Abstract
Low plasma B-vitamin levels and elevated homocysteine have been associated with cancer, cardiovascular disease and neurodegenerative disorders. Common variants in FUT2 on chromosome 19q13 were associated with plasma vitamin B12 levels among women in a genome-wide association study in the Nurses’ Health Study (NHS) NCI-Cancer Genetic Markers of Susceptibility (CGEMS) project. To identify additional loci associated with plasma vitamin B12, homocysteine, folate and vitamin B6 (active form pyridoxal 5′-phosphate, PLP), we conducted a meta-analysis of three GWA scans (total n = 4763, consisting of 1658 women in NHS-CGEMS, 1647 women in Framingham-SNP-Health Association Resource (SHARe) and 1458 men in SHARe). On chromosome 19q13, we confirm the association of plasma vitamin B12 with rs602662 and rs492602 (P-value = 1.83 × 10−15 and 1.30 × 10−14, respectively) in strong linkage disequilibrium (LD) with rs601338 (P = 6.92 × 10−15), the FUT2 W143X nonsense mutation. We identified additional genome-wide significant loci for plasma vitamin B12 on chromosomes 6p21 (P = 4.05 × 10−08), 10p12 (P-value=2.87 × 10−9) and 11q11 (P-value=2.25 × 10−10) in genes with biological relevance. We confirm the association of the well-studied functional candidate SNP 5,10-methylene tetrahydrofolate reductase (MTHFR) Ala222Val (dbSNP ID: rs1801133; P-value=1.27 × 10−8), on chromosome 1p36 with plasma homocysteine and identify an additional genome-wide significant locus on chromosome 9q22 (P-value=2.06 × 10−8) associated with plasma homocysteine. We also identified genome-wide associations with variants on chromosome 1p36 with plasma PLP (P-value=1.40 × 10−15). Genome-wide significant loci were not identified for plasma folate. These data reveal new biological candidates and confirm prior candidate genes for plasma homocysteine, plasma vitamin B12 and plasma PLP.
INTRODUCTION
One-carbon metabolism comprises a network of biochemical reactions involved in the transfer of single-carbon moieties essential for purine and thymidylate synthesis necessary for maintenance of genomic integrity and remethylation of homocysteine for S-adenosylmethionine (SAM)-dependent DNA methylation reactions (1,2). Folate, a B-complex nutrient from diet and supplements, donates methyl groups to generate methionine from homocysteine. Plasma homocysteine (Hcy), an integrated marker of one-carbon metabolism, is inversely related to folate, vitamin B6 (3) and vitamin B12 and positively related to high alcohol consumption (4,5). Vitamin B12 is a necessary co-factor for the methionine synthase catalyzed remethylation of Hcy into methionine. The biologically active form of vitamin B6, pyridoxal 5′-phosphate (PLP), catalyzes Hcy transulfuration (6) and is involved in over 100 enzyme reactions in the synthesis and catabolism of neurotransmitters (7), amino acids, glycogen and lipids (8).
Epidemiologic studies have shown that individuals with reduced intake of folate (found in fruits, vegetables and legumes) or vitamin B6 (found in poultry, fish, meat, legumes, nuts and potatoes) or vitamin B12 [found in liver, shellfish, fish, poultry, eggs and dairy products (9)] have an inverse relationship with Hcy levels. The accumulation of Hcy leads to an intracellular increase in S-adenosylhomocysteine (AdoHcy) and is an established risk predictor of cardiovascular diseases (6,10–16). In addition, individuals with inadequate intake of folate and vitamin B12 are at an elevated risk of uracil misincorporation into DNA and aberrant DNA methylation (17–20). Vitamin B12 deficiency (21), related to poor intestinal B12 absorption (9) or dietary deficiency, can lead to inactivation of methionine synthase and is associated with pernicious anemia (22), cardiovascular disease (23–25), cancer (26–32) and neurodegenerative disorders (5,33–35). Plasma PLP levels are significantly inversely associated with risk of colorectal cancer in men (36) and women (37).
Plasma Hcy is the integrated marker of the one-carbon metabolism pathway, with a heritability estimate of 44% (38) [heritability estimate range 8–57% (38–43)]. Until recently, common genetic variations reported to influence plasma folate, vitamin PLP, vitamin B12 and Hcy were focused on genes in the one-carbon metabolism pathway, specifically the Ala222Val polymorphism (23,44) (45) in 5,10 methylene-THF reductase (MTHFR), associated with reduced conversion of methylene-THF to methyl-THF (46,47). We previously identified a genome-wide significant association between common variants in FUT2 and plasma vitamin B12 levels in women (48). In a recent genome-wide analysis of data pooled from three genome-wide association study (GWASs) of 2931 persons and a replication study of 687 participants, Tanaka et al. (49) confirmed this loci and reported additional loci that met genome-wide significance criteria for PLP. In the current study, we report a meta-analysis of three GWASs with 4763 participants (1658 women in CGEMS, 1647 women in SHARe and 1458 men in SHARe) and test for the association between 2 404 675 genotyped or imputed SNPs with an R-sq > 0.3 (R-sq_hat estimates the squared correlation between imputed and true genotypes and allows us to assess imputation accuracy for markers with many different allele frequencies) in each study and plasma levels of Hcy, folate, vitamin B12 and PLP.
RESULTS
Genome-wide significant associations of P-value < 5 × 10−8 [corresponding to P < 0.05 corrected P-value after adjusting for approximately 1 million independent loci (50)] were identified in meta-analyses for log-transformed plasma Hcy, vitamin B12 and PLP. Age and the distributions of plasma vitamin B12 and homocysteine in the study populations are described in Table 1. As expected, plasma Hcy was found to be inversely related to plasma levels of folate, vitamin B12 and vitamin B6 in our study population (Supplementary Material, Table S1). No significant heterogeneity by sex or by study (Table 2) was detected for the genome-wide significant associations.
Table 1.
Mean and median plasma one-carbon metabolite levels in the NHS CGEMS, SHARe Women and SHARe Men study populations with GW data
| NHS CGEMSa, n = 1,658 | SHARe Women, Exam 6b, n = 1,647 | SHARe Men, Exam 6b, n = 1,458 | |
|---|---|---|---|
| Age, Mean, SD | 58.92, 6.20 | 58.60,9.71 | 58.57, 9.70 |
| Plasma HCYc, Mean, SD | 11.52, 5.41 | 9.06, 3.87 | 10.54, 3.99 |
| Plasma HCY Median (IQRd) | 10.62 (4.04) | 8.39 (3.32) | 9.82 (3.55) |
| Plasma Folatee, Mean, SD | 10.02, 9.03 | 7.85, 5.32 | 6.54, 4.23 |
| Plasma Folate Median (IQRd) | 7.69. (7.44) | 6.45 (5.80) | 5.68 (4.22) |
| Plasma B12f, Mean, SD | 473.90, 249.53 | 435.10, 181.80 | 398.59, 148.26 |
| Plasma B12 median (IQRd) | 436.31 (224.0) | 405.96 (211.91) | 377.34 (172.68) |
| Plasma PLPg, Mean, SD | 76.39, 90.55 | 83.14, 82.76 | 86.89, 84.05 |
| Plasma PLP median (IQRd) | 46.70 (47.77) | 57.55 (56.87) | 61.72 (52.61) |
aNHS blood collection was during 1989–1990.
bExam 6 refers to Framingham Heart Study laboratory exam 6 measurements (1995–1998). For all plasma, one-carbon metabolites exam 6 measurements were used in this analysis because this exam had the largest sample size, and laboratory measurements were conducted using assays comparable to NHS CGEMS.
cPlasma HCY measured in μmol/l; data were log-transformed for GWAS analysis.
dIQR, interquartile range.
ePlasma Folate measure in ng/ml; data were log-transformed for GWAS analysis.
fPlasma Vitamin B12 measured in pg/ml; data were log-transformed for GWAS analysis.
gPlasma PLP measured in pmol/ml; data were log-transformed for GWAS analysis.
Table 2.
Genome-wide significant associations for plasma one-carbon metabolites
| NHS CGEMS n = 1658 |
SHARe Women n = 1647 |
SHARe Men n = 1458 |
Meta-analysis n = 4763 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position (bp) | Gene | Alleles | MAFd | BETAa | S.E. | P-value | BETAb | S.E. | P-value | BETAb | S.E. | P-value | BETA | S.E. | Q | P-metac | P-het |
| Plasma homocysteine | |||||||||||||||||||
| rs12085006 | 1p36 | 11892989 | G,A | 0.43 | 0.04 | 0.01 | 1.21E−05 | 0.03 | 0.01 | 4.06E−03 | 0.04 | 0.01 | 6.49E−04 | 0.04 | 0.01 | 0.70 | 5.81E–10 | 0.72 | |
| rs1999594 | 1p36 | 11893482 | A,G | 0.43 | 0.04 | 0.01 | 1.32E−05 | 0.03 | 0.01 | 4.06E−03 | 0.04 | 0.01 | 6.49E−04 | 0.04 | 0.01 | 0.67 | 6.24E–10 | 0.72 | |
| rs1801133 | 1p36 | 11790644 | MTHFR | G,A | 0.33 | 0.04 | 0.01 | 5.12E−04 | 0.03 | 0.01 | 9.59E−03 | 0.04 | 0.01 | 1.51E–04 | 0.04 | 0.01 | 0.82 | 1.27E–08 | 0.65 |
| rs10986018 | 9q22 | 98202891 | GPR51 | C,T | 0.22 | −0.08 | 0.02 | 4.92E−05 | −0.04 | 0.02 | 4.85E−02 | −0.07 | 0.02 | 2.92E–04 | −0.06 | 0.01 | 2.10 | 2.06E–08 | 0.36 |
| Plasma Vitamin B12 | |||||||||||||||||||
| rs602662 | 19q13 | 53898797 | FUT2 | A,G | 0.44 | −0.08 | 0.01 | 3.09E−10 | −0.05 | 0.02 | 3.80E−04 | −0.05 | 0.01 | 2.80E–04 | −0.07 | 0.01 | 2.83 | 1.83E–15 | 0.23 |
| rs601338 | 19q13 | 53898486 | FUT2 | G,A | 0.45 | 0.09 | 0.01 | 4.25E−11 | 0.05 | 0.01 | 2.63E−03 | 0.05 | 0.01 | 4.02E–04 | 0.06 | 0.01 | 5.13 | 6.92E–15 | 0.08 |
| rs492602 | 19q13 | 53898229 | FUT2 | A,G | 0.44 | 0.09 | 0.01 | 5.39E−11 | 0.04 | 0.02 | 5.89E−03 | 0.05 | 0.01 | 2.36E–04 | 0.06 | 0.01 | 5.43 | 1.30E–14 | 0.06 |
| rs1801222 | 10p12 | 17196157 | CUBN | G,A | 0.28 | −0.05 | 0.01 | 9.04E−05 | −0.04 | 0.02 | 6.32E−03 | −0.05 | 0.02 | 3.56E–04 | −0.05 | 0.01 | 0.42 | 2.87E–09 | 0.81 |
| rs526934 | 11q11 | 59390069 | TCN1 | A,G | 0.27 | −0.05 | 0.01 | 1.27E−03 | −0.06 | 0.02 | 6.69E−05 | −0.06 | 0.02 | 1.64E–04 | −0.05 | 0.01 | 0.45 | 2.25E–10 | 0.82 |
| rs9473558e | 6p21 | 49520392 | MUT | C,T | 0.35 | −0.03 | 0.01 | 4.27E−02 | −0.03 | 0.01 | 1.87E−02 | −0.07 | 0.01 | 3.96E–07 | −0.04 | 0.01 | 5.48 | 4.05E–08 | 0.08 |
| rs9473555 | 6p21 | 49517446 | MUT | G,C | 0.35 | −0.03 | 0.01 | 4.27E−02 | −0.03 | 0.01 | 2.26E−02 | −0.07 | 0.01 | 3.71E–07 | −0.04 | 0.01 | 5.45 | 4.91E–08 | 0.08 |
| Plasma PLP | |||||||||||||||||||
| rs1256335 | 1p36 | 21635692 | ALPL | A,G | 0.21 | −0.15 | 0.03 | 6.44E−07 | −0.11 | 0.03 | 2.37E−04 | −0.16 | 0.03 | 2.36E–07 | −0.14 | 0.02 | 1.56 | 1.40E–15 | 0.46 |
| rs4654748 | 1p36 | 21531374 | NBPF3 | T,C | 0.48 | −0.12 | 0.03 | 1.40E−06 | −0.05 | 0.03 | 7.32E−02 | −0.12 | 0.02 | 2.21E–06 | −0.10 | 0.01 | 5.63 | 4.30E–11 | 0.12 |
aEstimates for NHS CGEMS data are from linear regression using log-transformed metabolite levels.
bEstimates SHARe data are from linear mixed-effects model using kinship information using log-transformed metabolite levels.
cMeta-analysis estimates and P-values for the NHS CGEMS, FHS SHARe Women and SHARe Men data sets were calculated using the fixed-effects model; between study variance is <0.001.
dMAF is averaged across the three GWA studies.
Plasma homocysteine
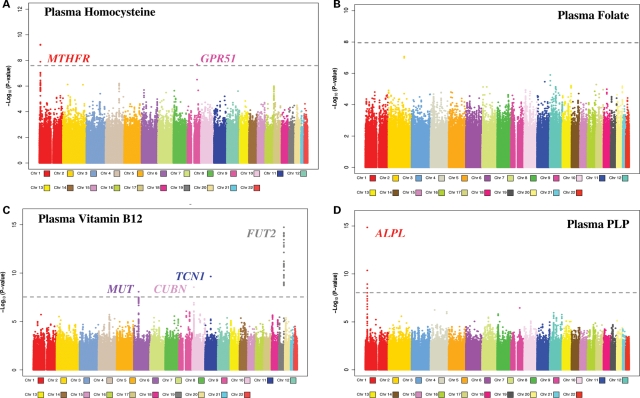
The well-studied functional polymorphism in MTHFR Ala222Val (also reported as MTHFR C677T), rs1801133, on chromosome 1 [position (bp): 11,790,644] was associated with plasma Hcy (P-value = 1.27 × 10−8; Fig. 1A; Table 2). Participants with the Ala/Ala (G/G) variant had lower plasma Hcy levels than variant carriers. Two other SNPs, rs12085006 [position (bp): 11,892,989] and rs1999594 [position (bp): 11,893,482], located 102 kb from MTHFR, had stronger associations with plasma Hcy (5.81 × 10−10 and 6.24 × 10−10, respectively; Fig. 1A; Table 2). Participants homozygous for the rs12085006 G and rs1999594 A alleles had higher Hcy levels compared with variant carriers for each SNP. The rs12085006 A allele is correlated with the MTHFR 222Val allele (composite r2 = 0.53; P < 0.0001) (51). After mutual adjustment in a univariate regression model, both rs1285006 and MTHFR Ala222Val remained nominally statistically significantly associated with plasma Hcy (P = 0.02 and 0.002, respectively). Variant carriers of both MTHFR 222Val and the A allele for rs12085006 had higher levels of plasma Hcy compared with non-carriers. Similar results were obtained for rs1999594. We also identified an association on chromosome 9 in the gamma-aminobutyric acid B-type receptor G-protein coupled receptor 51 (GPR51) gene, rs10986018, with plasma Hcy (P-value=2.06 × 10−8; Fig. 1A, Table 2). A SNP, rs11041321 in SYT9, reported by Tanaka et al. (49) with a P-value of 1.11 × 10−4 in their joint analysis of GWAS meta-analysis and replication study, had a P-value=0.44 in our meta-analysis data. The SNPs in CBS, CPS1, MUT and NOX4 reported in the Women's Genome Health Study by Paré et al. (52) were not genome-wide significant in our plasma homocysteine data (rs6586282 in CBS: P-value=6.30 × 10−1; rs7422339 in CPS1: P-value=4.18 × 10-4; rs4267943 in MUT: P-value 6.43 × 10−1; rs11018628 in NOX4: P-value=4.35 × 10−3).
Figure 1.
Meta-analysis of three GWA scans for plasma one-carbon metabolites. Manhattan plot of genome-significant associations (defined as P = 5 × 10−8 and depicted by the grey dashed line) for plasma homocysteine (A), plasma vitamin B12 (C) and plasma PLP (D) in a meta-analysis of participants from NHS CGEMS, Framingham SHARe Women and Framingham SHARe Men.
Plasma folate
Overall, there were no genome-wide significant associations for plasma folate in the meta-analysis (Fig. 1B). The strongest associations were in chromosome 2 in the FIGN gene, rs982393 (P-value=8.36 × 10−8; data not shown) and rs2119289 (P-value=1.03 × 10−7; data not shown). Although not genome-wide significant, a moderate signal for SNPs in MTHFR was observed (rs3737965, P-value=6.90 × 10−5; data not shown). As expected (53), the MTHFR Ala222Val variant carriers had lower plasma folate compared with the Ala/Ala variant (MTHFR Ala222Val P-value=1.14 × 10−3 from meta-analysis, data not shown). The composite linkage disequilibrium (LD) with MTHFR Ala222Val and rs3737965 was 0.15 in our data. The rs1999594 SNP associated with plasma folate in the report by Tanaka et al. (49) had P-value=3.01 × 10−3 in our analysis for plasma folate. The rs153734 SNP in the PRICKLE gene, reported by Tanaka et al. (49), was not associated with plasma folate in these data (P-value=0.44).
Plasma vitamin B12
Genome-wide significant associations were identified on chromosomes 6, 10, 11 and 19 for plasma vitamin B12 (Fig. 1C; Table 2). Two SNPs, in LD, in Methylmalonyl-CoA mutase (MUT) on chromosome 6p21—rs9473558 SNP and rs9473555 [chromosome position (bp): 49,520,392 and 49,517,446, respectively]—were genome-wide significant in the meta-analysis for plasma vitamin B12 (P-meta=4.91 × 10−8, 4.05 × 10−8 respectively, Fig. 1; Table 2). Participants homozygous for the rs9473558 C and rs9473555 G alleles had higher B12 levels compared with participants homozygous for the major allele. We identified an association (P = 2.87 × 10−9; Fig. 1C; Table 2) between rs1801222 in CUBN (Cubilin) on chromosome 10p12 and plasma vitamin B12 levels in the meta-analysis of 4763 participants. The SNP is in exon 8 of CUBN and codes for an amino acid change Ser253Phe [chromosome 10p position (bp): 17,196,157]. Participants homozygous for the rs1801222 G allele had higher B12 levels. The meta-analysis indicated several signals in the P < 10−5 range for other SNPs in CUBN and on chromosome 10p15 in the neighboring TRDMT1 gene (DNMT2), which may be explained by LD. A genome-wide significant association was also observed for rs526934 in the transcobalamin I (TCN1) gene P-value=2.25 × 10−10) located on chromosome 11q11 [position (bp): 59,390,069]. Variant carriers of the rs526934 G allele had lower vitamin B12 levels when compared with individuals with the homozygous AA variant.
The most significant association in the meta-analysis data (P-value=1.83 × 10−15; Fig. 1C; Table 2) was for the rs602662 SNP in FUT2. The FUT2 nonsense SNP W143X, rs601338 [chromosome 19q13 position (bp): 53,898,486], the determinant of FUT2 secretor status, was the second most significant association with a P-value=6.92 × 10−15. Participants homozygous for non-secretor variants had higher B12 levels than carriers of the secretor genotype. The rs492602 SNP reported to be in high LD (D′ = 1, r2 = 0.76) with FUT2 W143X had a P-value=1.30 × 10−14. We noted these associations despite the fact that the FUT2 SNPs were not well tagged in the 347 458 genotyped SNPs that passed Quality Control filters in the SHARe data set; the MACH imputation R-sq value for rs602662: was 0.45, and for rs492602 it was 0.41 in SHARe.
Plasma PLP
SNPs in alkaline phosphatase, ALPL, had genome-wide significant associations with plasma PLP in the meta-analysis. We observed the strongest association for rs1256335 (P-value=1.40 × 10−15; Fig. 1D; Table 2) in the ALPL gene. Participants homozygous for the A allele had higher plasma PLP levels compared with carriers of the G allele. The reported rs4654748 (49) SNP in NBPF3, upstream of ALPL, had a P-value=4.30 × 10−11 with plasma PLP in these data (composite r2 for rs1256335 and rs4654748 is 0.16, P < 0.0001).
DISCUSSION
We identified genome-wide significant associations for plasma homocysteine, plasma vitamin B12 and plasma PLP. SNPs in chromosome 6p21, 10p12, 11q11 and 19q13 showed strong signals for plasma vitamin B12. SNPs in chromosomes 1p36 and 9q22 were strongly associated with plasma homocysteine. We identified genome-wide significant associations between SNPs in chromosome 1p36 and plasma PLP.
Plasma homocysteine
Chromosome 1p36
MTHFR is a key enzyme required for DNA synthesis that catalyzes the irreversible transformation of 5,10-MTHF into 5-MTHF (54). The TT variant alleles of the well-known MTHFR 677 (amino acid change: MTHFR Ala222Val) polymorphism are related to a 30% reduction in enzyme activity (55). A decrease in MTHFR activity associated with the 222 Val/Val (677TT) polymorphism shifts the folate pool, leading to an elevation in 5,10-methyleneTHF. The MTHFR Ala222Val SNP results in a moderate (up to 15–19%) increase in mean Hcy levels between the Ala/Ala and Val/Val variants (23), and this effect is greatly attenuated in those individuals with mid-range to high folate levels (56). Rs12085006 and rs1999594, located 102 kb from MTHFR, had an even stronger association with plasma Hcy in our meta-analysis. RS1999594 had a genome-wide significant association (P = 8.5 × 10−12) with plasma Hcy in the Women's Genome Health Study (WGHS), but only rs180133 showed evidence for non-additive effects (52). The rs1999594 SNP was reported by Tanaka et al. (49) to be associated with plasma folate in a meta-analysis of 2931 participants (P = 1.12 × 10−7), although the association did not reach genome-wide significance in their report on plasma folate and was not reported for plasma Hcy. Given the strong correlation between rs1999594 (or rs12085006) and rs180133 and the modest evidence for association between rs1999594 (or rs12085006) and plasma Hcy after adjusting for MTHFR Ala222Val, we cannot conclude that these two SNPs mark distinct loci associated with plasma Hcy.
Chromosome 9q22
We identified a genome-wide significant association between plasma Hcy and a variant in GPR51. Carriers of the rs10986018 T allele had lower plasma vitamin B12 compared with carriers of the CC variant. Expression of GPR51 may be regulated by nicotine (57). Epidemiologic studies have reported increased homocysteine levels in smokers compared with non-smokers (58–62). The association with plasma Hcy and rs10986018 warrants future study on plasma Hcy and effect modification by smoking status.
Plasma folate
We did not identify any genome-wide significant association with plasma folate in this meta-analysis, although the MTHFR Ala222Val variant had the expected direction of association (53,56), with a modest P-value. FHS SHARe Exam 6 data available in dbGaP had the largest sample size for all four plasma one-carbon metabolites and laboratory assays comparable to the NHS CGEMS study population. All NHS CGEMS participants had plasma folate measured on blood drawn prior to folic acid fortification (1989–1990). However, the Framingham SHARe laboratory exam 6 measurements were taken during 1995–1998. While the majority of the SHARe participants included in this analysis had measurements prior to the US mandate for folic acid fortification in 1998, evidence suggests that the fortification may have started in 1996–1997. Pre-fortification plasma folate levels in the prior Framingham SHARe laboratory exam, available in dbGaP, were measured using a microbial assay and would not be equivalent to the HPLC assay used in the NHS CGEMS data. We also recognize that there may also be misclassification due to folic acid supplementation.
Plasma vitamin B12
Chromosome 6
MUT, located on chromosome 6p21, encodes the mitochondrial enzyme methylmalonyl Coenzyme A (CoA) mutase. MUT requires 5-prime-deoxyadenosylcobalamin (AdoCbl, a coenzyme form of vitamin B12) to catalyze the isomerization of L-methylmalonyl–coenzyme A (CoA) to succinyl-CoA, in the mitochondria, independent of folate status. Mutations in methylmalonyl-CoA mutase, especially in the C-terminus, result in methylmalonic aciduria (63–67). A patient with the mut(-) MMA phenotype due to the MUT Gly717Val variant was also homozygous for the His532Arg (rs9473558) polymorphism (68). The T variant of rs9473558 has reduced vitamin B12 levels in our data and may be associated with decreased conversion of methylmalonyl CoA to succinyl CoA. The rs9473558 SNP in MUT was associated with plasma homocysteine (P = 2.1 × 10–7) in the Women's Genome Health Study (WGHS) participants (52). The rs9473558 SNP was positively associated with plasma homocysteine in the WGHS study (β = 0.020), but inversely associated with plasma vitamin B12 in our meta-analysis (β = −0.04), which is consistent with the inverse relationship between homocysteine and vitamin B12. The expression of MUT was associated with the rs9473555 SNP with a LOD score = 7.095 [enzyme activity for the G-allele is reduced by 0.42 standard deviation; mRNA by SNP browser 1-0-1 database (69)]. The preliminary association with rs9473558 or rs9473555 in the MUT gene and plasma vitamin B12 should be replicated in other studies. Future studies on the relationship of rs9473555 or rs9473558 and MUT gene expression is warranted.
Chromosome 10
Cubilin (CUBN) is located on chromosome 10p12.31. The 460 kd multiligand hydrophobic CUBN protein binds to intrinsic factor-cobalamin (Cbl-IF) complexes with a high affinity and is expressed in both kidney and ileal epithelial cells. Mutations in CUBN are associated with megaloblastic anemia and characterized by selective intestinal vitamin B12 malabsorption (70). The P1297L mutation in CUBN causes a 5-fold decrease of the binding domain affinity for Cbl-IF (the IF-Cbl binding region includes amino acids 928–1386). The rs1801222 (Ser253Phe) variant was associated with lower vitamin B12 levels in this study and may decrease the binding and transport of vitamin B12 in the ileum. Another SNP in CUBN reported by Tanaka et al. (49) with a P-value of 1.11 × 10−6, rs11254363, had a P-value of 8.65 × 10−5 in our meta-analysis. The neighboring gene, TRDMT1, located on 10p15.1 is a methyltransferase that methylates the aspartic acid transfer RNA molecule (tRNA(Asp)) (71). A recent study on spina bifida and folate-related genes reported an association for different SNPs in CUBN and TRDMT1 with increased serum RBC folate levels in controls (72). Further research is warranted to understand the LD pattern across the 305.3 kb CUBN gene and the cis relationships of the genome-wide significant SNPs with CUBN and TRDMT1.
Chromosome 11
TCN1, on chromosome 11q11, is a vitamin B12 binding protein that promotes the cellular uptake of vitamin B12 by receptor-mediated endocytosis. The protein is a major constituent of secondary granules in neutrophils and facilitates the transport of cobalamin into cells (73). The meta-analysis and replication joint P-value for rs526934 from Tanaka et al., was P-value=1.51 × 10−6. The TCN1 rs526934 G variant may reduce transport of cobalamin, resulting in lower plasma vitamin B12 levels.
Chromosome 19
We previously reported an association with variants in FUT2 on chromosome 19q13 and plasma vitamin B12 [rs602662: P-value=3.52 × 10−15; rs492602: P-value=5.36 × 10−17 in the combined NHS CGEMS and colorectal neoplasia data sets; (48)]. In the SHARe women and the SHARe men data sets, we replicated our genome-wide significant finding for rs602662 (SHARe combined P = 5.07 × 10−8; Table 2 presents separate estimates and standard error, by SHARe Men and Women) and in this meta-analysis of CGEMS and SHARe data, we confirm the association of these SNPs with plasma vitamin B12. In this study of plasma vitamin B12, we confirm our previously reported hypothesis that FUT2 W143X, a nonsense mutation (74) and determinant of FUT2 secretor status, is the causal variant for the association with plasma B12 levels (rs601338: P-value=6.92 × 10−15). Individuals with the non-secretor status variant had higher plasma vitamin B12 levels compared with carriers of the secretor status genotypes. Reduced vitamin B12 absorption (75–79) in carriers of the secretor genotype may be a consequence of susceptibility to H. pylori infection compared with individuals with the non-secretor genotype. The association with rs602662 was replicated by an independent group (49) with a P-value of 2.43 × 10−12 in a meta-analysis of three GWASs (n = 2930 participants).
Plasma PLP
Mutations in the ALPL gene, on chromosome 1p36, are associated with hypophosphatasia. Increased circulating concentrations of PLP in all clinical forms of hypophosphatasia have been reported by Whyte et al. (80,81). The findings suggested that tissue-non-specific alkaline phosphatase is involved in vitamin B6 metabolism. The ALPL enzyme may function as an ectoenzyme to regulate extracellular concentrations of PLP (80). The rs1256335 G allele had lower plasma PLP levels compared with the homozygous A allele, possibly due to increased hydrolysis of PLP (82). The rs4654748 C allele, in the neuroblastoma breakpoint family, member 3, (NBPF3, alternate name AE2) gene had lower plasma PLP levels compared with the homozygous major allele.
Comparability of plasma phenotypes in the study population
The NHS and FHS study populations have comparable metabolite levels for conducting a meta-analysis of GWA data of plasma one-carbon metabolites and the biomarker measurement in these studies were all done in a single laboratory. However, one limitation is that the NHS CGEMS participants were all postmenopausal women, whereas SHARe Women participants included of both pre- and postmenopausal women. Plasma Hcy is lower in premenopausal women than men, but after menopause, homocysteine levels in women increased to approach those in men, consistent with our data. All NHS and the majority of the FHS blood samples for participants in this analysis were collected prior to the introduction of folic acid fortification of breads, cereals, flours, corn meal, pasta products, rice and other cereal grain products sold in the USA (83,84). Therefore, the genetic variability signal should not be attenuated by this recent environmental influence.
In summary, for plasma vitamin B12, we replicated our finding for FUT2 (48) and identified genome-wide significant SNPs in biological candidate genes: TCN1, a vitamin B12 binding protein; MUT, which converts methylmalonyl CoA to succinyl CoA; and CUBN, the receptor for intrinsic factor-vitamin B12 complexes. For plasma homocysteine, we observed genome-wide significant associations with the MTHFR functional SNP Ala222Val and a possible new independent locus 102 kb upstream of MTHFR. For plasma PLP, we noted genome-wide significant associations in ALPL. These data reveal new biological candidates and confirm prior candidate genes for plasma homocysteine, plasma vitamin B12 and plasma PLP.
MATERIALS AND METHODS
Ethics statement
All participants provided written informed consent, and local institutional review boards approved the study protocols.
Study populations
We conducted a meta-analysis of genome-wide data in 1658 women genotyped with the HumanHap500 as part of the Cancer Genetic Markers of Susceptibility Project (CGEMS) and in 1682 women genotyped with the Affymetrix 500K mapping array and the Affymetrix 50K Supplementary array as a part of the SNP Health Association Resource (SHARe data set, part of NHLBI FHS) to identify novel loci that influence plasma one-carbon metabolite levels. All participants were of self-reported European ancestry.
The Nurses’ Health Study (NHS)
The NHS was initiated in 1976, when 121 700 US registered nurses between the ages of 30 and 55 returned an initial questionnaire reporting medical histories and baseline health-related exposures. Every 2 years, follow-up questionnaires are mailed to the participants (questionnaire response rate >90% for all follow-up cycles). Diet was assessed in 1986, 1990, 1994 and 1996 with a semiquantitative food frequency questionnaire. Between 1989 and 1990, blood samples and relevant sample collection information were collected from 32 826 women.
The CGEMS project (http://www.cgems.cancer.gov) is an NCI initiative to conduct genome-wide association studies (GWAS) to identify genes involved in breast and prostate cancer. The initial CGEMS scan, designed and funded only to study the main effect of SNP variants on breast cancer risk in postmenopausal women, has been completed (85). A subset of the participants in the CGEMS project also has been assayed for plasma vitamin B12. The NHS CGEMS participants had similar laboratory measurement of the plasma one-carbon metabolites as well as host characteristics and blood draw features (fasting status and year and month of blood draw).
Framingham Heart Study (FHS)
The FHS began in 1948 with the enrollment of two-thirds of the adult population of Framingham, Massachusetts (86), including 2873 women aged 28–62 years. In 1971, 5124 offspring of the original cohort members and offspring spouses were enrolled in the Framingham Heart Study, including 2641 women ranging in age from 12 to 60 years. A data set consisting of repeated and highly accurate measurements of many different phenotypes collected over multiple exam biennial cycles is available for each participant. The NHLBI Framingham SNP-Health Association Resource (SHARe) is a large-scale GWAS on the FHS original, offspring and third-generation cohorts with data accessible to the research community (87). In the Framingham cohort, the SNP Health Association Resource (SHARe) project genotyped 9274 Caucasian participants with the Affymetrix 500K mapping array and the Affymetrix 50K supplemental array (Affymetrix, Santa Clara, CA, USA). The 8508 samples from these participants had a sample call rate ≥97% (88,89). Of the participants from the Framingham original and offspring cohorts, with measured plasma one-carbon metabolites at laboratory exam 6, between 1995 and 1998, the SHARe scan included 1647 (16) Caucasian women and 1458 Caucasian men. Exam 6 had the largest sample size for all four plasma one-carbon metabolites and laboratory assays comparable to the NHS CGEMS study population. Information on Framingham SHARe can be found at http://www.ncbi.nlm.nih.gov/projects/gap/cgi- bin/study.cgi?study_id=phs000007.v1.p1.
Laboratory assays
Plasma levels of homocysteine for both the NHS CGEMS and FHS SHARe studies were measured by high-performance liquid chromatography, with fluorescence detection (90). Plasma levels of folate and vitamin B12 for the NHS CGEMS and FHS SHARe participants were measured using by a radioassay kit (Bio-Rad, Richmond, CA, USA) according to the manufacturer's instructions. Plasma PLP levels were measured by an enzymatic procedure using radioactive tyrosine and the apo-enzyme tyrosine decarboxylase.
All assays were conducted at the Jean Mayer U.S. Department of Agriculture Human Nutrition Research Center on Aging at Tufts University. Plasma samples for matched case–control sets were always positioned next to each other, in random order, in boxes sent to the lab and were assessed in the same batch to minimize the impact of laboratory error due to batch drift. Quality control (QC) samples were also submitted with each batch and were randomly placed throughout the boxes (two assay batches were used for NHS CGEMS participants). Laboratory technicians were blinded to case, control and QC status of the samples. QC samples consisted of replicates of two pools of plasma. One QC sample was assayed per 10 study samples (27,37). No significant differences in mean levels of plasma homocysteine, folate, vitamin B12 or PLP were detected by breast cancer case–control status in the NHS. The mean coefficient of variation for 75 pairs of replicate 6.5% for folate, 7.9% for homocysteine, 7.3% for B12 and 7.2% for PLP (27).
Reliability of laboratory assays of plasma levels
In an analysis of 188 NHS participants, the correlation coefficients between folate intake calculated from the 1980 food frequency questionnaire and erythrocyte folate concentrations in 1987 were 0.55 for folate from foods and supplementation and 0.38 for folate from food only (18). Similar correlation coefficients were noted for plasma folate (0.55 for total folate; 0.35 for dietary folate) (18,27). The intraclass correlation coefficient over a 3-year period on 82 participants was 0.63 for plasma folate, suggesting reproducibility of these measurements over time.
Ilumina SNP genotyping and quality-control analysis
Genotyping in the CGEMS project was carried out using the Illumina Infinium Sentrix HumanHap550 bead chip, which contains 556 566 SNPs derived from the HapMap phase I and II data (85). The polymorphisms on the bead chip are selected to be direct surrogates (R2, a measure of pair-wise SNP correlation, >0.8) of more than 75% of the common polymorphisms genotyped as part of the HapMap Phase II project. Overall, genotyping success was 98.45% of the possible genotypes in the first attempt and 99.27% after failed samples were repeated. Quality control of genotyping reproducibility was 99.99%. SNPs with significant deviations from Hardy–Weinberg (HWE) proportions (5% of SNPs at the level of P = 0.05 and 1.28% at P = 0.01) were not excluded from the analysis, as the tests for association applied to these data are valid in the presence of departure from HW proportions. The NHS cases and controls included in CGEMS are of self-reported European ancestry (85). Four samples (three cases and one control) were removed because of intercontinental admixture. In the CGEMS data, we saw no evidence for systematic bias in the distribution of P-values for analyses with and without further adjustment for residual population structure using four principal components of genetic variation compatible with no confounding of SNP-metabolite associations due to population stratification (85,91). A total of 1658 NHS participants had GWAS and plasma vitamin B12 data.
Affymetrix SNP genotyping and quality-control analysis
Genotyping in the SHARe study (88,89) was carried out using the Affymetrix (Santa Clara, CA, USA) GeneChip Human Mapping 500K mapping array and supplemental 50K mapping array (HuGeneFocused50K) with gene-centric and coding SNPs. The Human Mapping 500K Array Set is comprised of two arrays, each capable of genotyping on average 250 000 SNPs (approximately 262 000 for Nsp arrays and 238 000 for Sty arrays).
Association analyses were restricted to individuals with ≥97% genotyping call-rate. SNP results were filtered on Hardy–Weinberg equilibrium P-value of 1 × 10−6, SNP call-rate of 95% and minor allele frequency (MAF) of 0.01. After cleaning the genotype data, 347 K SNPs were available for the analysis.
Statistical methods
We tested the association between log-transformed plasma one-carbon metabolite levels and each SNP using linear regression adjusted for age. SNP genotypes were coded as counts of minor alleles (additive coding) unless otherwise specified. We adopted an additive coding because it has nearly optimal power for a range of realistic genetic models, with the exception of rare recessive effects, which we have little power to detect (92). In CGEMS, we further adjusted for principal components of genetic variation, calculated with the EIGENSTRAT software (91).
The SHARe data were analyzed using linear mixed effects models to account for residual familial correlation; aside from fixed effects (age, SNP genotype), this model includes individual random effects with a correlation matrix equal to the kinship matrix times a scalar variance parameter. These analyses were implemented using the lmekin function in the R package kinship.
We imputed SNP genotypes using the hidden Markov model implemented in MACH 1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) for each CGEMS and SHARe separately. For the SHARe data, the hidden Markov model was trained using unrelated founders (922 women and 692 biologically unrelated men); the fitted model was then used to infer missing genotypes for the remaining subjects. The certainty of the imputation is dependent on the stretches of haplotypes shared between the reference panel (HapMap Phase II CEU population) and the study population. The imputation result at each of the imputed SNPs, per study participant, can be expressed as a set of three genotype probabilities, that vary between 0 and 1 per genotype and sum to 1, or as an ‘allele dosage’, which is defined as the expected number of copies of the minor allele at each SNP and varies between 0 and 2. In the CGEMS data, we evaluated both methods and obtained comparable results for the genotype probabilities and allele dosage methods. In the SHARe data, we evaluated both methods for the genome-wide significant associations and obtained comparable results for the genotype probabilities and allele dosage methods.
Genotyped and imputed SNPs with an R-sq_hat from MACH >0.30 in both the CGEMS Illumina and SHARe Affymetrix data were used for a meta-analysis (93) to examine the association between ∼2.5million polymorphisms and the plasma metabolites. A fixed-effect model was used to pool the results for the NHS CGEMS, SHARe Women and SHARe Men studies. Cochran's Q statistic and tau-squared were used to test for between-study heterogeneity, and the I-squared statistic to quantify the proportion of total variation due to heterogeneity will be calculated (94). We saw no evidence for systematic bias in the distribution of P-values from the meta-analyses (Supplementary Material, Fig. S1). The P-for-heterogeneity is reported in Table 2. SAS PROC GLM was used to test the independence of the estimates of multiple SNPs in a model. Composite LD was calculated according to the notation of Weir (51).
Our analyses do not explicitly account for the ascertainment of subjects on the basis of primary disease (breast cancer for the NHS CGEMS; Framingham SHARe data are from a cohort study and not ascertained on disease). This approach is appropriate, since our loci of interest are not associated with primary disease evaluated in the CGEMS NHS participants (85,95). All P-values are based on two-sided tests. All statistical analyses were performed with SAS (version 9.1; SAS Institute, Cary, NC, USA), R project (http://www.r-project.org) and PLINK (96).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health [research grant numbers. U54 CA100971, P01 CA87969, P01 CA55075, U01 CA098233, R01 CA 065725, R01 CA070817 and R03 CA133937].
The Framingham Heart Study is conducted and supported by the National Heart, Lung and Blood Institute (NHLBI) in collaboration with Boston University.
ACKNOWLEDGEMENTS
In this study, all plasma assays for CGEMS and SHARe were conducted at the Jean Mayer U.S. Department of Agriculture Human Nutrition Research Center on Aging at Tufts University. This manuscript does not necessarily reflect the opinions or views of Boston University or the NHLBI.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kim Y.I. Folate and DNA methylation: a mechanistic link between folate deficiency and colorectal cancer? Cancer Epidemiol. Biomarkers Prev. 2004;13:511–519. [PubMed] [Google Scholar]
- 2.Kim Y.I. Nutritional epigenetics: impact of folate deficiency on DNA methylation and colon cancer susceptibility. J. Nutr. 2005;135:2703–2709. doi: 10.1093/jn/135.11.2703. [DOI] [PubMed] [Google Scholar]
- 3.Bates C.J., Pentieva K.D., Prentice A., Mansoor M.A., Finch S. Plasma pyridoxal phosphate and pyridoxic acid and their relationship to plasma homocysteine in a representative sample of British men and women aged 65 years and over. Br. J. Nutr. 1999;81:191–201. [PubMed] [Google Scholar]
- 4.Selhub J., Miller J.W. The pathogenesis of homocysteinemia: interruption of the coordinate regulation by S-adenosylmethionine of the remethylation and transsulfuration of homocysteine. Am. J. Clin. Nutr. 1992;55:131–138. doi: 10.1093/ajcn/55.1.131. [DOI] [PubMed] [Google Scholar]
- 5.Selhub J. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J. Nutr. Health Aging. 2002;6:39–42. [PubMed] [Google Scholar]
- 6.Selhub J. Homocysteine metabolism. Annu. Rev. Nutr. 1999;19:217–246. doi: 10.1146/annurev.nutr.19.1.217. [DOI] [PubMed] [Google Scholar]
- 7.Clayton P.T. B6-responsive disorders: a model of vitamin dependency. J. Inherit. Metab. Dis. 2006;29:317–326. doi: 10.1007/s10545-005-0243-2. [DOI] [PubMed] [Google Scholar]
- 8.Bender D.A. Novel functions of vitamin B6. Proc. Nutr. Soc. 1994;53:625–630. doi: 10.1079/pns19940071. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe F. Vitamin B12 sources and bioavailability. Exp. Biol. Med. (Maywood) 2007;232:1266–1274. doi: 10.3181/0703-MR-67. [DOI] [PubMed] [Google Scholar]
- 10.Clarke R., Daly L., Robinson K., Naughten E., Cahalane S., Fowler B., Graham I. Hyperhomocysteinemia: an independent risk factor for vascular disease. N. Engl. J. Med. 1991;324:1149–1155. doi: 10.1056/NEJM199104253241701. [DOI] [PubMed] [Google Scholar]
- 11.Stampfer M.J., Malinow M.R., Willett W.C., Newcomer L.M., Upson B., Ullmann D., Tishler P.V., Hennekens C.H. A prospective study of plasma homocyst(e)ine and risk of myocardial infarction in US physicians. JAMA. 1992;268:877–881. [PubMed] [Google Scholar]
- 12.Selhub J., Jacques P.F., Bostom A.G., D'Agostino R.B., Wilson P.W., Belanger A.J., O'Leary D.H., Wolf P.A., Schaefer E.J., Rosenberg I.H. Association between plasma homocysteine concentrations and extracranial carotid-artery stenosis. N. Engl. J. Med. 1995;332:286–291. doi: 10.1056/NEJM199502023320502. [DOI] [PubMed] [Google Scholar]
- 13.Perry I.J., Refsum H., Morris R.W., Ebrahim S.B., Ueland P.M., Shaper A.G. Prospective study of serum total homocysteine concentration and risk of stroke in middle-aged British men. Lancet. 1995;346:1395–1398. doi: 10.1016/s0140-6736(95)92407-8. [DOI] [PubMed] [Google Scholar]
- 14.Mayer E.L., Jacobsen D.W., Robinson K. Homocysteine and coronary atherosclerosis. J. Am. Coll. Cardiol. 1996;27:517–527. doi: 10.1016/0735-1097(95)00508-0. [DOI] [PubMed] [Google Scholar]
- 15.Ridker P.M., Shih J., Cook T.J., Clearfield M., Downs J.R., Pradhan A.D., Weis S.E., Gotto A.M., Jr Plasma homocysteine concentration, statin therapy, and the risk of first acute coronary events. Circulation. 2002;105:1776–1779. [PubMed] [Google Scholar]
- 16.Selhub J. The many facets of hyperhomocysteinemia: studies from the Framingham cohorts. J. Nutr. 2006;136:1726S–1730S. doi: 10.1093/jn/136.6.1726S. [DOI] [PubMed] [Google Scholar]
- 17.Bird C.L., Swendseid M.E., Witte J.S., Shikany J.M., Hunt I.F., Frankl H.D., Lee E.R., Longnecker M.P., Haile R.W. Red cell and plasma folate, folate consumption, and the risk of colorectal adenomatous polyps. Cancer Epidemiol. Biomarkers Prev. 1995;4:709–714. [PubMed] [Google Scholar]
- 18.Giovannucci E., Stampfer M.J., Colditz G.A., Rimm E.B., Trichopoulos D., Rosner B.A., Speizer F.E., Willett W.C. Folate, methionine, and alcohol intake and risk of colorectal adenoma. J. Natl Cancer Inst. 1993;85:875–884. doi: 10.1093/jnci/85.11.875. [DOI] [PubMed] [Google Scholar]
- 19.Tseng M., Murray S.C., Kupper L.L., Sandler R.S. Micronutrients and the risk of colorectal adenomas. Am. J. Epidemiol. 1996;144:1005–1014. doi: 10.1093/oxfordjournals.aje.a008871. [DOI] [PubMed] [Google Scholar]
- 20.Giovannucci E., Stampfer M.J., Colditz G.A., Hunter D.J., Fuchs C., Rosner B.A., Speizer F.E., Willett W.C. Multivitamin use, folate, and colon cancer in women in the Nurses’ Health Study. Ann. Intern. Med. 1998;129:517–524. doi: 10.7326/0003-4819-129-7-199810010-00002. [DOI] [PubMed] [Google Scholar]
- 21.Andres E., Federici L., Affenberger S., Vidal-Alaball J., Loukili N.H., Zimmer J., Kaltenbach G. B12 deficiency: a look beyond pernicious anemia. J. Fam. Pract. 2007;56:537–542. [PubMed] [Google Scholar]
- 22.Hall C.A., Finkler A.E. Measurement of the amounts of the individual vitamin B12 binding proteins in plasma. II. Abnormalities in leukemia and pernicious anemia. Blood. 1966;27:618–622. [PubMed] [Google Scholar]
- 23.Klerk M., Verhoef P., Clarke R., Blom H.J., Kok F.J., Schouten E.G. MTHFR 677C–>T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA. 2002;288:2023–2031. doi: 10.1001/jama.288.16.2023. [DOI] [PubMed] [Google Scholar]
- 24.van Oijen M.G., Vlemmix F., Laheij R.J., Paloheimo L., Jansen J.B., Verheugt F.W. Hyperhomocysteinaemia and vitamin B12 deficiency: the long-term effects in cardiovascular disease. Cardiology. 2007;107:57–62. doi: 10.1159/000093746. [DOI] [PubMed] [Google Scholar]
- 25.Dhonukshe-Rutten R.A., de Vries J.H., de Bree A., van der Put N., van Staveren W.A., de Groot L.C. Dietary intake and status of folate and vitamin B12 and their association with homocysteine and cardiovascular disease in European populations. Eur. J. Clin. Nutr. 2009;63:18–30. doi: 10.1038/sj.ejcn.1602897. [DOI] [PubMed] [Google Scholar]
- 26.Dahlin A.M., Van Guelpen B., Hultdin J., Johansson I., Hallmans G., Palmqvist R. Plasma vitamin B12 concentrations and the risk of colorectal cancer: a nested case-referent study. Int. J. Cancer. 2008;122:2057–2061. doi: 10.1002/ijc.23299. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S.M., Willett W.C., Selhub J., Hunter D.J., Giovannucci E.L., Holmes M.D., Colditz G.A., Hankinson S.E. Plasma folate, vitamin B6, vitamin B12, homocysteine, and risk of breast cancer. J. Natl Cancer Inst. 2003;95:373–380. doi: 10.1093/jnci/95.5.373. [DOI] [PubMed] [Google Scholar]
- 28.Hultdin J., Van Guelpen B., Bergh A., Hallmans G., Stattin P. Plasma folate, vitamin B12, and homocysteine and prostate cancer risk: a prospective study. Int. J. Cancer. 2005;113:819–824. doi: 10.1002/ijc.20646. [DOI] [PubMed] [Google Scholar]
- 29.Xu X., Chen J. One-carbon metabolism and breast cancer: an epidemiological perspective. J. Genet. Genomics. 2009;36:203–214. doi: 10.1016/S1673-8527(08)60108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galvan-Portillo M.V., Cantoral A., Onate-Ocana L.F., Chen J., Herrera-Goepfert R., Torres-Sanchez L., Hernandez-Ramirez R.U., Palma-Coca O., Lopez-Carrillo L. Gastric cancer in relation to the intake of nutrients involved in one-carbon metabolism among MTHFR 677 TT carriers. Eur. J. Nutr. 2009;48:269–276. doi: 10.1007/s00394-009-0010-5. [DOI] [PubMed] [Google Scholar]
- 31.Johansson M., Van Guelpen B., Vollset S.E., Hultdin J., Bergh A., Key T., Midttun O., Hallmans G., Ueland P.M., Stattin P. One-carbon metabolism and prostate cancer risk: prospective investigation of seven circulating B vitamins and metabolites. Cancer Epidemiol. Biomarkers Prev. 2009;18:1538–1543. doi: 10.1158/1055-9965.EPI-08-1193. [DOI] [PubMed] [Google Scholar]
- 32.Maruti S.S., Ulrich C.M., White E. Folate and one-carbon metabolism nutrients from supplements and diet in relation to breast cancer risk. Am. J. Clin. Nutr. 2009;89:624–633. doi: 10.3945/ajcn.2008.26568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke R., Smith A.D., Jobst K.A., Refsum H., Sutton L., Ueland P.M. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch. Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- 34.Kang J.H., Irizarry M.C., Grodstein F. Prospective study of plasma folate, vitamin B12, and cognitive function and decline. Epidemiology. 2006;17:650–657. doi: 10.1097/01.ede.0000239727.59575.da. [DOI] [PubMed] [Google Scholar]
- 35.Selhub J., Morris M.S., Jacques P.F., Rosenberg I.H. Folate-vitamin B-12 interaction in relation to cognitive impairment, anemia, and biochemical indicators of vitamin B-12 deficiency. Am. J. Clin. Nutr. 2009;89:702S–706S. doi: 10.3945/ajcn.2008.26947C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.E., Li H., Giovannucci E., Lee I.M., Selhub J., Stampfer M., Ma J. Prospective study of plasma vitamin B6 and risk of colorectal cancer in men. Cancer Epidemiol. Biomarkers Prev. 2009;18:1197–1202. doi: 10.1158/1055-9965.EPI-08-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei E.K., Giovannucci E., Selhub J., Fuchs C.S., Hankinson S.E., Ma J. Plasma vitamin B6 and the risk of colorectal cancer and adenoma in women. J. Natl Cancer Inst. 2005;97:684–692. doi: 10.1093/jnci/dji116. [DOI] [PubMed] [Google Scholar]
- 38.Vermeulen S.H., van der Vleuten G.M., de Graaf J., Hermus A.R., Blom H.J., Stalenhoef A.F., den Heijer M. A genome-wide linkage scan for homocysteine levels suggests three regions of interest. J. Thromb. Haemost. 2006;4:1303–1307. doi: 10.1111/j.1538-7836.2006.01977.x. [DOI] [PubMed] [Google Scholar]
- 39.Cesari M., Burlina A.B., Narkiewicz K., Sartori M.T., Sacchetto A., Rossi G.P. Are fasting plasma homocyst(e)ine levels heritable? A study of normotensive twins. J. Investig. Med. 2000;48:351–358. [PubMed] [Google Scholar]
- 40.den Heijer M., Graafsma S., Lee S.Y., van Landeghem B., Kluijtmans L., Verhoef P., Beaty T.H., Blom H. Homocysteine levels—before and after methionine loading—in 51 Dutch families. Eur. J. Hum. Genet. 2005;13:753–762. doi: 10.1038/sj.ejhg.5201389. [DOI] [PubMed] [Google Scholar]
- 41.Jee S.H., Song K.S., Shim W.H., Kim H.K., Suh I., Park J.Y., Won S.Y., Beaty T.H. Major gene evidence after MTHFR-segregation analysis of serum homocysteine in families of patients undergoing coronary arteriography. Hum. Genet. 2002;111:128–135. doi: 10.1007/s00439-002-0757-8. [DOI] [PubMed] [Google Scholar]
- 42.Kullo I.J., Ding K., Boerwinkle E., Turner S.T., Mosley T.H., Jr, Kardia S.L., de Andrade M. Novel genomic loci influencing plasma homocysteine levels. Stroke. 2006;37:1703–1709. doi: 10.1161/01.STR.0000225929.96190.b3. [DOI] [PubMed] [Google Scholar]
- 43.Siva A., De Lange M., Clayton D., Monteith S., Spector T., Brown M.J. The heritability of plasma homocysteine, and the influence of genetic variation in the homocysteine methylation pathway. QJM. 2007;100:495–499. doi: 10.1093/qjmed/hcm054. [DOI] [PubMed] [Google Scholar]
- 44.Hubner R.A., Houlston R.S. MTHFR C677T and colorectal cancer risk: A meta-analysis of 25 populations. Int. J. Cancer. 2007;120:1027–1035. doi: 10.1002/ijc.22440. [DOI] [PubMed] [Google Scholar]
- 45.Larsson S.C., Giovannucci E., Wolk A. Folate and risk of breast cancer: a meta-analysis. J. Natl Cancer Inst. 2007;99:64–76. doi: 10.1093/jnci/djk006. [DOI] [PubMed] [Google Scholar]
- 46.Shan X., Wang L., Hoffmaster R., Kruger W.D. Functional characterization of human methylenetetrahydrofolate reductase in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:32613–32618. doi: 10.1074/jbc.274.46.32613. [DOI] [PubMed] [Google Scholar]
- 47.Goyette P., Rozen R. The thermolabile variant 677C–>T can further reduce activity when expressed in cis with severe mutations for human methylenetetrahydrofolate reductase. Hum. Mutat. 2000;16:132–138. doi: 10.1002/1098-1004(200008)16:2<132::AID-HUMU5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 48.Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet. 2008;40:1160–1162. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka T., Scheet P., Giusti B., Bandinelli S., Piras M.G., Usala G., Lai S., Mulas A., Corsi A.M., Vestrini A., et al. Genome-wide Association Study of Vitamin B6, Vitamin B12, Folate, and Homocysteine Blood Concentrations. Am. J. Hum. Genet. 2009;84:477–482. doi: 10.1016/j.ajhg.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pe'er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 51.Weir B.S. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Associates, Inc; 1996. [Google Scholar]
- 52.Paré G., Chasman D.I., Parker A.N., Zee R.R.Y., Mälarstig A., Seedorf U., Collins R., Watkins H., Hamsten A., Miletich J.P., et al. Novel Associations of CPS1, MUT, NOX4 and DPEP1 With Plasma Homocysteine in a Healthy Population: A Genome-Wide Evaluation of 13 974 Participants in the Women's Genome Health Study. Circ. Cardiovasc. Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q.H., Botto L.D., Gallagher M., Friedman J.M., Sanders C.L., Koontz D., Nikolova S., Erickson J.D., Steinberg K. Prevalence and effects of gene-gene and gene-nutrient interactions on serum folate and serum total homocysteine concentrations in the United States: findings from the third National Health and Nutrition Examination Survey DNA Bank. Am. J. Clin. Nutr. 2008;88:232–246. doi: 10.1093/ajcn/88.1.232. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y.I. Role of folate in colon cancer development and progression. J. Nutr. 2003;133:3731S–3739S. doi: 10.1093/jn/133.11.3731S. [DOI] [PubMed] [Google Scholar]
- 55.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J., den Heijer M., Kluijtmans L.A., van den Heuvel L.P., et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 56.Jacques P.F., Bostom A.G., Williams R.R., Ellison R.C., Eckfeldt J.H., Rosenberg I.H., Selhub J., Rozen R. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- 57.Sun D., Huang W., Hwang Y.Y., Zhang Y., Zhang Q., Li M.D. Regulation by nicotine of Gpr51 and Ntrk2 expression in various rat brain regions. Neuropsychopharmacology. 2007;32:110–116. doi: 10.1038/sj.npp.1301134. [DOI] [PubMed] [Google Scholar]
- 58.Refsum H., Nurk E., Smith A.D., Ueland P.M., Gjesdal C.G., Bjelland I., Tverdal A., Tell G.S., Nygard O., Vollset S.E. The Hordaland Homocysteine Study: a community-based study of homocysteine, its determinants, and associations with disease. J. Nutr. 2006;136:1731S–1740S. doi: 10.1093/jn/136.6.1731S. [DOI] [PubMed] [Google Scholar]
- 59.Stein J.H., Bushara M., Bushara K., McBride P.E., Jorenby D.E., Fiore M.C. Smoking cessation, but not smoking reduction, reduces plasma homocysteine levels. Clin. Cardiol. 2002;25:23–26. doi: 10.1002/clc.4950250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacques P.F., Bostom A.G., Wilson P.W., Rich S., Rosenberg I.H., Selhub J. Determinants of plasma total homocysteine concentration in the Framingham Offspring cohort. Am. J. Clin. Nutr. 2001;73:613–621. doi: 10.1093/ajcn/73.3.613. [DOI] [PubMed] [Google Scholar]
- 61.Targher G., Bertolini L., Zenari L., Cacciatori V., Muggeo M., Faccini G., Zoppini G. Cigarette smoking and plasma total homocysteine levels in young adults with type 1 diabetes. Diabetes Care. 2000;23:524–528. doi: 10.2337/diacare.23.4.524. [DOI] [PubMed] [Google Scholar]
- 62.Nygard O., Vollset S.E., Refsum H., Stensvold I., Tverdal A., Nordrehaug J.E., Ueland M., Kvale G. Total plasma homocysteine and cardiovascular risk profile. The Hordaland Homocysteine Study. JAMA. 1995;274:1526–1533. doi: 10.1001/jama.1995.03530190040032. [DOI] [PubMed] [Google Scholar]
- 63.Ogasawara M., Matsubara Y., Mikami H., Narisawa K. Identification of two novel mutations in the methylmalonyl-CoA mutase gene with decreased levels of mutant mRNA in methylmalonic acidemia. Hum. Mol. Genet. 1994;3:867–872. doi: 10.1093/hmg/3.6.867. [DOI] [PubMed] [Google Scholar]
- 64.Thoma N.H., Leadlay P.F. Homology modeling of human methylmalonyl-CoA mutase: a structural basis for point mutations causing methylmalonic aciduria. Protein Sci. 1996;5:1922–1927. doi: 10.1002/pro.5560050919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikami H., Ogasawara M., Matsubara Y., Kikuchi M., Miyabayashi S., Kure S., Narisawa K. Molecular analysis of methylmalonyl-CoA mutase deficiency: identification of three missense mutations in mut0 patients. J. Hum. Genet. 1999;44:35–39. doi: 10.1007/s100380050103. [DOI] [PubMed] [Google Scholar]
- 66.Acquaviva C., Benoist J.F., Pereira S., Callebaut I., Koskas T., Porquet D., Elion J. Molecular basis of methylmalonyl-CoA mutase apoenzyme defect in 40 European patients affected by mut(o) and mut- forms of methylmalonic acidemia: identification of 29 novel mutations in the MUT gene. Hum. Mutat. 2005;25:167–176. doi: 10.1002/humu.20128. [DOI] [PubMed] [Google Scholar]
- 67.Drennan C.L., Matthews R.G., Rosenblatt D.S., Ledley F.D., Fenton W.A., Ludwig M.L. Molecular basis for dysfunction of some mutant forms of methylmalonyl-CoA mutase: deductions from the structure of methionine synthase. Proc. Natl Acad. Sci. USA. 1996;93:5550–5555. doi: 10.1073/pnas.93.11.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ledley F.D., Crane A.M., Lumetta M. Heterogeneous alleles and expression of methylmalonyl CoA mutase in mut methylmalonic acidemia. Am. J. Hum. Genet. 1990;46:539–547. [PMC free article] [PubMed] [Google Scholar]
- 69.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 70.Aminoff M., Carter J.E., Chadwick R.B., Johnson C., Grasbeck R., Abdelaal M.A., Broch H., Jenner L.B., Verroust P.J., Moestrup S.K., et al. Mutations in CUBN, encoding the intrinsic factor-vitamin B12 receptor, cubilin, cause hereditary megaloblastic anaemia 1. Nat. Genet. 1999;21:309–313. doi: 10.1038/6831. [DOI] [PubMed] [Google Scholar]
- 71.Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 72.Franke B., Vermeulen S.H., Steegers-Theunissen R.P., Coenen M.J., Schijvenaars M.M., Scheffer H., den Heijer M., Blom H.J. An association study of 45 folate-related genes in spina bifida: involvement of cubilin (CUBN) and tRNA aspartic acid methyltransferase 1 (TRDMT1) Birth Defects Res. A Clin. Mol. Teratol. 2009;85:216–226. doi: 10.1002/bdra.20556. [DOI] [PubMed] [Google Scholar]
- 73.Johnston J., Yang-Feng T., Berliner N. Genomic structure and mapping of the chromosomal gene for transcobalamin I (TCN1): comparison to human intrinsic factor. Genomics. 1992;12:459–464. doi: 10.1016/0888-7543(92)90435-u. [DOI] [PubMed] [Google Scholar]
- 74.Kelly R.J., Rouquier S., Giorgi D., Lennon G.G., Lowe J.B. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 75.van Oijen M.G., Laheij R.J., de Jong C.A., Peters W.H., Jansen J.B. Vitamin B12 status and its association with Helicobacter pylori infection in alcohol dependent patients. J. Nutr. Sci. Vitaminol. (Tokyo) 2004;50:305–308. doi: 10.3177/jnsv.50.305. [DOI] [PubMed] [Google Scholar]
- 76.Dholakia K.R., Dharmarajan T.S., Yadav D., Oiseth S., Norkus E.P., Pitchumoni C.S. Vitamin B12 deficiency and gastric histopathology in older patients. World J. Gastroenterol. 2005;11:7078–7083. doi: 10.3748/wjg.v11.i45.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Annibale B., Capurso G., Delle Fave G. Consequences of Helicobacter pylori infection on the absorption of micronutrients. Dig. Liver Dis. 2002;34(Suppl. 2):S72–S77. doi: 10.1016/s1590-8658(02)80170-0. [DOI] [PubMed] [Google Scholar]
- 78.Tamura A., Fujioka T., Nasu M. Relation of Helicobacter pylori infection to plasma vitamin B12, folic acid, and homocysteine levels in patients who underwent diagnostic coronary arteriography. Am. J. Gastroenterol. 2002;97:861–866. doi: 10.1111/j.1572-0241.2002.05601.x. [DOI] [PubMed] [Google Scholar]
- 79.Kaptan K., Beyan C., Ural A.U., Cetin T., Avcu F., Gulsen M., Finci R., Yalcin A. Helicobacter pylori—is it a novel causative agent in Vitamin B12 deficiency? Arch. Intern. Med. 2000;160:1349–1353. doi: 10.1001/archinte.160.9.1349. [DOI] [PubMed] [Google Scholar]
- 80.Whyte M.P., Mahuren J.D., Fedde K.N., Cole F.S., McCabe E.R., Coburn S.P. Perinatal hypophosphatasia: tissue levels of vitamin B6 are unremarkable despite markedly increased circulating concentrations of pyridoxal-5′-phosphate. Evidence for an ectoenzyme role for tissue-nonspecific alkaline phosphatase. J. Clin. Invest. 1988;81:1234–1239. doi: 10.1172/JCI113440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whyte M.P., Mahuren J.D., Vrabel L.A., Coburn S.P. Markedly increased circulating pyridoxal-5′-phosphate levels in hypophosphatasia. Alkaline phosphatase acts in vitamin B6 metabolism. J. Clin. Invest. 1985;76:752–756. doi: 10.1172/JCI112031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson B.B., O'Brien H., Griffin G.E., Mollin D.L. Hydrolysis of pyridoxal-5′-phosphate in plasma in conditions with raised alkaline phosphate. Gut. 1980;21:192–194. doi: 10.1136/gut.21.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDowell M.A., Lacher D.A., Pfeiffer C.M., Mulinare J., Picciano M.F., Rader J.I., Yetley E.A., Kennedy-Stephenson J., Johnson C.L. Blood folate levels: the latest NHANES results. NCHS Data Brief. 2008;6:1–8. [PubMed] [Google Scholar]
- 84.Selhub J., Jacques P.F., Bostom A.G., Wilson P.W., Rosenberg I.H. Relationship between plasma homocysteine and vitamin status in the Framingham study population. Impact of folic acid fortification. Public Health Rev. 2000;28:117–145. [PubMed] [Google Scholar]
- 85.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kannel W.B. The Framingham Study: ITS 50-year legacy and future promise. J Atheroscler. Thromb. 2000;6:60–66. doi: 10.5551/jat1994.6.60. [DOI] [PubMed] [Google Scholar]
- 87.Herbert A., Lenburg M.E., Ulrich D., Gerry N.P., Schlauch K., Christman M.F. Open-access database of candidate associations from a genome-wide SNP scan of the Framingham Heart Study. Nat. Genet. 2007;39:135–136. doi: 10.1038/ng0207-135. [DOI] [PubMed] [Google Scholar]
- 88.Dehghan A., Kottgen A., Yang Q., Hwang S.J., Kao W.L., Rivadeneira F., Boerwinkle E., Levy D., Hofman A., Astor B.C., et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilk J.B., Chen T.H., Gottlieb D.J., Walter R.E., Nagle M.W., Brandler B.J., Myers R.H., Borecki I.B., Silverman E.K., Weiss S.T., et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Araki A., Sako Y. Determination of free and total homocysteine in human plasma by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. 1987;422:43–52. doi: 10.1016/0378-4347(87)80438-3. [DOI] [PubMed] [Google Scholar]
- 91.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 92.Lettre G., Lange C., Hirschhorn J.N. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet. Epidemiol. 2007;31:358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 93.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 95.Monsees G.M., Tamimi R.M., Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet. Epidemiol. 2009 doi: 10.1002/gepi.20424. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crane A.M., Martin L.S., Valle D., Ledley F.D. Phenotype of disease in three patients with identical mutations in methylmalonyl CoA mutase. Hum. Genet. 1992;89:259–264. doi: 10.1007/BF00220536. [DOI] [PubMed] [Google Scholar]
- 98.Crane A.M., Jansen R., Andrews E.R., Ledley F.D. Cloning and expression of a mutant methylmalonyl coenzyme A mutase with altered cobalamin affinity that causes mut- methylmalonic aciduria. J. Clin. Invest. 1992;89:385–391. doi: 10.1172/JCI115597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.