Abstract
Motivation: For the biologist, running bioinformatics analyses involves a time-consuming management of data and tools. Users need support to organize their work, retrieve parameters and reproduce their analyses. They also need to be able to combine their analytic tools using a safe data flow software mechanism. Finally, given that scientific tools can be difficult to install, it is particularly helpful for biologists to be able to use these tools through a web user interface. However, providing a web interface for a set of tools raises the problem that a single web portal cannot offer all the existing and possible services: it is the user, again, who has to cope with data copy among a number of different services. A framework enabling portal administrators to build a network of cooperating services would therefore clearly be beneficial.
Results: We have designed a system, Mobyle, to provide a flexible and usable Web environment for defining and running bioinformatics analyses. It embeds simple yet powerful data management features that allow the user to reproduce analyses and to combine tools using a hierarchical typing system. Mobyle offers invocation of services distributed over remote Mobyle servers, thus enabling a federated network of curated bioinformatics portals without the user having to learn complex concepts or to install sophisticated software. While being focused on the end user, the Mobyle system also addresses the need, for the bioinfomatician, to automate remote services execution: PlayMOBY is a companion tool that automates the publication of BioMOBY web services, using Mobyle program definitions.
Availability: The Mobyle system is distributed under the terms of the GNU GPLv2 on the project web site (http://bioweb2.pasteur.fr/projects/mobyle/). It is already deployed on three servers: http://mobyle.pasteur.fr, http://mobyle.rpbs.univ-paris-diderot.fr and http://lipm-bioinfo.toulouse.inra.fr/Mobyle. The PlayMOBY companion is distributed under the terms of the CeCILL license, and is available at http://lipm-bioinfo.toulouse.inra.fr/biomoby/PlayMOBY/.
Contact: mobyle-support@pasteur.fr; mobyle-support@rpbs.univ-paris-diderot.fr; letondal@pasteur.fr
Supplementary information:Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Over the last 10 years, an increasing number of bioinformatics tools, covering a growing spectrum of applications, including aspects of genomics, systems and structural biology have been made available to the community. The complexity of analyses undertaken in these domains makes the publication, integration and interconnection of these tools particularly challenging.
Several systems propose a solution to automate the access to bioinformatics resources. Web-based user interfaces, such as the Biology Workbench (Subramaniam, 1998), PISE (Letondal, 2001), wEMBOSS (Sarachu and Colet, 2005), Galaxy (Giardine et al., 2005), GenePattern (Reich et al., 2006), MOWServ (Navas-Delgado et al., 2006), the New Generation Biology Workbench (Rifaieh et al., 2007), BioManager (Cattley and Arthur, 2007) and BioExtract (Lushbough et al., 2008), simplify the access to powerful computer resources by providing a familiar graphical-based environment for inexperienced users and by saving them from installing software on their own computer. In contrast with these tools which focus on the execution of programs, and where the users can often interactively chain analyses, GBrowse MOBY (Wilkinson, 2006) and Seahawk (Gordon and Sensen, 2007b) propose data-centric solutions where the user can explore a set of BioMOBY (Wilkinson, 2004) services to analyse a set of given data, with edition, navigation and visualization components which fully exploit the composite nature of BioMOBY objects.
Standalone workflow systems such as Taverna (Oinn et al., 2004), Kepler (Altintas et al., 2004) or BioSide (Hallard et al., 2004) enable to combine tools within desktop applications, using graphically specified workflows. It is hence possible, for bioinformaticians, to organize and automate complex data processing. Such possibilities have also been offered on web interfaces, either in dedicated systems such as Remora (Carrere and Gouzy, 2006), or within some of the web-based systems cited above. PISE, Galaxy and BioExtract also offer the possibility to save interactively designed protocols.
Many of these systems now offer the possibility of accessing distributed resources, often using dedicated web-service solutions such as BioMOBY and SoapLab (Senger et al., 2003). While SoapLab offers a system to automate the distribution of asynchronous analytical services, the BioMOBY protocol provides in addition more detailed descriptions, including semantic metadata and a registration system to facilitate the discovery of relevant resources. A number of tools facilitates the publication and management of BioMOBY services, such as MoSeS (http://biomoby.open-bio.org/CVS_CONTENT/moby-live/Java/docs/Moses-generators.html) and the BioMOBY dashboard (http://biomoby.open-bio.org/CVS_CONTENT/moby-live/Java/docs/Dashboard.html). Many of the existing user interfaces rely on the descriptions provided by these systems to generate user interfaces: Ajax Command Definition from EMBOSS (Rice et al., 2000), BioMOBY or even pure WSDL definitions.
Mobyle is a generic web-based framework. While including advanced technologies such as web services, remote execution and dataflow mechanisms, it addresses major end-user concerns arising from the use of sophisticated bioinformatics systems. A recent study (Gordon and Sensen, 2007a) shows how simple issues in the user interface can impede the use of a system by scientists, causing them to waste time and even preventing them from using it further. To address this issue, the design of Mobyle's interface is user-centered, to provide a usable yet customizable access to a large panel of services, from genome analysis to structural bioinformatics. The description language it uses to generate user interfaces is sufficiently extensive and flexible for rich user interfaces to be generated. Its core partly relies on concepts previously embedded in PISE and the RPBS portal (Alland et al., 2005). Moreover, different Mobyle servers can be federated to integrate services distributed over various sites. This functionality enables the federation of a network of curated portals, combining the skills of each of them. The Mobyle program description language also enables to define web services, as shown by the PlayMOBY Mobyle companion tool, which automates the publication of BioMOBY web services, and has monitoring capabilities that provide the bases for service quality monitoring.
2 SYSTEM DESIGN
The Mobyle design process was based on a user-centered design process (Javahery et al., 2004; Letondal and Amanatian, 2004 Shachak et al., 2007), involving numerous interviews and a number of users workshops (see Section 4.1). We identified the major end-user concerns that needed to be addressed:
An integrated bioinformatics framework needs to provide end users with several important capabilities: (i) reuse of data and results without the burden of unsafe copy-and-paste operations; (ii) management of scientific studies (e.g. retrieving parameters of a job, comparing results or relaunching an analysis using different data); and (iii) access to a wide range of local or remote services within a unique user interface.
A bioinformatics framework should enable scientists to share knowledge: (i) on setting up analyses, through tutorials based on curated examples; (ii) by accessing protocols designed and validated by experts in the domain; and (iii) through the use of a confidence network, allowing the interconnection of distributed resources, taking advantages of local skills to improve service quality, instead of favouring centralized platforms.
Technical requirements for such criteria to be satisfied include: (i) a well-designed web user interface, enabling easy navigation and data management within a user workspace; (ii) a distributed architecture, enabling access to and combined use of local and remote services; (iii) workflow authoring and enacting tools to support protocols; (iv) sound software architecture and APIs, to simplify system maintenance, configuration and extension; (v) a flexible description language to enable domain-specific adaptation needs, including visualization of results using special-purpose graphics components.
3 SYSTEM OVERVIEW
In this section, we provide an overview of Mobyle. We describe the underlying concepts of the system, the design of the web user interface, the server components and the distributed architecture.
3.1 Concepts
3.1.1 Homogeneous user interfaces to heterogeneous programs
The steep learning curve involved in the use of command-line tools invocation is problematic for inexperienced users (Shneiderman, 1983). A classical solution involves wrapping each application in a custom CGI script and providing a web interface to reduce the burden of remembering the syntax of the command line and the name of the parameters. Given the sheer number of bioinformatics software programs available, we chose to automate the generation of web interfaces using an abstract definition of a program's parameters. This solution provides a homogeneous interface to all the programs, minimizing the syntactic complexity. It is also helpful in file management issues and program chaining.
3.1.2 Persistent user workspaces
One of the shortcomings of the PISE environment is its lack of support for a persistent user workspace (Gilbert, 2002). The new system, while still allowing a fast and temporary ‘guest’ access to the programs, also gives to the users the possibility to create registered accounts, which allows user data and jobs to be maintained and managed across multiple work sessions.
3.1.3 XML description of interfaces
Based on feedback from PISE server administrators, as well as the need to extend the capacity of functionalities such as web service wrapping or workflow management, we modified the schema that describes the system. All Mobyle data, including program descriptions, job definitions and user workspaces are stored in XML documents. The main element of the system, the program description, is inherited from the PISE system. It describes various aspects, in a language that targets simplicity as much as possible (experience shows that the authoring and maintenance of program descriptions is a complex and tedious task). Aspects covered include:
Program/service documentation: describes the accomplished task and guides users through the process. It also provides authoring and version informations.
Data typing: characterizes the parameters and results of the different programs. Further details are provided in Section 3.3.
Wrapping instructions: translates a user request into a valid execution of the program (e.g. in the case of a unix program call, the construction of the command line will be defined by rules included in the XML file).
User interface layout: the layout of the program invocation form and job result pages are by default automatically generated from the XML definition, but can be customized.
BioMoby integration: the XML files can be enriched with metadata such as BioMoby parameter types, which permits the publication of a program as a BioMoby service using PlayMOBY (see Section 3.6).
A formal description of the XML grammar used to describe Mobyle programs is available on the project webpage (see http://bioweb2.pasteur.fr/projects/mobyle/downloads.html), using Relax NG (Clark et al., 2001).
3.1.4 Network-enabled bioinformatics tools
The Mobyle system aims at acting as a ‘hub’ for a set of programs of interest. A program integrated in Mobyle can not only be local but also remote, using the Mobyle Net functionality (further described in Section 3.5). Additionally, current and future developments aim at providing gateways to web-service-based systems such as BioMOBY (see Section 3.6). The interest is to provide a unified framework for bioinformatics platforms maintainers, who have to publish and integrate various local or remote programs for both biologists and bioinformaticians.
3.2 User interface design
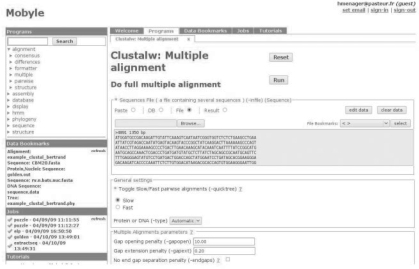
Mobyle provides the scientist with a global and integrated view of all the elements needed to perform his or her analyses. At one glance, the user can see which programs are available and which analyses have already been run. The portal is organized in three main parts (Fig. 1): a left navigation panel, a central panel displaying a selection of elements of the current work, such as forms and results, and horizontal tabs enabling the user to navigate between these elements.
Fig. 1.
Web user interface: portal overview, displaying the multiple alignment CLUSTALW submission form. The form in the main part displays a control, the databox, where the user can either paste data, enter a database ID or upload a file. The user can also select a previously uploaded file from the ‘File bookmarks’ menu. When available, results from previous jobs are also provided through a ‘Results’ menu.
3.2.1 Program search
Available programs are classified so that they can be searched, using program and parameter names, prompts and documentation, or bibliographic references. The page that displays the results of a given analysis also provides the user with the list of programs that can be run for further analysis. This list guides users, restricting them to a view of programs that are compatible with a given result file.
3.2.2 User workspace
At any time, the user can navigate between previously used forms and details of individual analyses, for instance to compare results. This view persists over separate uses of the portal during a given amount of time. This navigation model gives a ‘flat’ multi-directional access to several forms, which was not previously possible with the classical browser history mechanism. A list of bookmarked data is available: this provides the user with reuse-based access and information on previously submitted data.
3.2.3 Forms and reusability
Program forms are classical HTML forms. They contain a specific control element—the databox—designed to facilitate data reuse by biologists. This control allows users to set parameter values for biological data (e.g. DNA sequence or 3D protein structure) using various methods (pasting, file upload, databank entry retrieval, bookmark reuse).
3.2.4 Results and interactive chaining
The job status and results page provides a preview of the job data and metadata. It also includes an advanced control element—the resultbox—which lets users access results in plain browser windows, download them to their workstation, bookmark them or ‘pipe’ them to a new form, i.e. display a new form that is preloaded with the user data. This ability to pipe data into a new form, together with databox bookmark reuse, permits interactive program chainings within the portal.
3.2.5 Flexible layout design
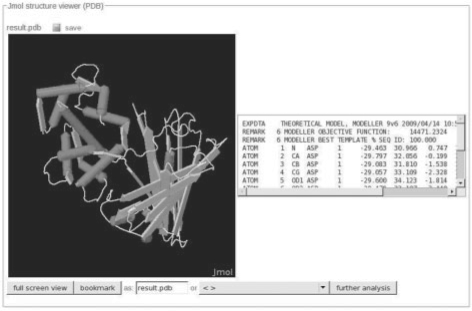
The portal, given a program definition, can automatically generate a form and a result page without any need for layout information. However, this process can be overridden to generate program-specific forms and job result pages with particular layouts. This possibility can prove extremely useful, for instance to fulfil custom program requirements, or to define a particular layout required for optimal use of a given program (see Fig. 2). This functionality relies on via specific layout tags or even HTML snippets embedded in the XML file, which can include inline javascript or Java applets.
Fig. 2.
Web user interface: example of customised interface, showing the simultaneous display of a program output in both a text box and in a Jmol applet.
3.3 Data typing and helpers
A major aspect of Mobyle is its capacity to facilitates automated data conversion and formatting for service integration, thus saving the user from tedious and non-scientific data manipulation tasks. The Mobyle typing system describes program parameters using a typing mechanism that aims to help users in such tasks. It modifies the interface display, the controls for user values (the input parameters), the possibility of chaining between programs and data reusability.
Similarly to SWAMI (Rifaieh et al., 2007), data and parameter characterization is multidimensional:
the biotype describes the biological object (e.g. nucleic acid, protein or drug);
the datatype describes the data ‘structure’ (e.g. sequence, alignment and matrix, but also more basic types for ‘non-bio’ parameters such as string or integer);
the format (blast html report, fasta sequence, phylip distance matrix).
The type of a data item or a parameter is used to:
detect, check and convert the format of the data provided by the user, based on external ancillary tools;
filter the available data sources (banks) that can be used to load data directly into a given tool;
filter the data bookmarks that can be reused in a new analysis;
filter the programs that a given result can be piped to;
offer specific visualization options in the interface (such as Java applets) (currently in development).
In contrast with other typing systems, such as the one offered by BioMOBY, the parameter compatibility between two tasks is based on a more abstract description. Since the Mobyle system can convert the data to the format which is accepted by the next task, there is no need to have service interoperability rely on a bioinformatics format description. For instance, in the Mobyle Pasteur server, the user can align a set of protein sequences using the clustalw-multialign interface, then pipe the clustalw-formatted result to Phylip protdist: although this program only accepts Phylip-formatted multiple alignments, the squizz program, which handles sequence and alignment conversions, will automatically detect and convert the multiple alignment into the accepted format. This approach simplifies the construction of the protocol: the user does not have to specify data transformation tasks, allowing optimal handling of the data (the format conversion information is directly accessible in the job result page). This typing system is configurable and relies on a ‘best-effort’ approach. Tasks such as format detection or conversion are handled by the system only if they are compatible with the local configuration.
This feature underlies the difference in what is considered to be a ‘service’ between Mobyle and BioMOBY: in Mobyle, data format conversions should be, as far as possible, considered as mere connectors in the analysis dataflow, whereas in BioMOBY they are considered to be on the same level as other analysis programs. Furthermore, since some programs accept multiple data formats as inputs, publishing them in BioMOBY involves registering multiple services on the registry (one per accepted format), whereas they are published as a single ‘interface’ in Mobyle.
3.4 Components
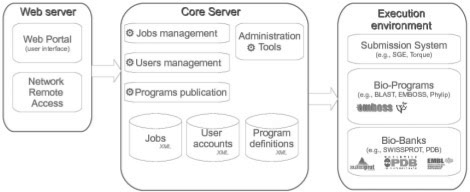
The Web portal provides a unified access to the services available on the server which is briefly described on Figure 3. We use Ajax (Garrett, 2005) to coordinate required information and to enable the user to explore various functionalities on a single page. For instance, a user action such as a job submission triggers an update of the user's job list and the data bookmarks list. This event triggers an update of the history data available in the program forms, without the page having to be completely reloaded.
Fig. 3.
Mobyle components overview.
The Mobyle server, based on a set of python modules, handles the various aspects of job, data and session managements. The most important of these, job submission, includes:
security validation of parameters and semantic validation (data types, range of value, compatibility between options);
command-line construction;
submission to an execution system, which could be local or on a cluster managed by SGE or torque;
job cancelling or status querying;
data management: all data needed to replay a job are stored (inputs, outputs, secondary parameters, etc.), ensuring the traceability of each job.
The server also has a substantial capacity for configuration, such as setting program access permissions based on request address, user limit for space size and handling user account types (registered and/or guest accounts). All of these features are based on server administrator needs.
3.5 The Mobyle network
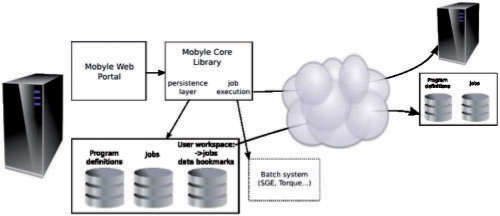
The Mobyle network allows the execution of programs that are available on different Mobyle servers from a single portal. This feature facilitates user access to services physically distributed over different Mobyle servers, within an integrated environment enabling data reuse and program chaining. As opposed to BioMOBY, Mobyle does not provide any central repository to register new services, relying instead on a distributed registration: each Mobyle server administrator chooses to publish a number of services, which can be upon his choice exported to or imported from remote Mobyle servers. Remote program invocation relies on the import of the remote program XML descriptions. Upon submission, this allows to generate a call to the remote server instead of generating and submitting a local batch call. In such a configuration, the user workspace, as well as the local jobs, remain stored on the initial server, but remotely invoked jobs are directly linked from their execution servers (Fig. 4).
Fig. 4.
Mobyle network architecture. The Mobyle server can invoke either local or remote programs, integrating them in a single work environment.
We demonstrate the use of this facility for the easy comparative modelling of the structure of a protein identified from analysis of the sunflower genome. A step by step tutorial is available on the MobyleNet help pages (http://mobyle.rpbs.univ-paris-diderot.fr/help/MobyleTutorials_network.html). This task can be broken down into several successive steps: inferring protein sequence from the genomic information, performing a search for a template structure, fitting the query sequence to the template and then generating a 3D model. Protein sequence inference can currently be performed through the LIPM Mobyle server, using the heliagene resource dedicated to sunflower genomic information. The next steps can be achieved through the RPBS Mobyle server using a two pass blast (PDBblast service) to identify candidate template structures from the query sequence. Sequence alignment can be performed through the Institut Pasteur Mobyle server using the clustalw service. Finally, the 3D modelling and model visualization/analysis can be performed through the RPBS site (Fig. 2). The Mobyle network allows efficient organization of the complete sequence of tasks as a pipeline, without having to leave the RPBS Mobyle portal, but making use of the resources and services at the three sites.
3.6 Deploying BioMOBY services from Mobyle program descriptions, using PlayMOBY
PlayMOBY is an external Mobyle companion tool allows the publication of BioMOBY-compliant web services, using Mobyle program descriptions. From the same XML description, bioinformatics programs can thus be both published on the Mobyle network and used by the code generator of PlayMOBY in order to implement, register and validate BioMOBY web services automatically. PlayMOBY also integrates a perl library to generate the corresponding Mobyle XML description file from program parameters. Thus, PlayMOBY reduces the overheads of publishing BioMOBY web services. To date, PlayMOBY handles more than 100 BioMOBY web services for providing generic sequence analysis tools and ensuring the interoperability of specialized databases (http://www.legoo.org, http://www.heliagene.org and http://narcisse.toulouse.inra.fr).
In addition to web service deployment, PlayMOBY provides daily monitoring tools for the deployed web services. During the Mobyle to PlayMOBY format conversion step, a service developer can provide a test dataset for each service input. These data are used to test web services availability and consistency by the provider himself. We made the choice to not allow web services users to launch these tests, which are instead automatically launched. The tests consist in verifying accessibility to the web service and consistency and stability of the results on test data. Their results are published as XML reports and RSS feeds. The PlayMOBY description for the QoS presently follows the specifications stated by the BioWorkflow group (Wessner et al., 2008). However, it could easily be extended to other frameworks such as the BioCatalogue project (Belhajjame et al., 2008). In the case of failures or unexpected results, alerts are sent by e-mail to the developers. Thus, PlayMOBY also provides the framework to evaluate Mobyle service reliability and curation. This problem is particularly important in bioinformatics, given the volatility of tools (especially web-based), impairing the reliability of biological data processing.
4 DISCUSSION AND PERSPECTIVES
4.1 System design: a user-centered approach
Over a period of 4 years leading up to Mobyle's first release, a series of about 30 user interviews and participatory design workshops involving brainstorming sessions and video prototyping were conducted. These were used to gain a deeper understanding of how a web portal could facilitate the use of bioinformatics tools by biologists. These studies are described in detail elsewhere (Letondal and Amanatian, 2004). The results showed the principal needs of biologists to be: (1) a stable and predictable set of known tools integrated in a portal designed for inexperienced users; (2) an overview of the analyses and careful organization of results; (3) reusability features to re-execute previous commands; and (4) user-defined and ready-to-use analysis pipelines, similar to benchmarked protocols. We also concluded that caution should be taken regarding advanced features. Indeed, (5) biologists are often sceptical about sophisticated tools that are difficult to understand: they have to be able to anticipate the benefits that such tools may provide.
Before opening the portal, we invited users to participate in test sessions. We selected 16 users among the users of the previous portal, ranging from beginners to frequent users, to participate in test sessions. We used the think-aloud method (Nielsen, 1994), involving users working in pair. Participants were told they could bring their own data. Most of them tried 2D structure analysis, alignment and phylogenetic programs. Several users perceived the new system as more complex, but more complete than the current one. They appreciated being able to retrieve jobs results, particularly from the databox menus. Most of the concerns, which were consequently solved, were related to (i) having to navigate back and forth between forms and results; (ii) user accounting; (iii) not fully understanding some English terms (such as ‘pairwise’ alignment) used in program classification; and (iv) ambiguities concerning data storage.
The Institut Pasteur, RPBS and LIPM Mobyle servers have been publicly available since January 2008, October 2008 and January 2009, respectively. The Pasteur server provides access to a wide range of bioinformatics tools mainly focused on sequence analysis and phylogeny; the LIPM server offers access to tools related to plant genomics; and the RPBS server publishes programs that mainly concern structural bioinformatics. Over 400 000 jobs have already been submitted by over 40 000 different users throughout the world on these servers.
4.2 Original contributions
The Mobyle project arose from the natural evolution of software such as PISE, offering a simple way to publish and to share bioinformatics software on the Web. Program chaining is limited to the most common usage patterns that we identified, and does not provide any of the advanced workflow patterns included in previously described systems (Taverna, Kepler). It is not either intended to handle analyses of very large quantities of data, data transfer over the network being too costly for web-based applications.
Several aspects make this approach novel:
the simplicity of the interface, aiming to provide advanced functionalities without imposing the steep learning curve generally associated with complex systems. For instance, data can be directly uploaded onto the program form (without having to navigate to a separate upload interface) and the data format is automatically detected and validated. This simplicity is in the continuity of the PISE approach, with the addition of new features such as a persistent workspace and multiple functionalities to facilitate data reuse.
flexibility in the design of the program user interfaces (submission forms and job results pages, as described in Section 3.2.5) and in the configuration of the system.
the Mobyle network enables the integration of local software with remote services, reducing the costs associated with the local maintenance of an exhaustive list of programs. Application of this strategy over several Mobyle sites should help to connect providers, generating a framework that emcompasses a wide spectrum of applications and covers complementary aspects of bioinformatics. This strategy needs to be tested for a large number of sites; however, we anticipate the generation of a confidence network combining specific tools offered by each platform and thus promoting quality management of the services.
deployment of BioMOBY web services from bioinformatics programs is made available through PlayMOBY. It thus benefits from the conceptual strength of the BioMOBY architecture, while avoiding the duplication of wrapper authoring effort, using a common format for both Mobyle and BioMOBY publications. Additionally, it encourages good practices such as services monitoring by integrating the design of tests into the publication process.
4.3 Current developments
Workflows and protocols: Mobyle already embeds a prototype dataflow-oriented workflow engine, enabling the chaining of successive or parallel tasks to be automated. It runs on top of the Mobyle core library, exploiting the Mobyle network to execute tasks distributed between local and remote servers, and orchestrates the tasks to synchronize their execution with the availability of all their input data.
Mobyle workflows modelize a set of tasks, but in contrast with existing workflow languages such as SCUFL in Taverna (Oinn et al., 2004) or MoML in Kepler (Altintas et al., 2004), do not specify explicitly some low-level data format detection and transformation tasks. Authors are, whenever possible according to local configuration, not asked to specify these steps. Hence, the storage of workflow definitions is planned to be based on an extension of the current Mobyle XML language, as they are based on a higher level perspective on the analyses they describe. An interesting outcome is the possibility to save an interactively designed program chaining as a reusable composite service. This perspective provides a gateway towards the publication of user-acknowledged protocols on a community-based platform such as myexperiment.org (De Roure et al., 2007). Future end-user access to the workflow definitions and workflow engine will be integrated to the current web interface.
Web services: the PlayMOBY system already allows the automated publication of BioMOBY web services using Mobyle program definitions. Future work also includes enabling the use of other approaches such as custom SOAP or REST interfaces, and SoapLab. Enabling the integration of such services as remote programs, for which the Mobyle Web Portal acts as a client will also be considered.
Didactic tutorials: further developments will encompass the design of tutorials. As explained by Cattley and Arthur (2007), integrated web portals are a very efficient learning and teaching tool. Mobyle additionally provides technical support for implementing interactive tutorials. A major issue to be incorporated in its design is to make more interactive components available: as shown by user studies, workflows should be available as semi-automatic protocols, with interactive components enabling the biologist to customize the visualization of the result from an analysis (e.g. a 3D molecule or an alignment).
5 CONCLUSIONS
The design of Mobyle, which provides an effective way to make a large panel of curated bioinformatics tools available in a homogeneous environment, has been driven with the concern to meet the requirements of different audiences—biologists and bioinformaticians, mostly. Our objective was thus to facilitate the access to complex features and advanced technologies by a design that suits the work of biologists, which we were able to observe and understand during a number of interviews and workshops.
Funding: the Agence Nationale de la Recherche (GPLA06026G ANR Genoplante, to PlayMOBY project ).
Conflict of Interest: none declared.
REFERENCES
- Alland C, et al. RPBS: a web resource for structural bioinformatics. Nucleic Acids Res. 2005;33:W44. doi: 10.1093/nar/gki477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altintas I, et al. SSDBM ′04: Proceedings of the 16th International Conference on Scientific and Statistical Database Management. Washington DC: IEEE Computer Society; 2004. Kepler: An extensible system for design and execution of scientific workflows; p. 423. [Google Scholar]
- Belhajjame K, et al. Proceedings of the Third International Biocuration Conference. Berlin, Germany: 2008. Biocatalogue: a curated web service registry for the life science community; pp. 16–19. April, 2009. [Google Scholar]
- Carrere S, Gouzy J. REMORA: a pilot in the ocean of BioMoby web-services. Bioinformatics. 2006;22:900–901. doi: 10.1093/bioinformatics/btl001. [DOI] [PubMed] [Google Scholar]
- Cattley S, Arthur J. BioManager: the use of a bioinformatics web application as a teaching tool in undergraduate bioinformatics training. Brief. Bioinform. 2007;8:457–465. doi: 10.1093/bib/bbm039. [DOI] [PubMed] [Google Scholar]
- Clark J, et al. RELAX NG Specification. 2001 Available at http://www.oasis-open.org/committees/relax-ng (last accessed date September 4, 2009) [Google Scholar]
- De Roure D, et al. eScience, Bangalore, December 10–13. IEEE Computer Society; 2007. Designing the myexperiment virtual research environment for the social sharing of workflows; pp. 603–610. [Google Scholar]
- Garrett J. Ajax: a new approach to web applications. Adapt. Path. 2005;18 [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. Pise: software for building bioinformatics webs. Brief. Bioinform. 2002;3:405–409. doi: 10.1093/bib/3.4.405. [DOI] [PubMed] [Google Scholar]
- Gordon PMK, Sensen CW. Technical Report #2007-874-26. Canada: University of Calgary; 2007a. A Pilot Study into the Usability of a Scientific Workflow Construction Tool. Department of Computer Science. [Google Scholar]
- Gordon P, Sensen C. Seahawk: moving beyond html in web-based bioinformatics analysis. BMC Bioinformatics. 2007b;8:208. doi: 10.1186/1471-2105-8-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallard M, et al. JOBIM 2004 : 5èmes journèes ouvertes biologie informatique mathèmatique, 27-30 juin, Montrèal, Canada. IASC - Dèpt. Intelligence Artificielle et Systèmes Cognitifs (Institut TELECOM; TELECOM Bretagne; 2004. Bioside: faciliter l'accès des biologistes aux ressources bioinformatiques; p. 64. [Google Scholar]
- Javahery H, et al. Beyond power: making bioinformatics tools user-centered. Commun. ACM. 2004;47:58–63. [Google Scholar]
- Letondal C, Amanatian O. Requirements Capture for Collaboration in e-Science Workshop. Edinburgh: National e-Science Center; 2004. Participatory design of pipeline tools and web services in bioinformatics. e-Science Institute. [Google Scholar]
- Letondal C. A web interface generator for molecular biology programs in Unix. Bioinformatics. 2001;17:73–82. doi: 10.1093/bioinformatics/17.1.73. [DOI] [PubMed] [Google Scholar]
- Lushbough C, et al. Bioextract server - an integrated workflow-enabling system to access and analyze heterogeneous, distributed biomolecular data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008;99 doi: 10.1109/TCBB.2008.98. [DOI] [PubMed] [Google Scholar]
- Navas-Delgado I, et al. Intelligent client for integrating bioinformatics services. Bioinformatics. 2006;22:106–111. doi: 10.1093/bioinformatics/bti740. [DOI] [PubMed] [Google Scholar]
- Nielsen J. Usability Engineering. San Francisco: Morgan Kaufmann; 1994. [Google Scholar]
- Oinn T, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rice P, et al. Emboss: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rifaieh R, et al. Swami: integrating biological databases and analysis tools within user friendly environment. In: Boulakia SC, Tannen V, editors. Data Integration in the Life Sciences (DILS) Vol. 4544. Philadelphia, PA: Springer; 2007. pp. 48–58. of Lecture Notes in Computer Science. [Google Scholar]
- Sarachu M, Colet M. wemboss: a web interface for emboss. Bioinformatics. 2005;21:540–541. doi: 10.1093/bioinformatics/bti031. [DOI] [PubMed] [Google Scholar]
- Senger M, et al. Proceedings of the UK e-Science All Hands Meeting 2–4th September, 2003. Nottingham: EPSRC; 2003. Soaplab - a unified sesame door to analysis tools; pp. 509–513. [Google Scholar]
- Shachak A, et al. Barriers and enablers to the acceptance of bioinformatics tools: a qualitative study. J. Med. Libr. Assoc. 2007;95:454–458. doi: 10.3163/1536-5050.95.4.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneiderman B. Direct manipulation: a step beyond programming languages. IEEE Comput. 1983;16:57–69. [Google Scholar]
- Subramaniam S. The biology workbench - a seamless database and analysis environment for the biologist. Proteins Struct. Funct. Genet. 1998;32:1–2. [PubMed] [Google Scholar]
- Wessner M, et al. Jobim (Journées Ouvertes Biologie Informatique Mathématiques. Lille France; 2008. BioWorkFlow: web services toolkit and workflow applications evaluation to deploy a confidence network. [Google Scholar]
- Wilkinson MD. BioMOBY - the MOBY-S Platform for Interoperable Data Service Provision. In: Grant RP, editor. Computational Genomics Theory and Application. Wymondham: Horizon Bioscience; 2004. [Google Scholar]
- Wilkinson M. Gbrowse Moby: a web-based browser for BioMoby services. Source Code Biol. Med. 2006;1:4. doi: 10.1186/1751-0473-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]