Abstract
Increased production and deposition of amyloid β-protein (Aβ) are believed to be key pathogenic events in Alzheimer's disease. As such, routes for lowering cerebral Aβ levels represent potential therapeutic targets for Alzheimer's disease. X11β is a neuronal adaptor protein that binds to the intracellular domain of the amyloid precursor protein (APP). Overexpression of X11β inhibits Aβ production in a number of experimental systems. However, whether these changes to APP processing and Aβ production induced by X11β overexpression also induce beneficial effects to memory and synaptic plasticity are not known. We report here that X11β-mediated reduction in cerebral Aβ is associated with normalization of both cognition and in vivo long-term potentiation in aged APPswe Tg2576 transgenic mice that model the amyloid pathology of Alzheimer's disease. Overexpression of X11β itself has no detectable adverse effects upon mouse behaviour. These findings support the notion that modulation of X11β function represents a therapeutic target for Aβ-mediated neuronal dysfunction in Alzheimer's disease.
INTRODUCTION
Neuritic plaques containing deposits of Aβ are hallmark pathologies of Alzheimer's disease. Aβ is a 40–42 amino acid peptide that is derived by proteolytic cleavage from a precursor, the amyloid precursor protein (APP). APP is a type-1 membrane-spanning protein and the Aβ sequence resides partly within the extracellular and membrane-spanning domains [see for review (1)].
A number of lines of evidence support the notion that altered APP processing leading to increased production, aggregation and deposition of Aβ are major pathogenic events in Alzheimer's disease [see for review (2)]. Indeed, elevated levels of Aβ have been linked to memory defects and changes in synaptic plasticity in a variety of animal models of Alzheimer's disease [see for reviews (3,4)]. One such model is the APPsweTg2576 (APPswe) transgenic mouse which expresses a familial Alzheimer's disease mutant APP gene that contains the ‘Swedish’ double mutation Lys670Asn/Met671Leu (5). APPswe mice are one of the most widely studied models of Alzheimer's disease and develop Aβ amyloid pathology and also age-related defects in spatial reference memory and long-term potentiation (LTP) that correlate with increasing levels of cerebral Aβ (6–9). Such studies have highlighted reduction of cerebral Aβ load as a prime target for therapeutic intervention in Alzheimer's disease.
X11β (also known as X11-like/munc-18-interacting protein-2; mint2) is a neuronal adaptor protein that binds to the intracellular domain of APP (10–13). This interaction is mediated by a central phospho-tyrosine-binding (PTB) domain in X11β and sequences surrounding the YENPTY motif in APP [see for reviews (14,15)]. Alterations to X11β expression influence APP processing and Aβ production. In particular, elevation of X11β inhibits Aβ production in cellular models and inhibits Aβ production and deposition in the brains of APPswe transgenic mice (12,16–20). Such observations suggest that modulation of X11β function might prove therapeutic in Alzheimer's disease. Here, we show that X11β not only inhibits cerebral Aβ production, but additionally corrects age-dependent defects in both cognition and in vivo LTP in aged Alzheimer's disease APPswe Tg2576 mice.
RESULTS
Overexpression of X11β has no detrimental effect on sensorimotor function in APPswe Tg2576 mice
To investigate the effect of X11β on cerebral Aβ levels, memory function and LTP, we crossed X11β overexpressing transgenic mice (16) with APPswe Tg2576 (APPswe) transgenic mice (5). APPswe mice show age-related defects in spatial reference memory and LTP that correlate with increasing levels of cerebral Aβ (6–9). Analyses of the offspring from the cross revealed that their genotypes approximated to that expected of Mendelian inheritance; 1/4 each non-transgenic (NTg); APPswe; X11β; APPswe/X11β. Mice were analyzed at 3–5 months and at 12–16 months of age. Up to 6 months of age, APPswe mice have normal memory function and no Aβ pathology but then develop memory and LTP defects that correlate with increasing Aβ load (6,8,21). There were no significant age differences between the four genotypes in the 3–5 months age group; likewise there were no significant differences between the four genotypes in the 12–16 months age group. Finally, no significant differences in sex ratios between genotypes were detected in either age group (chi-square analyses). Male and female mice were thus pooled for analyses.
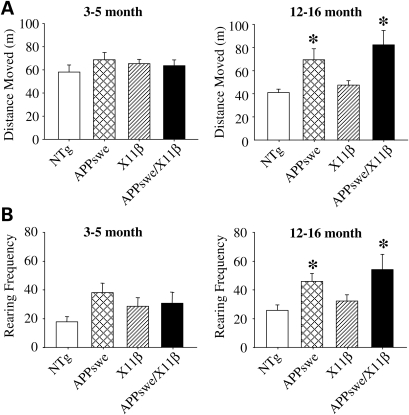
Both the 3–5 and 12–16 months mice were analyzed for sensorimotor defects using the SHIRPA primary screen (22). No differences between the four genotypes were detected at either age demonstrating that overexpression of X11β has no overt detrimental effect on sensorimotor function in either the absence or presence of the APPswe transgene. However, on open-field testing 12–16 months but not 3–5 months APPswe and APPswe/X11β mice both displayed significant 1.5–2-fold increases in distance travelled and rearing frequencies compared with both NTg and X11β animals (Fig. 1). In contrast, no differences between genotype in either the 3–5 or 12–16-month-old mice in the total time spent within, and number of entries into the central zone of the open-field arena were detected (one-way Anova, 3–5-month-old mice, n = 12–20; 12–16 months mice, n = 17–21). Previous studies of motor activity in APPswe mice have produced inconsistent results with reports of hyperactivity in three but not 9-month-old animals, hyperactivity in 17-month-old mice and no differences between APPswe and their NTg littermates at 16–18 months (6,23,24). Recently, an increase in locomotor activity has been linked to behavioural disinhibition in APPswe mice (25). Whatever, our finding that X11β overexpression does not influence locomotor activity in either the presence or absence of APPswe, suggests that the presence of the X11β transgene is unlikely to confound interpretation of assays of memory function in the Morris water maze which rely in part on locomotor function. Moreover, since X11β overexpression reduces cerebral Aβ levels (16) (and see below) the effect of APPswe on locomotor activity in 12–16 months mice may not be linked to cerebral Aβ load. Rather, the effect may be a functional consequence of APP (or APPswe) overexpression per se.
Figure 1.
Twelve to 16 months but not 3–5-month-old APPswe mice travel more and have increased rearing frequencies in the open-field test and these are unaffected by X11β overexpression. The total distances moved by each of the four genotypes in a 10 min session in the open-field arena are shown (A) along with rearing frequencies displayed as total rears per 10 min session (B). * Indicates significant differences (P < 0.05 one-way ANOVA with LSD post hoc test). n = 12–20 for 3–5 months mice and n = 17–21 for 12–16 months mice. Error bars are ± SEM.
X11β reduces cerebral Aβ levels in APPswe Tg2576 mice
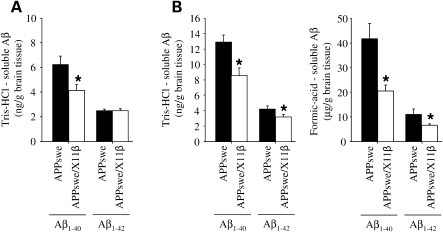
We analyzed brain Aβ1–40 and Aβ1–42 levels in APPswe and APPswe/X11β littermates. In young APPswe mice (up to 6 months), Aβ species are mainly soluble in aqueous buffers (Tris–HCl-soluble Aβ) but in older mice, Aβ levels increase dramatically and require solubilisation in agents such as formic acid (formic acid-soluble Aβ) (7,16). We therefore prepared Tris–HCl-soluble and formic acid-soluble fractions for analyses. In 3–5 months animals, X11β significantly reduced Tris–HCl-soluble Aβ1–40 but not Aβ1–42 levels. The lack of an effect of X11β on Aβ1–42 levels may be partly a consequence of the relatively low levels of this species in the brains of APPswe mice at this young age and the consequent difficulties in its robust detection. Indeed, this lack of effect of X11β on Aβ1–42 levels in young APPswe mice is consistent with previous reports in these mice (16). Formic acid-soluble Aβ1–40 and Aβ1–42 levels in 3–5-month-old mice were below the level of accurate detection by the commercial ELISA kits used here (Fig. 2A). Again this is in agreement with previous studies that also reported very low levels of formic acid-soluble Aβ1–40 and Aβ1–42 in mice up to 6 months of age and which were difficult to robustly detect (7,16). However, in 12–16-month-old APPswe mice, formic acid-soluble Aβ levels increase by over a thousand-fold (7,16). At this age, X11β induced significant decreases in both Tris–HCl-soluble and formic acid-soluble Aβ1–40 and Aβ1–42 levels (Fig. 2B). Of particular interest, X11β reduced total Aβ1–40 and Aβ1–42 levels by approximately 50 and 40%, respectively, in the 12–16 months mice. We have previously shown that X11β also markedly reduces Aβ plaque numbers in APPswe transgenic mice (16).
Figure 2.
X11β reduces brain Aβ levels in APPswe mice. (A) Shows Tris–HCl-soluble Aβ1–40 and Aβ1–42 levels in 3–5-month-old APPswe and APPswe/X11β transgenic mice (n = 12–14). X11β significantly lowered Aβ1–40 levels. (B) Shows Tris–HCl-soluble and formic acid-soluble Aβ1–40 and Aβ1–42 levels in 12–16 months APPswe and APPswe/X11β transgenic mice (n = 17–21). X11β significantly lowered Aβ1–40 and Aβ1–42 levels in both biochemical fractions. * Indicates significant differences (P < 0.05 t-test). Error bars are ± SEM.
X11β rescues spatial reference memory deficits in aged APPswe mice
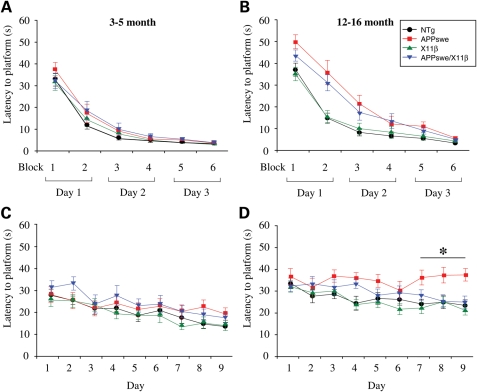
To determine the effect of X11β on age-related memory loss in APPswe mice, we tested spatial reference memory function in NTg, APPswe, X11β and APPswe/X11β mice using the Morris water maze at both 3–5 and 12–16 months of age. For these studies, we performed visible platform training followed by hidden platform testing with three rounds of probe trials as described by others for APPswe Tg2576 mice (8,21). All genotypes in both age groups were able to learn the location of the visible platform by 3 days of training (Fig. 3A and B) and within each age group were similarly proficient swimmers (data not shown). However, APPswe and APPswe/X11β 12–16 months but not 3–5-month-old mice both had increased escape latencies (time in seconds to reach the platform) compared with NTg and X11β littermate mice in the first three training blocks (Fig. 3A and B). This is consistent with previous studies that likewise demonstrated an initial lag in performance to reach the visible platform in middle aged (12–18 months) but not young (4–5 months) APPswe Tg2576 mice (8). Despite this lag in older APPswe and APPswe/X11β mice, the finding that there were no significant differences between any genotype in the last three training blocks mitigates against sensorimotor deficits as a potential explanation for any impaired performance in acquisition or retention of the location of the hidden platform in the Morris water maze. Others have reached the same conclusion with highly similar data on APPswe Tg2576 mice (8). Moreover, our finding that there are no significant differences in escape latencies to the visible platform between NTg and X11β, and between APPswe and APPswe/X11β mice at any time studied (Fig. 3A and B) argues that the phenotype is due to APPswe and is not influenced by the presence of the X11β transgene. Thus, X11β overexpression has no detectable effect on escape latency to the visible platform in the Morris water maze.
Figure 3.
X11β improves acquisition of water maze platform location in 12–16 months APPswe mice. Escape latencies in seconds (s) for NTg, APPswe, X11β and APPswe/X11β mice were measured during visible and hidden platform training in the Morris water maze test. (A) And (B) shows escape latencies to the visible platform in 3–5 months (A) and 12–16 months (B) mice. No significant differences were detected between any of the genotypes in visible platform training at 3–5 months of age (two-way ANOVA). In 12–16 months animals both APPswe and APPswe/X11β transgenics showed increased latencies in the first three blocks (day 1 and first block of day 2) of visible training compared with both X11β and NTg (P < 0.05); however, no differences were detected between NTg and X11β and between APPswe and APPswe/X11β at any time point (two-way Anova with LSD post hoc test). (C) And (D) shows escape latencies to the hidden platform in 3–5 months (C) and 12–16 months (D) mice. No significant differences were detected between any genotype in 3–5 months mice (two-way ANOVA) and all genotypes showed significant improvement in escape latencies over the testing period (both days 1 and 2 versus days 5–9, P < 0.05; two-way ANOVA with LSD post hoc test). Twelve to 16 months NTg, X11β and APPswe/X11β likewise showed significant improvement in escape latencies over the testing period (day 1 versus days 7, 8 and 9, P < 0.05; two-way ANOVA with LSD post hoc test). However, in 12–16 months mice, latencies were specifically increased in APPswe compared with all other genotypes on days 7, 8 and 9 of testing and this effect was rescued in APPswe/X11β mice (two-way ANOVA with LSD post hoc test; P < 0.01 day 7; P < 0.001 days 8 and 9). In addition, APPswe (but not APPswe/X11β) mice showed a significant increase in escape latency compared NTg and X11β mice on day 4 (P < 0.01 two-way ANOVA). * Indicates significant differences between APPswe and all other genotypes. n = 12–20 for 3–5 months mice; n = 17–21 for 12–16 months mice. Error bars are ± SEM.
We next analyzed escape latencies to reach the hidden platform in the Morris water maze. APPswe 12–16 months but not 3–5 months mice displayed significant deficits in spatial reference learning and memory as compared with NTg littermates. This was revealed by an increase in the escape latencies by APPswe mice on days 7–9 of training (Fig. 3C and D) and by analyses of swimming patterns in probe trials conducted on days 4, 7 and 10 of testing where the platform was removed. In addition, APPswe but not APPswe/X11β transgenics also showed an increase in latency to reach the platform compared with NTg and X11β littermates on day 4 of testing. This may be related to the first probe trial that occurred prior to testing on day 4. Indeed, others using these same water maze methods for testing have also shown an increase in escape latency by similarly aged (12–18 months) APPswe mice on day 4 of testing following the first probe trial (8). However, platform seeking behaviour was not noticeably different after the probe trial (no indications of thigmotaxis, circling or non-swimming/floating behaviour was observed) which supports the notion that the day 4 effect on APPswe mice is transient. Indeed, the data we present on escape latencies for APPswe and NTg littermates in the 12–16 months mice are remarkably similar to those described by others in similarly aged APPswe Tg2576 mice (8).
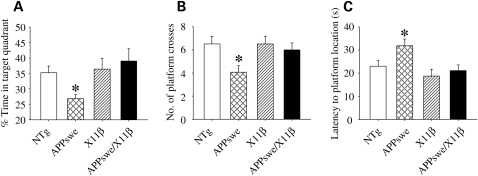
Concordant with analyses of escape latencies, the probe trial data demonstrated significant reductions in mean time spent in the target quadrant and number of target crosses, and a significant increase in mean latency to swim to the platform location by APPswe mice (Fig. 4A–C). These results are consistent with previous reports (8,21). There were no detectable differences between X11β and NTg mice in any of the tests. However, APPswe/X11β mice displayed significant improvements in both escape latencies and mean probe trial scores as compared with APPswe mice such that their performances were not significantly different from NTg or X11β animals (Figs 3C, D and 4). Thus, overexpression of X11β reduces Aβ levels in the brains of APPswe mice and improves spatial reference learning and memory in 12–16-month-old APPswe mice.
Figure 4.
X11β improves retention of water maze platform location in 12–16 months APPswe mice. Spatial memory retention in 12–16 months NTg, APPswe, X11β and APPswe/X11β mice were assessed in probe trials that determined the % of time spent in target quadrant (A), mean number of times the platform location was crossed (B) and latency to first reach the platform location (C). APPswe mice performed less well compared with all other genotypes and this effect was rescued in APPswe/X11β mice [one-way ANOVA with LSD post hoc test; P = 0.029 (A); P = 0.026 (B); P = 0.009 (C) n = 17–21]. * Indicate significant differences. Error bars are ± SEM.
X11β rescues age-dependent deficits in LTP in APPswe mice
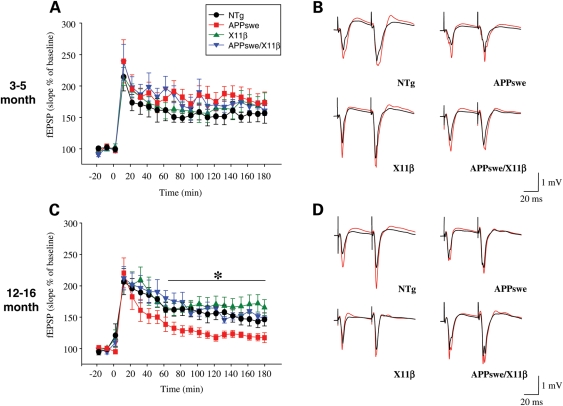
Defective learning and memory in APPswe mice has been associated with LTP deficits (6). LTP was therefore analyzed at Schaffer-collateral pyramidal cell synapses in the CA1 of anaesthetized 3–5 and 12–16 months NTg, APPswe, X11β and APPswe/X11β mice. Tetanic stimulation produced LTP in all four genotypes at both ages (Fig. 5). In 3–5-month-old mice, there were no significant differences between any of the genotypes during the induction (10–70 min), maintenance (70–130 min) and late (130–180 min) phases (Fig. 5A and B). However, in 12–16 months APPswe mice, LTP declined more rapidly than in the other groups and was significantly less in both the maintenance and late phases (Fig. 5C and D). These data are consistent with earlier reports (6). Notably, LTP in 12–16 months APPswe/X11β and X11β mice were not significantly different to NTg animals in any phase. Thus, expression of X11β alone has no discernable effect on LTP in either the 3–5 or 12–16 months mice but rescues LTP deficits in APPswe/X11β animals.
Figure 5.
X11β improves LTP in 12–16 months APPswe mice. LTP in 3–5 months (A) and 12–16 months (C) NTg, APPswe, X11β and APPswe/X11β mice are shown. fEPSP slopes are expressed as the % of the pre-tetanus baseline. Representative examples of stimulus responses both before (black) and after (red) tetanus are also shown for both age groups (B; 3–5 months, D; 12–16 months). (A) No significant differences in fEPSPs between genotypes were detected during either induction-phase (10–70 min), maintenance-phase (70–130 min) or late-phase (130–180 min) of LTP in 3–5 months mice (one-way ANOVA; n = 7–9). (C) No significant differences in fEPSPs between genotypes were detected in the induction phase in 12–16 months mice. Likewise, no differences in fEPSPs were detected between NTg, X11β and APPswe/X11β in the maintenance and late phases of LTP. In contrast LTP was significantly less well maintained in the maintenance- and late-phases in APPswe mice compared with all other genotypes (one-way ANOVA with LSD post hoc test P < 0.05; n = 6–8). * Indicate significant differences. Error bars are ± SEM.
DISCUSSION
X11β (mint2) is a member of a small family of related proteins which include X11α (also known as mint1) and X11γ (also known as mint3) that bind to the intracellular domain of APP via PTB domains; X11α and X11β are the two neuronal-specific isoforms [see for reviews (14,15)]. Numerous studies have shown that overexpression of either X11α, X11β or X11γ all inhibit Aβ production (12,16,17,26–30). In contrast, the effect of loss of X11s on in vivo Aβ production is less clear (17,31–33). However, of the three, X11β is expressed highest in the cerebral cortex, entorhinal cortex and hippocampus, regions of the brain most acutely affected in Alzheimer's disease (34). Thus, our findings that overexpression of X11β reduces Aβ levels and rescues learning, memory and LTP defects in APPswe mice has particular relevance for Alzheimer's disease.
X11β function is not properly understood but there is evidence that the X11s have synaptic roles. Thus, X11α and X11α/X11β double knockout mice have presynaptic defects involving altered neurotransmitter release (35–37). In apparent contrast to the above, other studies provide evidence for post-synaptic functions for the X11s and in particular in the trafficking of NMDA receptors (38–40). Finally, the X11s may be involved in synaptic targeting of N-type calcium channels and Kir2 potassium channels (41,42). However, as yet there are no reports on electrophysiological characterization of X11β single knockout mice and this confounds the proper interpretation of the role of X11β in synaptic function. This is especially so since there is now strong evidence that the different X11s have unique functions. For example, X11α and X11β but not X11γ bind munc-18, and X11α but not X11β or X11γ binds to the CASK/mLin-2 and Veli/mLin7 complex (43–45). There is even some evidence that the effects of X11α and X11β on APP and Aβ production may involve different underlying mechanisms (33,46).
What we show here is that overexpression of X11β has no discernible effect on learning, memory and LTP in X11β single transgenics and this is highly complementary to those of a recent study on X11β knockout mice which likewise demonstrate no role for X11β in learning and memory (also using the Morris water maze) (47). Thus, neither overexpression nor loss of X11β influences hippocampal dependent learning and memory functions. A large body of evidence now demonstrates that learning, memory and associated LTP defects in APPswe Tg2576 mice and other Alzheimer's APP mice are due to increased cerebral levels of Aβ and/or specific Aβ assemblies (5–9,21,48–52). Thus, the most likely interpretation for the restoration of learning, memory and LTP in APPswe/X11β mice is X11β-mediated reduction in brain Aβ levels. However, we cannot exclude the possibility that there are learning mechanisms (as opposed to simply locomotor ones) that are impaired in APPswe mice that are Aβ-independent, X11β-resistant or both.
The precise molecular mechanisms by which the X11s inhibit Aβ production are not properly understood. Aβ is produced by consecutive processing of APP by β- and γ-secretases. β-secretase (BACE1) cleaves APP at the amino-terminus of the Aβ sequence and γ-secretase (comprising Presenilin, Nicastrin, APH-1 and PEN-2) at the carboxy-terminus of the Aβ sequence (53,54). Some studies suggest that the X11s inhibit γ-secretase cleavage while others propose they inhibit β-secretase cleavage of APP (55,56). Recently, the X11s have been shown to regulate BACE1 processing of APP by altering APP compartmentalization into detergent-resistant domains within membranes (lipid rafts) or microdomains such that APP and BACE1 are in different membranous compartments (31,57). This may involve altered trafficking of APP and indeed the X11s are linked to kinesin family molecular motors. X11α binds to the kinesin KIF17 and X11β binds to alcadein/calsyntenin, a scaffolding protein that interacts with kinesin-1 (20,38,58,59). In particular, the X11s and/or alcadein/calsyntenin may be involved in trafficking APP from the Golgi (17,60). Finally, the X11s also interact with the copper chaperone for superoxide dismutase-1 (CCS) and CCS additionally binds BACE1 (61,62). The roles of CCS on APP processing and Aβ production are not known but its interaction with both BACE1 and X11 provides a further route whereby the X11s might inhibit brain Aβ production. Whatever the precise mechanism, the data presented here which demonstrate that X11β not only reduces cerebral Aβ load but also corrects the associated cognitive and synaptic plasticity defects, validate modulation of X11β function as a therapeutic target for Alzheimer's disease.
MATERIALS AND METHODS
Transgenic mice
APPswe Tg2576 mice obtained from Taconic farms (Germantown, NY, USA) were maintained by breeding males with C57Bl/6/SJL F1 females as recommended by the suppliers. X11β transgenic line 42 mice generated from C57Bl/6/SJL embryos have been described previously and were backcrossed onto C57Bl/6 eight times prior to crossing with APPswe Tg2576 animals. Male APPswe mice were mated with female X11β mice and offspring genotyped by PCR using primer sets 5′-CGACTCGACCAGGTTCTGGGT-3′, 5′-ATAACCCCTCCCCCAGCCTAGA-3′ (APP) and 5′-TGAAGAACCAAACCCAGGTAAAGC-3′, 5′-CTACAGATCCTCTTCTGAGATGAG-3′ (X11β). Mice were housed on 12 h light:12 h dark cycles. All experiments were performed under the terms of the UK Animals (Scientific Procedures) Act 1986.
Behavioural testing
Experimenters were blind to genotype for all behavioural testing. Mice were initially assessed for sensorimotor function using a modified SHIRPA primary screen (22). For open field testing of locomotor function and exploratory activity, we used a white circular arena (80 cm diameter) with a 60 cm high wall, surrounded by a visually uniform environment. For single trials, mice were placed into the edge of the arena, facing the wall, and allowed to explore for 10 min. Trials were monitored and analyzed using the Ethovision package (Noldus, The Netherlands). We measured total locomotor activity plus time spent in, and visits to, the central (60 cm diameter) more anxiogenic zone. Rearing frequencies were monitored using Ethovision software.
Hippocampus-dependent spatial reference learning and memory were tested using a version of the Morris water maze task that has been specifically designed for APPswe Tg2576 mice, the Alzheimer's disease mouse model used here (8,21). This involved visible platform training followed by hidden platform testing with three rounds of probe trials (8,21). We used a white 1.4 m pool filled with water (22–24°C) opacified using Opacifier E308 (Rohm and Haas, UK). Mice underwent visible platform (10 cm diameter) training for three consecutive days with each days training split into two blocks of four trials. During visual training, a curtain was drawn around the pool to remove any visual cues. The platform location (NE, SE, SW or NW) and start positions (N, NE, E, SE, S, SW, W or NW) were varied pseudorandomly between these trials although start positions were never immediately adjacent to the platform. A black and white striped flagpole was situated in the centre of the platform to mark its location. Hidden platform training was conducted over the next 9 days (four trials per day) where mice searched for the platform now submerged 1.5 cm below the water surface. The location of the platform remained constant during hidden platform trials and the mice entered the pool in one of the pseudorandomly selected locations excluding the positions immediately adjacent to the platform. Mice that were unable to find the platform within 60 s were led to it with an escape scoop. In aged mice, probe trials were performed on the beginning of the 4th, 7th and 10th days of hidden platform training, when the platform was removed and mice allowed 60 s search time. Trials were recorded and analyzed using Ethovision. Time taken to reach the platform (escape latency) during each trial was monitored. During probe trials, time in target quadrant, latency to platform location and number of platform crosses were measured and mean performance across the three trials determined as previously described (63,64).
Aβ assays
Human Aβ1–40 and Aβ1–42 levels in the brains of mice were determined using ELISA kits (TKHS-Set, The Genetics Company, Switzerland) essentially following the manufacturers instructions. Samples for analyses were prepared as Tris–HCl-soluble and formic acid-soluble fractions as described previously (16). Briefly, brains were prepared for assay by homogenization as 20% homogenates in 20 mm Tris–HCl pH 8.0 containing 5 mm EDTA plus Complete protease inhibitor cocktail (Roche) (Assay buffer) using a Dounce homogenizer. Thereafter, the samples were spun at 100 000 g(av) for 1 h and the supernatant containing soluble Aβ then removed and diluted as appropriate in Assay buffer for analyses (Tris–HCl-soluble Aβ). To assay for insoluble Aβ, the remaining pellet was extracted as a 15% homogenate in 70% formic acid by sonication at level 4 for 35 s using a Vibra Cell Disruptor (Sonic & Materials Inc.) and the mixture then spun at 100 000 g(av) for 1 h and the supernatant removed and diluted 1:20 with 1 m Tris to neutralize the pH. The samples were then diluted as appropriate in Assay buffer for analyses (formic acid-soluble Aβ). Signals from ELISA were quantified at 450 nm using a microplate reader (Victor 3 1420, Perkin Elmer).
Electrophysiology
Mice underwent in vivo electrophysiological testing to investigate changes in hippocampal LTP essentially as previously described (65). Mice were anaesthetized with urethane (1.2–1.8 g/kg i.p.) followed by supplemental injections as required and fixed in a stereotaxic frame with bregma and lambda in the same horizontal plane. Body temperature was maintained at 37°C using an electrical heating pad. Bipolar stimulating electrodes were placed in the Schaffer-commissural input to CA1 of the left cerebral hemisphere (coordinates: 1.0 mm posterior to bregma; 0.5 mm lateral; 2.0 mm below brain surface), and the recording electrodes were sited in the left stratum oriens (2.2 mm posterior to bregma; 1.5 mm lateral; 1.2 mm below brain surface). After optimization and stabilization of evoked, negative-going, field excitatory postsynaptic potentials (fEPSPs), pulse pairs (200 µs pulse width, 50 ms interpulse interval, 2 pulse pairs per minute) were delivered at an intensity producing fEPSPs with ∼40% of the maximal amplitude to obtain 20 min of baseline responses (−20 to 0 min). For analytical purposes, slopes of four consecutive fEPSPs were averaged to provide one mean of fEPSP over each 2 min period. The data analysis sections were divided into induction phase (10–70 min), maintenance phase (70–130 min) and late phase LTP (130–180 min). Input–output curves, expressed as percentages of the maximum fEPSP slopes against stimulating current, were obtained for the four genotypes in both the 3–5 and 12–16-month-old mice. The curves were not affected by genotype (P = 0.516, F3,418 = 0.761; three-way ANOVA) or age (P = 0.249, F1,418 = 1.334; three-way ANOVA). Nor was there any interaction between age and genotype (age × genotype: P = 0.327, F3,418 = 1.155; three-way ANOVA). This indicates that basal synaptic transmission was unaffected by genotype or age. LTP was then evoked using high frequency tetanic stimulation (single pulses at 100 pulses/sec for 1 s, repeated three times at 20 s intervals), after which paired-pulse stimulation was resumed for a further 180 min. LTP experiments were conducted blind to genotype.
FUNDING
This work was supported by grants from the Wellcome Trust, the Alzheimer's Society Medical Research Council, Alzheimer's Association, Alzheimer's Research Trust, European Union NeuroNE and The Health Foundation.
ACKNOWLEDGEMENTS
We thank Gerald Finnerty (IoP King's College London) for helpful discussions on electrophysiology.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thinakaran G., Koo E.H. Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh D.M., Selkoe D.J. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004;44:181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D.J. Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior. Behav. Brain. Res. 2008;192:106–113. doi: 10.1016/j.bbr.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 5.Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F.S., Cole G. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 6.Chapman P.F., White G.L., Jones M.W., Cooper-Blacketer D., Marshall V.J., Irizarry M., Younkin L., Good M.A., Bliss T.V., Hyman B.T., et al. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nat. Neurosci. 1999;2:271–276. doi: 10.1038/6374. [DOI] [PubMed] [Google Scholar]
- 7.Kawarabayashi T., Younkin L.H., Saido T.C., Shoji M., Ashe K.H., Younkin S.G. Age-dependent changes in brain, CSF, and plasma amyloid beta-protein in the Tg2576 transgenic mouse model of Alzheimer's disease. J. Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westerman M.A., Cooper-Blacketer D., Mariash A., Kotilinek L., Kawarabayashi T., Younkin L.H., Carlson G.A., Younkin S.G., Ashe K.H. The relationship between Abeta and memory in the Tg2576 mouse model of Alzheimer's disease. J. Neurosci. 2002;22:1858–1867. doi: 10.1523/JNEUROSCI.22-05-01858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotilinek L.A., Bacskai B., Westerman M., Kawarabayashi T., Younkin L., Hyman B.T., Younkin S., Ashe K.H. Reversible memory loss in a mouse transgenic model of Alzheimer's disease. J. Neurosci. 2002;22:6331–6335. doi: 10.1523/JNEUROSCI.22-15-06331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLoughlin D.M., Irving N.G., Brownlees J., Brion J.-P., Leroy K., Miller C.C.J. Mint2/X11-like co-localises with the Alzheimer's disease amyloid precursor protein and is associated with neuritic plaques in Alzheimer's disease. Eur. J. Neurosci. 1999;11:1988–1994. doi: 10.1046/j.1460-9568.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- 11.McLoughlin D.M., Miller C.C.J. The intracellular cytoplasmic domain of the Alzheimer's disease amyloid precursor protein interacts with phosphotyrosine binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;397:197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 12.Tomita S., Ozaki T., Taru H., Oguchi S., Takeda S., Yagi Y., Sakiyama S., Kirino Y., Suzuki T. Interaction of a neuron-specific protein containing PDZ domains with Alzheimer's amyloid precursor protein. J. Biol. Chem. 1999;274:2243–2254. doi: 10.1074/jbc.274.4.2243. [DOI] [PubMed] [Google Scholar]
- 13.Lau K.-F., McLoughlin D.M., Standen C.L., Irving N.G., Miller C.C.J. Fe65 and X11b co-localize with and compete for binding to the amyloid precursor protein. NeuroReports. 2000;16:3607–3610. doi: 10.1097/00001756-200011090-00041. [DOI] [PubMed] [Google Scholar]
- 14.Rogelj B., Mitchell J.C., Miller C.C., McLoughlin D.M. The X11/Mint family of adaptor proteins. Brain Res. Brain Res. Rev. 2006;52:305–315. doi: 10.1016/j.brainresrev.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Miller C.C., McLoughlin D.M., Lau K.F., Tennant M.E., Rogelj B. The X11 proteins, Abeta production and Alzheimer's disease. Trends Neurosci. 2006;29:280–285. doi: 10.1016/j.tins.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.H., Lau K.F., Perkinton M.S., Standen C.L., Rogelj B., Falinska A., McLoughlin D.M., Miller C.C. The neuronal adaptor protein X11beta reduces Abeta levels and amyloid plaque formation in the brains of transgenic mice. J. Biol. Chem. 2004;279:49099–49104. doi: 10.1074/jbc.M405602200. [DOI] [PubMed] [Google Scholar]
- 17.Shrivastava-Ranjan P., Faundez V., Fang G., Rees H., Lah J.J., Levey A.I., Kahn R.A. Mint3/X11gamma is an Arf-dependent adaptor that regulates the traffic of the Alzheimer's Precursor Protein from the TGN. Mol. Biol. Cell. 2008;19:51–64. doi: 10.1091/mbc.E07-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita S., Fujita T., Kirino Y., Suzuki T. PDZ domain-dependent suppression of NF-kB/p65-induced Ab42 production by a neuron-specific X11-like protein. J. Biol. Chem. 2000;275:13056–13060. doi: 10.1074/jbc.c000019200. [DOI] [PubMed] [Google Scholar]
- 19.Lee D.-S., Tomita S., Kirino Y., Suzuki T. Regulation of X11L-dependent APP metabolism by XB51, a novel X11L-binding protein. J. Biol. Chem. 2000;275:23134–23138. doi: 10.1074/jbc.C000302200. [DOI] [PubMed] [Google Scholar]
- 20.Araki Y., Tomita S., Yamaguchi H., Miyagi N., Sumioka A., Kirino Y., Suzuki T. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta -protein precursor metabolism. J. Biol. Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- 21.Lesne S., Koh M.T., Kotilinek L., Kayed R., Glabe C.G., Yang A., Gallagher M., Ashe K.H. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 22.Rogers D.C., Fisher E.M., Brown S.D., Peters J., Hunter A.J., Martin J.E. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome. 1997;8:711–713. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- 23.King D.L., Arendash G.W., Crawford F., Sterk T., Menendez J., Mullan M.J. Progressive and gender-dependent cognitive impairment in the APP(SW) transgenic mouse model for Alzheimer's disease. Behav. Brain Res. 1999;103:145–162. doi: 10.1016/s0166-4328(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 24.Lalonde R., Lewis T.L., Strazielle C., Kim H., Fukuchi K. Transgenic mice expressing the betaAPP695SWE mutation: effects on exploratory activity, anxiety, and motor coordination. Brain Res. 2003;977:38–45. doi: 10.1016/s0006-8993(03)02694-5. [DOI] [PubMed] [Google Scholar]
- 25.Gil-Bea F.J., Aisa B., Schliebs R., Ramirez M.J. Increase of locomotor activity underlying the behavioral disinhibition in tg2576 mice. Behav. Neurosci. 2007;121:340–344. doi: 10.1037/0735-7044.121.2.340. [DOI] [PubMed] [Google Scholar]
- 26.Lee J.H., Lau K.F., Perkinton M.S., Standen C.L., Shemilt S.J., Mercken L., Cooper J.D., McLoughlin D.M., Miller C.C. The neuronal adaptor protein X11alpha reduces Abeta levels in the brains of Alzheimer's APPswe Tg2576 transgenic mice. J. Biol. Chem. 2003;278:47025–47029. doi: 10.1074/jbc.M300503200. [DOI] [PubMed] [Google Scholar]
- 27.Ho C.S., Marinescu V., Steinhilb M.L., Gaut J.R., Turner R.S., Stuenkel E.L. Synergistic effects of Munc18a and X11 proteins on amyloid precursor protein metabolism. J. Biol. Chem. 2002;277:27021–27028. doi: 10.1074/jbc.M201823200. [DOI] [PubMed] [Google Scholar]
- 28.Mueller H.T., Borg J.P., Margolis B., Turner R.S. Modulation of amyloid precursor metabolism by X11alpha/mint-1: a deletion analysis of protein-protein interaction domains. J. Biol. Chem. 2000;275:39302–39306. doi: 10.1074/jbc.M008453200. [DOI] [PubMed] [Google Scholar]
- 29.Sastre M., Turner R.S., Levy E. X11 interaction with B-amyloid precursor protein modulates its cellular stabilization and reduces amyloid B-protein secretion. J. Biol. Chem. 1998;273:22351–22357. doi: 10.1074/jbc.273.35.22351. [DOI] [PubMed] [Google Scholar]
- 30.Borg J.P., Yang Y.N., De Taddéo-Borg M., Margolis B., Turner R.S. The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J. Biol. Chem. 1998;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 31.Saito Y., Sano Y., Vassar R., Gandy S., Nakaya T., Yamamoto T., Suzuki T. X11 proteins regulate the translocation of APP into detergent resistant membrane and suppress the amyloidogenic cleavage of APP by BACE in brain. J. Biol. Chem. 2008;283:35763–35771. doi: 10.1074/jbc.M801353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho A., Liu X., Sudhof T.C. Deletion of Mint proteins decreases amyloid production in transgenic mouse models of Alzheimer's disease. J. Neurosci. 2008;28:14392–14400. doi: 10.1523/JNEUROSCI.2481-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z., Romano D.M., Tanzi R.E. RNAi-mediated silencing of X11alpha and X11beta attenuate Abeta levels via differential effects on APP processing. J. Biol. Chem. 2005;280:15413–15421. doi: 10.1074/jbc.M414353200. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima Y., Okamoto M., Nishimura H., Obata K., Kitano H., Sugita M., Matsuyama T. Neuronal expression of mint1 and mint2, novel multimodular proteins, in adult murine brain. Mol. Brain Res. 2001;92:27–42. doi: 10.1016/s0169-328x(01)00126-7. [DOI] [PubMed] [Google Scholar]
- 35.Ho A., Morishita W., Atasoy D., Liu X., Tabuchi K., Hammer R.E., Malenka R.C., Sudhof T.C. Genetic Analysis of Mint/X11 Proteins: Essential Presynaptic Functions of a Neuronal Adaptor Protein Family. J. Neurosci. 2006;26:13089–13101. doi: 10.1523/JNEUROSCI.2855-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho A., Morishita W., Hammer R.E., Malenka R.C., Sudhof T.C. A role for Mints in transmitter release: Mint 1 knockout mice exhibit impaired GABAergic synaptic transmission. Proc. Natl. Acad. Sci. USA. 2003;100:1409–1414. doi: 10.1073/pnas.252774899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori A., Okuyama K., Horie M., Taniguchi Y., Wadatsu T., Nishino N., Shimada Y., Miyazawa N., Takeda S., Niimi M., et al. Alteration of methamphetamine-induced striatal dopamine release in mint-1 knockout mice. Neurosci. Res. 2002;43:251–257. doi: 10.1016/s0168-0102(02)00041-x. [DOI] [PubMed] [Google Scholar]
- 38.Setou M., Nakagawa T., Seog D.-H., Hirokawa N. Kinesin superfamiy motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 39.Guillaud L., Setou M., Hirokawa N. KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J. Neurosci. 2003;23:131–140. doi: 10.1523/JNEUROSCI.23-01-00131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillaud L., Wong R., Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat. Cell Biol. 2007;10:19–29. doi: 10.1038/ncb1665. [DOI] [PubMed] [Google Scholar]
- 41.Maximov A., Bezprozvanny I. Synaptic targeting of N-type calcium channels in hippocampal neurons. J. Neurosci. 2002;22:6939–6952. doi: 10.1523/JNEUROSCI.22-16-06939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonoudakis D., Conti L.R., Radeke C.M., McGuire L.M., Vandenberg C.A. A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J. Biol. Chem. 2004;279:19051–19063. doi: 10.1074/jbc.M400284200. [DOI] [PubMed] [Google Scholar]
- 43.Butz S., Okamoto M., Sudhof T.C. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in the brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 44.Borg J.-P., Straight S.W., Kaech S.M., De Taddéo-Borg M., Kroon D.E., Karnak D., Turner R.S., Kim S.K., Margolis B. Identification of an evolutionarily conserved hetrotrimeric protein complex involved in protein targeting. J. Biol. Chem. 1998;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- 45.Borg J.P., Lopez-Figueroa M.O., De Taddéo-Borg M., Kroon D.E., Turner R.S., Watson S.J., Margolis B. Molecular analysis of the X11-mLin-2/CASK complex in brain. J. Neurosci. 1999;19:1307–1316. doi: 10.1523/JNEUROSCI.19-04-01307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biederer T., Cao X., Sudhof T.C., Liu X. Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms. J. Neurosci. 2002;22:7340–7351. doi: 10.1523/JNEUROSCI.22-17-07340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sano Y., Ornthanalai V.G., Yamada K., Homma C., Suzuki H., Suzuki T., Murphy N.P., Itohara S. X11-like protein deficiency is associated with impaired conflict resolution in mice. J. Neurosci. 2009;29:5884–5896. doi: 10.1523/JNEUROSCI.5756-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobsen J.S., Wu C.C., Redwine J.M., Comery T.A., Arias R., Bowlby M., Martone R., Morrison J.H., Pangalos M.N., Reinhart P.H., et al. Early-onset behavioral and synaptic deficits in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2006;103:5161–5166. doi: 10.1073/pnas.0600948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cleary J.P., Walsh D.M., Hofmeister J.J., Shankar G.M., Kuskowski M.A., Selkoe D.J., Ashe K.H. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 50.Janus C., Pearson J., McLaurin J., Mathews P.M., Jiang Y., Schmidt S.D., Chishti M.A., Horne P., Heslin D., French J., et al. Ab peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer's disease. Nature. 2000;408:979–982. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- 51.Morgan D., Diamond D.M., Gottschall P.E., Ugen K.E., Dickey C., Hardy J., Duff K., Jantzen P., DiCarlo G., Wilcock D., et al. Ab peptide vaccination prevents memory loss in an animal model of Alzheimer's disease. Nature. 2000;408:982–985. doi: 10.1038/35050116. [DOI] [PubMed] [Google Scholar]
- 52.Lee E.B., Leng L.Z., Zhang B., Kwong L., Trojanowski J.Q., Abel T., Lee V.M. Targeting amyloid-beta peptide (Abeta) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Abeta precursor protein (APP) transgenic mice. J. Biol. Chem. 2006;281:4292–4299. doi: 10.1074/jbc.M511018200. [DOI] [PubMed] [Google Scholar]
- 53.Steiner H., Fluhrer R., Haass C. Intramembrane proteolysis by gamma-secretase. J. Biol. Chem. 2008;283:29627–29631. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole S.L., Vassar R. The role of amyloid precursor protein processing by BACE1, the beta-secretase, in Alzheimer disease pathophysiology. J. Biol. Chem. 2008;283:29621–29625. doi: 10.1074/jbc.R800015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King G.D., Cherian K., Turner R.S. X11alpha impairs gamma- but not beta-cleavage of amyloid precursor protein. J. Neurochem. 2004;88:971–982. doi: 10.1046/j.1471-4159.2003.02234.x. [DOI] [PubMed] [Google Scholar]
- 56.Sano Y., Syuzo-Takabatake A., Nakaya T., Saito Y., Tomita S., Itohara S., Suzuki T. Enhanced amyloidogenic metabolism of APP in the X11L-deficient mouse brain. J. Biol. Chem. 2006;281:37853–37860. doi: 10.1074/jbc.M609312200. [DOI] [PubMed] [Google Scholar]
- 57.Sakurai T., Kaneko K., Okuno M., Wada K., Kashiyama T., Shimizu H., Akagi T., Hashikawa T., Nukina N. Membrane microdomain switching: a regulatory mechanism of amyloid precursor protein processing. J. Cell Biol. 2008;183:339–352. doi: 10.1083/jcb.200804075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Araki Y., Kawano T., Taru H., Saito Y., Wada S., Miyamoto K., Kobayashi H., Ishikawa H.O., Ohsugi Y., Yamamoto T., et al. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konecna A., Frischknecht R., Kinter J., Ludwig A., Steuble M., Meskenaite V., Indermuhle M., Engel M., Cen C., Mateos J.M., et al. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol. Biol. Cell. 2006;17:3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ludwig A., Blume J., Diep T.M., Yuan J., Mateos J.M., Leuthauser K., Steuble M., Streit P., Sonderegger P. Calsyntenins mediate TGN exit of APP in a Kinesin-1-dependent manner. Traffic. 2009;10:572–589. doi: 10.1111/j.1600-0854.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 61.McLoughlin D.M., Standen C.L., Lau K.-F., Ackerley S., Bartnikas T.P., Gitlin J.D., Miller C.C.J. The neuronal adaptor protein X11a interacts with the copper chaperone for SOD1 and regulates SOD1 activity. J. Biol. Chem. 2001;276:9303–9307. doi: 10.1074/jbc.M010023200. [DOI] [PubMed] [Google Scholar]
- 62.Angeletti B., Waldron K.J., Freeman K.B., Bawagan H., Hussain I., Miller C.C., Lau K.F., Tennant M.E., Dennison C., Robinson N.J., et al. BACE1 cytoplasmic domain interacts with the copper chaperone for superoxide dismutase-1 and binds copper. J. Biol. Chem. 2005;280:17930–17937. doi: 10.1074/jbc.M412034200. [DOI] [PubMed] [Google Scholar]
- 63.Billings L.M., Oddo S., Green K.N., McGaugh J.L., LaFerla F.M. Intraneuronal Abeta causes the onset of early Alzheimer's disease-related cognitive deficits in transgenic mice. Neuron. 2005;45:675–688. doi: 10.1016/j.neuron.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 64.Cheng I.H., Scearce-Levie K., Legleiter J., Palop J.J., Gerstein H., Bien-Ly N., Puolivali J., Lesne S., Ashe K.H., Muchowski P.J., et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 65.Hooper C., Markevich V., Plattner F., Killick R., Schofield E., Engel T., Hernandez F., Anderton B., Rosenblum K., Bliss T., et al. Glycogen synthase kinase-3 inhibition is integral to long-term potentiation. Eur. J. Neurosci. 2007;25:81–86. doi: 10.1111/j.1460-9568.2006.05245.x. [DOI] [PubMed] [Google Scholar]