Abstract
Born-Oppenheimer ab initio QM/MM molecular dynamics simulation with umbrella sampling is a state-of-the-art approach to calculate free energy profiles of chemical reactions in complex systems. To further improve its computational efficiency, a mass-scaling method with the increased time step in MD simulations has been explored and tested. It is found that by increasing the hydrogen mass to 10 amu, a time step of 3 fs can be employed in ab initio QM/MM MD simulations. In all our three test cases, including two solution reactions and one enzyme reaction, the resulted reaction free energy profiles with 3 fs time step and mass scaling are found to be in excellent agreement with the corresponding simulation results using 1 fs time step and the normal mass. These results indicate that for Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, the mass-scaling method can significantly reduce its computational cost while has little effect on the calculated free energy profiles.
Keywords: molecular dynamics simulation, ab initio QM/MM method, time step, free energy profile, umbrella sampling
Introduction
To reliably simulate chemical reactions in enzymes and in solution not only requires a reasonably accurate potential energy surface, but also needs extensive sampling due to the complexity of the energy landscape. The free energy changes associated with the reactions are not only better-defined for such systems, but also characterize the reactions better than potential energies. One of the most promising approaches to meet both daunting tasks is Born-Oppenheimer molecular dynamics (BO-MD) simulation with ab initio QM/MM potential and umbrella sampling: a first principle quantum mechanical method is used to describe a small region where the chemical reaction takes place, while a molecular mechanical force field is employed for the rest of the system; at each time step, the atomic forces as well as the total energy of the whole system come from the ab initio QM/MM approach [1-12] on-the-fly with a converged SCF calculation, and Newton equations of motion are integrated; from a series of biased simulations, the potential of mean force (PMF) along the reaction coordinate is obtained with the weighted histogram analysis method (WHAM) [13-16]. This direct ab initio QM/MM BO-MD approach provides an ab initio description of chemical bond formation and breaking process, properly and explicitly models the rest of the system, and takes accounts of dynamics of reaction center and its environment on an equal footing. In spite of the conventional wisdom that such simulations would be computationally too expensive to be applicable for enzyme reactions, this state-of-the-art approach has been demonstrated to be feasible and successful in elucidating the catalytic power of histone lysine methyltransferase SET7/9 [17], providing insights into the methylation state specificity of histone lysine methylation [18] and characterizing the Sir2 catalyzed nicotinamide cleavage reaction [19]. In the latter case, 720 ps B3LYP(6-31G*) QM/MM BO-MD simulations have been employed to study the SIR2 enzyme, in which the QM subsystem has 65 QM atoms and 560 basis functions while the MM sub-system has more than 9000 atoms. This would represent a typical application that we would be able to carry out with the currently available computational resources.
To further enhance the efficiency of ab initio QM/MM BO-MD simulations, it is of significant interest to increase the time step so that the number of force evaluations can be reduced to achieve the same simulation length. In classical MD simulations of molecular systems, the employed time step typically is no larger than 1.0 fs, which is limited by the highest frequency motion of bond variations in the system. One widely employed approach to increase the time step to 2.0 fs is to remove bond-stretching variations by using a bond constraint algorithm, such as SHAKE [20]. However, to constrain covalent bonds is not an option in QM/MM BO-MD simulations of chemical reactions, which involve bond forming and breaking. Since atomic motions are dependent on their masses, another intriguing approach is to scale atomic masses to slow down the high frequency motions [21-24]. In principle, mass-scaling would not affect the thermodynamic properties of the system although it would certainly lead to significant change in its kinetic properties. In molecular systems, hydrogen atoms are the lightest and are invariably involved in the fastest motions. For MD simulations with classical MM force field, several mass-scaling schemes have been tested in combination with bond constraints [21-24].
In this work, we have explored the mass-scaling idea in ab initio QM/MM BO-MD simulations and demonstrated that it is a promising approach to improve the computational efficiency in calculating free energy barriers of chemical reactions in complex systems. Our results showed that increasing the hydrogen mass to 10 amu, a simple mass scaling scheme which was introduced by Pomes and McCammon [22], would allow a time step of 3 fs in ab initio QM/MM MD simulations. For two solution reactions and one enzyme reaction, including Cl− + CH3Cl, → ClCH3 + Cl− and in aqueous solution, and the monomethylation reactions catalyzed by the enzyme SET7/9, the calculated reaction free energy profiles with 3 fs time step and mass scaling are found to be in excellent agreement with the corresponding simulation results using 1 fs time step and the normal mass.
Methods
ab initio QM/MM BO-MD Simulation
In the QM/MM framework [1-3], the total energy of the system can be expressed as following:
| (1) |
where Eqm(QM) is the quantum mechanical energy of the QM subsystem and Emm(MM) is the standard molecular mechanical interactions involving entirely atoms in the MM region. Eqm/mm(QM/MM) is the QM/MM interaction between the QM region and MM region, which can be casted as:
| (2) |
where three terms refer to the electrostatic, van der Waals and MM-bonded interactions between the QM and MM regions respectively.
In ab initio QM/MM calculations with electrostatic embedding, EMM-bonded(QM/MM), Evdw(QM/MM) and Emm(MM) are typically calculated with a MM force field; the sum of Eqm(QM) and Eele(QM/MM) are calculated with ab initio quantum mechanical calculations with an effective Hamiltonian Heff,
| (3) |
| (4) |
where the last two terms describe the QM/MM electrostatic interactions, and Neff is the total number of the electrons in the QM sub-system. In the pseudobond approach for the treatment of QM/MM covalent interface [25, 26], the effective core potential of boundary atoms would be an additional term in Heff.
In ab initio QM/MM BO-MD simulations, the atomic forces as well as the total energy of the whole system are calculated at each time step with the above formalism on-the-fly with a converged SCF calculation, and Newton equations of motion are integrated.Thus it takes accounts of dynamics of reaction center and its environment on an equal footing. The free energy profile F(η) along a chosen reaction coordination can be obtained from the probability distribution ρ(η) [27],
| (5) |
where kB is the Boltzmann constant, T is the absolute temperature, and ρ(η) is the probability distribution, which represents the occurring frequency of configurations with a given value of η among a canonical ensemble of structures. Due to the presence of large energy barriers along η, it is impractical to directly carry out the ordinary MD or MC simulations to obtain an adequate sampling of the probability distribution ρ(η). One of the most widely employed approach to meet this challenge is the umbrella sampling method [28, 29], which was initially introduced by Patey, Valleau, and Torrey [30, 31].
Umbrella Sampling and WHAM
In the umbrella sampling method [28-31], an artificial biasing potential ωi along a chosen reaction coordinate would be added to the total energy of the system in order to either constrain the simulation to sample a particular range more efficiently or flatten problematic barriers. The biasing function often takes the form ωi(η) = ½k(η - ηi)2, where ηi denotes the value of successive reaction coordinate that each biasing potential centers on. For each window, an independent equilibrium simulation would be performed, spanning the relevant range of the reaction coordinate. These simulations would result in a series of probability distribution profiles (e.g. histograms) along the reaction coordinate, one from each window. However, these histograms have been obtained using different biasing potentials, thus each of them needs to be unbiased and subsequently can be combined together to obtain the unbiased free energy profile. For this data analysis task, a widely employed approach is the weighted histogram analysis method (WHAM) [32-38].
In a canonical ensemble, the probability distribution along a reaction coordinate η can be written as:
| (6) |
where R represents all the the spatial coordinates of the system, U(R) is the corresponding energy, δ is the Dirac delta function, η’[R] is the reaction coordinate in the configuration R, β = (kBT)−1, and the integrals are over all values of R.
For the ith umbrella simulation with the presence of a biased potential ωi(η), the corresponding biased probability distribution can be casted into the following form:
| (7) |
Here by defining 〈e-βωi(η)〉 = e-βfi, where fi is an unknown constant for the umbrella simulation i, the unbiased probability density from the biased simulation i can be written as
| (8) |
We can see that can be calculated from the simulation trajectory, ωi(η) is the known potential function, and the only unknown parameter is the constant fi. In a typical umbrella sampling simulation which employs N biased windows, a most widely employed and successful approach to determine fi for each window and to combine all data together is the weighted histogram analysis method (WHAM) [32-38]. In order to provide an optimal estimate for the unbiased probability distribution as a η-dependent weighted sum over the N individual unbiased probability distribution, WHAM employs an iterative method to solve the following equation:
| (9) |
where N is the number of the umbrella windows and ni is the total configuration numbers of the i-th window. Typically, by first giving an initial estimate of fi, where i ranges from 1 to N, the above equation can be solved iteratively until all fi do not change. The resulted fi can be employed to obtain the total unbiased probability distribution along the reaction coordinate using the following equation:
| (10) |
Mass Scaling Molecular Dynamics
The time step employed in molecular dynamics simulations is limited by the highest frequency motion in the molecular system. One simple way to slow it down is to increase the mass of light atoms. Although mass-scaling certainly would lead to significant change in its kinetic properties, in principle it should not affect the thermodynamic properties of the system. Take the free free energy profile as an example, Eq. 5 and 6 indicate that it only depends on the potential energy of the system, and is independent of the kinetic part of the Hamiltonian. Thus, it should be able to be obtained from mass-scaling molecular dynamics. For MD simulations with classical MM force field, several mass-scaling schemes have been tested with bond constraint [21-24], including mass tensor molecular dynamics, increasing the mass of hydrogen atom, reducing the mass of non-hydrogen atoms, etc. In this work, by experimenting several previously proposed mass-scaling schemes, it is found that increasing the mass of hydrogen to 10 amu [22] is a very simple and efficient way for ab initio QM/MM BO-MD simulations of chemical reactions with umbrella sampling. It would allow a time step of 3 fs and lead to very little change in the simulated free energy profiles and structural properties in comparison with corresponding simulations using 1 fs time step and the normal mass.
Computation and Results
We have implemented ab initio QM/MM BO-MD simulations with umbrella sampling and mass scaling based on modified GAUSSIAN03 package [39] and TINKER4.2 [40]. Simulations have been carried out on two solution reactions and one enzyme reaction, including Cl− + CH3Cl, → ClCH3 + Cl− and in aqueous solution, and the mono-methylation reaction catalyzed by the enzyme SET7/9. For each test system, the only difference between the mass scaling simulation and the corresponding conventional one is that in mass-scaling simulation the time step is 3 fs and the mass for hydrogen is 10 amu while the conventional one uses 1 fs time step and the normal mass 1 amu for the hydrogen atom. Other simulation details are the same which allows a fair comparison. All umbrella simulations were performed in the NVT ensemble with the Berendsen thermostat method for controlling the system temperature at 300 K. For each umbrella window, 30 ps simulations were carried out and the last 20 ps trajectory has been used for data analysis.
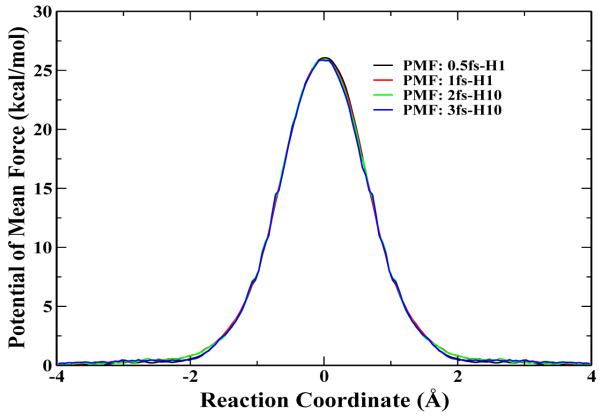
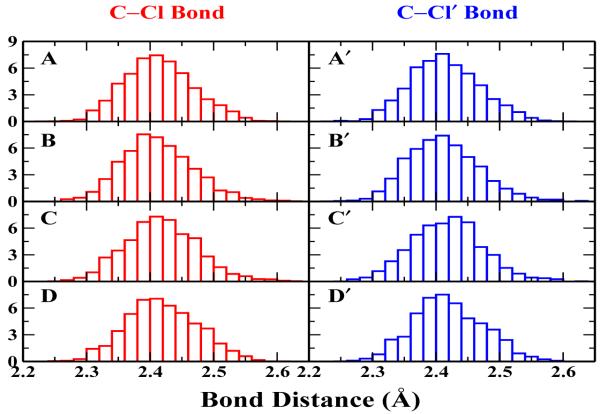
First, we carried out HF/6-31G* QM/MM BO-MD simulations of the Cl− + CH3Cl → ClCH3 + Cl− reaction in aqueous solution. This reaction has been extensively studied both experimentally and theoretically [41-51], which has been an excellent system to test our implementation and computational protocol. HF/6-31G* method has been employed, which has been known to describe such methyl-transfer reactions well with a reasonable computational cost [41,44-47,50]. The simulation protocol is very similar to the one in our recent studies [52]. In the QM/MM calculations, the solutes were solvated with 796 water molecules in a sphere of 15 Å radius and the TIP3P [53] water model was used. A spherical boundary condition has been employed, in which solvent molecules within 13 Å sphere of the sphere center were allowed to move during simulations. To prevent the solutes from moving to the boundary, their center of mass was constrained to the center of the sphere. The reaction coordinate Rc was defined as Rc = rCCl’ - rCCl, where Cl’ is the leaving atom. A total of 43 umbrella windows have been employed from -3.8 to 3.8 Å, with a harmonic force constant 50 kcal mol−1 Å−2. For this typical system with a relatively small QM sub-system, besides 1 fs time step with the normal mass (1fs-H1) and 3 fs time step with 10 amu for H (1fs-H10), we have also carried out simulations using 0.5 fs time step with normal mass (0.5fs-H1) and 2 fs time step with 10 amu for H (2fs-H10). The computed free energy profiles are presented in Fig. 1, and the four curves are almost indistinguishable. The calculated free energy barriers, as listed in Table I, are also very consistent with each other. It is ranging from 26.1 ±0.1 kcal/mol for the 0.5fs-H1 simulation to 25.7 ± 0.3 kcal/mol for the 3fs-H10 simulation, all of which is in excellent agreement with the experimental value (26.6 kcal/mol [49]), as well as with results from other calculation methods such as RISM/MD by Truong’s group (27.1 kcal/mol [54]), and the Monte Carlo simulation with umbrella sampling by Jorgensen’s group (26.3±0.5 kcal/mol [50]). Meanwhile, as expected, the timing data in Table I indicates that the use of the longer time step leads to the significantly less computational cost for accumulating the same length of the ab initio QM/MM BO-MD simulation trajectory. We have also checked the distribution of bond lengths at the transition state for all four simulations. As shown in Fig. 2, both Cl-C and C-Cl’ bond distributions are very consistent among these four simulations, which is reassuring that mass-scaling MD simulations would lead to very similar structural properties as the corresponding simulations with the normal mass.
Figure 1.
The computed free energy profile for Cl− + CH3Cl → ClCH3 + Cl− reaction in water. Four individual PMF curves obtained from different hydrogen atom mass and time steps have been plotted, which are 1 amu with 0.5 fs, 1 amu with 1 fs, 10 amu with 2 fs, 10 amu with 3 fs, respectively.
Table 1.
The summary of the free energy barrier (in kcal/mol) in solvent and computational cost for a 30 ps trajectory ( wall time in hours on running on a IBM PowerPC 970 computer cluster ) for the tested reactions
Figure 2.
The C-Cl and C-Cl’ bond distributions at the transition state for Cl− + CH3Cl → ClCH3 + Cl− reaction in water. Graph A, B, C, D indicate the C-Cl bond distributions for 0.5 fs and 1 fs with standard mass, 2 fs and 3 fs with modified hydrogen mass of 10 amu; Graph A’, B’, C’, D’ point to the C-Cl’ bond distributions for the same set of simulations respectively.
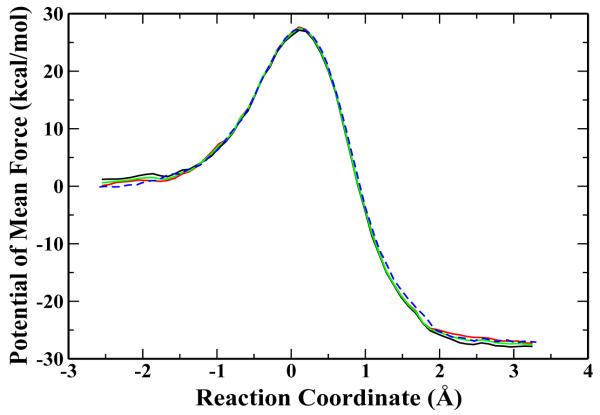
The second reaction we studied is the methyl transfer between and NH(CH3)2 in water. This reaction can be considered as a model for the methyl-transfer reaction catalyzed by histone lysine methyltransferases (HKMTs) in water, and the free energy reaction barrier has been measured experimentally [55]. To our best knowledge, there is no previous theoretical study on this reaction. Our simulation protocol is very similar to the one for the first reaction. The radius of the solvent water box is 15 Å, which contains 552 water molecules. The solutes are treated with HF/6-31G* and water molecules are described by the TIP3P water model. The spherical boundary condition (13 Å) was also applied. This methyl transfer reaction involves the breaking of S - C bond C - N, and therefore the reaction coordinate Rc was defined by Rc = rS-C - rN-C, where C is the carbon atom of the transfered methyl group. We have employed 22 umbrella windows, centered at -2.5, ... ,3.2 Å, with the harmonic force constant 70 kcal/mol−1Å−2. The calculated free energy profiles are shown in Figure 3, which have converged reasonably well and are very consistent with each other. The calculated free energy barriers are 27.4±0.7 kcal/mol for the 1fs-H1 simulation and 27.0 ±0.8 for the 3fs-H10 simulation, both of which are in good agreement with the experimental value of 28.1 kcal/mol [55].
Figure 3.
The free energy profile for reaction in condensed phase at 300 K. The three solid curves represent the PMF obtained from the mass scaling MD using the hydrogen mass of 10 amu with time step 3 fs from different time interval, which are from 10 ps to 20 ps; 20 ps to 30 ps; 10 ps to 30 ps respectively. The dashed curve shows the PMF obtained from the standard MD using the hydrogen mass of 1 amu with 1 fs time step.
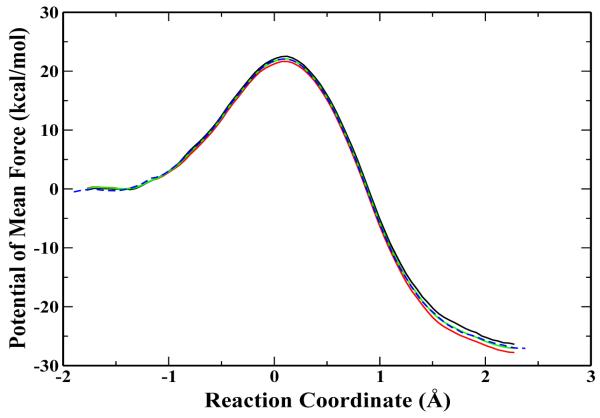
Finally, we tested the mass-scaling simulation on the mono-methylation reaction catalyzed by the histone lysine methyltransferase SET7/9, which involves the transfer of a methyl group from AdoMet to the histone-lysine residue H3-K4. The corresponding ab initio QM/MM BO-MD simulation with 1 fs time step and atomic normal mass has already been presented previously [17]. Here the same simulation protocol has been employed for the 3fs-H10 simulation, and the resulted free energy profile is shown in Fig. 4. Comparing the curves between the 3fs-H10 and 1fs-H1 simulations, again we can see that they are overlapping very well. The calculated free energy barriers are 22.1±0.6 kcal/mol and 22.5±0.5 kcal/mol from 3fs-H10 and 1fs-H1 simulations respectively, and their difference is within the statistical error of the sampling. These results indicate that the mass-scaling method is also applicable to ab initio QM/MM BO-MD simulations of enzyme reactions.
Figure 4.
The potential of mean force for the mono-methylation catalyzed by SET7/9. All the solid curves are obtained from the simulations by applying hydrogen mass of 10 amu pair with 3 fs step length. And corresponding to the black, red and green curves, the time period are from 10 ps to 20 ps; 20 ps to 30 ps; 10 ps to 30 ps. The dashed blue curve is from the standard mass coupled with 1 fs time step.
Conclusion
In order to further improve the computational efficiency of ab initio QM/MM BO-MD simulations, a mass-scaling method with the increased time step has been explored and tested. Our results showed that increasing the hydrogen mass to 10 amu would allow a time step of 3 fs in ab initio QM/MM BO-MD simulations and lead to very little change in the simulated free energy profiles and structural properties in comparison with corresponding simulations using 1 fs time step and the normal mass. Certainly, mass scaling would affect the dynamics of the system. Take water as an example, the increase of the hydrogen mass to 10 amu would decrease the diffusion constant of water by 30 % [22]. However, it would not be a main concern as long as the thermodynamic and structural properties are the main interests of the simulation. Meanwhile, it is also worth to note that the diffusion constant of TIP3P water model is about twice as large as the experimental value. These results suggest that the mass scaling method is a simple and quite effective approach to improve the computational efficiency of ab initio QM/MM BO-MD simulations.
Acknowledgments
We acknowledge support from National Science Foundation (CHE-CAREER-0448156), National Institute of Health (R01-GM079223) and NYSTAR (James D. Watson Young Investigator Award). We appreciate the computational resources provided by the NYUITS.
References
- [1].Warshel A, Levitt M. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- [2].Singh UC, Kollman PA. J. Comput. Chem. 1986;7:718–730. [Google Scholar]
- [3].Field MJ, Bash PA, Karplus M. J. Comput. Chem. 1990;11:700–733. [Google Scholar]
- [4].Gao J, Truhlar DG. Annu. Rev. Phys. Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- [5].Cui Q, Karplus M. Adv. Protein Chem. 2003;66:315–372. doi: 10.1016/s0065-3233(03)66008-0. [DOI] [PubMed] [Google Scholar]
- [6].Shurki A, Warshel A. Adv. Protein Chem. 2003;66:249–313. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]
- [7].Friesner RA, Guallar V. Annu. Rev. Phys. Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- [8].Mulholland AJ. Drug Discov. Today. 2005;10:1393–1402. doi: 10.1016/S1359-6446(05)03611-1. [DOI] [PubMed] [Google Scholar]
- [9].Zhang Y. Theor. Chem. Acc. 2006;116:43–50. [Google Scholar]
- [10].Senn HM, Thiel W. Top. Curr. Chem. 2007;268:173–290. [Google Scholar]
- [11].Hu H, Yang W. Annu. Rev. Phys. Chem. 2008;59:573–601. doi: 10.1146/annurev.physchem.59.032607.093618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Senn HM, Thiel W. Angew. Chem. Int. Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- [13].Patey GN, Valleau JP. J. Chem. Phys. 1975;63:2334–2339. [Google Scholar]
- [14].Boczko EM, Brooks CL. J. Phys. Chem. 1993;97:4509–4513. [Google Scholar]
- [15].Roux B. Comput. Phys. Commun. 1995;91:275–282. [Google Scholar]
- [16].Ferrenberg AM, Swendsen RH. Phys. Rev. Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- [17].Wang S, Hu P, Zhang Y. J. Phys. Chem. B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hu P, Wang S, Zhang Y. J. Am. Chem. Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hu P, Wang S, Zhang Y. J. Am. Chem. Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- [21].Bennett CH. J. Comput. Phys. 1975;19:267–279. [Google Scholar]
- [22].Pomes R, Mccammon JA. Chem. Phys. Lett. 1990;166:425–428. [Google Scholar]
- [23].Feenstra KA, Hess B, Berendsen HJC. J. Comput. Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [24].Stocker U, Juchli D, van Gunsteren WF. Mol. Simul. 2003;29:123–138. [Google Scholar]
- [25].Zhang Y, Lee T, Yang W. J. Chem. Phys. 1999;110:46–54. [Google Scholar]
- [26].Zhang Y. J. Chem. Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- [27].Landau LD, Lifshiz EM. Statistical Physics. 2nd ed. Pergamon; New York: 1969. pp. 343–353. [Google Scholar]
- [28].Sheinerman FB, Brooks CL. J. Mol. Biol. 1998;278:439–456. doi: 10.1006/jmbi.1998.1688. [DOI] [PubMed] [Google Scholar]
- [29].Allen MP, Tildesley DJ. Computer Simulation of Liquid. Clarendon Press; Oxford: 1989. [Google Scholar]
- [30].Patey GN, Valleau JP. J. Chem. Phys. 1975;63:2334–2339. [Google Scholar]
- [31].Torrie GM, Valleau JP. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- [32].Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- [33].Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. J. Comput. Chem. 1995;16:1339–1350. [Google Scholar]
- [34].Souaille M, Roux B. Comput. Phys. Commun. 2001;135:40–57. [Google Scholar]
- [35].Ferrenberg AM, Swendsen RH. Phys. Rev. Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- [36].Ferrenberg AM, Swendsen RH. Phys. Rev. Lett. 1989;63:1195–1198. doi: 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- [37].Roux B. Comput. Phys. Commun. 1995;91:275–282. [Google Scholar]
- [38].Boczko EM, Brooks CL. J. Phys. Chem. 1993;97:4509–4513. [Google Scholar]
- [39].Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheese-man JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision D.01. [Google Scholar]
- [40].Ponder JW. TINKER, Software Tools for Molecular Design, Version 3.6. Jun, 2004. [Google Scholar]
- [41].Bento AP, Sola M, Bickelhaupt FM. J. Comput. Chem. 2005;26:1497–1504. doi: 10.1002/jcc.20261. [DOI] [PubMed] [Google Scholar]
- [42].Bogdanov B, Mcmahon TB. Int. J. Mass Spectrom. 2005;241:205–223. [Google Scholar]
- [43].Chabinyc ML, Craig SL, Regan CK, Brauman JI. Science. 1998;279:1882–1886. doi: 10.1126/science.279.5358.1882. [DOI] [PubMed] [Google Scholar]
- [44].Hase WL. Science. 1994;266:998–1002. doi: 10.1126/science.266.5187.998. [DOI] [PubMed] [Google Scholar]
- [45].Mohamed AA, Jensen F. J. Phys. Chem. A. 2001;105:3259–3268. [Google Scholar]
- [46].Morokuma K. J. Am. Chem. Soc. 1982;104:3732–3733. [Google Scholar]
- [47].Truong TN, Stefanovich EV. J. Phys. Chem. 1995;99:14700–14706. [Google Scholar]
- [48].Yang SY, Fleurat-lessard P, Hristov I, Ziegler T. J. Phys. Chem. A. 2004;108:9461–9468. doi: 10.1021/jp045943n. [DOI] [PubMed] [Google Scholar]
- [49].Mclennan DJ. Aust. J. Chem. 1978;31:1897–1909. [Google Scholar]
- [50].Chandrasekhar J, Smith SF, Jorgensen WL. J. Am. Chem. Soc. 1984;106:3049–3050. [Google Scholar]
- [51].Gould IR, Moody R, Farid S. J. Am. Chem. Soc. 1988;110:7242–7244. [Google Scholar]
- [52].Zheng H, Zhang Y. J. Chem. Phys. 2008;128:204106. doi: 10.1063/1.2920476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- [54].Freedman H, Truong TN. J. Phys. Chem. B. 2005;109:4726–4730. doi: 10.1021/jp044944c. [DOI] [PubMed] [Google Scholar]
- [55].Callahan BP, Wolfenden R. J. Am. Chem. Soc. 2003;125:10481–10481. doi: 10.1021/ja021212u. [DOI] [PubMed] [Google Scholar]
- [56].Trievel RC, Beach BM, Dirk LMA, Houtz RL, Hurley JH. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]