Abstract
The Golgi apparatus is a central hub for both protein and lipid trafficking/sorting and is also a major site for glycosylation in the cell. This organelle employs a cohort of peripheral membrane proteins and protein complexes to keep its structural and functional organization. The conserved oligomeric Golgi (COG) complex is an evolutionary conserved peripheral membrane protein complex that is proposed to act as a retrograde vesicle tethering factor in intra-Golgi trafficking. The COG protein complex consists of eight subunits, distributed in two lobes, Lobe A (Cog1–4) and Lobe B (Cog5–8). Malfunctions in the COG complex have a significant impact on processes such as protein sorting, glycosylation, and Golgi integrity. A deletion of Lobe A COG subunits in yeasts causes severe growth defects while mutations in COG1, COG7, and COG8 in humans cause novel types of congenital disorders of glycosylation. These pathologies involve a change in structural Golgi phenotype and function. Recent results indicate that down-regulation of COG function results in the resident Golgi glycosyltransferases/glycosidases to be mislocalized or degraded.
Keywords: Golgi, COG complex, Protein glycosylation, Glycosyltransferases, Lectin, Vesicle tethering
1. The Golgi apparatus and glycosylation
The Golgi apparatus is a hub of vesicular protein trafficking and is a major site for the glycosylation of proteins and lipids.1 In eukaryotic cells, both secretory and plasma membrane proteins enter the secretory pathway and are core-glycosylated in the ER and then transported in vesicular coat complex II (COPII)-coated membrane trafficking intermediates to the Golgi complex. The Golgi is a dynamic polarized organelle that receives cargo vesicles from the ER compartment at its cis-compartment and moves cargo from cis- to trans- through the medial compartment.2 Within the Golgi, proteins and lipids are further modified by glycosyltransferases prior to their export to the plasma membrane, the endosomal-lysosomal compartments, or back to ER for retrieval. The Golgi is responsible for the organization of both anterograde and retrograde trafficking (see Ref. 3 for a review). Golgi enzymes and associated substrate transporters are non-uniformly distributed within Golgi stack allowing sequential modification of passing glycoproteins. For instance, N-acetylglucosaminyltransferase I (GlcNAcTI) and man-nosidase II (ManII) are preferentially localized in cis/medial cisternae, while galactosyltransferase (GalT) and sialyltransferase (ST) are mostly found in trans-Golgi. 4,5 To generate and maintain this non-uniform distribution, the enzymes must first be transported to specific Golgi cisternae. Their consequent steady-state localization is maintained then through a combination of retention and retrieval.6
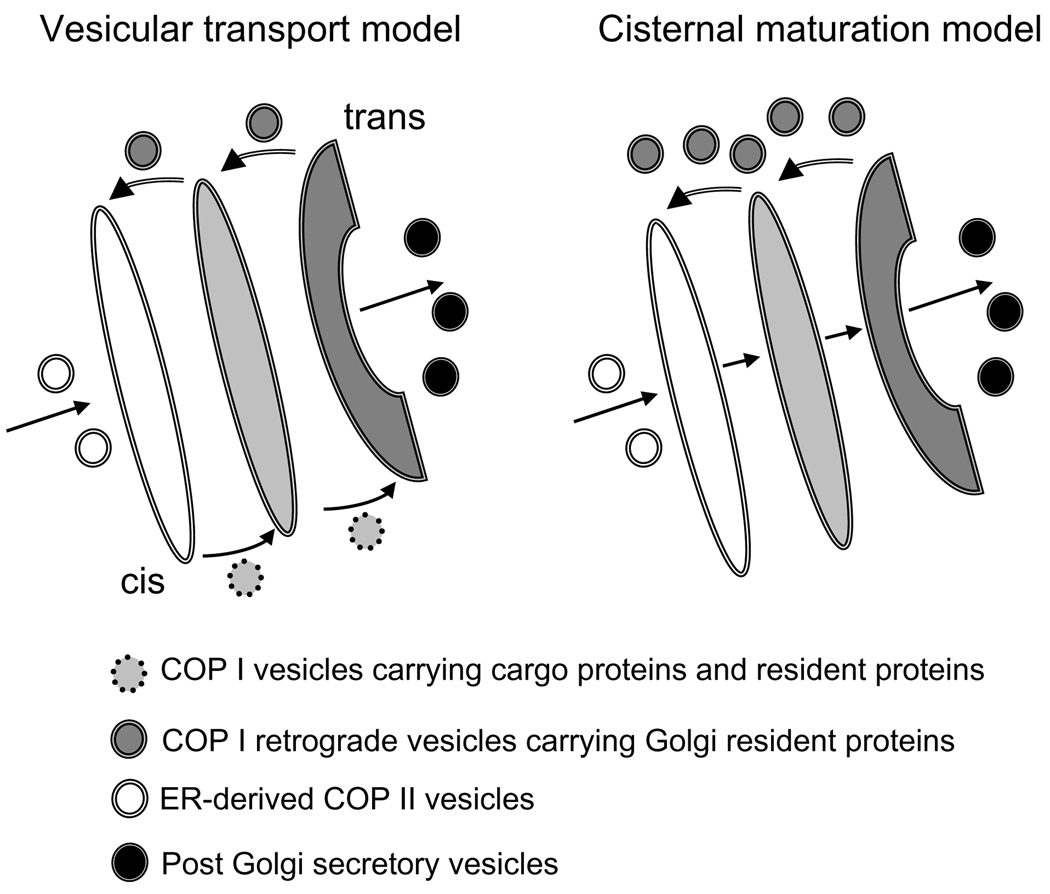
Vesicular coat complex I (COPI)-coated proteins are involved in both intra-Golgi trafficking and maintaining the normal structure of the Golgi complex.7 Anterograde vesicular transport and cisternal maturation are two alternative models of intra-Golgi transport.8 A fundamental difference between these two models (Fig. 1) is the differential movement of resident and cargo proteins. The anterograde vesicular transport model predicts that cargo molecules will move forward in transport vesicles, while resident proteins are specifically retained. The cisternal maturation model predicts that cargo molecules are moving through the stack passively as the cisternae move forward, while resident proteins are recycled by retrograde transport to establish differential concentrations across the stack. These two models are not mutually exclusive and may occur simultaneously.9 Inherent in both models is the dependence of recycling to localize glycosyltransferases, SNAREs, and other resident Golgi proteins.
Figure 1.
Models for intra-Golgi transport.
It has been demonstrated in both yeast and mammalian systems that Golgi enzymes and cargo receptors constantly recycle in a retrograde trans-to-cis fashion by the means of COPI-coated vesicles and COPI-independent tubular intermediates.10,11 Recycling transport vesicles fuses with the acceptor compartment in a SNARE-dependent fashion (for a review, see Ref. 12). To coordinate proper vesicle targeting and fusion, the Golgi apparatus employs a cohort of the so-called vesicle tethering factors. These tethering factors comprise two families of coiled-coil and oligomeric proteins that serve to establish a link between vesicle and appropriate Golgi compartment prior to formation of fusogenic SNARE complexes (for a review, see Refs. 13 and 14).
2. The COG complex
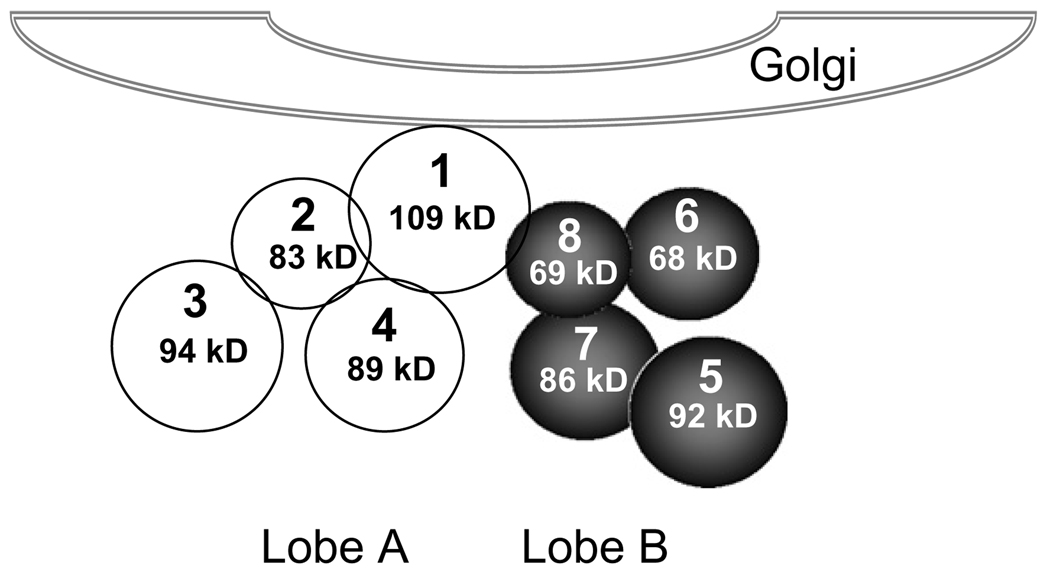
The COG protein complex consists of eight subunits named COG1 through COG8.15–20 Based on yeast genetic studies and observations with electron microscopy, 19 the COG subunits have been grouped into two lobes consisting of Cog1p–Cog4p and Cog5p–Cog8p, respectively (Fig. 2). The two lobes appear to be interconnected by thin rods and/or globules. Cog1p is likely a bridging subunit between the two COG lobes in yeast cell.21 The bridge that joins Lobe A and Lobe B in the mammalian COG complex is similarly composed of Cog1 and Cog8.17
Figure 2.
Subunit composition of the COG complex. Apparent molecular weights and known subunit interactions for the human COG complex are shown. Cog1p is responsible for both the COG complex contact with the Golgi membrane and for the Lobe A–Lobe B communication.
The COG complex is evolutionary conserved; homologues of its most conserved subunits, Cog3p, Cog4p, Cog6p, and Cog8p, are found in different genera, including yeast, plants, insects, worms, and mammals.22 The COG complex appears structurally related to other tethering complexes.15 Iterative database searches using N-terminal domains of several COG components revealed similarities in the components of the exocyst (for a recent review, see Ref. 23) and the GARP24 complex. The COG, exocyst, and GARP complexes turned out to be distantly related, multimeric assemblies that evolved to tether membranes at distinct stages of the secretory or endocytic pathways. The solute structure of a large portion of yeast Cog2p was recently reported.25 The structure reveals a six-helix bundle with few conserved surface features but a general resemblance to a crystal structure of exocyst subunits.
COG subunits form a peripheral Golgi protein complex. 18,19,26 Immunogold electron microscopy was used to visualize the intra-Golgi distribution of a functional, hemagglutinin epitope-labeled COG subunit, Cog1-HA. COG was found to be localized primarily on or in close proximity to the tips and rims of the Golgi cisternae and their associated vesicles.27 None of the COG subunits has either a transmembrane or lipid-binding domain and most likely they attach to the membrane via specific and highly regulated protein–protein interactions. Although it was proposed that the Cog1p subunit is responsible for the Golgi localization/attachment of the COG complex,28,29 data from other and our laboratories suggest that both Golgi localization and membrane attachment of the COG complex are probably more multifaceted and involved several COG subunits. We have found that the absence of one subunit of the yeast COG complex does not significantly affect membrane association of others.21 Both Golgi-localized lipid-modified Rab proteins and transmembrane SNARE proteins were implicated as factors that facilitate COG–membrane binding.
Protein trafficking in eukaryotic cells is controlled by the members of the Rab family of small GTPases. Rab proteins act as molecular switches that shuttle on/off membrane while attracting a diverse group of effectors. 30 There are more than 20 Rab proteins associated with the mammalian Golgi apparatus.31 Both coiled-coil and oligomeric tethering factors are known to interact with Rab proteins.14 In yeast cells, the COG complex interacts with two Golgi Rab GTPases, Ypt1 and Ypt6.15,17,18 In vitro experiments indicate that yeast COG complex binds preferentially to GTP-loaded Ypt1.18 Overexpression of Ypt1 suppresses growth defects associated with the loss of the COG functions.32,33 On the other hand, the simultaneous loss of one of the non-essential COG subunits and another Golgi Rab, Ypt6, produces a synthetic growth lethality.34 Recent yeast two-hybrid assays identified several mammalian Golgi Rabs that interact with COG complex subunits in a GTP-dependent manner (D. Ungar, University of York, personal communication). Epistatic depletion of Rab6 inhibited the Golgi disruptive effects of COG complex inactivation by siRNA or antibodies, indicating that the COG complex operates downstream of the Rab6 in Golgi trafficking.35
A number of oligomeric tethering complexes co-localize and directly interact with SNARE molecules,18,36,37 but the mechanism of this tether–SNARE interaction is not well understood. Sec6p, a subunit of the exocyst complex, binds in vitro to the plasma membrane t-SNARE Sec9p where it modulates the interaction between Sec9p and Sso1p.38 Vps51p, a subunit of the trans-Golgi-localized GARP complex, specifically binds to the conserved N-terminal domain of the t-SNARE Tlg1p. However, Vps51/Tlg1 interaction is not essential for GARP-mediated endosome-trans-Golgi network (TGN) trafficking.36
Numerous genetic interactions have been detected between the COG complex and SNARE proteins.17,32,33,39 Subunits of the yeast COG complex co-immunoprecipitates (co-IP) with the Golgi t/v-SNAREs Sed5p, Gos1p, Ykt6p, and Sec22p18 and the mammalian COG complex co-IPs with the GS28, mammalian homologue of Gos1p.40 The data also indicate that the localization and stability of Golgi SNAREs are altered in cells expressing a defective COG complex.21,40–42 These findings led to a hypothesis that the COG complex regulates the formation and/or stability of the intra-Golgi SNARE complex.
A third class of protein to which COG complex has been linked is the COPI coat.17,18,42 COPI coats are found on trafficking vesicles originating within the Golgi, and have an established role in retrograde trafficking.43 Oka et al.42 have employed short interfering RNA (siRNA)-mediated ζ-COP depletion and established that COG and ζ-COP contribute to similar functions necessary for normal Golgi organization. These data are in a good agreement with the proposal that in yeast cells COG complex mediates tethering of COPI-coated trafficking intermediates.18 Thus, the COG complex ‘interactome’ includes SNAREs, Rabs, and COPI coat proteins, all proteins with well-established roles in vesicular transport (Table 1). Further analysis of interactions between the COG complex subunits and other Golgi-localized components of vesicle trafficking machinery will help in understanding the COG complex cellular function.
Table 1.
COG interactome
| COG subunits | Species | Interacting protein |
Comments |
|---|---|---|---|
| COG complex | Budding yeasts | Ypt1-GTP | In vitro binding18 |
| Ypt6 | In vitro binding18 | ||
| Sed5p | In vitro binding18 | ||
| Cog1p | Budding yeasts | Sed5p | Co-IP18 |
| COPI | Co-IP18 | ||
| Cog2p | Mammals | P115 | Y2H, in vitro binding, co-IP59 |
| Budding yeasts | γ-COPI | Y2H18 | |
| Sed5p | Co-IP18 | ||
| Cog3p | Mammals | GS28 | Co-IP40 |
| β-COPI | Co-IP40 | ||
| P115 | Co-IP59 | ||
| GM130 | Co-IP59 | ||
| Giantin | Co-IP59 | ||
| Budding yeasts | Sed5p | Co-IP, in vitro binding18 |
|
| Ykt6p | Co-IP18 | ||
| Gos1p | Co-IP18 | ||
| Sec22p | Co-IP18 | ||
| COPI | Co-IP18 | ||
| Cog4p | Budding yeasts | Sed5p | Co-IP18 |
| COPI | Co-IP18 | ||
| Cog6p | Budding yeasts | Sed5p | Co-IP18 |
| COPI | Co-IP18 |
3. From yeast to humans
Mutation or deletion of the individual COG subunits gives rise to different phenotypes, suggesting that each member protein plays a distinct role within the complex. In yeast Saccharomyces cerevisiae, deletion of COG1–COG4 results in severe growth defects,17,18,32,33,39,44 accumulation of vesicles,15,45 reduced glycosylation of proteins,15,18 and an altered distribution of SNAREs.18,15 In other multicellular organisms, mutations in COG subunits affect the structure and the function of the Golgi apparatus (Table 2).17–19,46,47
Table 2.
Phylogenetic distribution of COG complex subunits and pathology/glycosylation defects associated with mutations/deletions of COG complex subunits
| Budding yeasts S. cerevisiae |
Worms C. elegans |
Fruit fly D. melanogaster |
Humans mammals |
Pathology/glycosylation defects |
|---|---|---|---|---|
| Cog1 Sec36 Cod3 |
Cogc-1 Mig30 |
dCog1 | Cog1 ldlB |
|
| Cog2 Sec35 |
Cogc-2 | dCog2 | Cog2 ldlC |
|
| Cog3 Sec34 Grd20 |
Cogc-3 Mig29 |
dCog3 | Cog3 hSec34 |
|
| Cog4 Sec38 Cod1 Sgf1 |
Cogc-4 | dCog4 | Cog4 Cod1 |
Glycosylation defects in yeast cells lacking COG4.18 |
| Cog5 Cod4 |
Cogc-5 | dCog5 fws |
hCog5 GTC-90 |
|
| Cog6 Sec37 Cod2 |
Cogc-6 | dCog6 | Cog6 hCod2 |
Glycosylation defects in yeast cells lacking COG6.15,18,21 |
| Cog7 Cod5 |
Cogc-7 | dCog7 | Cog7 | |
| Cog8 Dor1 |
Cogc-8 | dCog8 | Cog8 hDor1 |
In Caenorabditis elegans, the COG complex is involved in biogenesis of Mig17, an ADAM (a disintegrin and metalloprotease) protease, which is glycosylated and plays a role in the development of distal tip cells needed for gonad morphogenesis.48 The deletion of Cogc-1 or Cogc-3 leads to underglycosylation of Mig17. This deletion caused the distal tip cells to be mis-directed which is observed in the Mig17 deletion. Therefore, either a deletion in Mig17 or Cogc-1/Cogc-3 result in mislocalization of the gonads.
In Drosophila melanogaster, the gene four way stop (fws) encodes a Golgi-localized protein homologous to Cog5 protein.46 This fws when deleted in males causes the spermatids to contain two to four nuclei which is caused by failure of cytokineses during the first stage of meiosis (meiosis I). The authors proposed that fws may directly or indirectly facilitate efficient vesicle traffic through the Golgi to support rapid and extensive increases in cell surface area during spermatocyte cytokinesis and polarized elongation of differentiating spermatids.
Monty Krieger’s laboratory at MIT created several mutant Chinese hamster ovary (CHO) cell lines that display severe defects in the low-density lipoprotein (LDL) receptor.49 It was subsequently shown that ldlB (COG1 knockout19,50) and ldlC (COG2 knockout19,47) are deficient in glycosylation of both N-linked and O-linked glycans,16 and that the altered glycosylation of LDLR results in rapid down-regulation from the cell surface and intracellular degradation.
Both ldlB and ldlC cells also displayed N-linked dysglycosylation of the vesicular stomatitis virus G (VSV-G) protein.16 Because there are defects in N- and O-glycosylation for LDL receptor and VSV-G protein, all glycoproteins passing through the Golgi may be dysglycosylated. Additionally, lipid-linked sugars (glycolipids) are also affected in ldlB and ldlC cells. Although there are no major alterations in the structure of the Golgi for both ldlB and ldlC cells, both Cog1 and Cog2 are required for normal trafficking through the Golgi.47
Toxic lectins have been used as a tool to test sensitivity of CHO, ldlB, and ldlC cells: WGA (wheat germ agglutinin, which specifically binds to sialic acid or N-acetylglucosamine), ricin (RIC; specifically binds to terminal galactose and N-acetylgalactosamine), PHA (phytohemagglutinin, which specifically binds to galactose in a β1–4 branch), Con A (concanavalin A, which specifically binds to mannose), and Lens culinaris agglutinin (LCA, which specifically binds to mannose residues in fucosylated N-linked glycoproteins) to test their sensitivities of these CHO and mutant cells.16 The LD10 (lectin sensitivity) was measured between control and COG1 and COG2 knockout cells. For WGA, PHA, and LCA the LD10 was increased (=less sensitive); for RIC and Con A, the LD10 was decreased (more sensitive).
Recent studies of human COG7-deficient fibroblasts51 (see also next section) revealed that retrograde transport of multiple Golgi proteins via brefeldin A-induced tubules was significantly slower in mutant cells, whereas anterograde protein trafficking was much less affected. After prolonged treatment with brefeldin A, several Golgi proteins were detected in clusters that co-localize with the microtubule-organizing center in patient cells. All of these abnormalities were normalized in COG7-corrected patient fibroblasts.
4. COG complex defects and congenital disorders of glycosylation
Mutations in a COG subunit, which can cause their truncation or mutation, lead to a congenital disorder of glycosylation (CDG) in humans.52 Most CDGs are caused by deletions in glycosyltransferases/glycosidases or nucleotide-sugar transporters responsible for glycan synthesis or modification. All CDGs are homozygous recessive that cause multisystem disorders. Type I CDGs are caused by defects in the assembly or transfer of the dolichol-bound 14 sugar oligosaccharide precursor for N-linked glycosylation. Type II CDGs involve defects in the subsequent processing of protein-bound N-linked oligosaccharides within the ER and Golgi and may be associated with alterations in O-glycosylation as well.53 Isoelectric focusing (IEF) of serum transferrin can determine if a patient is diagnosed with CDG. Once an IEF of serum transferrin has been performed, another IEF of Apolipoprotein C-III will show whether or not O-glycosylation is defective.53
In 2004, patients (three siblings) were described from healthy father and mother from the Mediterranean area that had a reduction in total serum sialic acid representing a new type of CDG (CDG-IIe).54 All three patients possessed homozygous intronic mutation of the COG7 gene that resulted in a truncated version of this protein that was reduced in the patients. All patients died either at birth or 10 weeks after birth. The patients displayed perinatal asphyxia, low-set dysplastic ears, micrognathia, short neck, and loose wrinkled skin and exhibited severe epilepsy and later died of infection and cardiac insufficiency. Mutation of Cog7p resulted in significant reduced activities of nucleotide-sugar transporters (CMP-Sia and UDP-Gal) and glycosyltransferases (Core1 GalT and ST3Gal-I) of the Golgi. There was also reduced retrograde trafficking for ST3Gal-I in COG7 mutant fibroblast compared to control cells. Normal trafficking and glycosylation was restored with re-introduction of COG7.
Three more Cog7p-deficient patients were of Moroccan decent.55 All patients displayed dysmorphic facial features, microcephaly, adducted thumbs, skeletal anomalies, wrinkled skin, ventricular septal defect, failure to thrive, and liver disease. Both IEF of serum transferrin and ApoC-III showed a type II CDG (both a defect in N- and O-glycosylation).
CDG-II/Cog1 is caused by a homozygous nonsense mutation of COG1 inserting a stop codon at positive 900 causing the last 80 amino acid residues to be deleted. 53 A 3-month old girl displayed hypotonia, small hands and feet, straightened bitemporal space, and anti-mongoloid eyelids. She later on developed growth and psychomotor retardation. This mutation in Cog1p also lead to a decrease in Cog2p, Cog3p, Cog4p, and Cog8p levels and lead to a down-regulation of Man II and GalT. Staining of PNA Alexa-Fluor 488 showed staining of the COG1-deficient fibroblast when compared to a control fibroblast. Placing neuraminidase on the patient’s fibroblast increased the staining of the fibroblast showing that there in a defect in the sialylation. Mass spectroscopy of the patient along with control showed differences in glycans profiles. When tritiated UDP-galactose or CMP-neuraminic acid were added to the patient’s cells, there was a reduced incorporation of these activated sugars suggesting a defect in both sialylation and galactosylation in CDG-II/Cog1 cells.
CDG-IIh (or CDG-II/Cog8) is also caused by a homozygous nonsense mutation of COG8 in which the C is changed to a G causing the threonine residue to be a stop codon.56 The Cog8p molecule was truncated 76 amino acids in the C-terminus. This protein had an increase in the mobility of the Western blot and causes a down-regulation of Cog1p (~75%). The patient of Spanish origin with deleted Cog8p displayed acute encephalopathy, hypotonia, minor dysmorphic characters, and mental retardation. IEF of serum transferrin again showed a CDG. IEF of ApoC-III also indicated a disorder of O-glycosylation. PNA showed a positive result indicating a defect in sialylation. MALDI-TOF of a patient with Cog8p mutations revealed increases in the fucosylation of agalacto-asialo glycans with hyposialylation of N-glycans.57 Furthermore, there was a 2-fold increase in the number of non-sialylated proteins, which is consistent with a decrease on ST3Gal-I in this patient.
All of the known COG deletions in humans cause microcephaly (‘small brain’), hypotonia (low muscle tone due to defect in cerebellar growth), and failure to thrive.54 Among several COG mutations that have been described as type II CDGs, the COG7 deletion is having the highest mortality within the first year of life.
5. Cellular defects on COG knockdown cells
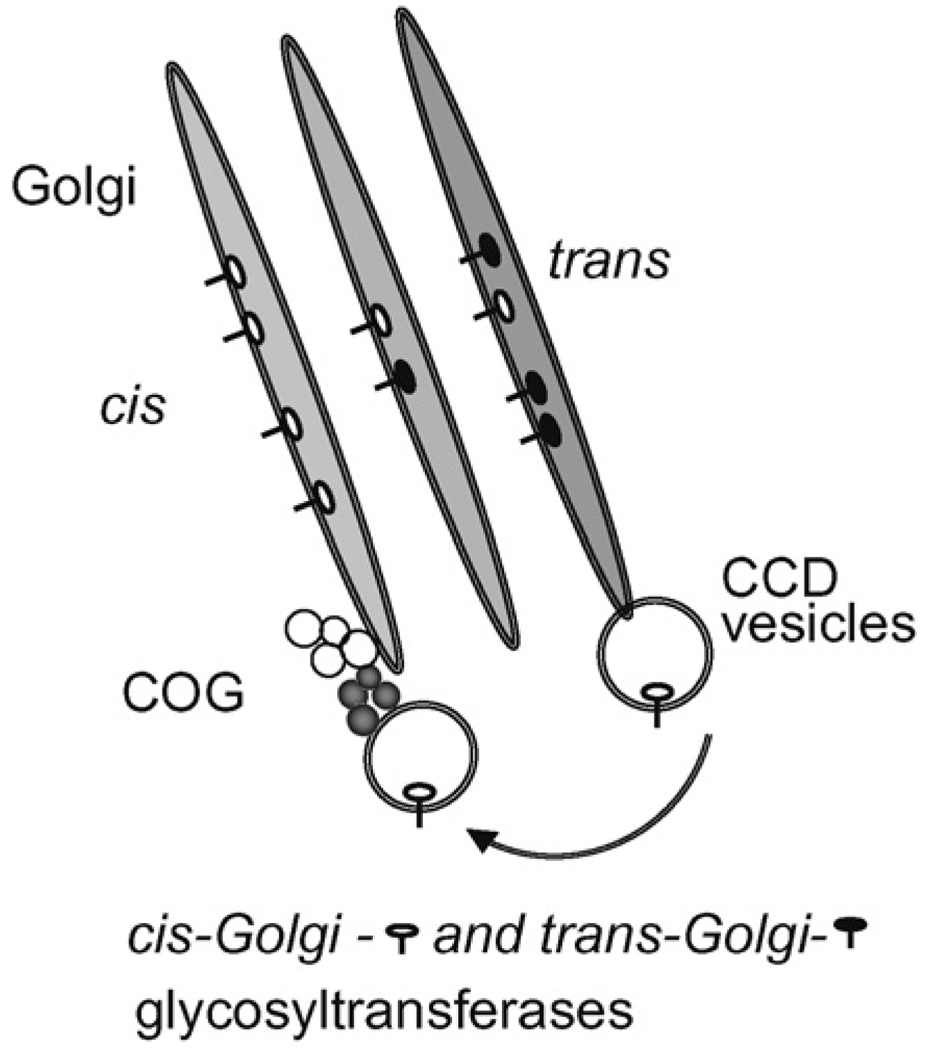
Our laboratory has recently demonstrated that the acute knockdown (KD) of the COG3 in HeLa cells was accompanied by reduction in Cog1, 2, and 4 protein levels and resulted in accumulation of COG complex-dependent (CCD) vesicles carrying Golgi v-SNARE molecules.40 Vesicle accumulation as a response for the COG complex dysfunction is evolutionary conserved, since previous electron microscopy experiments revealed that yeast cog2 and cog3 temperature-sensitive mutants accumulated 50 nm vesicles.45 COG complex-dependent docking of isolated CCD vesicles was reconstituted in vitro supporting their role as functional trafficking intermediates.41 Prolonged block in CCD vesicle tethering is accompanied by substantial fragmentation of the Golgi ribbon. Fragmented Golgi membranes maintain their juxtanuclear localization, cisternal organization, and competence for anterograde protein trafficking to the plasma membrane. These findings led to the hypothesis that the COG complex acts as a tether which connects COPI vesicles with cis-Golgi membranes during retrograde intra-Golgi traffic.18 Additional evidence that COG plays a role in the retrograde vesicular transport of Golgi proteins, including the glycosylation enzymes required for normal Golgi function, came from surveying the steady-state levels of Golgi proteins in wild-type and COG-deficient mammalian cells.42 Seven Golgi membrane proteins (called ‘GEAR’s by the authors), including processing enzyme Man II, were found to exhibit reduced steady-state levels in both ΔCOG1 and ΔCOG2 CHO cells. Detailed analysis of cellular phenotypes at different stages of COG3 KD was very instrumental to uncover the molecular link between COG function and glycosylation disorders. It was found that in COG-depleted cells, two medial-Golgi enzymes, GlcN-AcTI and Man II, are transiently relocated into CCD vesicles. As a result, Golgi modifications of both plasma membrane (CD44) and lysosomal (Lamp2) glycoproteins are distorted. Final intracellular localization of these proteins was not altered indicating that COG complex is not required for anterograde trafficking and accurate sorting. COG7 KD and double COG3/COG7 KD caused similar defects with respect to both Golgi traffic and glycosylation suggesting that the entire COG complex orchestrates the recycling of medial-Golgi resident proteins.41 Similar defects in localization of cis-Golgi mannosyltransferase Och1p were found in yeast cog3 temperature-sensitive mutant cells58 supporting the hypothesis that COG complex-dependent recycling of Golgi glycosylation enzymes is evolutionary conserved. Altogether, the data suggest that constantly cycling medial-Golgi enzymes are transported from distal compartments to cis/medial-Golgi in CCD vesicles (Fig. 3). Dysfunction of the COG complex leads to separation of glycosyltransferases from anterograde cargo molecules passing along secretory pathway thus affecting normal protein glycosylation.
Figure 3.
Model for the COG complex functions in recycling of Golgi glycosyltransferases. The COG complex specifically tethers vesicles that retrieve recycling cis/medial-Golgi proteins from trans-Golgi compartments.
6. Concluding remarks
The Golgi apparatus is playing the major role in both N-and O-linked glycosylation of proteins and lipids in eukaryotic cells. The proper intra-Golgi localization of glycosylation enzymes is achieved by a combination of various retentions and retrieval mechanisms. The importance of COG complex-mediated trafficking of components of Golgi glycosylation machinery is highlighted by a recent discovery of several novel CDG-II human glycosylation disorders. Many lines of evidence now suggest that COG complex is orchestrating the accurate tethering of COPI-coated retrograde vesicles that transport Golgi enzymes. Future research is needed to elucidate the functional interplay between the COG complex and other components of vesicle tethering/fusion machinery (Golgi-localized Rabs, SNAREs and coiled-coil tethering factors). COG complex-related human CDG-II disorders are not currently treatable. Understanding the molecular underpinning of glycosylation disorders will hopefully provide a foundation for potential future therapies.
Acknowledgments
This study was supported by grants from the NSF (MCB-0234822 and MCB-0645163) and the Mizutani Foundation for Glycoscience.
Abbreviations
- CCD vesicles
COG complex-dependent vesicles
- CDG
congenital disorders of glycosylation
- CHO
Chinese hamster ovary
- COG
conserved oligomeric Golgi
- Con A
concanavalin A
- fws
four way stop
- GalNAcT2
N-acetylgalactosaminyltransferase-2
- GalT
galactosyltransferase
- GlcNAcT1
N-acetylglucosaminyltransferase I
- IEF
isoelectric focusing
- LCA
Lens culinaris agglutinin
- Ldl
low-density lipoprotein
- PHA
phytohemaglutinin
- PNA
peanut agglutinin
- siRNA
small interfering RNA
- WGA
wheat germ agglutinin
Footnotes
This work was supported by grants from the National Science Foundation (MCB-0234822; MCB-0645163) and Mizutani Foundation for Glycoscience.
References
- 1.Warren G, Malhotra V. Curr. Opin. Cell Biol. 1998;10:493–498. doi: 10.1016/s0955-0674(98)80064-1. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Glick BS. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 3.Shorter J, Warren G. Annu. Rev. Cell Dev. Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson T, Pypaert M, Hoe MH, Slusarewicz P, Berger EG, Warren G. J. Cell Biol. 1993;120:5–13. doi: 10.1083/jcb.120.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. J. Cell Sci. 1995;108:1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- 6.Opat AS, van Vliet C, Gleeson PA. Biochimie. 2001;83:763–773. doi: 10.1016/s0300-9084(01)01312-8. [DOI] [PubMed] [Google Scholar]
- 7.Duden R. Mol. Membr. Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- 8.Pelham HR, Rothman JE. Cell. 2000;102:713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 9.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Söllner TH, Rothman JE. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 10.Harris SL, Waters MG. J. Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opat AS, Houghton F, Gleeson PA. Biochem. J. 2001;358:33–40. doi: 10.1042/0264-6021:3580033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahn R, Scheller RH. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 13.Lupashin V, Sztul E. Biochim. Biophys. Acta. 2005;1744:325–339. doi: 10.1016/j.bbamcr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Sztul E, Lupashin V. Am. J. Physiol. 2006;290:C11–C26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 15.Whyte JR, Munro S. Dev. Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 16.Kingsley DM, Kozarsky KF, Segal M, Krieger M. J. Cell Biol. 1986;102:1576–1585. doi: 10.1083/jcb.102.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ram RJ, Li B, Kaiser CA. Mol. Biol. Cell. 2002;13:1484–1500. doi: 10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suvorova ES, Duden R, Lupashin VV. J. Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. J. Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suvorova ES, Kurten RC, Lupashin VV. J. Biol. Chem. 2001;276:22810–22818. doi: 10.1074/jbc.M011624200. [DOI] [PubMed] [Google Scholar]
- 21.Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. J. Biol. Chem. 2005;280:27613–27623. doi: 10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- 22.Koumandou VL, Dacks JB, Coulson RM, Field MC. BMC Evol. Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu SC, TerBush D, Abraham M, Guo W. Int. Rev. Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- 24.Liewen H, Meinhold-Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, Jung M, Pfreundschuh M, Stenner-Liewen F. Exp. Cell Res. 2005;306:24–34. doi: 10.1016/j.yexcr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Cavanaugh LF, Chen X, Richardson BC, Ungar D, Pelczer I, Rizo J, Hughson FM. J. Biol. Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 26.Walter DM, Paul KS, Waters MG. J. Biol. Chem. 1998;273:29565–29576. doi: 10.1074/jbc.273.45.29565. [DOI] [PubMed] [Google Scholar]
- 27.Vasile E, Oka T, Ericsson M, Nakamura N, Krieger M. Exp. Cell Res. 2006;312:3132–3141. doi: 10.1016/j.yexcr.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M. J. Biol. Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- 29.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. J. Biol. Chem. 2005;280:32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 30.Zerial M, McBride H. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 31.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 32.VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. J. Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG. J. Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Ménard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Shestakova A, Hunt L, Sehgal S, Lupashin V, Storrie B. Mol. Biol. Cell. 2007;18:4129–4142. doi: 10.1091/mbc.E07-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fridmann-Sirkis Y, Kent HM, Lewis MJ, Evans PR, Pelham HR. Traffic. 2006;7:182–190. doi: 10.1111/j.1600-0854.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- 37.Price A, Seals D, Wickner W, Ungermann C. J. Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sivaram MV, Saporita JA, Furgason ML, Boettcher AJ, Munson M. Biochemistry (Mosc) 2005;44:6302–6311. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- 39.Kim DW, Sacher M, Scarpa A, Quinn AM, Ferro-Novick S. Mol. Biol. Cell. 1999;10:3317–3329. doi: 10.1091/mbc.10.10.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zolov SN, Lupashin VV. J. Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shestakova A, Zolov S, Lupashin V. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- 42.Oka T, Ungar D, Hughson FM, Krieger M. Mol. Biol. Cell. 2004;15:2423–2435. doi: 10.1091/mbc.E03-09-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letourneur F, Gaynor EC, Hennecke S, Démollière C, Duden R, Emr SD, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 44.Kim DW, Massey T, Sacher M, Pypaert M, Ferro-Novick S. Traffic. 2001;2:820–830. doi: 10.1034/j.1600-0854.2001.21111.x. [DOI] [PubMed] [Google Scholar]
- 45.Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. Genetics. 1996;142:393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farkas RM, Giansanti MG, Gatti M, Fuller MT. Mol. Biol. Cell. 2003;14:190–200. doi: 10.1091/mbc.E02-06-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Podos SD, Reddy P, Ashkenas J, Krieger M. J. Cell Biol. 1994;127:679–691. doi: 10.1083/jcb.127.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubota Y, Sano M, Goda S, Suzuki N, Nishiwaki K. Development. 2006;133:263–273. doi: 10.1242/dev.02195. [DOI] [PubMed] [Google Scholar]
- 49.Kingsley DM, Krieger M. Proc. Natl. Acad. Sci. U.S.A. 1984;81:5454–5458. doi: 10.1073/pnas.81.17.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterton JE, Hirsch D, Schwartz JJ, Bickel PE, Rosenberg RD, Lodish HF, Krieger M. Proc. Natl. Acad. Sci. U.S.A. 1999;96:915–920. doi: 10.1073/pnas.96.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steet R, Kornfeld S. Mol. Biol. Cell. 2006;17:2312–2321. doi: 10.1091/mbc.E05-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeze HH. Curr. Mol. Med. 2007;7:389–396. doi: 10.2174/156652407780831548. [DOI] [PubMed] [Google Scholar]
- 53.Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, Jaeken J, Mills P, Winchester B, Krieger M, Annaert W, Matthijs G. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH. Nat. Med. 2004;10:518–523. doi: 10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- 55.Morava E, Zeevaert R, Korsch E, Huijben K, Wopereis S, Matthijs G, Keymolen K, Lefeber DJ, De Meirleir L, Wevers RA. Eur. J. Hum. Genet. 2007;15:638–645. doi: 10.1038/sj.ejhg.5201813. [DOI] [PubMed] [Google Scholar]
- 56.Foulquier F, Ungar D, Reynders E, Zeevaert R, Mills P, Garc±a-Silva MT, Briones P, Winchester B, Morelle W, Krieger M, Annaert W, Matthijs G. Hum. Mol. Genet. 2007;16:717–730. doi: 10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- 57.Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Winkel RD, Cipollo JF, Costello CE, Loh E, Hong W, Freeze HH. Hum. Mol. Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- 58.Bruinsma P, Spelbrink RG, Nothwehr SF. J. Biol. Chem. 2004;279:39814–39823. doi: 10.1074/jbc.M405500200. [DOI] [PubMed] [Google Scholar]
- 59.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. Traffic. 2007;8:270–284. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 60.Shite S, Seguchi T, Yoshida T, Kohno K, Ono M, Kuwano M. J. Biol. Chem. 1988;263:19286–19289. [PubMed] [Google Scholar]
- 61.Ng BG, Kranz C, Hagebeuk EE, Duran M, Abeling NG, Wuyts B, Ungar D, Lupashin V, Hartdorff CM, Poll-The BT, Freeze HH. Mol. Genet. Metab. 2007;91:201–204. doi: 10.1016/j.ymgme.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]