Abstract
IL-13 is a central effector of Th2-mediated allergic inflammation and is critical for the induction of IgE synthesis. Common IL13 variants are associated with allergy phenotypes in populations of distinct ethnic background. We recently demonstrated that IL13 expression by human CD4+ T cells is paralleled by extensive IL13 locus remodeling, which results in the appearance of multiple DNase I hypersensitive sites. Among these, HS4 in the distal promoter is constitutive in both naïve and polarized Th1 and Th2 cells, and spans a common single nucleotide polymorphism, IL13-1512A>C (rs1881457), strongly associated with total serum IgE levels. We recently characterized HS4 as a novel cis-acting element that upregulates IL13 transcription in activated human and murine T cells. Here we show that IL13-1512A>C is a functional polymorphism that significantly enhances HS4-dependent IL13 expression by creating a binding site for the transcription factor Oct-1. Of note, endogenous Oct-1 was preferentially recruited to the IL13-1512C risk allele in primary CD4+ T cells from IL13-1512A>C heterozygous subjects. Moreover, the IL13-1512C allele was overexpressed in transfected Th2 cells from Oct1+/+ mice, but not from Oct1+/− mice, demonstrating that increased activity was exquisitely dependent on physiologic levels of Oct-1. Our results illustrate how a functional variant in a regulatory element enhances transcription of an allergy-associated gene, thereby modulating disease susceptibility.
INTRODUCTION
The cytokine IL-13 is a central effector of Th2 responses and is necessary and sufficient to induce all the cardinal features of allergic lung inflammation and experimental asthma. IL-13 promotes allergen-induced airway hyperresponsiveness, epithelial cell damage, goblet cell metaplasia with mucus hyperproduction and eosinophilia (1,2). Furthermore, IL-13 shares with another cytokine, IL-4, a unique capacity to stimulate human B lymphocytes to synthesize IgE (3), which further amplifies allergen-specific inflammatory responses. Long-term lung inflammation is sustained by the IL-13-dependent activation of resident lung cells, which release chemokines, cytokines (4) and other effector molecules (5) and are involved in airway fibrosis (6,7). Of note, IL-13 is readily detectable in the human placenta (8) and is vigorously secreted by neonatal T cells (9), in line with the notion that early life immunoregulatory events play a critical role in the inception of asthma and allergy (10).
Because of its critical role as an effector of Th2 immunity, IL13 is a strong candidate gene for allergy and asthma. Indeed, the associations between allergy-related phenotypes (such as IgE levels, asthma and atopy) and individual IL13 variants (particularly IL13-1112C>T, rs1800925 and IL13+2044G>A, rs20541) are robust and widely replicated (reviewed in 11). However, dissection of IL13 genotype/phenotype associations and identification of causative variants require functional studies, because the IL13 locus harbors numerous common single nucleotide polymorphisms (SNPs) and complex patterns of linkage disequilibrium (LD). We recently showed that IL13-1112C>T in the promoter increases IL13 transcription in transfected murine and human Th2 cells by creating a YY1 binding site and relieving STAT6-mediated repression (12). IL13+2044G>A, a non-synonymous SNP in the fourth exon in modest LD with IL13-1112C>T, leads to the expression of an IL-13 variant with increased biological activity (13).
Studies in mouse models have shown that expression of IL13 and the other Th2 cytokine genes, IL5 and IL4, is orchestrated by multiple conserved and developmentally regulated nuclease hypersensitive (HS) sites that are scattered throughout the 160 kb Th2 locus (14) and include gene-specific enhancers as well as a locus control region (15). HS sites typically map to cis-regulatory elements and reflect the DNA binding activity of sequence-specific trans-acting factors that induce destabilization or displacement of local nucleosomes (16). We extensively analyzed chromatin remodeling at the human IL13 locus in naïve and differentiating CD4+ Th cells (17). Among the novel HS sites we identified was HS4, which resides in the distal IL13 promoter and co-localizes with CpG hypomethylation in naïve and in vitro polarized Th1 and Th2 cells. Early and persistent accessibility at HS4 suggests that occupancy by constitutive transcription factors may poise the promoter for rapid IL13 expression upon T cell activation and/or differentiation.
We recently showed that HS4 marks the location of a cis-regulatory region which is highly conserved in primates and enhances IL13 promoter activity in transfected human Jurkat T cells and murine Th2 cells (P. Kiesler et al., submitted). Interestingly, HS4 harbors a common SNP, IL13-1512A>C (rs1881457, minor allele frequency >20%), which is strongly associated with total serum IgE levels in unselected populations from the United States and Germany (18). Moreover, an analysis of the association between several IL13 SNPs and total IgE levels in the British 1958 Birth Cohort showed IL13-1512A>C accounted for 85% of the phenotypic variance attributable to IL13 polymorphisms (19). An association between IL13-1512A>C and IgE levels was also found in atopic asthmatic children from Korea (20). These strong genetic associations, and the localization of IL13-1512A>C within a cis-acting element, prompted us to investigate whether IL13-1512A>C alters the IL13 regulatory properties of HS4.
RESULTS
IL13-1512A>C is a functional polymorphism that enhances HS4 activity
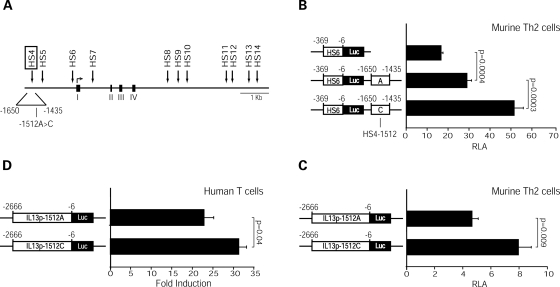
We recently provided evidence that the IL13 promoter region marked by HS4 (-1650 to -1435: numbering is relative to the IL13 ATG) acts as a position-independent, positive regulator of IL13 promoter activity in transfected human Jurkat T cells and primary, in vitro differentiated murine Th2 cells (P. Kiesler et al., submitted). IL13-1512A>C lies in the central portion of HS4 (Fig. 1A). To directly test the impact of IL13-1512A>C on the regulatory properties of this cis-acting element, we took a reductionist approach that relied on two sets of reporter vectors: one in which HS4 was linked to the IL13 proximal promoter (HS6/Luc), and one in which HS4 resided at its genomic location within the distal IL13 promoter. In order to characterize HS4 function in a significant nuclear environment, we compared the ability of the HS4 variants to upregulate the IL13 proximal promoter in transiently transfected, in vitro differentiated primary murine CD4+ Th2 cells. These cells are programmed for high-rate IL13 expression in vivo and are endowed with a transcriptional machinery ideal for investigating allele-specific differences in human IL13 expression (12). Figure 1B shows that both HS4-1512A and HS4-1512C significantly enhanced IL13 promoter activity. However, the HS4 variant carrying the -1512C risk allele was significantly (P = 0.0003) more active than the A allele. Moreover, a full length (2.7 kb) IL13 promoter construct carrying −1512C was significantly more active than the A variant in both murine Th2 cells (Fig. 1C) and human Jurkat T cells (Fig. 1D). Collectively, these results demonstrate that IL13-1512A>C is a functional polymorphism that enhances HS4 activity and HS4-driven IL13 expression.
Figure 1.
IL13-1512C enhances HS4-dependent IL13 expression. (A) Schematic representation of DNase I HS sites (arrows) within the human IL13 locus. Exons I–IV are marked by black boxes. The location of IL13-1512A>C within HS4 is depicted. (B and C) Actively proliferating murine Th2 cells polarized in vitro for 9–10 days were nucleofected with HS4/Luc constructs (B) or -2666IL13p/Luc constructs (C) carrying an A or a C at position -1512. Cells were harvested 16 h post-transfection. Results are expressed as RLA (mean±SE) measured in 10 (B) or 6 (C) independent experiments. (D) Jurkat T cells were transiently transfected with -2666IL13p/Luc constructs carrying an A or a C at position -1512 and harvested after 16 h of culture in the presence or absence of PMA (20 ng/ml) and ionomycin (1 µM). Results are expressed as the mean±SE of fold induction values, i.e. the ratio between RLA in stimulated and unstimulated cells measured in six independent experiments. Statistical significance was calculated using the Wilcoxon two-sample test.
The IL13-1512C allele selectively binds Oct-1 in vitro
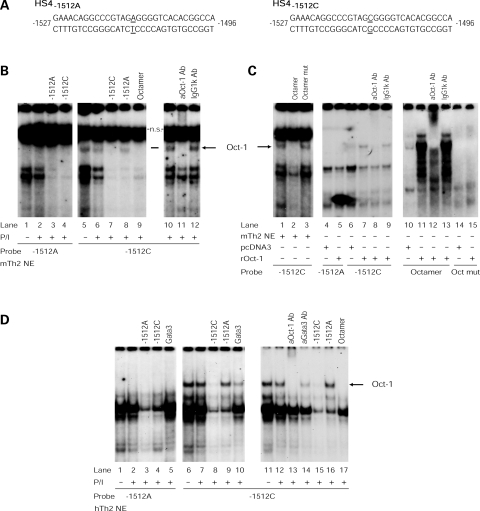
In order to start characterizing the molecular mechanisms underlying the differential activity of the IL13-1512 alleles in CD4+ T cells, we searched for allele-specific patterns of human IL13 promoter/nucleoprotein interactions. Figure 2B shows that EMSA analysis with -1512 allele-specific probes and competitors (Fig. 2A) and murine Th2 cell nuclear extracts identified a specific complex, which constitutively bound HS4-1512C (lanes 5–6) but not HS4-1512A (lanes 1–2) and was cross-competed by -1512C (lane 7) but not -1512A (lane 8) unlabeled oligonucleotides. This complex contained Oct-1, because it was specifically competed by an oligonucleotide carrying a canonical octamer motif (21) (Fig. 2C, lane 2), but not by an oligonucleotide in which the motif had been disrupted (22) (lane 3). IL13-1512C allele-specific binding of Oct-1 was further confirmed by supershifting experiments with Oct-1-specific and control antibodies (Fig. 2B, lanes 11–12), as well as by using in vitro translated recombinant Oct-1 (Fig. 2C, lane 7). The ability of recombinant Oct-1 to interact with a -1512C and a bona fide octamer probe (lane 11) also suggests that Oct-1 may form a complex directly with DNA. Of note, EMSA analysis with nuclear extracts from primary, in vitro differentiated human Th2 cells (Fig. 2D) or human Jurkat T cells (data not shown) also showed that the -1512C (lanes 6–7), but not the -1512A (lanes 1–2), allele bound Oct-1. Collectively, these analyses show that the replacement of an A with a C at position -1512 of the distal IL13 promoter creates an Oct-1 binding motif.
Figure 2.
HS4-1512C selectively binds Oct-1. Radiolabeled HS4 probes carrying an A or a C at position –1512 (A) were incubated with nuclear extracts from in vitro polarized murine Th2 cells (mTH2 NE) (B and C) or human Th2 cells (hTh2 NE) (D) cultured for 3 h in the presence or absence of PMA (20 ng/ml) and ionomycin (1 µM) (P/I) or in vitro translated recombinant Oct-1 (rOct-1) (C). EMSA probes and experimental conditions are noted below the gels. Competitors (100-fold molar excess) and supershifting antibodies added to each reaction are noted above the relevant lanes. A Gata3 consensus oligonucleotide and IgG1κ or an anti-Gata3 antibody were used as unrelated controls. Ns: non-specific.
Oct-1 is preferentially recruited to the IL13-1512C allele in vivo
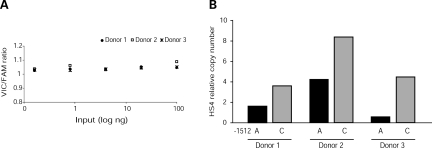
To validate the preferential interaction between Oct-1 and the IL13-1512C allele, allele-specific ChIP was used to test the ability of Oct-1 to bind the endogenous HS4 variants in vivo. Cross-linked chromatin harvested from primary, activated CD4+ T cells from IL13-1512A>C heterozygotes was immunoprecipitated with anti-Oct-1 or IgG isotype control antibodies. Real-time PCR with a custom TaqMan SNP genotyping assay, which was designed to amplify a 68 bp region of the human IL13 distal promoter spanning IL13-1512A>C (nucleotides -1547 to -1479), tested for enrichment of the -1512A or -1512C alleles using two specific reporter probes: VIC recognized the A allele, whereas FAM recognized the C allele. Results were expressed as HS4 relative copy number, i.e. the ratio between HS4 copies immunoprecipitated by anti-Oct-1 and IgG isotype control. Figure 3A shows that this approach resulted in accurate quantification of both HS4 alleles, because the VIC (-1512A)/FAM (-1512C) signal ratio was consistently equal or close to 1 over several orders of magnitude of input DNA (Fig. 3A). Figure 3B shows that endogenous Oct-1 was preferentially recruited to the IL13-1512C promoter allele in three independent experiments, each relying on chromatin from a different IL13-1512A>C heterozygous donor. These results support a critical role of Oct-1 in the differential activity of the IL13-1512 alleles.
Figure 3.
Allele-specific recruitment of Oct-1 to HS4. Chromatin from primary human CD4+ T cells heterozygous at IL13-1512A>C and activated with PMA (20 ng/ml) and ionomycin (2 µM) for 3 h was crosslinked, sonicated and immunoprecipitated with an anti-Oct-1 antibody or an IgG isotype control antibody. Target enrichment was assessed by allele-specific, real time PCR using a custom TaqMan SNP genotyping assay that amplified a 68 bp region spanning IL13-1512A>C (nucleotides -1547 to -1479) and discriminated between the A (VIC) and the C (FAM) alleles with reporter probes. (A) VIC:FAM signal ratios obtained from standard curves for each allele generated with serial dilutions of sonicated genomic DNA (input) from each experiment. (B) Ratio between number of HS4 targets immunoprecipitated with anti-Oct-1 Ab and control IgG Ab in three independent experiments, each with chromatin from a different donor.
The increased activity of HS4-1512C requires physiologic levels of Oct-1 in the nuclear environment
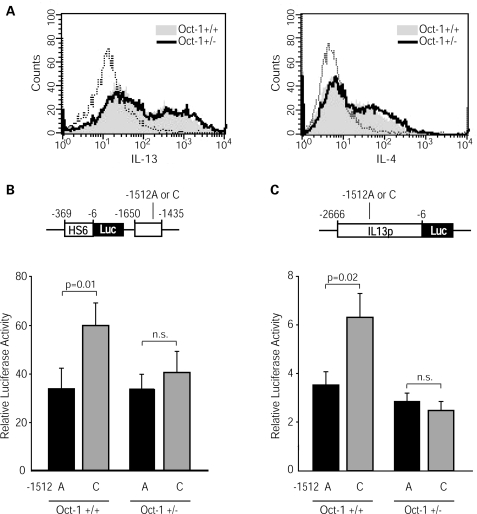
Because Oct-1 binds selectively the -1512C allele, and this allele exhibits higher activity, we hypothesized that Oct-1 is required for enhanced HS4 activity. To test our hypothesis, we turned to genetically modified mice in which the nuclear environment contains different levels of Oct-1. Oct1 deletion by gene targeting is embryonically lethal because of defective erythropoiesis, but heterozygous Oct1+/− mice are fully viable (21). We compared the activities of HS4-1512A and HS4-1512C reporter constructs following transfection into Th2 cells from Oct1+/+ and Oct1+/− C57BL/6 mice. Figure 4A shows that endogenous Th2 cell differentiation, as revealed by intracellular IL-4 and IL-13 protein expression, was fully preserved in Oct1+/− cells. Of note, HS4-1512C was significantly (P = 0.01) more active than HS4-1512A in upregulating the human IL13 promoter in Oct1+/+ Th2 cells. In contrast, the partial decrease in Oct-1 binding activity caused by heterozygosity for the Oct1 null mutation (≈60% as assessed by EMSA, data not shown) was sufficient to abrogate the increased activity of the -1512C allele (Fig. 4B). Comparable results were obtained by transfecting Oct1+/+ and Oct1+/− Th2 cells with -1512 allelic variants of the full-length IL13 promoter construct (Fig. 4C). These results show that the enhanced activity of HS4-1512C was exquisitely dependent on physiologic levels of Oct-1.
Figure 4.
The increased activity of HS4-1512C requires physiologic levels of Oct-1 in the nuclear environment. (A) Th2 cells from C57BL/6 wild-type or Oct1+/− mice were generated by in vitro polarization for 9–10 days, and analyzed by immunofluorescence for intracellular Th2 cytokine expression. The dotted lines correspond to the negative control. Th2 cells were nucleofected with HS4/Luc constructs (B) or -2666IL13p/Luc constructs (C) carrying an A or a C at position -1512. Cells were harvested 16 h after transfection. Results are expressed as RLA (mean±SE) measured in 8 (B) or 11 (C) independent experiments. Statistical significance was calculated using the Wilcoxon two-sample test.
DISCUSSION
Extensive studies in mouse models have shown that the expression of Th2 cytokines, including IL13, is controlled by an array of conserved DNase I HS regulatory elements that are positioned throughout the Th2 locus (reviewed in 23). Few of these elements have been functionally characterized in mice (24–30) and virtually none in humans. Our current analysis focused on HS4, an element located in the distal human IL13 promoter which is detectable in naïve CD4+ T cells and persists throughout T cell differentiation. Our group has shown that HS4 is a functional regulatory element that strongly upregulates IL13 transcription in response to T cell activation cues (P. Kiesler et al., submitted).
The IL13-1512C allele creates an Oct-1 binding site that is essential for the enhanced activity of the -1512C HS4 variant. Although the site contains a non-canonical octamer motif, its ability to specifically bind Oct-1 was supported by functional studies in murine Th2 cells and by DNA/protein interaction analysis in vitro and in vivo. To our knowledge, this is the first time that allele-specific binding of a transcription factor to a polymorphic site in the human IL13 locus is demonstrated in vivo and shown to have a significant transcriptional impact. Oct-1 belongs to a family of transcription factors characterized by the POU domain, whose peculiar bipartite structure confers to these proteins great flexibility in their interactions with DNA and other transcriptional regulators (31). Because of this functional versatility, the role played by Oct proteins in the regulation of individual genes varies with the sequence of the octamer motif, the promoter context and the identity of the regulators that engage in interactions with the POU domain. In T lymphocytes, Oct-1, a ubiquitously expressed protein, has been shown to activate tissue-specific gene transcription by cooperatively interacting with lineage-selective factors, particularly NFAT family proteins, within DNase I HS regions (32–34). Interestingly, HS4 contains several putative NFAT binding sites, one of which lies immediately upstream of IL13-1512A>C and may provide a platform for productive NFAT/Oct-1 interactions. Further studies are in progress to characterize the molecular events responsible for Oct-1-dependent IL13 upregulation. It is also noteworthy that Oct-1 had not been previously linked to IL13 regulation (21), and our experiments showed endogenous IL13 expression was fully preserved in Th2 cells from Oct1+/− mice. The low degree of conservation of the human distal IL13 promoter, and the absence of IL13-1512A>C from the mouse IL13 promoter, may explain the latter finding. Our results illustrate how creation of a transcription factor binding site within an important cis-element may redirect the regulatory program, thereby offering evolutionarily novel opportunities for modulating gene expression and disease susceptibility.
To what extent do the gain-of-function properties of IL13-1512A>C explain the previously reported (18–20) associations between this polymorphism and allergy-related phenotypes, particularly elevated IgE levels? The answer to this question depends on both LD patterns and magnitude of the variant's functional effects. In Caucasians, IL13-1512A>C is in strong, albeit not perfect, LD with IL13-1112C>T (12), which also resides within the distal IL13 promoter. Indeed, our LD analysis, which focused on a population of non-Hispanic white children from Tucson, AZ (n = 558), and relied on a widely used algorithm (Haploview: ref. 35), yielded an r2 value of 0.804 for these polymorphisms (D. Stern and M. Halonen, unpublished), and a comparable LD estimate (r2 = 0.71) was recently reported for the ISAAC cohort (n = 526). Because of strong LD, the phenotypic effects of IL13-1512A>C and IL13-1112C>T are not readily separable at the population level, at least in Caucasians. LD estimates for large populations of other ethnicities are currently unavailable. In terms of transcriptional effects, both IL13-1512A>C and IL13-1112C>T are gain-of-function polymorphisms. However, the increases in IL13 transcription dependent on the -1512C or the -1112T allele (12) were statistically significant but admittedly modest, a functional outcome which is quite typical of common regulatory variants (36–38). Therefore, taken in isolation, neither IL13-1512A>C nor IL13-1112C>T are likely to dysregulate IL13 expression intensely enough to cause an allergic disease phenotype. On the other hand, both IL13-1512A>C and IL13-1112C>T lie within an LD block that extends ≈87 kb upstream of the IL13 promoter, reaching beyond the Th2 locus control region located in RAD50 (39). Remarkably, several of the SNPs in this block fall within highly conserved HS sites that have been shown to be critical for the coordinated regulation of Th2 cytokine gene expression in mice (15). It is therefore tempting to speculate that, by acting in concert, linked variants of individually modest effects may profoundly alter Th2 cytokine expression, thereby modifying susceptibility to Th2-mediated allergic inflammation.
MATERIALS AND METHODS
Mice
Oct1+/− mice on a C57BL/6 background (21) and C57BL/6 control mice (The Jackson Laboratory) were maintained under specific pathogen-free conditions. All experiments were performed according to institutional and federal guidelines.
T-cell isolation and Th2 differentiation
To generate murine Th2 cells, CD4+ T lymphocytes were isolated from splenocyte suspensions using the CD4+ T cell isolation kit (Miltenyi) as recommended by the manufacturer. Cells (∼5 × 106) were resuspended in DMEM supplemented with FCS (10%), HEPES (10 mm), 2-β-ME (0.1 mm), penicillin (100 U/ml), streptomycin (100 µg/ml) and l-glutamine (2 mm), and stimulated for 3 days with plate-bound anti-CD3 mAb (clone 145-2C11, 1 µg/ml) and anti-CD28 mAb (clone 37.51, 1 µg/ml) in the presence of IL-4 (1000 U/ml) and neutralizing antibodies against IFN-γ (clone R4-6A2, 5 µg/ml) and IL-12 (clone C17.8, 3 µg/ml) (40). After expansion in the presence of IL-4, neutralizing antibodies and IL-2 (20 U/ml) for 4 days, cells were re-stimulated with plate-bound anti-CD3 and anti-CD28 mAb, and nucleofected at day 9–10. Th2 cell polarization was assessed by intracellular cytokine staining (17).
Human primary CD4+ T cells were isolated from peripheral blood samples using the RosetteSep human T cell enrichment cocktail (Stemcell technologies) following the manufacturer's directions and resuspended in RPMI 1600 supplemented with FBS (10%), penicillin (100 U/ml), streptomycin (100 µg/ml) and l-glutamine (2 mm). In vitro polarized human Th2 cells were generated as previously described (12). Briefly, naïve cord blood CD45RO−CD4+ T cells were obtained by negative selection and depletion of memory T cells (Miltenyi). Cells were stimulated for 4 days with plate-bound anti-CD3 mAb (UCHT1, 2 µg/ml) and soluble anti-CD28 mAb (1 µg/ml) in the presence of IL-2 (5 ng/m), IL-4 (4 ng/ml) and neutralizing antibodies to IL-12 (2 µg/ml) and IFN-γ (2 µg/ml) and then rested for 3 days in IL-2, IL-4 and anti-IL-12. Subsequently, cells were restimulated for 4 days on anti-CD3-coated plates with IL-4 and anti-IL-12 mAb. Experiments were performed on day 16. Th2 cell polarization was assessed by intracellular cytokine staining (17).
Jurkat T cells (ATCC clone E6-1) were cultured in RPMI 1640 supplemented with fetal bovine serum (10%), penicillin (100 units/ml), streptomycin (100 µg/ml) and l-glutamine (2 mm). These cells exhibit DNase I hypersensitivity at HS4 (data not shown) and vigorously up-regulate IL13 mRNA upon activation (12).
Reporter constructs
2666IL13p/Luc, a luciferase reporter construct driven by the major IL13-1512A promoter variant (12), was used as a template to generate -2666IL13p-1512C/Luc through QuickChange site-directed mutagenesis (Stratagene). HS6/Luc consisted of a 363 bp region encompassing the human IL13 proximal promoter (−369 to −6) cloned into pGL3 Basic (Promega) (41). PCR amplification of HS4 using -2666IL13p/Luc or -2666IL13p-1512C/Luc as templates and primers HS4-1650 (5′-ATACTCGTCGACATAAGGGGCGTTGACTCAC) and HS4-1435 (5′-TTGATGTCGACTCTGACTCCCAGAAGTCTG) and cloning of these amplicons into the SalI restriction site of HS6/Luc in the forward orientation generated HS4-1512A/Luc and HS4-1512C/Luc, respectively. All constructs were verified by sequencing.
Transient transfections
Anti-CD3/anti-CD28 mAb-activated murine Th2 cells (1 × 106) were nucleofected with equimolar amounts of endotoxin-free reporter constructs (1 µg) and pRL-TK (50 ng: Promega) using the Amaxa nucleofector and the mouse T-cell nucleofector kit (Amaxa). Firefly and renilla luciferase activities in cell lysates were assessed 16 h after nucleofection using the Dual Luciferase Assay System (Promega). Total protein concentrations in cell lysates were determined using the BCA assay (Pierce). Results were expressed as RLA (relative luciferase activity) units, i.e. firefly luciferase activity corrected for renilla luciferase (transfection efficiency) and protein content of each sample.
Endotoxin-free plasmids (5 µg) were electroporated into Jurkat T cells (5 × 106) in log phase of growth using the BTX ECM830 square wave electroporator (1 pulse, 250 V, 50 ms). Cells were co-transfected with pRLTK (25 ng, Promega) to normalize for transfection efficiency. After electroporation, cells were incubated in the presence or absence of PMA, phorbol 12-myristate 13-acetate, (20 ng/ml, Sigma) and ionomycin (1 µm, Sigma). Results were expressed as fold induction values, i.e. the ratio between RLA in stimulated and unstimulated cells.
EMSA
Murine Th2 cells, human Th2 cells or Jurkat T cells (15 × 106) were incubated in the presence or absence of PMA (20 ng/ml) and ionomycin (1 µm) for 3 h. After stimulation, cells were collected by centrifugation and washed twice with PBS. Cell pellets were resuspended (1:5 v/v) in buffer A (3 mm MgCl2, 10 mm NaCl, 10 mm Tris pH 7.5, 0.1 mm EGTA, 0.5 mm DTT, 1 µg/ml aprotinin, 2 µm leupeptin, 1 mm PMSF) and incubated on ice for 10 min. NP40 (0.5%) was added and nuclei were pelleted by centrifugation. Nuclear pellets were resuspended (1:2.5 v/v) in buffer C (1.5 mm MgCl2, 20 mm HEPES, pH 7.0, 420 mm NaCl, 25% glycerol, 0.2 mm EDTA, 0.5 mm DTT, 1 µg/ml aprotinin, 2 µm leupeptin, 0.5 mm PMSF) and incubated on ice for 30 min. After centrifugation, supernatants were collected, snap-frozen in liquid nitrogen and stored at −80°C. A pcDNA3.1-Oct-1 expression vector or empty pcDNA3.1 control (Invitrogen) were in vitro translated using the TNT Quick Coupled Transcription/Translation System (Promega) following the manufacturer's directions.
Nuclear extracts (5 µg) or in vitro translated Oct-1 were incubated with 32P-labeled HS4-1512A or HS4-1512C probes in binding buffer (10 mm Tris-Cl, 1 mm EDTA, 1 mm DTT, 0.5 mm MgCl2, 100 mm NaCl, 10% glycerol) and poly dI-dC (50 ng/µl) for 30 min at 4°C. Oligonucleotide competitors (Octamer: 5′-GGATGCAAATATGCAAATATGCAAATGG (21) and Octamer mut: 5′-GGcgGCAAATcgGCAAATcgGCAAATGG (22), 100-fold molar excess) or antibodies (anti-Oct-1, C-21X, anti-Gata3, B-10X, from Santa Cruz Biotechnologies, or IgG1κ, PMP48, from Serotec: 2 µg) were pre-incubated with nuclear extracts for 30 min at 4°C before addition of radiolabeled probe. DNA/protein complexes were separated on 5% (w/v) non-denaturing polyacrylamide gels in TBE (0.5×) for 5 h at 4°C.
Allele-specific ChIP
Freshly isolated peripheral blood CD4+ T cells (∼7 × 106/condition) from IL13-1512A>C heterozygotes were stimulated in the presence of PMA (20 ng/ml) and ionomycin (2 µm) for 3 h. Chromatin immunoprecitation experiments were performed using the ChIP assay kit (Millipore) following the manufacturer's directions with few modifications. Specifically, paraformaldehyde fixation was stopped by the addition of 125 mm glycine (RT, 5 min). Crosslinked chromatin was sonicated six times for 10 s each time with a Microson XL200 sonicator (Misonix). An aliquot of chromatin (1/20th) was set aside and used as input DNA. After preclearing, chromatin samples were immunoprecipitated with a rabbit anti-Oct-1 antibody (20 µg, sc-232X, Santa Cruz Biotechnology) or a rabbit IgG isotype control antibody (20 µg, 12-370, Millipore).
Allele-specific enrichment of the polymorphic HS4 region in immunoprecipitated samples was quantified using a custom TaqMan SNP genotyping assay (Applied Biosystems) on an ABI Prism 7900 Sequence Detection System. PCR primers (5′-GGCCCTCTACTACAGATTAGGAAAC and 3′-CCTGGAGTGCCGCTACTTG) were designed to amplify a 68 bp region of the human IL13 distal promoter that spans IL13-1512A>C (nucleotides -1547 to -1479). Two TaqMan probes discriminated between the two alleles. The -1512A allele was recognized by the reporter dye VIC (5′-CGTGTGACCCCTCTAC), whereas the -1512C allele was recognized by the reporter dye FAM (5′-CGTGTGACCCCGCTAC). Real-time PCR amplifications were performed in triplicate with the following cycling conditions: 10 min at 95°C followed by 40 cycles of 15 s at 92°C and 30 s at 60°C. Serial dilutions of input DNA were used to generate standard curves for each allele in each experiment and PCR efficiency was determined with the formula: E = 10(−1/k)− 1, where k represents the slope of each standard curve. Efficiency values were essentially equal to 1 and closely comparable between reporter dyes. Moreover, as expected for heterozygous genomic DNA, VIC:FAM signal ratios were essentially 1:1 and remained constant across several orders of magnitude of input DNA. Copy number values were calculated with the formula: N = (1 + E)[40 −Ct(sample)], where E represents the PCR efficiency. For each allele, results were expressed as relative copy numbers, that is, as the ratio between the numbers of HS4 targets immunoprecipitated with anti-Oct-1 and the numbers of HS4 targets immunoprecipitated with control IgG.
Human subjects
Cord blood was obtained from anonymous cesarean deliveries. Peripheral blood was drawn from healthy non-allergic subjects of known IL13-1512A>C genotype after written informed consent was obtained. All protocols were approved by the Institutional Review Board of the University of Arizona.
FUNDING
This work was supported by NIH grant RO1 HL66391 (to D.V.).
ACKNOWLEDGEMENTS
We thank our colleagues Debra A. Stern and Marilyn Halonen for linkage disequilibrium analyses in the Tucson population, and Robin B. Webster for providing in vitro differentiated human Th2 cells.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wills-Karp M., Luyimbazi J., Xu X., Schofield B., Neben T.Y., Karp C.L., Donaldson D.D. Interleukin-13: Central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 2.Grünig G., Warnock M., Wakil A.E., Venkayya R., Brombacher F., Rennick D.M., Sheppard D., Mohrs M., Donaldson D.D., Locksley R.M., et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punnonen J., Aversa G., Cocks B.J., Mckenzie A.N.J., Menon S., Zurawski G., de Waal Malefyt R., de Vries J.E. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc. Natl Acad. Sci. USA. 1993;90:3730–3734. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J.H., Kaminski N., Dolganov G., Grunig G., Koth L., Solomon C., Erle D.J., Sheppard D. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am. J. Respir. Cell. Mol. Biol. 2001;25:474–485. doi: 10.1165/ajrcmb.25.4.4522. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Z., Zheng T., Homer R.J., Kim Y.-K., Chen N.Y., Cohn L., Hamid Q., Elias J.A. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 6.Lee C., Homer R., Zhu Z., Lanone S., Wang X., Koteliansky V., Shipley J., Gotwals P., Noble P., Chen Q., et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor β-1. J. Exp. Med. 2001;194:809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanone S., Zheng T., Zhu Z., Liu W., Lee C.G., Ma B., Chen Q., Homer R.J., Wang J., Rabach L.A., et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13-induced inflammation and remodeling. J. Clin. Invest. 2002;110:463–474. doi: 10.1172/JCI14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams T.J., Jones C.A., Miles E.A., Warner J.O., Warner J.A. Fetal and neonatal IL-13 production during pregnancy and at birth and subsequent development of atopic symptoms. J. Allergy Clin. Immunol. 2000;105:951–959. doi: 10.1067/mai.2000.106211. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro-do-Couto L.M., Boeije L.C., Kroon J.S., Hooibrink B., Breur-Vriesendorp B.S., Aarden L.A., Boog C.J. High IL-13 production by human neonatal T cells: neonate immune system regulator? Eur. J. Immunol. 2001;31:3394–3402. doi: 10.1002/1521-4141(200111)31:11<3394::aid-immu3394>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 10.Martinez F.D. Toward asthma prevention–does all that really matters happen before we learn to read? N. Engl. J. Med. 2003;349:1473–1475. doi: 10.1056/NEJMe030041. [DOI] [PubMed] [Google Scholar]
- 11.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat. Rev. Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 12.Cameron L., Webster R.B., Strempel J.M., Kiesler P., Kabesch M., Ramachandran H., Yu L., Stern D.A., Graves P.E., Lohman I.C., et al. Th2-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J. Immunol. 2006;177:8633–8642. doi: 10.4049/jimmunol.177.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vladich F.D., Brazille S.M., Stern D., Peck M.L., Ghittoni R., Vercelli D. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J. Clin. Invest. 2005;115:747–754. doi: 10.1172/JCI22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loots G.G., Locksley R.M., Blankenspoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13 and 5 by cross-species sequence comparison. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 15.Wilson C.B., Rowell E., Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 16.Boyes J., Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 17.Webster R.B., Rodriguez Y., Klimecki W.T., Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J. Biol. Chem. 2007;282:700–709. doi: 10.1074/jbc.M609501200. [DOI] [PubMed] [Google Scholar]
- 18.Graves P.E., Kabesch M., Halonen M., Holberg C.J., Baldini M., Fritzsch C., Weiland S., Erickson R.P., von Mutius E., Martinez F.D. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J. Allergy Clin. Immunol. 2000;105:506–513. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 19.Maier L.M., Howson J.M.M., Walker N., Spickett G.P., Jones R.W., Ring S.M., McArdle W.L., Lowe C.E., Bailey R., Payne F., et al. Association of IL13 with total IgE: Evidence against an inverse association of atopy and diabetes. J. Allergy Clin. Immunol. 2006;117:1306–1313. doi: 10.1016/j.jaci.2005.12.1354. [DOI] [PubMed] [Google Scholar]
- 20.Kim H.B., Lee Y.C., Lee S.Y., Jung J., Jin H.S., Kim J.H., Kim B.S., Kang M.J., Jang S.O., Kim J., et al. Gene-gene interaction between IL-13 and IL-13Ralpha1 is associated with total IgE in Korean children with atopic asthma. J. Hum. Genet. 2006;51:1055–1062. doi: 10.1007/s10038-006-0061-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang V.E., Schmidt T., Chen J., Sharp P.A., Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 2004;24:1022–1032. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleary M.A., Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol. Cell. Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ansel K.M., Djuretic I., Tanasa B., Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal S., Avni O., Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 25.Ansel K.M., Greenwald R.J., Agarwal S., Bassing C.H., Monticelli S., Interlandi J., Djuretic I.M., Lee D.U., Sharpe A.H., Alt F.W., et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 26.Solymar D.C., Agarwal S., Bassing C.H., Alt F.W., Rao A. A 3′ enhancer in the IL-4 gene regulates cytokine production by TH2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 27.Lee G., Fields P., Griffin T., Flavell R. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 28.Lee G.R., Spilianakis C.G., Flavell R.A. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 29.Mohrs M., Blankespoor C.M., Wang Z.E., Loots G.G., Afzal V., Hadeiba H., Shinkai K., Rubin E.M., Locksley R.M. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 30.Fields P.E., Lee G.R., Kim S.T., Bartsevich V., Flavell R.A. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Herr W., Cleary M.A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes. Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 32.Duncliffe K.N., Bert A.G., Vadas M.A., Cockerill P.N. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 1997;6:175–185. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- 33.Bert A.G., Burrows J., Hawwari A., Vadas M.A., Cockerill P.N. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 2000;165:5646–5655. doi: 10.4049/jimmunol.165.10.5646. [DOI] [PubMed] [Google Scholar]
- 34.Zabel M.D., Wheeler W., Weis J.J., Weis J.H. Yin Yang 1, Oct1 and NFAT-4 form repeating, cyclosporin-sensitive regulatory modules within the murine CD21 intronic control region. J. Immunol. 2002;168:3341–3350. doi: 10.4049/jimmunol.168.7.3341. [DOI] [PubMed] [Google Scholar]
- 35.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.LeVan T.D., Bloom J.W., Bailey T.J., Karp C.L., Halonen M., Martinez F.D., Vercelli D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J. Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 37.Tokuhiro S., Yamada R., Chang X., Suzuki A., Kochi Y., Sawada T., Suzuki M., Nagasaki M., Ohtsuki M., Ono M., et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- 38.Silverman E.S., Palmer L.J., Subramaniam V., Hallock A., Mathew S., Vallone J., Faffe D.S., Shikanai T., Raby B.A., Weiss S.T., et al. Transforming growth factor-β1 promoter polymorphism C-509T is associated with asthma. Am. J. Respir. Crit. Care Med. 2004;169:214–219. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 39.Weidinger S., Gieger C., Rodriguez E., Baurecht H., Mempel M., Klopp N., Gohlke H., Wagenpfeil S., Ollert M., Ring J., et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avni O., Lee D., Macian F., Szabo S.J., Glimcher L.H., Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 41.Strempel J.M., Vercelli D. Functional dissection Identifies a CNS-1 core that mediates IL13 and IL4 transcriptional enhancement. J. Biol. Chem. 2007;282:3738–3746. doi: 10.1074/jbc.M606615200. [DOI] [PubMed] [Google Scholar]