Abstract
Motivation: High-throughput protein identification experiments based on tandem mass spectrometry (MS/MS) often suffer from low sensitivity and low-confidence protein identifications. In a typical shotgun proteomics experiment, it is assumed that all proteins are equally likely to be present. However, there is often other evidence to suggest that a protein is present and confidence in individual protein identification can be updated accordingly.
Results: We develop a method that analyzes MS/MS experiments in the larger context of the biological processes active in a cell. Our method, MSNet, improves protein identification in shotgun proteomics experiments by considering information on functional associations from a gene functional network. MSNet substantially increases the number of proteins identified in the sample at a given error rate. We identify 8–29% more proteins than the original MS experiment when applied to yeast grown in different experimental conditions analyzed on different MS/MS instruments, and 37% more proteins in a human sample. We validate up to 94% of our identifications in yeast by presence in ground-truth reference sets.
Availability and Implementation: Software and datasets are available at http://aug.csres.utexas.edu/msnet
Contact: miranker@cs.utexas.edu, marcotte@icmb.utexas.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
High-throughput protein identification in biological samples aids our understanding of complex cellular systems and their behavior. Mass spectrometry (MS)-based shotgun proteomics offers fast, high-throughput characterization of complex protein mixtures. Several thousand proteins may be identified in a sample using high-resolution MS/MS instruments and/or extensive biochemical fractionation (Brunner et al., 2007; Graumann et al., 2007), but standard approaches only identify a fraction of the expected proteins.
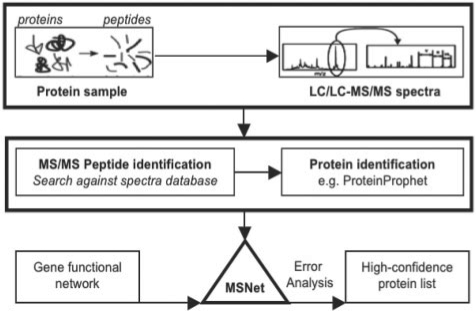
A shotgun proteomics experiment typically proceeds by MS/MS analysis of peptides from proteolytically digested proteins, followed by in silico matching of the MS/MS spectra against a database of theoretical peptide spectra derived from protein sequences (Fig. 1). Proteins are identified using combined evidence from constituent peptides, resulting in a list in which each protein is associated with a score signifying the confidence of correct identification. We refer to this score as the MS/MS protein score, e.g. ProteinProphet's protein probability (Nesvizhskii et al., 2003). Proteins with scores that satisfy an error threshold are labeled present by the MS analysis software.
Fig. 1.
Integrative analysis of MS-based shotgun proteomics and gene functional networks. A complex protein sample, e.g. cellular extract, is enzymatically digested into peptides and subjected to tandem mass spectrometry. Experimental spectra are searched against a database of theoretical spectra generated from protein sequences, or identified via de novo sequencing, using a peptide and protein identification software pipeline that produces a confidence score per protein [e.g. PeptideProphet (Keller et al., 2002) and Protein-Prophet (Nesvizhskii et al., 2003)] and a list of high-confidence proteins with scores that satisfy an error threshold (e.g. 5% FDR). We introduce a next stage of computational analysis which places proteins in a broader systems biological framework. MSNet uses protein-protein links from a functional network to identify proteins that may not be identified with high confidence by MS evidence alone, but are nevertheless highly likely to be present as demonstrated by the combination of MS evidence with functional links to other MS identified proteins. We find that the integrated analysis of mass spectrometry experiments and gene functional networks can improve the precision and sensitivity of protein identification at acceptable error rates.
Effective MS/MS protein identification is hindered by factors such as noisy spectra, low-concentration proteins, post-translational modifications and chemical properties that interfere with peptide ionization. For complex samples such as cell lysates, current MS search algorithms typically only match a small percentage (<20%) of all MS/MS spectra to real peptides, resulting in higher error rates and low recall at the protein level. As a result, only a percentage of the expected proteins are identified with confidence despite presence in the biological sample, and the MS/MS identification scores of many other proteins fall below acceptable confidence thresholds.
MS/MS protein identification scoring schemes, such as BioWorks (ThermoFinnegan) and ProteinProphet (Nesvizhskii et al., 2003), assume that all proteins are equally likely to be present. In reality, other information may be available and can be used to influence the inferred probability of protein presence thereby rescuing proteins that fall below confidence thresholds.
We use gene functional networks (Marcotte et al., 1999) as an external information source to analyze proteins in a sample in the context of the biological processes that are active in the cell. Given a list of proteins identified in an MS experiment (M), we determine a more complete list (M′) by considering the proteins that are expected to be present (or absent) based on their functional linkages to proteins in M. Each protein receives a revised identification score with contributions both from direct MS-based evidence, and MS evidence of neighbors in the gene functional network. Since current gene networks can be incomplete, we intend for M′ to serve as a complement to M, rather than replace it as the authoritative list of expressed proteins.
Our data integration approach has the potential to enable pathway-based interpretation of high-throughput MS/MS experiments that are otherwise run in isolation. For instance, by integrating mass spectrometry data from yeast grown in rich medium with a published yeast functional network (Lee et al., 2007), we were able to confidently identify many proteins from ribosomal complexes and proteins involved in RNA binding, processing and degradation, thereby increasing the protein coverage in several active pathways (Section 4). When our method was applied to yeast grown in minimal medium, we increased the number of proteins identified in the reductive carboxylate cycle pathway (Ogata et al., 1999). In both cases, we expect the newly identified proteins to be present in the sample, but they were not identified with confidence by the MS analysis software, despite having at least one peptide identified per protein.
We demonstrate the applicability of MSNet to data from different organisms, mass spectrometers, MS analysis pipelines, and experimental conditions. We identify 8–29% more proteins on different yeast datasets at the same error rate, and evaluate the quality of protein identifications via ROC and precision–recall plots. In yeast grown in rich medium, analyzed on a high-resolution mass spectrometer, we identify 29% more proteins than the original MS analysis, 97% of which are present in a reference set derived from independent identification experiments. We also demonstrate direct applicability to the human proteome using a human functional gene network, reporting 37% more proteins than the original MS analysis.
2 METHODS
2.1 MSNet algorithm
MSNet introduces an additional stage of computational analysis to MS/MS shotgun protein identification (Fig. 1). In this section, we introduce the MSNet protein identification score. Specifically, if two proteins are known to be ‘functionally linked’ i.e. proteins p1 and p2 are known to physically interact, be co-expressed or co-regulated across several biological conditions, and p1 has been observed in a MS experiment, we propose that p1 should be assigned a revised identification score that depends not only on its own MS-based identification score c1, but also on the MS identification of its functional neighbor p2 and the strength of belief in the functional link between p1 and p2. The concept can be extended from two genes to pathways of co-functioning genes, generating revised identification scores for every protein encoded in the genome. Note that the confidence score c1 represents protein presence, and not protein abundance.
We use the yeast gene functional network developed by Lee et al. (Lee et al., 2004, 2007) which spans >95% of the yeast genes. The network forms a graph G = (V, E) with |V| = N genes and |E| weighted edges (wij) between nodes. The weight wij of an edge between two genes i and j is defined as the log of the likelihood odds ratio that there exists a link, and is determined by Bayesian integration of thousands of diverse experiments that estimate functional association e.g. mRNA co-expression, phylogenetic profiles, protein interaction experiments and co-citation in published literature (Lee et al., 2007). Intuitively, wij denotes the strength of a functional link between two genes. For human samples, we use a similarly constructed human gene network (Lee and Marcotte, manuscript in preparation).
MSNet computes a score yi for each protein i, which represents how likely it is for i to be present in the sample given MS evidence for i and its functionally related proteins j. The MSNet score for protein i (Equation 2) is the convex combination of two terms: (i) the probability that the protein is present in the sample given evidence from a MS experiment (oi) and (ii) the weighted average of MSNet scores of i's immediate network neighbors j (Equation 4). We set oi to the MS protein probability generated by ProteinProphet (Nesvizhskii et al., 2003), but any posterior probability of protein presence given sample-specific experimental data may be used instead (see discussion in Section 4). Since yi is defined in terms of yj, we update scores iteratively. At each iteration t, the algorithm includes evidence from neighbors at path length=t.
| (1) |
| (2) |
| (3) |
| (4) |
The MSNet score can be rewritten in vector notation using the weighted adjacency matrix UN×N and MS protein probability vector ON×1 to generate score vector YN×1 (Equation 2).
The MSNet algorithm is closely related to diffusion algorithms like Google's PageRank (Langville and Meyer, 2006; Page et al., 1999). PageRank has been successfully used to determine a relevancy ranking of webpages based on the hyperlink structure of the web (Langville and Meyer, 2006). MSNet generates a ranking of proteins that is based not only on the link structure of a gene functional network, but also on per-protein relevance to a given sample. In Supplementary Appendix I, we show that MSNet is equivalent to a personalized (Page et al., 1999) or topic-sensitive variant of PageRank (Haveliwala, 2003) with two differences. First, PageRank is defined on a directed graph. Gene functional networks are undirected, so each edge must be interpreted as being bi-directional. A second related difference is that PageRank uses a column-normalized weight matrix H = UT. We justify the use of U in Supplementary Appendix I, and show that it performs better in our domain in Supplementary Figure S6.
MSNet can be shown to converge to a unique solution irrespective of starting vector Y(0) (proof of convergence is in Supplementary Appendix I). In practice, MSNet converges within 10−6 tolerance in tens of iterations (Equation 3). In our experiments, we initialize Y(0) = O. Parameter (1 − γ)/γ weights the network's contribution to the MSNet score. We optimize γ in yeast by maximizing the area under the ROC curve (AUC) while maintaining similar error rates as the MS analysis across multiple datasets. AUC is not very sensitive to (1 − γ)/γ in the range [5,50] (see Supplementary Fig. S3). We set (1 − γ)/γ = 6 for yeast.
2.2 Evaluation methodology
In this section, we describe the MSNet evaluation framework, introduce the error measures used and describe how they are computed. For a given mass spectrometry experiment and gene functional network, we calculate the MSNet protein identification score for every protein on a genome-wide scale. To test robustness to missing network links, the reported MSNet score is averaged across 10 runs of 10-fold cross-validation. We restrict our evaluation to proteins with at least one peptide identified in the MS experiment.
We use a 5% false discovery rate (FDR) (Storey and Tibshirani, 2003) to determine a high-confidence list of proteins. The FDR at a score t is the fraction of false instances among all identifications with score ≥t. We employ two approaches to estimate the FDR: (i) using a protein reference set as ground-truth to categorize proteins as true or false instances; (ii) generating true and false (null) score distributions independent of ground truth as described in detail below.
We conducted functional analysis of yeast proteins using SGD (Nash et al., 2007), FunSpec (Robinson et al., 2002) and FuncAssociate (Berriz et al., 2003), applying Bonferroni corrections.
2.2.1 Evaluation against a protein reference set
When a protein reference dataset is available, we use it to label a protein as a true instance (T) if it is present in the reference set, and as a false instance (F) otherwise. We estimate the FDR at score threshold s as FDRref = F/(T + F), the percentage of false instances that have score ≥s. We also plot receiver operator characteristic (ROC) and precision-recall curves using the reference set to determine true and false instances. A ROC curve plots true positive rate (TPR) versus the false positive rate (FPR). A precision-recall curve plots (1-FDR) (precision) versus TPR (recall). TPR at a score threshold t is the fraction of true instances with score ≥t. FPR at score threshold t is the fraction of false instances with score ≥t. FDR is defined above. We also report the ROC AUC, the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one (Fawcett, 2006).
2.2.2 Evaluation independent of a protein reference set
When protein reference sets are unavailable, it is standard to compute error estimates by generating a null distribution of scores, and using the ratio of the areas of null and true distributions at scores ≥s as an estimate of the FDR at score threshold s. Though there has been extensive recent work on the estimation of FDRs at the peptide-level (Choi and Nesvizhskii, 2008; Kall et al., 2008), there is no consensus at the protein identification level (Tabb, 2008). Our purpose however is to develop an error model for MSNet, and we do not address the reliability of MS error models in this article. We generate an error model using a method we refer to as network-shuffling, similar to randomization or permutation tests used in statistical hypothesis testing. For a given dataset, we generate a null distribution of MSNet scores by running MSNet on a network where the labels on the nodes (protein names) are shuffled, such that proteins maintain features such as the MS protein identification score, but have a different set of network neighbors. This label-shuffling destroys any biological gene–gene association signal, while maintaining the total node degree (topology). We repeat the shuffling process multiple times and pool all generated scores to estimate the null score distribution. The true score distribution is generated by running MSNet on the original network. We plot density distributions for null and true scores (Supplementary Fig. S2) and estimate FDR as FDRshuff = Ns/Ts, where Ns is the area under the null distribution for scores ≥s and Ts is the area under the true distribution for scores ≥s. In this article, FDR refers to FDRshuff unless stated otherwise.
2.3 Datasets
We evaluated MSNet on different organisms, experimental conditions and mass spectrometers (Table 1). MS/MS data was collected on low and high-resolution mass spectrometers: ThermoFinnigan's Surveyor/DecaXP+(LCQ) and LTQ-OrbiTrap (ORBI). MS/MS protein identification was conducted using Bioworks 3.3 (ThermoFinnigan), PeptideProphet (Keller et al., 2002) and ProteinProphet (Nesvizhskii et al., 2003). We considered the entire yeast genome except for proteins annotated as ‘dubious’, since these proteins were not considered in the yeast network (Lee et al., 2007). All MS yeast experiments were the result of combined MS analysis of multiple injections of the sample. An identified protein was labeled as a true instance if it was present in the corresponding protein reference set (Table 1).
Table 1.
Datasets and experimental setup
| Dataset | MS/MS experiment | Protein reference set | Number of proteins |
|---|---|---|---|
| YPD-ORBI | Cell lysate from yeast BY4742 wild-type grown in rich medium (YPD) analyzed on LTQ- ORBItrap (8inj) | YPD*: Proteins identified in ≥ 1 of three non-mass spectrometry experiments (Futcher et al., 1999; Ghaemmaghami et al., 2003; Newman et al., 2006) or ≥ 2 of four MS experiments (Chi et al., 2007; de Godoy et al., 2006; Peng et al., 2003; Washburn et al., 2001). Total 4264 proteins (67% of yeast genes) | 3816 |
| YPD-LCQ | Cell lysate from yeast BY4742 wild-type grown in rich medium (YPD) analyzed on LCQ (5inj) | YPD* defined above | 4385 |
| YPD-LCQ-Fraction | Cell lysate, fractionated in polysomal gradient from yeast grown in rich medium (YPD) analyzed on LCQ (3inj) | Known ribosomal, translation and ribosome biogenesis proteins (Nash et al., 2007; Planta and Mager, 1998) | 1393 |
| YMD-LCQ | Cell lysate from yeast BY4742 wild-type grown in minimal medium (YMD) analyzed on LCQ (6inj) | YMD*: Proteins identified in at least one of three experiments (de Godoy et al., 2006; Newman et al., 2006; Zybailov et al., 2005). | 4651 |
| Human-293T, ORBI | HEK293T kidney embryonic cells transfected with GFP lenti-virus vector | No comprehensive reference set available | 1860 |
The protein sample undergoes MS/MS analysis to generate a list of proteins identified by MS/MS identification software. We generate MSNet protein identification scores, on a genome-wide scale, for each protein that has at least one peptide identified in the MS experiment (Number of proteins). When available, we use a protein reference set as ground-truth to determine true and false identifications for evaluation. Inj—injection, i.e. technical replicate during MS/MS experiment; LCQ—LCQ DecaXP+MS/MS instrument; ORBI—LTQ-OrbiTrap MS/MS instrument).
2.3.1 Yeast (rich medium)
Cell lysate from wild-type yeast grown in rich medium was analyzed on both LCQ and ORBI mass spectrometers. The LCQ data has been published previously (Lu et al., 2007).
2.3.2 Yeast (rich medium, polysomal fraction)
Cellular lysate was separated in 7–47% sucrose gradient and fractions were monitored by UV absorbance for RNA content (Li et al., 2009). We chose the fraction containing 80S ribosomes for LC–MS/MS analysis on the LCQ.
2.3.3 Yeast (minimal medium)
We used MS/MS data on wild-type yeast grown in minimal medium (MOPS9), previously published in (Lu et al., 2007), with cell lysate analyzed on an LCQ mass spectrometer.
2.3.4 Human
Protein extracts from human HEK293T cell lines were prepared for MS/MS analysis as described in the Supplement. We evaluated results using the shuffled network approach, since no comprehensive protein reference set was available for this dataset.
2.3.5 Availability
Yeast LCQ data has been previously published (Lu et al., 2007). Software and datasets are available at http://aug.csres.utexas.edu/msnet. Further details about sample preparation and protein reference sets are in the Supplement.
3 IMPLEMENTATION AND RESULTS
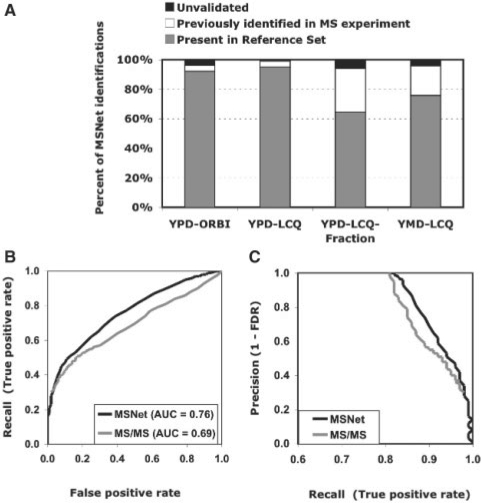
We demonstrate that incorporating functional association information can substantially boost correct identification of proteins in a shotgun proteomics experiment, across a range of sample conditions and mass spectrometers. For each dataset in Table 1, we measured the number of proteins identified by MSNet at 5% FDR as compared to the original MS experiment at its 5% FDR. ProteinProphet (Nesvizhskii et al., 2003) computes FDR directly from protein probabilities, which the authors empirically show to be good estimates of the true posterior probability of protein presence. MSNet consistently increased the number of identified proteins by 8–29% across yeast experiments (Table 2) and at least 94% of MSNet proteins were validated—either by presence in the reference set, or previous identification in the MS experiment (Fig. 2A). When protein reference sets were available, MSNet increased the number of identifications at 5% FDRref by 12–100% across datasets (Supplement Table S3) and increased ROC-AUC by up to 24% (Table 2). We also demonstrate MSNet's applicability to data generated from different MS pipelines. We describe these results in detail below.
Table 2.
MSNet performance evaluated with and without a protein reference set
| AUC (using reference set) |
Number of proteins at 5% FDR (using network shuffling) |
|||||
|---|---|---|---|---|---|---|
| Experiment | MS | MSN | % Increase | MS | MSN | % Increase |
| YPD-ORBI | 0.69 | 0.76 | 10 | 1420 | 1835 | 29 |
| YPD-LCQ | 0.55 | 0.68 | 24 | 548 | 591 | 8 |
| YPD-LCQ-Fraction | 0.78 | 0.91 | 17 | 246 | 285 | 16 |
| YMD-LCQ | 0.59 | 0.69 | 17 | 644 | 699 | 9 |
| Human-293T | – | – | – | 877 | [870–1233] | [0–40] |
First, we evaluated the performance of MSNet and the MS experiment using protein reference sets (Table 1), marking an identified protein as a true instance if it was present in the reference set and false otherwise. MSNet increased the AUC by 10–24% across datasets. Next, we evaluated MSNet independent of protein reference sets using a network-shuffling procedure (Section 2.2.2). We computed FDRshuff as the ratio between the cumulative null and true score densities at each score x. MSNet reported 8–29% more protein identifications at 5% FDRshuff in yeast and up to 40% more in human than ProteinProphet (Nesvizhskii et al., 2003) at its 5% FDR. MSN—MSNet, MS—ProteinProphet.
Fig. 2.
Performance of MSNet on yeast grown in rich medium analyzed on a high-resolution mass spectrometer. (A) At least 94% of proteins identified by MSNet at 5% FDR can be validated either by presence in the protein reference set or by identification in the MS analysis; (B) ROC curves using a protein reference set to determine true and false identifications: MSNet identifies more true instances over a range of FPRs than original MS experiment and results in 10% higher AUC; (C) precision–recall curves: MSNet identifies more proteins at high precision (i.e. low FDR) than the MS analysis.
3.1 Yeast grown in rich medium
We tested the applicability of our method to whole-cell lysate samples using yeast grown in rich medium analyzed on high and low-resolution mass spectrometers. In Table 2, we report the number of proteins identified by MSNet for the yeast rich medium sample analyzed on the high resolution LTQ-Orbitrap (Table 1, YPD-ORBI). MSNet reported 1835 identifications at 5% FDR, a 29% increase over the original MS experiment.
We validated 96% of MSNet's 5% FDR proteins—92% were present in the reference set and a further 4% were previously identified in the original MS experiment (Fig. 2B). There were 460 new MSNet proteins not previously identified in the MS experiment. They were enriched for ribosome or translation-associated functions when compared against a background of the whole genome, and for proteins of unknown function compared to a background of MSNet 5% FDR proteins (P < 0.001). Eighty-five percent of the 460 new identifications were present in the reference set and the remaining 15% were not enriched for any functional category—thus there were no obvious false-positive identifications based on protein function analysis.
We generated ROC and precision–recall plots for both MSNet and the original MS experiment, marking protein as a true instance if it was present in the YPD* reference set (Table 1), and false otherwise. In a ROC plot (Fig. 2B), MSNet identified more true instances (proteins present in the reference set) than the original MS experiment over a range of FPRs. Similarly, in a precision–recall plot (Fig. 2C) MSNet identified more true instances over a range of FDRs (1–precision), e.g. identifying 12% more proteins at 5% FDRref (Supplement Table S3). MSNet also resulted in a 10% increase in ROC-AUC (Table 2), i.e. MSnet is 10% more likely than MS analysis to rank a randomly chosen true instance higher than a randomly chosen negative instance (Fawcett, 2006).
MSNet improved performance even when the original MS experiment was limited by instrument resolution, as we observed on the same sample re-analyzed on a low-resolution mass spectrometer (Table 1, YPD-LCQ). MSNet reported 8% more proteins than the original MS experiment (Table 2) and increased AUC by 24% (Table 2, Supplementary Fig. S1). The new MSNet identifications were enriched for ribosomal proteins (P <0.001).
3.2 Yeast grown in minimal medium
We expect our method to be applicable to yeast in different sample conditions, since the gene network was constructed by integrating diverse biological experiments. Indeed, when applied to yeast grown in minimal medium (Table 1, YMD-LCQ), MSNet identified 9% more proteins at 5% FDR (Table 2). The new MSNet identifications were enriched for ribosomal proteins (P < 0.001) as in the rich-medium yeast experiment, but also for proteins of small molecule biosynthesis (P < 0.001) e.g. carboxylic acid, amine or folate metabolism, which is expected for growth in minimal medium. MSNet increased AUC by 17% when evaluated against the YMD* reference set (Table 2, Supplementary Fig. S1).
3.3 Yeast polysomal fraction
We expect MSNet to be especially effective on smaller, focused protein preparations. Accordingly, we tested MSNet on a polysomal fraction of yeast grown in rich medium, fractionated on a sucrose density gradient (Table 1, YPD-LCQ-Fraction). Proteins in this sample were restricted to those co-fractionating with 80S ribosomes and were expected to be associated with ribosomal and translation functions.
MSNet identified 16% more proteins at 5% FDR than the original MS experiment (Table 2). Ninety-four percent of MSNet identifications were validated, either by presence in the fractionation reference set or by previous identification in the MS experiment (Fig. 2A). In a function analysis, all but three new MSNet proteins were found to be associated with the ribosome, ribosomal functions or translation. The three proteins might represent false positives: inosine monophosphate dehydrogenase IMD2 which catalyzes the first step of GMP biosynthesis; ADK2, a mitochondrial adenylate kinase which catalyzes the reversible synthesis of GTP and AMP from GDP and ADP; and FLC1, a putative FAD transporter (Nash et al., 2007). MSNet increased AUC by 17% when evaluated against the fractionation protein reference set (Table 1). The corresponding ROC and precision–recall curves are plotted in Supplementary Figure S1.
3.4 Applicability to higher organisms
Finally, we tested MSNet in higher organisms by evaluating proteins expressed in human HEK293T cells analyzed on a high-resolution mass spectrometer (Table 1, Human-293T). We used a human gene functional network (Lee and Marcotte, manuscript in preparation). We considered 18 514 protein-coding genes present in the network, and reported up to 40% increase in the number of identified proteins at 5% FDR. We present a range of results in Table 2 with parameter (1 − γ)/γ varying in [6,10]. As in yeast (Section 2.1), this parameter may be optimized as reference sets for human data become available. The new 5% FDR MSNet proteins were not enriched for any functional category.
3.5 Performance on different MS/MS pipelines
We tested the applicability of MSNet to MS/MS data analyzed using different software pipelines. There are several issues with systematic testing and comparison of different MS pipelines. First, there is currently only one published, freely available analysis pipeline that generates protein-level probabilities and FDRs i.e. the TransProteomicPipeline [TPP, (Keller et al., 2002; Nesvizhskii et al., 2003)], which we used for our main results. Second, a systematic comparison is non-trivial since each pipeline makes different statistical assumptions and the hypotheses are not independent. Third, any such effort also entails significant development to accommodate different data formats (Prince and Marcotte, 2008).
Nevertheless, we tested four pipelines: (i) TPP with SEQUEST (Bioworks) for spectral matching (used for main results); (ii) TPP with X!Tandem (Craig and Beavis, 2004) for spectral matching; (iii) CRUX for spectral matching (Park et al., 2008), Percolator (Kall et al., 2007) for peptide-matching and DTASelect (Tabb et al., 2002) for protein reports; and finally (iv) a simple average of protein probabilities from the above pipelines. Since DTASelect does not generate protein scores or FDR, we implemented a simple protein probability as the probability that at least one constituent peptide's identification was correct as described in (Nesvizhskii et al., 2003).
MSNet showed comparable performance across pipelines, with 10–12% higher AUC, and 7–12% more proteins at 5% FDR than the original analysis. The percentage increase in reported proteins depended on the coverage of the MS analysis software. As expected, the more the proteins confidently identified at the MS stage, the fewer the new MSNet identifications (details are in Supplementary Tables S4–S5 and Supplementary Fig. S5).
4 DISCUSSION AND CONCLUSIONS
We have presented a method that improves the sensitivity and precision of protein identification by integrating functional linkage information into the computational analysis of MS shotgun proteomics experiments. Our methodology places MS experiments in a larger biological framework, where proteins expressed in a given cellular state may be readily analyzed in the context of their functionally related neighbors.
We have shown that integrating data sources from outside an MS experiment can improve the protein identification rate of current MS technology and software. We increased the number of proteins identified at 5% FDR by 8–40%. We also improved performance against the original MS analysis in ROC and precision–recall plots, using our compilation of protein reference sets, showing 10–24% increase in ROC-AUC. We also presented an evaluation methodology to generate null distributions and FDRs for MSNet using network-shuffling, independent of gold-standard reference sets. These null distributions may be used to compute any other desired error estimate (e.g. p- and q-value).
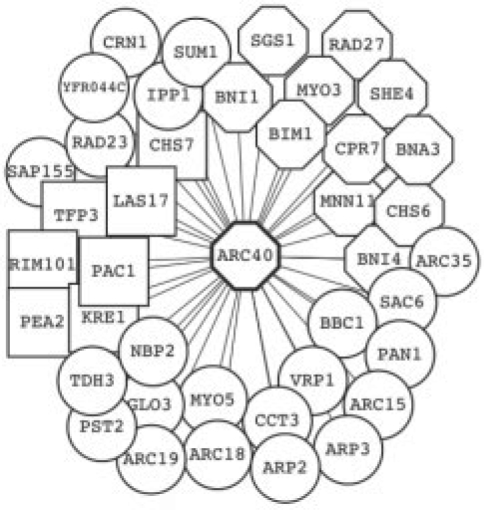
In two specific examples, we examine the immediate neighbors of two proteins identified by MSNet at 5% FDR in the proteome for yeast grown in rich medium. ARC40 is an essential subunit of the ARP2/3 complex (Fig. 3), and RPS29B is a member of the 40S ribosomal complex (Supplementary Fig. S4). Both proteins had multiple peptides identified in the MS experiment, but their MS protein scores fell below the error threshold of the MS software, and they were not identified with confidence. Both proteins have functions appropriate for yeast growing in rich medium, and have previously been identified with high confidence in the YPD* reference set. Moreover, deletion of either gene causes notable growth defects (Giaever et al., 2002); strongly supporting their expression in the sample. MSNet effectively rescues both proteins and gives them higher scores, based on the their MS evidence and their functional associations to other proteins that were confidently identified in the MS analysis. MSNet improved protein recall in several active pathways in rich-medium yeast e.g. glycolysis/gluconeogenesis, fatty acid metabolism, RNA biosynthesis, amino-acid biosynthesis and degradation (Dennis et al., 2003) (EASE-value=0.05). MSNet may be viewed as a quantitative complement to graphical tools that map ‘omics’ experiment results onto known functional pathways (Dennis et al., 2003; Paley and Karp, 2006).
Fig. 3.
Protein YBR234C (ARC40) and its immediate neighbors from the yeast gene functional network (Lee et al., 2007). The protein was identified with high confidence by MSNet, but not by the original MS analysis. YBR234C is an essential subunit of the ARP2/3 complex required for the motility and integrity of cortical actin patches, and involved in cell growth and polarity. Deletion of the gene causes notable growth defects (Giaever et al., 2002), a fact that strongly supports its expression. It is also present in the yeast reference set (Table 1, YPD*). MSNet gave YBR234C a high score because it had multiple neighbors that were either confidently identified in the MS experiment (circle) or had some MS evidence (hexagon, ≥1 peptide identified). The other neighbors (square) had no peptides identified. Figures were created using Cytoscape (Shannon et al., 2003).
MSNet improves protein identification by both increasing the number of true identifications and reducing false identifications. Since MSNet produces a revised ranking of MS-identified proteins, some proteins can receive lower ranks than in the MS analysis and fall below MSNet's 5% FDR threshold, despite satisfying the MS 5% FDR threshold. There is some evidence that these demoted proteins might be false positive MS identifications: in yeast, the percentage of demoted proteins that can be validated by presence in the reference set is much smaller than the percentage of new MSNet proteins that can be validated similarly (Supplementary Table S6). In human, all demoted proteins were network singletons i.e. they had no network neighbors. We list the demoted proteins for all experiments, as well as the union of MS and MSNet identifications in Supplementary Table S6. Using the high-confidence list of MSNet identifications as a starting point, one may narrow the range of additional experiments that are run to validate the existence of computationally predicted proteins.
To the best of our knowledge our method is the first to use gene networks to improve protein identification in shotgun proteomics. Gene functional networks have been widely used for predicting gene function. For example, Deng et al. (2003) modeled functional linkages as a Markov network, predicting a gene's function based on the functions of its neighbors. More recently, Wei and pan (2008) used functional associations to learn per-gene mixing proportions in a spatially correlated mixture model to improve large-scale studies such as differential gene expression. We have shown that MSNet is able to exploit a single organism-wide gene functional network to improve protein identification across different sample conditions, including different growth media and ranging from proteome-wide analysis to subcellular fractions.
In contrast to previous approaches using MS and mRNA expression data (Ramakrishnan et al., 2009), MSNet is easily applicable across datasets and experimental conditions, and does not depend on the availability of matching sample-specific data. MSNet is also directly applicable to smaller, focused protein preparations (Section 3.3) and to higher organisms, as we show for the proteome of cultured human cells. It is also possible to incorporate other sample-specific data when available by replacing the mass-spectrometry specific term oi (Equation 1) by a probability conditioned on other data sources e.g. LC separation profiles. ‘Omics’ integration approaches like MSNet will become increasingly powerful as functional association networks become broadly available, as for C.elegans (Lee et al., 2008), mouse (Guan et al., 2008; Kim et al., 2008; Pena-Castillo et al., 2008) and other organisms (Bowers et al., 2004; von Mering et al., 2003).
ACKNOWLEDGEMENTS
The authors thank Dan Boutz for mass-spectrometry assistance, Insuk Lee for pre-publication access to the human gene network, and Prof. William Noble and Lukas Kall for assistance with Percolator. They also thank Prof. Inderjit Dhillon, Prateek Jain and Raghu Meka for feedback on algorithm convergence, Martin Blom for discussions on network-shuffling, Lee Thompson for proofreading the convergence proofs and Lilyana Mihalkova, Prof. Raymond Mooney and Prof. William Press for useful discussions on relational learning.
Funding: National Science Foundation grant (DBI-0640923); the National Institutes of Health grants (GM067779, GM076536); the Welch (F-1515) and Packard Foundations grant; International Human Frontier Science Program support (to C.V.).
Conflict of Interest: none declared.
REFERENCES
- Berriz GF, et al. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Bowers PM, et al. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- Chi A, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl Acad. Sci. USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Nesvizhskii AI. False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res. 2008;7:47–50. doi: 10.1021/pr700747q. [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, et al. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, et al. Prediction of protein function using protein-protein interaction data. J. Comput. Biol. 2003;10:947–960. doi: 10.1089/106652703322756168. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006;27:861–874. [Google Scholar]
- Futcher B, et al. A sampling of the yeast proteome. Mol. Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Graumann J, et al. SILAC-labeling and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol. Cell Proteomics. 2007;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- Guan Y, et al. A genomewide functional network for the laboratory mouse. PLoS Comput. Biol. 2008;4:e1000165. doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveliwala TH. Topic-sensitive PageRank: a context-sensitive ranking algorithm for web search. IEEE Trans. Knowledge Data Eng. 2003;15:784–796. [Google Scholar]
- Kall L, et al. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kall L, et al. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- Keller A, et al. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim WK, et al. Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol. 2008;9(Suppl. 1):S5. doi: 10.1186/gb-2008-9-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langville Meyer . Google's PageRank and Beyond: The Science of Search Engine Rankings. Princeton, NJ, USA: Princeton University Press; 2006. [Google Scholar]
- Lee I, et al. A probabilistic functional network of yeast genes is accurate, extensive, and highly modular. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Lee I, et al. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat. Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLOS Biol. 2009 doi: 10.1371/journal.pbio.1000213. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, et al. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, et al. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Nash R, et al. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L, et al. Technical Report. Stanford Digital Libraries; 1999. The PageRank citation ranking: bringing order to the web. Available at http://ilpubs.stanford.edu:8090/422/ (Previous number = SIDL-WP- 1999-0120) [Google Scholar]
- Paley SM, Karp PD. The pathway tools cellular overview diagram and omics viewer. Nucleic Acids Res. 2006;34:3771–3778. doi: 10.1093/nar/gkl334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, et al. Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 2008;7:3022–3027. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Castillo L, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9(Suppl. 1):S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LCMS/ MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Prince JT, Marcotte EM. mspire: mass spectrometry proteomics in Ruby. Bioinformatics. 2008;24:2796–2797. doi: 10.1093/bioinformatics/btn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SR, et al. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics. 2009;25:1397–1403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, et al. FunSpec: a webbased cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL. What's driving false discovery rates? J. Proteome Res. 2008;7:45–46. doi: 10.1021/pr700728t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, et al. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, et al. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wei pan Incorporating gene networks into statistical tests for genomic data via a spatially correlated mixture model. Bioinformatics. 2008;24:404–411. doi: 10.1093/bioinformatics/btm612. [DOI] [PubMed] [Google Scholar]
- Zybailov B, et al. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]