. DctD | 0.00746 | ||
| Q | K | Y | B. sub. Spo0F | 0.00447 | CheYSpo0F | 0.085 ± .00922 |
| E | N | R | E. coli ArcA | 0.00548/0.0249 | CheYArcA | 0.40 ± 0.10 |
All response regulators on the same line of the table have the indicated combination of variable active site amino acids.
Species abbreviations: E. coli, Escherichia coli; S. mel., Sinorhizobium meliloti; B. sub., Bacillus subtilus.
Superscript indicates literature source of previously published data. Values measured in our laboratory include the mean ± standard deviation for at least two independent measurements. Measurements from other laboratories are rounded to one significant figure.
Not available. We are unaware of any kdephos measurement for CusR.
Autodephosphorylation kinetics of CheY mutants
All of the CheY mutants demonstrated a marked decrease in autodephosphorylation rate relative to wild type CheY (Table 1). CheYCusR and CheYFixJ DctD had the slowest rate constants (kdephosph = 0.060 min−1), down more than 40-fold from wild type CheY. CheYArcA displayed a rate constant only modestly (six-fold) decreased from the wild type value. However, the reduced rate constants did not reach the values of the response regulators mimicked by the CheY mutants, with discrepancies ranging from 12- to 80-fold. The inability of the CheY mutants to achieve the autodephosphorylation rates characteristic of other response regulators was also observed in our previous study22 and suggests that although positions '59' and '89' substantially modulate autodephosphorylation rate, additional factors exist. To gain insight into the structural bases for the observed decreases in autophosphorylation rates of the CheY mutants relative to wild type CheY and to potentially ascertain other structural factors that may affect autodephosphorylation, we determined and analyzed the structures of the five CheY mutants.
General structural features of CheY mutants
All of the CheY mutants were crystallized as a complex with Mn2+ and the phosphoryl analogue BeF3−. This structure represents the phosphorylated (activated) form of the protein, which is the initial ground state for the autodephosphorylation reaction. CheY complexed with BeF3− exhibits all of the structural changes observed for phosphorylated derivatives of response regulators11. Particularly important for the current study is that response regulator-BeF3− complexes tightly bind divalent cation, which is an absolute requirement for catalysis of autodephosphorylation. Although Mn2+ was used as divalent cation here in order to produce diffraction quality crystals, other response regulator-BeF3− structures [e.g. PhoB (pdb 1ZES) and ArcA (pdb 1XHF)] containing the more physiologically relevant Mg2+ show virtually identical locations and coordination properties for the metal ion, indicating that the identity of the cation (Mg2+ or Mn2+) does not affect the structure. Crystallization conditions for all of the CheY mutants were essentially identical to those used for the wild type CheY•BeF3−•Mn2+ structure (pdb 1FQW)11. Consequently, all six CheY structures (the five mutants plus wild type CheY) shared the same space group and crystal contacts, and the crystal contacts did not involve the active site region. Crystallography data and statistics are summarized in Table 2.
Table 2.
Data collection and refinement statistics
| Data set | CheYPhoB (pdb 3FGZ) | CheYCusR (pdb 3F7N) | CheYFixJ DctD (pdb 3FFX) | CheYSpo0F (pdb 3FFW) | CheYArcA (pdb 3FFT) |
|---|---|---|---|---|---|
| A. Data collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | a = b =53.5, c =161.9 | a = b =53.5, c =161.8 | a = b =53.5, c =161.8 | a = b =53.5, c =161.6 | a = b =53.5, c =161.7 |
| Resolution (Å) | 2.00 | 2.00 | 2.01 | 2.00 | 2.21 |
| No. of unique reflections | 31410 | 31381 | 31825 | 31409 | 23551 |
| Rsyma,b (%) | 6.5 (41.6) | 7.3 (33.1) | 7.9 (42.7) | 5.8 (33.3) | 9.9 (35.6) |
| Average redundancy | 6.6 | 4.6 | 4.7 | 4.0 | 3.7 |
| Completenessb | 97.2 (95.3) | 97.2 (88.7) | 99.7 (99.7) | 97.6 (99.5) | 97.5 (99.2) |
| Average I/(σ I)b | 16.8 (5.0) | 13.1 (5.0) | 11.5 (3.4) | 13.3 (3.4) | 8.1 (3.4) |
| B. Refinement | |||||
| No. of monomersc | 2 | 2 | 2 | 2 | 2 |
| No. of protein atoms in asymmetric unitd | 1979 (5) | 1984 (5) | 2026 (8) | 2016 (6) | 1964 (1) |
| No. of ligand atomse | 23 (0/2/2/2) 1 NH4 | 44 (2/4/2/2) | 55 (3/5/2/2) | 38 (2/3/2/2) | 44 (2/4/2/2) |
| No. of solvent molecules | 169 | 371 | 348 | 388 | 344 |
| Resolution range (Å) | 30 - 2.00 | 30 - 2.00 | 30 - 2.01 | 30 - 2.00 | 30 - 2.21 |
| No. of used reflections | 29778 | 29747 | 30163 | 29767 | 22311 |
| Rfactor (%)/Rfree (%)f,g | 19.6/22.0 | 17.1/21.9 | 17.8/21.0 | 16.2/20.6 | 16.1/20.3 |
| Average B factor (Å2) | 25.4 | 18.4 | 20.5 | 18.6 | 17.0 |
| RMS deviation from ideality bond length (Å)/angles (°) | 0.015/1.432 | 0.015/1.002 | 0.015/1.546 | 0.015/1.470 | 0.018/1.595 |
| Ramachandran plot Preferred/Allowed/Outliers (%) | 97.62/1.59/0.79 | 97.10/2.49/0.41 | 96.64/2.94/0.42 | 97.01/2.56/0.43 | 96.81/3.19/0.00 |
Wavelength (Å) for all data sets: 1.5418
Rsym = Σ|Iobs − Iavg|/ΣIavg.
Values in parentheses correspond to the highest resolution shell. Highest shell resolution range: CheYPhoB:2.07-2.00, CheYCusR:2.07-2.00, CheYFixJ DctD:2.08-2.01, CheYSpo0F:2.07-2.00, CheYArcA:2.29-2.21
Protein monomers per asymmetric unit
Values in parentheses correspond to number of residues with multiple conformations
In parentheses are numbers of sulfates/glycerols/BeF3−/Mn2+
R = Σ||Fobs|(hkl) − |Fcalc|(hkl)|Σ|Fobs|(hkl)
Test set is 5% of data in all structures
All of the CheY mutants displayed the full activating conformational change observed for wild type CheY•BeF3−•Mn2+. Superimpositions of the CheY mutant structures over the wild type structure (pdb 1FQW) showed that the β3α3 and β4α4 loop backbones, and side chains that undergo rotations upon activation (e.g. Trp58, Val86, and Phe111), were in their fully active conformations. There was a hydrogen bond present between the backbone carbonyl oxygen atom of the residue at position 89 and the side chain hydroxyl group of Tyr106 in all of the structures, a hallmark of complete activation that is absent in some partially activated structures26,27. The locations of the BeF3− and other conserved active site moieties were all virtually superimposable on their counterparts in the wild type CheY•BeF3−•Mn2+ structure (pdb 1FQW) with root mean square deviations (rmsd) less than 0.1 Å.
Potential structural influences on autodephosphorylation kinetics
Response regulator autodephosphorylation involves in-line attack by a water molecule on a tetrahedral phosphoryl group (the ground state), passage through a planar transition state with inversion of stereochemistry, release of inorganic phosphate, and change of the receiver domain from the active to inactive conformation. Consideration of this reaction pathway led us to five different hypotheses concerning potential mechanisms by which receiver domain structure might affect autodephosphorylation kinetics:
Hypothesis 1
Interactions of amino acid side chains with a water molecule that orient and/or activate the water for nucleophilic attack are known to accelerate the rate of dephosphorylation reactions14,20. Therefore, differences in interactions of residues with appropriately positioned water molecules could affect reaction rate. Examination of ordered water molecules in the CheY mutant structures could suggest specific mechanisms to directly influence reaction rate.
Hypothesis 2
Because a water molecule must take a specific in-line route to attack the phosphorus atom, differences in accessibility to the phosphoryl group could affect reaction rate.
Hypothesis 3
Response regulator dephosphorylation is accompanied by a return to the inactive conformational state. If the dephosphorylation reaction is complete before the structure rearranges, then conformational change cannot affect the reaction. However, if phosphoryl group hydrolysis and conformational change are coupled, then perturbations to the kinetics of conformational change could affect dephosphorylation reaction rate. Coupling between the hydrolysis event and the conformational change could occur if the phosphorylated response regulator is able to occasionally access the inactive state and only the inactive state is capable of hydrolysis. Alternatively, coupling could occur if the hydrolysis and conformational change occur in a single step with a shared transition state. Regardless of the exact mechanism, structural features that might alter the stability of the active conformation relative to the inactive might affect the kinetics of the conformational change, and hence the autodephosphorylation rate, if the steps are coupled.
Hypothesis 4
The phosphoryl group specifically interacts with multiple side chain and backbone atoms in the ground state11. Although the dephosphorylation transition state has not been directly observed for response regulators, models of the planar intermediate in closely related members of the haloacid dehalogenase family show interactions between the same protein and phosphoryl group atoms that are observed in the ground state19,20. In principle, the presence or absence of particular interactions between the protein and the phosphoryl group, or differences in the interaction distances and/or geometries in different response regulators, could affect the energy of the ground and/or transition states and in so doing alter activation energy and hence reaction kinetics.
Hypothesis 5
A specific subset of hypothesis 4 is inspired by the previously noted correlation between the introduction of positively charged residues at positions '59' or '89' and a reduction in autodephosphorylation rate22. The positively charged side chains might interact directly with the negatively charged phosphoryl group to lower the ground state energy, thus increasing activation energy (the difference in energy between the transition and ground states) and slowing the reaction.
Comparisons between the structures of CheY, the CheY mutants, and the model response regulators were used to test our five hypotheses outlined above. The hypotheses are not mutually exclusive and multiple factors might act simultaneously to determine response regulator autodephosphorylation rate.
Analysis of water molecules in the CheY mutant structures
In phosphoserine phosphatase, an enzyme with an active site structurally similar to response regulators, orientation/activation of water by Asp'59' leads to a dephosphorylation rate two orders of magnitude faster than that of CheY-P20,28. However, residue 59 does not appear to play a catalytic role in autodephosphorylation of CheY-P, because replacement of the wild type Asn59 with Asp (to mimic phosphoserine phosphatase) accelerates autodephosphorylation only two-fold22 and replacement with Ala does not change the reaction rate24. Nevertheless, we examined the structures with hypothesis 1 in mind, that interactions between the protein and a nucleophilic water molecule could affect reaction rate. Analysis of the locations of ordered water molecules in the CheY structures (mutants and wild type) revealed that only one of the structures (CheYArcA) had an ordered water molecule in a location consistent with a nucleophilic water, and this water, in fact, formed a hydrogen bond with Asn59. However, the autodephosphorylation rate of CheYArcA is slower than wild type CheY (Table 1). Therefore, in this set of CheY structures, the locations of ordered water molecules do not appear to reveal relevant information about catalytic mechanism or rate, providing no support for hypothesis 1.
CheYPhoB and CheYCusR mutants share obstructive Met'59' with model protein PhoB
In wild type CheY•BeF3−•Mn2+, the Asn59 and Glu89 side chains point away from the BeF3− and do not obstruct access to the active site (Fig. 2a). In contrast, for both CheYPhoB and CheYCusR, which exhibited autodephosphorylation rates that were reduced 20- to 40-fold from wild type CheY (Table 1), the Met59 side chain projects out into the active site with the final bond rotated such that the terminal methyl group is located over the beryllium atom (Figs. 2ab). This conformation of Met59 appears to be stabilized by van der Waals interactions with the residue at position 89 - the Leu89 side chain in CheYCusR or the hydrocarbon portion of the Arg89 side chain in CheYPhoB - both of which, in turn, form additional van der Waals contacts with Trp58 (Fig. 2c). Furthermore, attempts to model other common rotamers of methionine29 into the structure resulted in either steric clash with the residue 89 side chain or the loss of stabilizing van der Waals interactions (data not shown). The Met59 Cε atom is close to the beryllium atom (3.3 Å and 4.1 Å separation for CheYPhoB and CheYCusR, respectively) and occupies the same location in space as would a water molecule positioned for in-line nucleophilic attack on the phosphoryl group (Fig. 2b). This apparent steric blockage of the electrophilic phosphorous is consistent with hypothesis 2 and provides a straightforward explanation for the reduction in autodephosphorylation rates observed in CheYPhoB and CheYCusR mutants compared to wild type CheY (Table 1).
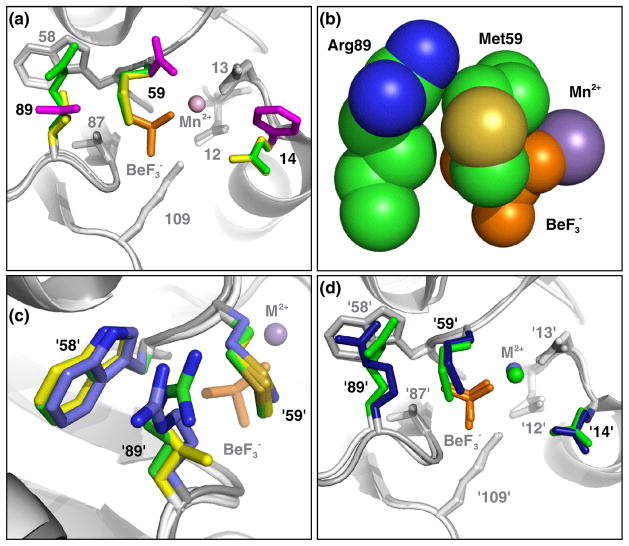
Fig. 2.
CheYPhoB•BeF3−•Mn2+ and CheYCusR•BeF3−•Mn2+ display the same obstructive conformation of Met59. Panels a, b, and d are from the perspective of a nucleophilic water molecule, looking straight down the line connecting the beryllium atom and Asp57 Oδ1 atom. (a) Superimposition of CheYPhoB•BeF3−•Mn2+, CheYCusR•BeF3−•Mn2+, and CheY•BeF3−•Mn2+ (pdb 1FQW). Residues 14, 59, and 89, are shown as colored sticks with CheYPhoB (green), CheYCusR (yellow), and CheY (magenta). Conserved residues Asp12, Asp13, Trp58, Thr87, and Lys109 are shown as sticks with CheYPhoB and CheYCusR (light gray) and wild type CheY (dark gray). The backbone is shown in ribbon representation with CheYPhoB and CheYCusR (light gray) and wild type CheY (dark gray). BeF3− is orange and Mn2+ is marked by a sphere in the color of the parent molecule. (b) Space-filling representation of CheYPhoB•BeF3−•Mn2+ active site demonstrating steric blockage of the BeF3− by Met59 and van der Waals interactions between Met59 and Arg89. (c) Superimposition of PhoB•BeF3−•Mg2+ (blue, pdb1ZES), CheYPhoB•BeF3−•Mn2+ (green), and CheYCusR•BeF3−•Mn2+ (yellow) demonstrating that residues at positions '58', '89', and '59' (PhoB Met54, Arg85, and Trp55, respectively) pack similarly. (d) Superimposition of CheYPhoB•BeF3−•Mn2+ and PhoB•BeF3−•Mg2+ (pdb 1ZES). The same coloring scheme as panel (a) applies with PhoB colored blue. The divalent cation (represented as a sphere) is Mn2+ for the CheY structures and Mg2+ for PhoB.
Superimposition of the CheYPhoB structure over PhoB•BeF3−•Mg2+ showed that Met'59' in PhoB has a similar conformation as Met59 in CheYPhoB and CheYCusR (Fig. 2cd). Furthermore, like Met59 in the CheY mutants, the PhoB Met'59' side chain is involved in van der Waals interactions with the hydrocarbon portion of the Arg'89' side chain and Trp'58' (Fig. 2c). Because residues '59' and '89' in PhoB are in nearly the same conformations as observed in CheYPhoB, it is reasonable to conclude that residues '59' and '89' affect the autodephosphorylation rate by a similar mechanism (steric obstruction of the phosphoryl group) in both CheYPhoB and PhoB.
CheYFixJ DctD mutant shares obstructive His'89' with model proteins DctD and FixJ
The autodephosphorylation rate of CheYFixJ DctD was reduced 40-fold from wild type CheY, similar to CheYPhoB and CheYCusR (Table 1). CheYFixJ DctD•BeF3−•Mn2+ also contained features consistent with steric obstruction of the BeF3− from water attack. In CheYFixJ DctD, both Arg59 and His89 assumed different conformations in the two molecules within the asymmetric unit (Fig. 3a). The side chain electron density for Arg59 required two conformations to be modeled into the structure of each monomer, consistent with high mobility and the absence of interactions with other side chains (Fig. 3a). The electron density for His89 was well defined and indicated different orientations in each monomer (Fig. 3a). In monomer A, His89 extends to interact with Trp58, as seen for Arg89 and Leu89 in CheYPhoB and CheYCusR respectively. In monomer B, the His89 side chain is oriented so that the imidazole group is located in a position that causes steric obstruction of the BeF3−. Taken together with the structures of CheYPhoB and CheYCusR, it is apparent that the side chains of residues at both positions '59' and '89' are capable of assuming conformations that fill the space directly in front of the BeF3−, and may act to obstruct an incoming water molecule during autodephosphorylation. Thus, steric obstruction of the phosphoryl group (hypothesis 2) also provides a potential explanation for the 40-fold slower autodephosphorylation rate of CheYFixJ DctD relative to CheY.
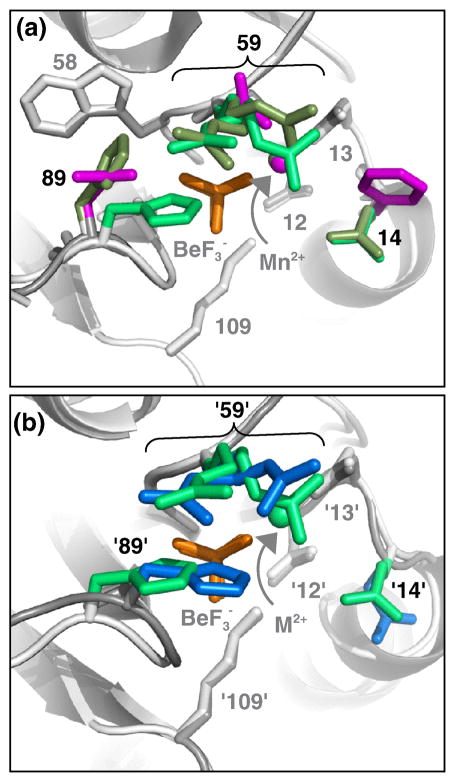
Fig. 3.
CheYFixJ DctD•BeF3− displays two conformations of His89, one of which is in a sterically obstructive position. Both panels are from the perspective of a nucleophilic water molecule, looking straight down the line connecting the beryllium atom and Asp57 Oδ1 atom. The same general modeling scheme described in Fig. 2a, with only residues 14, 59, and 89 colored, applies. BeF3− is orange and Mn2+ is marked by a sphere in the color of the parent molecule. (a) Overlay of CheYFixJ DctD•BeF3−•Mn2+ chain A (olive green), CheYFixJ DctD•BeF3−•Mn2+ chain B (green), and wild type CheY•BeF3−•Mn2+ (magenta, pdb 1FQW). Note both chains A and B of CheYFixJ DctD contain two rotamers of Arg59. (b) Overlay of CheYFixJ DctD•BeF3−•Mn2+ chain B (green) and DctD•BeF3−•Mg2+ (blue, pdb 1L5Y).
Comparison of the active sites of CheYFixJ DctD, DctD, and FixJ (Fig. 3b), like the CheYPhoB/PhoB pair (Fig. 2cd), shows remarkable similarity in the positions of the residues at positions '59' and '89'. The crystal structures of FixJ-PO32−•NH4+ (pdb 1D5W) and DctD•BeF3−•Mg2+ (pdb 1L5Y), like CheYFixJ DctD •BeF3−•Mn2+, contain multiple conformations of Arg'59'. However, for both DctD and FixJ, all of the monomers within the asymmetric unit share one orientation of His'89' that blocks the BeF3−. DctD His'89' (Fig. 3b) and FixJ His'89' (data not shown) are oriented similarly to His89 in CheYFixJ DctD monomer B but display decidedly more obstructive positions over the phosphoryl/BeF3− group. Like CheYPhoB and CheYCusR, the slow autodephosphorylation rate for CheYFixJ DctD can largely be explained in terms of steric obstruction of the phosphoryl group. However, in this case, it appears that different degrees of steric obstruction (hypothesis 2) can also explain the ~10-fold difference between the autodephosphorylation rates of CheYFixJ DctD and the FixJ/DctD models.
Does energetic linkage of protein conformational change and phosphoryl group hydrolysis affect autodephosphorylation rate?
In contrast to the previously described structures, the CheYSpo0F•BeF3−•Mn2+ structure revealed no obvious obstruction of the BeF3− group (Fig. 4a), even though the autodephosphorylation rate of CheYSpo0F was 30-fold reduced from that of wild type CheY (Table 1). The Lys59 side chain of CheYSpo0F extends straight out towards solvent and does not interact with other protein atoms. Strikingly for a surface Lys residue, the electron density for Lys59 was clear for the entire length of the side chain, leaving no ambiguity about the ability of Lys59 to occupy a nonobstructive conformation. The Tyr89 side chain appears to be held out of the path of an attacking water molecule through van der Waals interactions with Trp58. Therefore, in CheYSpo0F, residues 59 and 89 must influence autodephosphorylation by a mechanism other than obstruction of water.
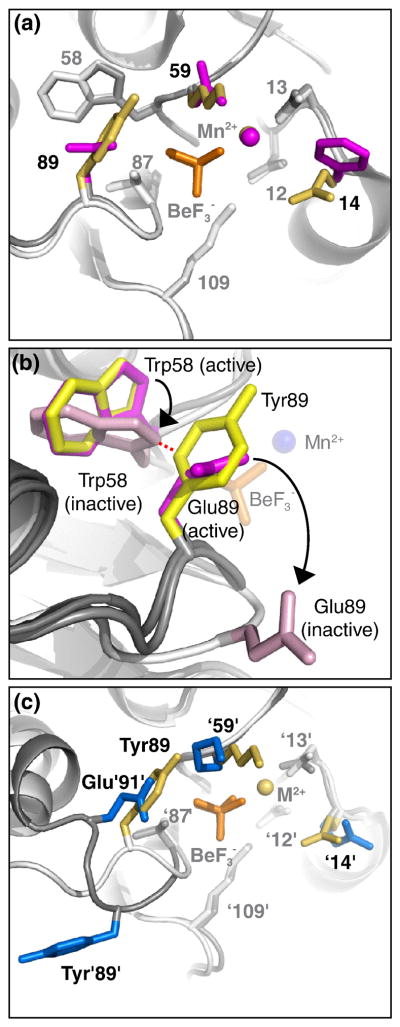
Fig. 4.
CheYSpo0F•BeF3−•Mn2+ does not display steric obstruction of the BeF3−. Panels a and c are from the perspective of a nucleophilic water molecule, looking straight down the line connecting the beryllium atom and Asp57 Oδ1 atom. The same general modeling scheme described in Fig. 2a, with only residues 14, 59, and 89 colored, applies to panels a and c. In panel b, the viewpoint is different and only residues 58 and 89 are colored. BeF3− is orange and Mn2+ is marked by a sphere in the color of the parent molecule. (a) Overlay of CheYSpo0F•BeF3−•Mn2+ (yellow side chains, white ribbon) and wild type CheY•BeF3−•Mn2+ (magenta side chains, gray ribbon). (b) Superimposition of CheY•BeF3−•Mn2+ (magenta side chains, gray ribbon, pdb 1FQW), apo-CheY (light pink side chains, white ribbon, pdb 3CHY), and CheYSpo0F•BeF3−•Mn2+ (yellow side chains, gray ribbon) reveals steric clash between CheYSpo0F Tyr89 and apo-CheY Trp58. CheY•BeF3−•Mn2+ is a model for the initial state of the autodephosphorylation reaction and apo-CheY is a model for the final state. BeF3− is orange sticks and Mn2+ is marked by a blue sphere. The distance marked by the red dashed line is <1.4 Å, indicative of a steric clash. Black arrows indicate movement of Trp58 and Glu89 from active to inactive conformation. (c) Overlay of CheYSpo0F•BeF3−•Mn2+ (yellow) and Spo0F•BeF3−•Mg2+ (blue, pdb 1FTK).
The observation of van der Waals interactions between Tyr89 and Trp58 in CheYSpo0F, absent in wild type CheY, led to consideration of a possible role of conformational change in autodephosphorylation kinetics (hypothesis 3). Removal of the phosphoryl group from a response regulator necessarily results in the reversal of the well established activating conformational change11. The wild type CheY Trp58 side chain undergoes a rotation of ~60° when changing from the active to the inactive conformation (Fig. 4b). Additionally, the β4α4 loop (containing residue 89) moves 4 Å away from the active site. If the dephosphorylation chemistry and conformation reversal are coupled, then the energetics of the conformational change might impact the autodephosphorylation rate. Interactions that stabilize the activated conformation and/or destabilize the transition state of the conformational change would retard the autodephosphorylation reaction. In CheYSpo0F, the van der Waals interactions between Tyr89 and Trp58 may help hold the β4α4 loop in the active conformation (Fig. 4b). Furthermore, introduction of the bulky Tyr89 residue creates a steric clash with the position that Trp58 occupies in the inactive conformation (Fig. 4b). The structural evidence for an effect on conformational change is consistent with hypothesis 3, which might be a mechanism by which variable active site residues influence autodephosphorylation rate.
Additional indirect evidence consistent with coupled autodephosphorylation and conformational change (hypothesis 3) can be seen in other structures. The autodephosphorylation rate of CheYArcA was about six-fold reduced from wild type CheY (Table 1), but the active site of CheYArcA displayed no obstruction of the BeF3− (Fig. 5a). Instead, the Asn59 and Arg89 side chains are both oriented towards Trp58 and their conformations are stabilized by cation-π interactions (between Arg89 and Trp58) and hydrogen bond formation (between Arg89 and Asn59) (Fig. 5b). This network of interactions may retard the movement of residue 89 (on the β4α4 loop) away from residues 58 and 59 as the receiver domain changes from the active to the inactive conformation. In another example, PhoB autodephosphorylated 12-fold slower than CheYPhoB despite similar occlusion of the active sites in the two proteins (Fig. 2d). Although the conformations of Arg'89' are similar in PhoB and CheYPhoB, the position of Arg'89' in PhoB is additionally stabilized by cation-π interactions30 between the guanidinium group of Arg'89' and Trp'58' (Fig. 2c) and by a salt bridge between Arg'89' and Asp'94'31. Again, these interactions might stabilize the active conformation and so reduce the rate of a coordinated autodephosphorylation process.
Fig. 5.
CheYArcA•BeF3−•Mn2+ does not display steric obstruction of the BeF3−. Both panels are from the perspective of a nucleophilic water molecule, looking straight down the line connecting the beryllium atom and Asp57 Oδ1 atom. The same general modeling scheme described in Fig. 2a, with only residues 14, 59, and 89 colored, applies. BeF3− is orange and Mn2+ is marked by a sphere in the color of the parent molecule. (a) Overlay of CheYArcA•BeF3−•Mn2+ (cyan) and wild type CheY•BeF3−•Mn2+ (magenta, pdb 1FQW). (b) Close-up view illustrating the packing of CheYArcA•BeF3−•Mn2+ residues Trp58, Asn59, and Arg89. Cation-π interactions are represented with red dotted lines and the hydrogen bond between Asn59 and Arg89 is illustrated as a green dotted line. (c) Overlay of CheYArcA•BeF3−•Mn2+ (cyan) and ArcA•BeF3−•Mg2+ (blue, pdb 1XHF).
Finally, recall that although steric occlusion (hypothesis 2) appears sufficient to account for the diminished autodephosphorylation rates of CheYPhoB and CheYCusR compared to wild type CheY, the mutants contain interactions between residue 89 and Trp58 that are not present in wild type CheY (Fig. 2a). Thus, all five of the CheY mutants described here (but not wild type CheY) displayed direct interactions between Trp58 and the side chains at position 59 and/or 89 that could increase the stability of the active conformation and result in a slower autodephosphorylation rate.
Although hypothesis 3, that the autodephosphorylation reaction and change to the inactive conformation are coupled, provides an intellectually satisfying interpretation of much of our structural and kinetic data, it should be noted that some published data is consistent with dephosphorylation occurring prior to, rather than during, the conformational change. In particular, binding of CheZ or FliM peptides to CheY-P or binding of ompC DNA to OmpR-P should result in stabilization of the active conformation of the receiver domain and hence might be predicted by hypothesis 3 to slow the autodephosphorylation rate. However, these binding events actually have no effect or slightly increase (up to two-fold) autodephosphorylation rates32,33. Also, in the structurally related phosphoserine phosphatase, Ser'87' remains bound to the phosphoryl group analog AlF3 in a model of the autodephosphorylation transition state20. If this was also the case for response regulator autodephosphorylation, it would follow that phosphoryl group hydrolysis must precede the conformational change because Ser/Thr'87' undergoes substantial movement away from the active site to reach the inactive conformation. However, this data is indirect, multiple interpretations are possible, and additional experiments will be required to determine whether response regulator autodephosphorylation rate is coupled to the kinetics of conformational change.
Do small differences in geometry of conserved active site residues affect autodephosphorylation rates?
Autodephosphorylation of ArcA is 20- to 80-fold slower than that of CheYArcA (Table 1). However, superimposition of CheYArcA and ArcA shows similarly unobstructed active sites due to similar, but not identical, conformations of Asn'59' and Arg'89' (Fig. 5c). Therefore steric hindrance (hypothesis 2) cannot account for the autodephosphorylation rate difference. Although the Asn'59' side chains of CheYArcA and ArcA superimpose well, the Arg'89' side chains diverge after Cδ, perhaps due to the absence of a tryptophan at position '58' in ArcA. Instead of interacting with residue '58' as in CheYArcA, the guanidinium group of ArcA Arg'89' is oriented towards solvent. Thus the features proposed to inhibit conformational change and hence retard autodephosphorylation (hypothesis 3) in CheYArcA are also absent in ArcA, leaving us with no obvious explanation for the slow autodephosphorylation rate in ArcA based on these structures.
Likewise, autodephosphorylation of Spo0F is 20-fold slower than that of CheYSpo0F (Table 1) even though the active sites of the two proteins appear similarly unobstructed (Fig. 4c). Furthermore, unlike Tyr89 in CheYSpo0F, Spo0F Tyr'89' is far removed from the active site (>10 Å) due to large differences in the β4α4 loops between the two proteins. Therefore, Tyr'89' does not appear to be in a position to affect the conformational change of Spo0F, and the reason for the reduced autodephosphorylation rate of Spo0F is not apparent.
We considered hypothesis 4 (differences in interactions between the active site and the transition state that could potentially affect activation energy) as a possible explanation for the slower autodephosphorylation rates of ArcA and Spo0F compared to their respective CheY mutants. Several strictly conserved features of receiver domain active sites, including Lys109, Thr87, and Mg2+, play a catalytic role in autodephosphorylation (Fig. 1b). Could subtle positional variations in these or other conserved elements of the active sites for different response regulators contribute to changes in autodephosphorylation rate despite identical residues at positions '59' and '89’? Autodephosphorylation rate differences of 20- to 80-fold, such as observed between CheYArcA and ArcA or between CheYSpo0F and Spo0F (Table 1), correspond to a difference in activation energy of only ~2 kcal/mol, comparable to the energy of a weak hydrogen bond.
The precise nature of the bipyramidal transition state for autodephosphorylation (Fig. 1b) is not known. An “associative” phosphotransfer mechanism would be expected to give a tight transition state (short apical bonds) with significant positive charge on the oxygen atom of the attacking water molecule (Fig. 1b). A dissociative mechanism would give a more extended transition state (longer apical bonds) with significant amount of negative charge on the leaving group Asp’57’ carboxylate oxygen34. We overlaid a set of 13 conserved active site atoms (Be, three F, Mn2+, Asp'57'Oδ1, Asp'57'Cγ, Lys'109'Nζ, Lys'109'Cε, Thr'87'Oγ1 Thr'87'Cβ, Res'58'N, Res'59'O) in each CheY mutant/model response regulator pair to obtain a quantitative indicator of the degree of similarity of their geometries. The chosen atoms represent the phosphoryl group, plus side chain and backbone atoms that interact with the phosphoryl group in the ground state11 or could potentially interact with atoms in the transition state19,20, whether it proceeds by an associative or dissociative mechanism. Three of the pairs (CheYPhoB/PhoB, CheYFixJ DctD/DctD, and CheYArcA/ArcA) gave excellent overlays (rmsd < 0.25 Å). Overlay of CheYSpo0F and Spo0F gave a slightly higher rmsd of 0.45 Å, perhaps because the Spo0F•BeF3− structure was solved in the context of a co-crystal with the Spo0B phosphotransferase. The rmsd values are low compared to the resolution of the crystal structures and the significance, if any, of an rmsd of 0.25 Å or even 0.45 Å is difficult to assess. The comparisons confirmed the high degree of structural conservation between response regulator active sites and did not provide compelling evidence of interaction differences between the phosphoryl group and conserved active site features (hypothesis 4) that could account for the disparity in autodephosphorylation rates.
Positively charged side chains at positions '59' and '89' do not stabilize the phosphoryl (BeF3−) group by direct interaction
No direct interactions are observed between side chains at positions '59' or '89' and the phosphoryl group or BeF3− analog in any of the CheY mutant or model response regulator structures examined in this work (Figs. 2, 3, 4, and 5). Thus, the crystal structures do not support hypothesis 5, which suggested that positively charged side chains at positions '59' or '89' might slow autodephosphorylation by stabilizing the ground state of the negatively charged phosphoryl group.
Summary of structural evidence and hypotheses
To summarize our findings, we characterized the autodephosphorylation rates and three-dimensional structures of five CheY mutants in an attempt to test five hypotheses about the possible influence of receiver domain structure on autodephosphorylation rate. The structures of five mutants containing the CheY core combined with the variable active site residues of a model response regulator were each compared to wild type CheY, and four of the mutants were compared to a matching model response regulator structure (two models in the case of CheYFixJ DctD), for a total of ten pairwise comparisons (Table 3). Interpretation of the structures is necessarily tempered by the knowledge that X-ray crystallography provides static pictures of dynamic protein structures, but nevertheless provided the following insights:
Table 3.
Summary of hypothesized mechanisms of autodephosphorylation rate modulation based on structural comparisons between CheY mutants and wild type CheY or model response regulators
| Compared to | CheY Mutant |
||||
|---|---|---|---|---|---|
| CheYPhoB | CheYCusR | CheYFixJ DctD | CheYSpo0F | CheYArcA | |
| Wild type | Obstructiona | Obstruction | Obstruction | Conformation | Conformation |
| CheY | (Conformation)b | (Conformation) | (Conformation) | ||
| Model response regulator | Conformation | NAc | Obstructiond | Unknown | Unknown |
“Obstruction” means steric obstruction of attacking water molecule in hydrolysis reaction (hypothesis 2 in the text). “Conformation” means interference with the change from active to inactive conformation coupled to autodephosphorylation (hypothesis 3 in the text).
Secondary explanation in parentheses
Not applicable. No structure of CusR is available for comparison.
CheYFixJ DctD was compared to both DctD and FixJ but the latter comparison was not shown in a figure.
None of the CheY mutant or model response regulator structures provided evidence in support of hypothesis 1, that differences in interaction with the attacking water molecule might account for the observed differences in reaction rates.
Five comparisons (CheY vs. CheYPhoB, Fig. 2ab; CheY vs. CheYCusR, Fig. 2a; CheY vs. CheYFixJ DctD, Fig. 3a; CheYFixJ DctD vs. DctD, Fig. 3b; CheYFixJ DctD vs. FixJ, not shown) provided direct evidence in support of hypothesis 2, that reduction of response regulator autodephosphorylation rate could be attributed to varying degrees of obstruction of the phosphoryl group from the attacking water molecule.
Three comparisons (CheY vs. CheYSpo0F, Fig. 4ab; CheY vs. CheYArcA, Fig. 5ab; CheYPhoB vs. PhoB, Fig. 2cd) revealed structural features consistent with hypothesis 3, that perturbation of the conversion of the receiver domain from the active to the inactive conformation might affect the rate of the autodephosphorylation reaction, if the two processes are coupled. Three additional comparisons (CheY vs. CheYPhoB, Fig. 2ab; CheY vs. CheYCusR, Fig. 2a; CheY vs. CheYFixJ DctD, Fig. 3a), in which hypothesis 2 already provided a sufficient explanation of reaction rate differences, also showed structural differences consistent with hypothesis 3. The evidence in support of hypothesis 3 is necessarily indirect because we did not actually measure the rate of conformational change.
None of the 10 pair wise comparisons provided convincing evidence in support of hypothesis 4, that differences in the distances and/or geometry between the phosphoryl group and the side chain, backbone, or metal ion atoms with which the phosphoryl group interacts might affect autodephosphorylation rate.
The structures of the four CheY mutants (all except CheYCusR) and five model response regulators with positively charged side chains at positions '59' and/or '89' provided no evidence in support of hypothesis 5, that reduced autodephosphorylation rate in the presence of positively charged residues at variable active site positions might be due to direct interaction with the negatively charged phosphoryl group.
Finally, two comparisons (CheYSpo0F vs. Spo0F, Fig. 4c; CheYArcA vs. ArcA, Fig. 5c) did not reveal an obvious structural basis for observed differences in autodephosphorylation rate, which suggests that additional contributing factors remain to be discovered.
We anticipated that comparisons between the structures of the CheY mutants and wild type CheY (first row of Table 3) might reveal mechanisms by which residues '59' and '89' affect autodephosphorylation rate, and this approach indeed was fruitful in providing support for hypotheses 2 and 3 while ruling out hypothesis 5. We also anticipated that comparison of the CheY mutants with the model response regulators might reveal features that influence autodephosphorylation rate other than the eight matched active site residues. However, the latter strategy (second row of Table 3) had little success at identifying other contributing factors. In two such comparisons (CheYFixJ DctD vs. DctD, Fig. 3b; CheYFixJ DctD vs. FixJ, not shown), it appears that residues '59'/'89' alone influenced the reaction rate, and in two other cases (CheYSpo0F vs. Spo0F, Fig. 4c; CheYArcA vs. ArcA, Fig. 5c) no plausible causes were identified. Only one case (CheYPhoB vs. PhoB, Fig. 2cd) of comparison between a CheY mutant and the model response regulator it was designed to mimic pointed to involvement of a previously unidentified factor, which was the residue at position '58'. However, the potential involvement of residue '58' was further highlighted by the observation of interactions between Trp58 and the residue at position 89 (Arg, His, Leu, Tyr) in all five CheY mutants examined, in contrast to the lack of interaction between Trp58 and Glu89 exhibited by wild type CheY. Thus, the structural comparisons described here suggest that the variable active site residue at position '58' might contribute to modulation of response regulator autodephosphorylation rate, a hypothesis that is amenable to future direct experimental test.
Can response regulator phosphoryl group stability be predicted from amino acid sequence?
A plethora of protein sequence information is currently available. Combined with the rapid increase in protein sequence database content, a current goal is to develop tools to utilize the data to more fully predict aspects of protein function. Previous studies22 established a correlation between autodephosphorylation rates and the identities of residues at positions '59' and '89' of response regulators. In this study, we initiated investigation of the mechanisms by which dephosphorylation rate modulation occurs. Mechanistic information potentially enhances predictive power because it allows extrapolation of general principles without empirically testing every combination of amino acids.
The structures analyzed here provided strong evidence for steric obstruction of the phosphoryl group by side chains at positions '59' or '89' as one structural mechanism for the rate modulation. Phosphorous substitution reactions have the stringent requirement that the incoming nucleophile attacks from a position that is co-linear with the bond that breaks in the reaction. Many of the structures described here had side chain atoms that occupied space required for the hydrolytic water. Consideration of the identities of residues at both positions '59' and '89' is essential because the two residues often interact with each other22, thereby mutually affecting conformation. The identity of the residue at position '58' may also be important based on the presence of direct interactions between residue '59' and/or '89' and Trp'58' in all five CheY mutants examined here as well as PhoB. Finally, the consistent similarity of the conformations of identical residues at positions '59' and '89' in all of the CheY mutants and their associated model response regulators considered here (amino acid sequence identity of only ~20–25%) strongly suggests that a given residue has the same effect in different response regulators, greatly enhancing the potential to predict phosphoryl group stability and perhaps other functions that vary in magnitude between response regulators from amino acid sequence information.
As an example of a specific prediction, the structures of CheYPhoB, CheYCusR, and PhoB all display direct interaction between Met'59' and the hydrocarbon portion of Arg'89' (CheYPhoB and PhoB) or Leu89 (CheYCusR), suggesting that a side chain with some hydrophobic portion is required at position '89' to stabilize the obstructive position of Met'59'. Interaction between residues 59 and 89 may explain a previous observation that four CheY mutants with methionine at position 59 [CheYPhoB (Glu14/Met59/Arg89), CheY Phe14/Met59/Arg89, CheYOmpR (Asp14/Met59/Lys89), CheY Phe14/Met59/Lys89] all had autodephosphorylation rates ~30-fold slower than wild type CheY whereas CheY Phe14/Met59/Glu89 had a rate that was two-fold faster22. It is likely that Arg89 or Lys89 provide nonpolar atoms to stabilize Met59 in its extended obstructive positions and the terminal positive charge may provide an additional attractive interaction with the Trp58 indole group30. The shorter and anionic Glu89 in CheY Phe14/Met59/Glu89 may not provide these stabilizing forces, resulting in a different (nonobstructive) conformation for Met59. Therefore, the presence of a methionine at position '59' coupled with a Lys, Arg, or hydrophobic side chain at position '89' may indicate a slow response regulator autodephosphorylation rate. Indeed, three response regulators with among the slowest known autodephosphorylation rates (E. coli OmpR32, Enterococcus faecium VanR35, and Thermotoga maritima DrrA21) all contain Met'59'/Lys'89'. About one-third of response regulators belong to the OmpR/PhoB subclass, based on a recent census of more than 4,600 response regulator amino acid sequences36. It may be noteworthy that among the 37 receiver domains (32 in response regulators and five in hybrid sensor kinases) from one organism (E. coli), Met'59' is found exclusively in the OmpR/PhoB subclass and half (7/14) of the OmpR/PhoB subclass response regulators contain the Met'59' and Arg/Leu/Lys'89' combination of residues predicted to result in slow autodephosphorylation due to steric hindrance of the water molecule necessary for hydrolysis.
Some response regulators with the same combination of amino acids at positions '59' and '89' exhibit very similar autodephosphorylation rates, whereas other matched pairs have kdephos values that differ by up to two orders of magnitude (see Table S5 of Ref. 22). Although the instances with similar rates are encouraging, the discrepant cases clearly indicate that additional determinants of response regulator phosphoryl group stability remain to be identified, and stability cannot yet be confidently predicted from amino acid sequence. We previously found that changing CheY residues 59 and 89 simultaneously had a greater effect than changing either residue alone22, consistent with interactions between residues '59' and '89' observed in some of the structures reported here. The structural results described here further suggest that interaction of residue '58' with residues '89' and/or '59' depend on the particular amino acids involved, and that networks of interactions involving multiple active site residues might influence autodephosphorylation reaction rate. Therefore, future attempts to mimic the kinetic characteristics of one response regulator in the context of another may benefit from expanding the set of variable active site residues that are matched between the two proteins, especially by including position '58'.
Materials and Methods
Site directed mutagenesis and protein purification
The Quikchange mutagenesis (Stratagene) procedure was used to make cheY mutants encoding CheY F14E E89R (CheYArcA), CheY F14E N59M E89L (CheYCusR), and CheY F14E N59R E89H (CheYFixJ DctD) using the pRS3 plasmid37 as template. CheY F14E N59M E89R (CheYPhoB) and CheY F14Q N59K E89Y (CheYSpo0F) were from a previous study22. The CheY mutants were overexpressed and purified as previously described22. CheY concentrations were determined using an extinction coefficient of 0.727 (mg/ml)−1 cm−1 for all mutants38.
Measurement of autodephosphorylation kinetics
Rates of CheY autodephosphorylation were measured by quantitation of the loss radioactivity from [32P]CheY-P by gel electrophoresis and phosphorimaging as previously described22. Briefly, [32P]CheA-P (28 pmol) and excess CheY (400 pmol) were mixed in buffer containing 100 mM Tris, pH 7.5, 10 mM MgCl2 and aliquots were removed and quenched in an equal volume of 2x SDS sample buffer at various times. Samples were then subjected to SDS polyacrylamide gel electrophoresis (18% acrylamide Tris-glycine gel). The gels were dried and analyzed by quantitative phosphorimaging to determine the amount of radioactivity associated with protein at each time point. Phosphotransfer from CheA-P to CheY was complete by the first (10 s) time point for all of the CheY mutants. Subsequent time points showed the decay of the 32P signal associated with CheY due to autodephosphorylation. The data was fit to a single exponential decay using Prism software to determine the rate constant. Each rate was measured twice, and the average and standard deviation reported.
Crystallization and structure determination
Protein crystals were grown at room temperature using the hanging drop vapor diffusion method. Each drop consisted of 1 μL of reservoir buffer (100 mM Tris, pH 8.4, 1.8 –2.4 M ammonium sulfate, and 5–7% (v/v) glycerol) and 1 μL of a mix containing CheY (at 1.5, 2.6, 4.3, 7.4, and 7.8 mg/mL for CheYFixJ DctD, CheYSpo0F, CheYArcA, CheYCusR, and CheYPhoB, respectively), 1 mM BeCl2, 10 mM NaF, and 1 mM MnCl2. Crystals were visible within 12 hours. Crystals were cryoprotected by gradual exposure to reservoir buffer supplemented with increasing concentrations of glycerol (5–15 % (v/v)) as described11 and were transferred immediately from the equilibrated drop into a cryogenic nitrogen stream (100 K, Oxford Cryo System). Diffraction data were collected using a Ru-H3R generator and R-Axis IV++ detector. Data was processed using Denzo and Scalepack in the HKL2000 suite39 (see Table 2 for data statistics).
The structures of all the CheY mutants were determined using molecular replacement methods. Specifically, we used Phaser in the CCP4 program suite40, with the E. coli wild type CheY•BeF3−•Mn2+ structure (pdb 1FQW) as our search model. Initial rigid body refinement, was applied to refine the position and orientation of the molecule in the asymmetric unit and initial density maps created. Manual model building, alteration to the correct amino acids in positions 14, 59 and 89, and refinement were performed using Coot41, followed by restrained refinement using the Refmac5 program in CCP440. After several cycles of refinement and dropping of Rwork and Rfree values, water molecules were added with ARP/wARP in CCP440. Water molecules were confirmed by visually checking their environment for valid distances and angles. Finally, sulfate ions and glycerol molecules were modeled into appropriate densities. Final refinement statistics are shown in Table 2. In all of the CheY mutant structures, over 95% of the residues were in the generously allowed region in the Ramachandran plot, and the only residue in the disallowed region was Asn62, as previously observed42.
Structural comparisons and analysis
For all the crystal structures considered here, the individual chains comprising the asymmetric unit were first analyzed so that a representative chain could be chosen for comparative purposes. For structures where all active site residues (including the residues '14', '59', and '89') were in identical conformations in each monomer, a chain was chosen randomly. If there were noted differences in conformation, chains were eliminated based on the presence of intermolecular crystal contacts involving residues '14', '59', or '89'. CheYPhoB, CheYCusR, CheYArcA, and CheYSpo0F displayed no heterogeneity in conformations of active site residues and chain A was chosen randomly. For CheYFixJ DctD, there were different conformations for both Arg59 and His89 in the two monomers but neither could be eliminated based on involvement in intermolecular crystal contacts so both were considered (see Results). For the model response regulators, the following chains were selected based on the above criteria: PhoB•BeF3−•Mg2+ (pdb 1ZES chain B), FixJ-PO32−•NH4+ (pdb 1D5W chain B), Spo0F•BeF3−•Mg2+•Spo0B (pdb 2FTK chain H), ArcA•BeF3−•Mg2+ (pdb 1XHF chain A), and DctD•BeF3−•Mg2+ (pdb 1L5Y chain B). For CheY•BeF3−•Mn2+ (pdb 1FQW), there were different conformations of Glu89 in the two monomers. The two conformations of Glu89 superimposed until Cβ and then diverged, with monomer A tilted towards the BeF3−. Monomer A was chosen because the orientation of the Cβ-Cγ bond matched that for the residue at position 89 of the CheY mutants.
Superimpositions were carried out using the PyMOL43 “align” command with a specified set of atoms. For overlays of different CheY mutants, all of the backbone atoms were used. For overlays of CheY mutants with a receiver domain from a different response regulator, a portion of the backbone roughly corresponding to the β-strands plus the βα loops (except β4α4) with half of the ensuing α helix, (CheY residues 7–22, 34–43, 53–69, 83–87, 105–121) were used. These criteria gave reasonable overlays of the active site regions of CheY mutants with all of the response regulators except FixJ and Spo0F. For FixJ, the discrepancy may have been at least partially due to the absence of a metal ion in the structure because overlays using a small set of active site atoms gave reasonable alignments of the active site except for the metal binding region. The structure for Spo0F•BeF3−•Mg2+ was solved as a complex with Spo0B (pdb 2FTK), a histidyl phosphotransferase that interacts with the active site of Spo0F. In Spo0F•BeF3−•Mg2+, the β4α4 loop is far removed from the active site perhaps due to conformational rearrangement upon binding Spo0B (the β4α4 loop interacts directly with Spo0B). Furthermore, Spo0F does not undergo the complete activating conformational change because Spo0F His101 (analogous to CheY Tyr106) does not change rotameric state upon activation. Thus, the utility of the Spo0F•BeF3−•Mg2+ structure for comparison to CheYSpo0F may be limited.
Deposition of structure coordinates
The coordinates and structure factors for the five CheY mutants described here have been deposited in the Protein Data Bank. The accession number(s) are pdb 3FGZ (CheYPhoB = CheY F14E N59M E89R), pdb 3F7N (CheYCusR = CheY F14E N59M E89L), pdb 3FFX (CheYFixJ DctD = CheY F14E N59R E89H), pdb 3FFT (CheYArcA = CheY F14E E89R), and pdb 3FFW (CheYSpo0F = CheY F14Q N59K E89Y).
Acknowledgments
We thank Brenda Temple for expert advice in interpretation of structures. This work was supported by National Institutes of Health grant GM050860 (to R.B.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Abbreviations
- rmsd
root mean square deviation
- '59'
the residue in a response regulator that corresponds to position 59 (or whatever number is enclosed by single quotation marks) in Escherichia coli CheY
References
- 1.Jordan JD, Landau EM, Iyengar R. Signaling networks: the origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downward J. The ins and outs of signalling. Nature. 2001;411:759–762. doi: 10.1038/35081138. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 4.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 7.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Stephenson K, Hoch JA. Virulence- and antibiotic resistance-associated two-component signal transduction systems of Gram-positive pathogenic bacteria as targets for antimicrobial therapy. Pharmacol Ther. 2002;93:293–305. doi: 10.1016/s0163-7258(02)00198-5. [DOI] [PubMed] [Google Scholar]
- 9.Stock AM, Guhaniyogi J. A new perspective on response regulator activation. J Bacteriol. 2006;188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA, Wemmer DE. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 11.Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- 12.Park SY, Chao X, Gonzalez-Bonet G, Beel BD, Bilwes AM, Crane BR. Structure and function of an unusual family of protein phosphatases: the bacterial chemotaxis proteins CheC and CheX. Mol Cell. 2004;16:563–574. doi: 10.1016/j.molcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Muff TJ, Ordal GW. The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem. 2007;282:34120–34128. doi: 10.1074/jbc.M706432200. [DOI] [PubMed] [Google Scholar]
- 14.Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- 15.Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 16.Lukat GS, Stock AM, Stock JB. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 17.Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 18.Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts A, Lee SY, McCullagh E, Silversmith RE, Wemmer DE. YbiV from Escherichia coli K12 is a HAD phosphatase. Proteins. 2005;58:790–801. doi: 10.1002/prot.20267. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HT, Grigoriev IV, Wemmer DE, Kim SH. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319:421–431. doi: 10.1016/S0022-2836(02)00324-8. [DOI] [PubMed] [Google Scholar]
- 21.Goudreau PN, Lee PJ, Stock AM. Stabilization of the phospho-aspartyl residue in a two-component signal transduction system in Thermotoga maritima. Biochemistry. 1998;37:14575–14584. doi: 10.1021/bi980869i. [DOI] [PubMed] [Google Scholar]
- 22.Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagadeesen S, Mann P, Schink CW, Higgs PI. A novel “four-component” two component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J Biol Chem. 2009 doi: 10.1074/jbc.M109.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silversmith RE, Smith JG, Guanga GP, Les JT, Bourret RB. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J Biol Chem. 2001;276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
- 25.Silversmith RE, Guanga GP, Betts L, Chu C, Zhao R, Bourret RB. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol. 2003;185:1495–1502. doi: 10.1128/JB.185.5.1495-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonovic M, Volz K. A distinct meta-active conformation in the 1.1 Å resolution structure of wild-type apoCheY. J Biol Chem. 2001;276:28637–28640. doi: 10.1074/jbc.C100295200. [DOI] [PubMed] [Google Scholar]
- 27.Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. BeF3− acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3− complex with phosphoserine phosphatase. Proc Natl Acad Sci U S A. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbrack RL, Jr, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dougherty DA. Cation-pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 31.Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc Natl Acad Sci U S A. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maegley KA, Admiraal SJ, Herschlag D. Ras-catalyzed hydrolysis of GTP: a new perspective from model studies. Proc Natl Acad Sci U S A. 1996;93:8160–8166. doi: 10.1073/pnas.93.16.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright GD, Holman TR, Walsh CT. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32:5057–5063. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- 36.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silversmith RE. High mobility of carboxyl-terminal region of bacterial chemotaxis phosphatase CheZ is diminished upon binding divalent cation or CheY-P substrate. Biochemistry. 2005;44:7768–7776. doi: 10.1021/bi0501636. [DOI] [PubMed] [Google Scholar]
- 38.Silversmith RE, Appleby JL, Bourret RB. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Schuster M, Zhao R, Bourret RB, Collins EJ. Correlated switch binding and signaling in bacterial chemotaxis. J Biol Chem. 2000;275:19752–19758. doi: 10.1074/jbc.M909908199. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL molecular graphics system. 2002 www.pymol.org.
- 44.Fisher SL, Kim SK, Wanner BL, Walsh CT. Kinetic comparison of the specificity of the vancomycin resistance VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- 45.Weinstein M, Lois AF, Ditta GS, Helinski DR. Mutants of the two-component regulatory protein FixJ of Rhizobium meliloti that have increased activity at the nifA promoter. Gene. 1993;134:145–152. doi: 10.1016/0378-1119(93)90088-k. [DOI] [PubMed] [Google Scholar]
- 46.Meyer MG, Park S, Zeringue L, Staley M, McKinstry M, Kaufman RI, Zhang H, Yan D, Yennawar N, Yennawar H, Farber GK, Nixon BT. A dimeric two-component receiver domain inhibits the sigma54-dependent ATPase in DctD. Faseb J. 2001;15:1326–1328. doi: 10.1096/fj.00-0516fje. [DOI] [PubMed] [Google Scholar]
- 47.Zapf J, Madhusudan M, Grimshaw CE, Hoch JA, Varughese KI, Whiteley JM. A source of response regulator autophosphatase activity: the critical role of a residue adjacent to the Spo0F autophosphorylation active site. Biochemistry. 1998;37:7725–7732. doi: 10.1021/bi9729615. [DOI] [PubMed] [Google Scholar]
- 48.Georgellis D, Kwon O, De Wulf P, Lin EC. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 49.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]