Abstract
Cadherin-11/Cdh11 is expressed through early development and strongly during inner ear development (otic placode and vesicle). Here we show that antisense knockdown of Cdh11 during early zebrafish development interferes with otolith formation. Immunofluorescence labeling showed that Cdh11 expression was concentrated on and within the otolith. Cdh11 was faintly detected at the lateral surface (sites of cell-cell contact) of otic epithelial cells and in the cytoplasm. Strongly labeled Cdh11 containing puncta were detected within the otolymph (the fluid within the otic vesicle) and associated with the otolith surface. BODIPY-ceramine labeled vesicular structures detected in the otolymph were larger and more numerous in Cdh11 knockdown embryos. We present evidence supporting a working model that vesicular structures containing Cdh11 (perhaps containing biomineralization components) are exported from the otic epithelium into the otolymph, adhere to one another and to the surface of the growing otolith, facilitating otolith growth.
Keywords: Cadherin-11, otolith, zebrafish, ear development, antisense oligonucleotide
Introduction
Cadherins are homotypic cell adhesion molecules, which provide strong, dynamic adhesion activity in cells. Cadherins are typically associated with an adhesive, actin-associated structure in the cell, called the adherens junction. These adhesion molecules control morphogenesis, cell migration and cell shape changes during a variety of developmental processes (Gumbiner, 2005; Halbleib and Nelson, 2006). Cadherin activity also participates in differentiation events during embryogenesis, for example, in bone formation (Mbalaviele et al., 2006). We have initiated a series of studies to determine the functions of cadherin cell adhesion molecules during inner ear and lateral line system development using zebrafish as a model system (Novince et al., 2003; Kerstetter et al., 2004; Babb-Clendenon et al., 2006; Wilson et al., 2007). Several cadherins are expressed in the developing inner ear (Novince et al., 2003). Cadherin-11/Cdh11 (previously called VN-cadherin and OB-cadherin) expression was detected early during gastrulation, during neural development, and in the forming inner ear (Franklin and Sargent, 1996). Cdh11 knockout mice display defects in bone formation, bone growth and arthritis pathogenesis (Kawaguchi et al., 2001; Lee et al., 2007), indicating that Cdh11 plays a role in biomineralization.
After the otic placode forms in fish and frogs, these cells delaminate from the ectoderm and form a fluid filled otic vesicle or cyst from a solid cell mass by cavitation (Nicolson, 2005). In other vertebrates, the otocyst forms by invagination, pinching off a cystic structure from the otic placode cells (Barald and Kelley, 2004). Zebrafish otic induction and ectodermal thickening (otic placode) is evident at 16 hours postfertilization (hpf). By 18 hpf, cavitation has occurred, and hair cells differentiate soon afterwards. Otolith nucleation occurs around 20 hpf in association with kinocilia of the newly differentiated hair cells. Neuroblasts delaminate and begin to form the statoacoustic ganglion during this time period. Later, semicircular canal formation initiates, forming epithelial bridges around 45 hpf that fuse to separate the three fluid canals by 55 hpf. Semicircular canal growth proceeds for several days, and by 96 hpf ears are fully functional (Nicolson, 2005). Genetic analysis in the zebrafish and other vertebrates has identified genes that control these various processes (Malicki et al., 1996; Whitfield et al., 1996; Riley and Grunwald, 1996).
The inner ear detects gravitation, linear acceleration, perceives body orientation in free space, and detects sound. Inner ear development is conserved among vertebrates (Nicolson, 2005). Otic placode induction is initiated by signals from the hindbrain and/or periotic mesoderm, and hindbrain signaling helps shape the developing otic vesicle (Barald and Kelley, 2004). The otic vesicle undergoes a morphogenetic transformation to produce three semicircular canals (Malicki et al., 1996; Whitfield et al., 1996; Babb-Clendenon et al., 2006; Blasiole et al., 2006), the sacculus, the utricle and in most vertebrates the basilar papilla or the cochlea (Barald and Kelley, 2004). Within the sensory organs, differentiation produces sensory epithelial patches containing hair cells that detect and transduce fluid movement forces (Malicki et al., 1996; Whitfield et al., 1996; Barald and Kelley, 2004; Nicolson, 2005). To heighten sensitivity to acceleration and to detect gravitation, inertial structures, otoliths (“ear stones” in fish and frogs: homologous structures in the mammalian sacculus and utricle are called otoconia) are assembled and attached to the sensory patches via the kinocilia (Riley and Grunwald, 1996; Riley et al., 1997; Hughes et al., 2006).
Normal otolith (and otoconia) formation requires that a series of developmental processes to occur (Riley et al., 1997; Nicolson, 2005; Hughes et al., 2006). First, the otic placode is induced, and otic structures differentiate, including the sensory patches (Barald and Kelley, 2004; Nicolson, 2005). Otolith assembly initiates in association with the sensory patch, particularly associated with the developing kinocilium (Riley et al., 1997; Fekete, 2003; Murayama et al., 2005). Appropriate ionic transport processes develop to establish the proper ionic environment for otolith growth (Ibsch et al., 2004; Hardison et al., 2005; Blasiole et al., 2006). Several proteins, including otolith matrix proteins are secreted by the otic epithelium that control biomineralization or control attachment of otoliths to the sensory patch (Sollner et al., 2003; Hughes et al., 2004; Sollner et al., 2004; Murayama et al., 2005). Rapid growth may require efficient delivery of assembly materials to the surface of the growing otolith (Hughes et al., 2006). Many mechanistic details of otolith nucleation and assembly are unknown in any species. The zebrafish provides a useful model to study otolith formation (Sollner and Nicolson, 2005; Hughes et al., 2006).
Vesicular structures within otolymph were hypothesized to participate in fish otolith (and mammalian otoconia) formation (Hughes et al., 2006). Riley et al. (1997) observed particles (termed “seeding particles”) that moved rapidly and coalesce during otolith assembly. Here, we document an inner ear defect resulting from cadherin-11 gene (cdh11) loss-of-function: reduced and absent otoliths. We present evidence that membrane structures are released from zebrafish otic epithelial cell surfaces that may contain Cdh11. These membrane structures are found within the otolymph fluid that fills the otic vesicle and are associated with (presumably adhering to) the surfaces of growing otoliths. Cdh11 loss-of-function reduces otolith assembly. Our evidence supports a working model: Cdh11 on the surface of membrane structures that are exported from the otic vesicle epithelium facilitates their adhesion to one another and to the surface of the growing otolith. We speculate that other components required for otolith assembly may be found in Cdh11 containing vesicles, and our proposed adhesive mechanism would facilitate efficient delivery and concentration of assembly components at the growing surface of the otolith.
Results
Cdh11 knockdown phenotype: reduced or absent otolith formation
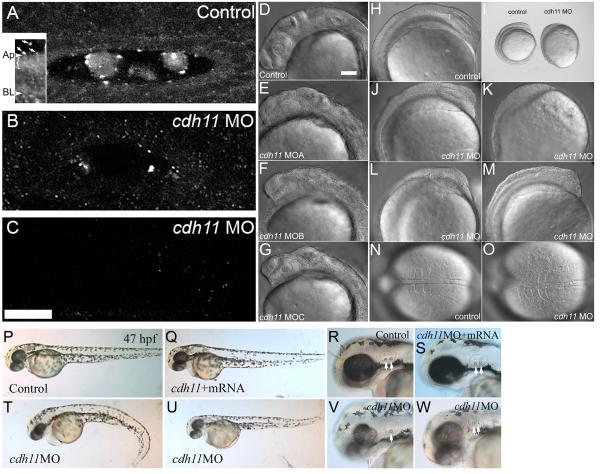
To examine cdh11 loss-of-function, we designed splice junction-blocking morpholino oligonucleotides (Draper et al., 2001; Morcos, 2007; Eisen and Smith, 2008). Injecting these morpholino oligonucleotides into 1-4 cell stage embryos reduced cadherin-11 protein expression throughout the embryo (not shown), and of particular interest for these studies, in the inner ear (Fig. 1A-C). Immunoblot analysis using our Cdh11 specific antiserum (see Supplemental Data, Fig. S1A-C) showed reduced expression in cdh11 morpholino oligonucleotide injected (knockdown) embryos as compared to control morpholino oligonucleotide injected embryos (see Supplemental Data, Fig. S1D). We categorized the morphant phenotype into graded series of effects, noting a gradient of severity in effects on body length, tail curl, pigmentation, hindbrain, eyes, ears and otoliths. Using these observations, we examined whether phenotypic severity correlated with Cdh11 protein expression. Immunofluorescence is semi-quantitative when using a confocal microscope at equal laser power, gain settings and sub-saturation conditions for the photomultiplier tube. We found that expression levels correlated with severity of the phenotype (Fig. 1A-C). Cdh11 expression was reduced in slightly affected embryos (near normal body length, no tail curl, reduced pigmentation, hindbrain defects; Fig. 1B); Cdh11 expression was nearly undetectable in moderately affected embryos (short body length, slight tail curl, small eyes, small otoliths; Fig. 1C); and Cdh11 was undetectable in severely affected embryos (very short body length, tail extremely curled, very small eyes, very small and sometimes absent otoliths; data not shown). In immunofluorescence image volumes that contain the entire otic vesicle of a slightly affected embryo showed a small amount of labeling (see Supplemental Data, Fig. S2A), and a moderately affected embryo showed little or no labeling except a background level of fine, punctate staining (see Supplemental Data, Fig. S2B; higher background is due to summation of the background from 25 optical sections). Increasing amounts of Cdh11 morpholino oligonucleotide produced more severely affected embryos (Table I), indicating that Cdh11 knockdown phenotype strength was concentration dependent. Finally, three independent morpholino oligonucleotides induced the identical phenotype (Fig. 1D-G; Table II), indicating that the phenotype is specifically due to cdh11 loss-of-function and not non-specific effects of the morpholino oligonucleotide reagent injection.
Figure 1. Morpholino oligonucleotide knockdown reduces Cdh11 expression in zebrafish embryos, causing defects in epiboly, gastrulation and neural development.
Zebrafish were injected with control morpholino oligonucleotide (A) or cdh11 splice-site blocking antisense morpholino oligonucleotide (B, C), and these embryos (30 hpf) were processed for wholemount immunofluorescence using antibodies raised to Cdh11 peptide sequences. Morpholino oligonucleotide knockdown produces a series of hypomorph phenotypes that were categorized as slight (near normal body length, no tail curl, reduced pigmentation, hindbrain defects), moderate (short body length, slight tail curl, small eyes, small otoliths) and severe (very short body length, tail extremely curled, very small eyes, very small or absent otoliths). Small amounts of Cdh11 staining were detected in slightly affected phenotype embryos (B), but no Cdh11 staining above background was detected in moderate (C) and severe (not shown) embryos. Panels A, B and C have identical orientation, with rostral to the left and lateral toward the top. All are projection images of five optical sections (0.3 μm thick) through the middle of the otic vesicle. Otoliths were not minimized by choice of plane in knockdown embryos. Laser power, gain settings, filters and image processing were identical for panels A, B and C. Two-photon imaging was performed using Zeiss LSM510 Meta NLO, C-Apochromatic 40×W 1.2NA objective, TRITC-labeled secondary antibody, 800 nm excitation, and mounted in PBS. Scale bar in panel C represents 25 microns, and applies to panels A-C. Inset shows an image that was processed to highlight abundant Cdh11 labeling in the apical cytoplasm (bracket) of otic epithelial cells. Apical (Ap) and basal (BL) plasma membrane is indicated. Arrows indicate Cdh11 labeled puncta outside the otic epithelial cells, in the otolymph. Specificity of the Cdh11 peptide antibody was validated by competition of the immunoreaction with the immunizing peptide detected by immunoblot and immunofluorescence (see Supplementary Figure S1). Three morpholino oligonucleotides, cdh11MOA, cdh11MOB and cdh11MOC, are described in Materials and Methods, which were designed to hybridize with intron-exon splicing border sequences. These morpholino oligonucleotides were tested for effects on Cdh11 expression and their ability to induce aberrant phenotypes. The three morpholino oligonucleotides produced identical phenotypes (compare control, panel D, with experimental morpholino injected, panels E-G) and with similar efficacy (not shown). DIC and stereomicroscope images of 8-somite control morpholino oligonucleotide injected embryos (H, I, N) with cdh11 antisense morpholino oligonucleotide injected embryos at the same developmental age (15 hpf; I-M, O) were compared. Scale bar in D represents 100 microns for panels D-H, J-O. Defects in epiboly were evident: panel I shows that the embryo proper completely engulfs the large yolk cell in the control embryo, but the embryo proper is only at around 80-90% epiboly in the cdh11 antisense morpholino oligonucleotide injected embryo shown in panel I. The phenotype varied in severity and examples are shown in panels J-M. Dorsal DIC images of the somites are shown in panels N and O. In the cdh11 antisense morpholino oligonucleotide injected embryo shown in panel O, the somites are compressed in the anterior-posterior axis (left to right) and expanded in the medial-lateral axis, indicative of convergence and extension cell movement defects. (P-W) Rescue experiment showing that synthetic cdh11 mRNA injection compensates for morpholino-induced defects. (P-R) Embryos (47 hpf) injected with control morpholino closely resemble (nearly indistinguishable from) embryos co-injected with cdh11 morpholino oligonucleotide and synthetic cdh11 mRNA (Q-S). Morphant phenotypes produced by injecting cdh11 morpholino oligonucleotide are shown (V-W): curled tail, reduced neural development, smaller eyes, etc. Higher magnification images show reduced inner ear size, reduced otolith size (arrows highlight otoliths; compare R and S with V and W). Also, a Cdh11 morphant embryo (V) had one small and one missing otolith.
Table I. Dose response to different amounts of cdh11 morpholino oligonucleotide.
Zebrafish embryos at 1-4 cell stage were injected with control morpholino or cdh11MOA at the volumes and concentrations shown. At 42-48 hpf, embryo phenotype was evaluated. These data are from a representative experiment. Using the following criteria, phenotype severity was evaluated and categorized into three groups. Normal embryos showed distinguishable phenotype. Slightly affected embryos displayed near normal body length, no tail curl, reduced pigmentation and hindbrain defects. Moderately affected embryos displayed short body length, slight tail curl, small eyes and small otoliths. Severely affected embryos displayed very short body length, extremely curled tail, very small eyes, very small and sometimes absent otoliths. Most embryo death may be due to disastrous effects of Cdh11 depletion on epiboly and early gastrulation. Statistically significant differences (P=0.0001 level) were detected among groups. Significantly more severe effects were detected in cdh11 MO 5 nL, 0.25 mM than all other groups. The effects of cdh11 MO 1-2 nL 0.5 mM were more severe than cdh11 MO 1-2 nL, 0.25 mM; Control MO 5 nL, 0.25 mM; and Control MO 1-2 nL, 0.25 mM. Also, cdh11 MO 1-2 nL 0.25 mM was significnantly worse than Control MO 5 nL, 0.25 mM, but cdh11 MO 1-2 nL 0.25 mM was not different from Control MO 1-2 nL, 0.25 mM. The two controls were also not significantly different from each other.
| Normal | Slight MO effect |
Moderate MO effect |
Severe MO effect |
Dead | |
|---|---|---|---|---|---|
| Control MO 1-2 nL, 0.25 mM | 2 | 0 | 0 | 0 | 0 |
| Control MO 5 nL, 0.25 mM | 6 | 0 | 0 | 0 | 0 |
| cdh11 MO 1-2 nL, 0.25 mM | 4 | 7 | 0 | 0 | 0 |
| cdh11 MO 1-2 nL, 0.5 mM | 0 | 6 | 10 | 1 | 0 |
| cdh11 MO 5 nL, 0.25 mM | 0 | 0 | 0 | 2 | 8 |
Table II. Phenotype severity comparison using three independent morpholino oligonucleotides.
As with all data figures, zebrafish embryos at 1-4 cell stage was injected with ~1-2 nL of 0.5 mM morpholino oligonucleotide (MO). For most data figures cdh11MOB was used inhibit Cdh11 expression. Here, standard control MO (n=41), cdh11MOA (n=17), cdh11MOB (n=93), or cdh11MOC (n=28) were used. At 42-48 hpf, embryo phenotype was evaluated. Using the following criteria, phenotype severity was evaluated and categorized into three groups. Slightly affected embryos displayed near normal body length, no tail curl, reduced pigmentation, hindbrain defects; moderately affected embryos displayed short body length, slight tail curl, small eyes, small otoliths; and severely affected embryos displayed very short body length, extremely curled tail, very small eyes, very small and sometimes absent otoliths. The three cdh11 specific MOs produced similar or identical phenotypes, but proportions of different severity levels varied (for cdh11MOC total percentage only adds to 99 because slight and severe were 8 individuals of 28 total, at two significant figures). Statistically significant differences were detected among the groups (P=0.0001 level). All cdh11 MO groups had significantly more severe effects than Control MO group. The three cdh11 MO groups did not have significantly different severity from each other.
| Normal | Slight | Moderate | Severe | |
|---|---|---|---|---|
| Control MO | 100% | 0 | 0 | 0 |
| cdh11MOA | 0 | 35% | 59% | 6% |
| cdh11MOB | 0 | 28% | 46% | 26% |
| cdh11MOC | 0 | 28% | 43% | 28% |
Additional confirmation of the cdh11 morpholino oligonucleotide induced phenotype specificity was provided by rescuing cdh11 loss-of-function phenotype with the co-injection of synthetic cdh11 mRNA. PCR cloning and subcloning cdh11 cDNA is described in Materials and Methods. Because we used splice blocking morpholino oligonucleotides, synthetic mRNA injection bypasses the splicing block. Injecting embryos with 0.5 mM cdh11 morpholino oligonucleotide produced mostly moderate or severe cdh11 knockdown phenotypes. Increasing numbers of normal/rescue embryos were produced when 0.5 mM cdh11 morpholino oligonucleotide was co-injected with increasing concentration (100 or 300 pg) of synthetic cdh11 mRNA (Table III). Very complete gross morphological rescue was observed (Fig. 1P-W). This finding, together with controls outlined above, strongly support the contention that cdh11 morpholino oligonucleotides produces a series of phenotypes (slight-to-severe) that result from increasing cdh11 loss-of-function.
Table III. Rescue of cdh11 knockdown phenotype by co-injecting synthetic cdh11 mRNA.
Zebrafish embryos at 1-4 cell stage were injected with ~1-2 nL of 0.5 mM morpholino oligonucleotide (MO); either, cdh11MOB alone (n=42); cdh11MOB and 100 pg mRNA (n=50); or cdh11MOB and 300 pg mRNA (n=50). At 42-48 hpf, embryo phenotype was evaluated. Using the following criteria, phenotype severity was evaluated and categorized into three groups. Normal embryos were indistinguishable from “rescue” embryos, i. e., embryos injected with morpholino oligonucleotide and synthetic mRNA. Embryos that were categorized as “partial rescue” were indistinguishable from slightly affected embryos that displayed near normal body length, no tail curl, reduced pigmentation and hindbrain defects. We used a morpholino concentration that produced few if any slightly affected embryos. Normal embryos in the ‘0.5 mM cdh11 MO’ row may have been injected incompletely, delivering a very low amount of morpholino. The moderately affected embryos displayed short body length, slight tail curl, small eyes, and small otoliths. Severely affected embryos displayed very short body length, extremely curled tail, very small eyes, very small and sometimes absent otoliths. ‘Non-specific effects’ were due to mechanical damage that severely disrupted development (e.g. little or no head, truncated body). This type of embryo is seen in other morpholino injection experiments targeting other genes and control injections (control morpholino or buffer alone). Statistically significant differences were detected among the groups (P=0.0001 level). Increasing levels of cdh11 mRNA were associated with decreased severity (P=0.0001 for 0.5 mM cdh11 MO vs. 0.5 mM cdh11 MO and 100 pg cdh11 mRNA; P=0.0001 for 0.5 mM cdh11 MO vs. 0.5 mM cdh11 MO and 300 pg cdh11 mRNA; and P=0.0325 for 0.5 mM cdh11 MO and 100 pg cdh11 mRNA vs. 0.5 mM cdh11 MO and 300 pg cdh11 mRNA).
| Normal/ Rescue |
Partial rescue |
Moderate MO effect |
Severe MO effect |
Non- Specific |
|
|---|---|---|---|---|---|
| 0.5 mM cdh11 MO | 14% | 0 | 12% | 67% | 7% |
|
0.5 mM cdh11 MO and
100 pg cdh11 mRNA |
50% | 36% | 6% | 0 | 8% |
|
0.5 mM cdh11 MO and
300 pg cdh11 mRNA |
84% | 10% | 0 | 0 | 6% |
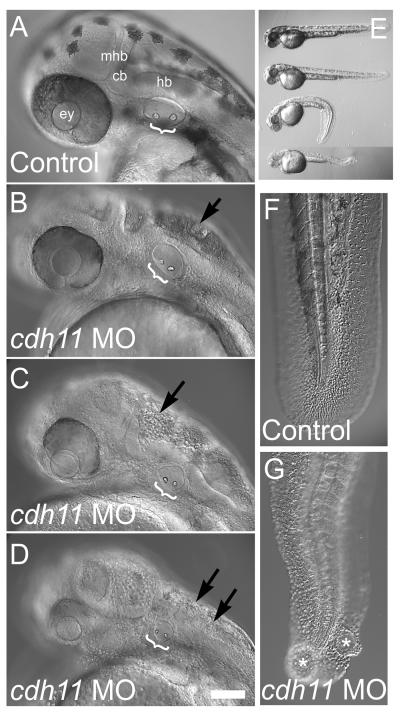
Embryonic development was affected in cdh11 morphants, including epiboly, gastrulation, eye, ear, brain, pigment cell and tail development. Cdh11 is expressed in the early gastrulating embryo, and early embryonic phenotypes were detected (Fig. 1H-O). We detected epiboly delay (Fig. 1I) and somite dysmorphogenesis in morphants (Fig 1N-O). Reduced somite anterior-posterior length and increased medial-lateral width suggests that Cdh11 knockdown reduces cell movements during gastrulation. Body length was also reduced in Cdh11 knockdown embryos (Fig. 2), consistent with an effect on convergence and extension cell movements. Reduced body length may be due to cell division or apoptosis rate changes, but this was not measured. Tail axis development was also affected. Tails were curled and had cysts in Cdh11 knockdown embryos (Fig. 2E-G). In addition, brain development defects were seen in Cdh11 knockdown embryos, showing considerable cell-cell dysadhesion (Fig. 2A-D; note: many rounded, cellular structures, which appear to be non-adherent cells, fill the fluid space of the hindbrain, fourth ventricle of morphants).
Figure 2. cdh11 loss-of-function neural and tail phenotype.
Zebrafish were injected with control morpholino oligonucleotide (A, E, F) or cdh11 antisense morpholino oligonucleotide (B, C, D, E, G). Embryos (48 hpf) were imaged using DIC microscopy (A, B, C, D, F, G; Nikon diaphot 200, Plan 20× 0.4NA objective using DIC) or a stereo microscope (E; Leica MZ12) using a SPOT Camera (Model 2.2.0 Diagnostic Instruments and Spot software). Increasing severity of cdh11 loss-of-function phenotype is illustrated: panel B, slightly affected embryo; panel C, moderately affected embryo; panel D, severely affected embryo. Brackets highlight otoliths within the inner ears. Effects on melanocytes, the neural axis and eye development were also evident. In panel E, a control-injected embryo is at the top, and the three embryos below are cdh11 morphants with slight, moderate and severe phenotypes. Higher magnification images of tails (F, G) are shown to illustrate that severely affected cdh11 loss-of-function embryos have significant tail curling, cyst formation (labeled with asterix in G) and rounded cells indicative of a dysadhesion effect during tail formation. Also, evidence of cellular disorganization and possibly dysadhesion was detected in the brains of all affected cdh11 antisense morpholino oligonucleotide-injected embryos (arrows indicate many rounded, cellular structures fill the fluid space of the hindbrain, fourth ventricle; see panels B, C, D). Scale bar in D represents 100 microns for panels A-D, F-G. Abbreviations: ey=eye, mhb=midbrain-hindbrain boundary, cb=cerebellum, hb=hindbrain.
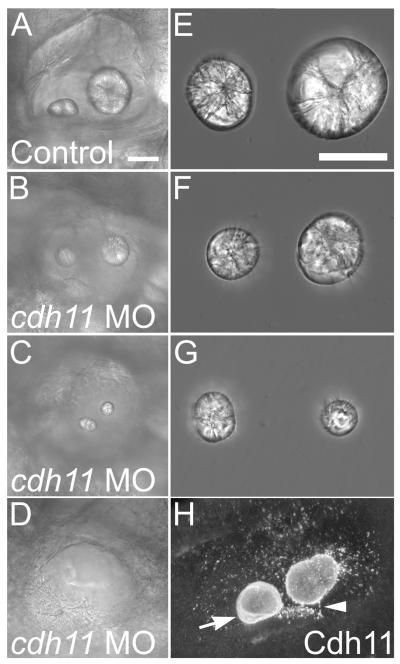
In the ear, we noted small or absent otoliths in the Cdh11 knockdown embryos (Fig. 2A-D and Fig. 3A-D). Otoliths were dissected from control and Cdh11 knockdown embryos to directly visualize effects on otolith growth (Fig. 3E-G). Reduced otolith size suggests that cdh11 loss-of-function reduces biocrystallization rate. Normal otoliths have a flat surface facing the sensory patch (Sollner and Nicolson, 2005). We noted that most knockdown embryos had otoliths that were more rounded on the surfaces facing sensory patches (not shown) similar to the surfaces normally seen on the opposite face, not facing the sensory patch. In some knockdown embryos, the otolith associated with the posterior macula was smaller than that associated with the anterior macula (Fig. 3G), which is the opposite of what is seen in control embryos where the otolith associated with the posterior macula is the larger of the two. Otic vesicles in some severely affected Cdh11 knockdown embryos lacked otoliths (Fig. 3D), which presumably represents the null phenotype. We conclude that otolith assembly was blocked by Cdh11 knockdown.
Figure 3. Otolith morphology in Cdh11 knockdown embryos.
Zebrafish were injected with control morpholino oligonucleotide (A, E) or cdh11 antisense morpholino oligonucleotide (B, C, D, F, G), and these embryos (7 dpf) (A-D) and their dissected otolith crystals (E-G) were imaged by DIC microscopy (Nikon diaphot 200: panels A-D, Plan 20× 0.4 NA objective; panels E-G 40× 0.55 NA objective). The otoliths shown in E, F and G were the same otoliths seen in the inner ears in A, B and C, respectively. Live embryos mounted in embryo medium (A-D) and otoliths were mounted in PBS. Note that the crystal diffraction patterns are similar, comparing control and morphant otoliths, but the sizes of morphant otolith crystals are reduced (F, G). The more caudal otolith (associated with the posterior macula) was smaller than the more rostral otolith in some morphants (for example, in C and G). Cdh11 immunofluorescence of 3 dpf wholemount labeled embryos mounted in PBS (H) shows annular patterns in the growing otolith (arrow), and abundant vesicular structures, many associated with the surface of the growing otolith (note: speckle pattern on otolith surface). Two photon microscopy using a BioRad MRC1024, 60× W 1.2 NA, 800 nm illumination, pixel size 0.39 micron, step size 0.4 micron. A projection image of 50 optical sections was made using Voxx Software in alpha blending mode. Arrowhead highlights a linear array of punctate Cdh11 immunolabeling that may represent association with a kinocilium. Scale bar in A represents 50 microns for panels A-D, and the scale bar in E represents 50 microns for panels E-G. For panels A-D and H, rostral is left, and dorsal is up. Figure was made with Photoshop 6, applying contrast stretch and unsharp mask.
Cdh11 immunofluorescence microscopy of the inner ear showed the most intense staining within the otolith crystal (Fig. 1A and 3H), showing that Cdh11 is part of the otolith matrix. Incubating antiserum with the immunizing peptide eliminated otolith and all other staining (see Supplementary Data, Fig. S1B-C). Also, prominent punctate structures within the otolymph were detected by immunofluorescence using Cdh11 antiserum, which may be vesicles since cadherins are integral membrane proteins; similar punctate structures were detected on the surface of the forming otoliths (Fig. 1A and 3H). Cdh11 was only faintly detected at the lateral domain (sites of cell-cell contact) and in cytoplasmic structures (possibly vesicles) within the cytoplasm of otic vesicle epithelial cells (Fig. 1A, inset), which appeared to be concentrated in a sub-apical location of the epithelial cells (Fig. 1A, bracket in inset). This sub-apical vesicular appearance may be due to exclusion from more basal cytoplasm by the nucleus. We also noted bright, punctate structures associated with the apical plasma membrane domains of otic epithelial cells (Fig. 1A, arrow inset). Cdh11 immunofluorescence staining was occasionally associated with linear structures with similar size and location as sensory patch kinocilia (Fig. 3H, arrowhead). It is unclear whether this Cdh11 protein was produced in a hair cell or another epithelial cell type, that is, if you consider that Cdh11 may be expressed and exported from otic epithelial cells.
Evidence for vesicle export from otic epithelial cells and fusion with otolith
Loss-of-function experiments indicated that otolith assembly is Cdh11-dependent. Franklin and Sargent (1996) showed that cdh11 message is abundantly expressed in otic epithelial cells. Immunofluorescence experiments showed that Cdh11 protein was detected in all otic epithelial cells, including sensory patches, but cellular staining did not accumulate (Fig. 1A). Punctate structures within the otolymph and associated with the surface otoliths were more intensely labeled (Fig. 1A). This staining was eliminated by morpholino oligonucleotide knockdown of Cdh11 (Fig. 1A-C). In summary, Cdh11 containing membrane structures accumulate in the otolymph and adhere with the otolith, and blocking Cdh11 expression reduces otolith growth.
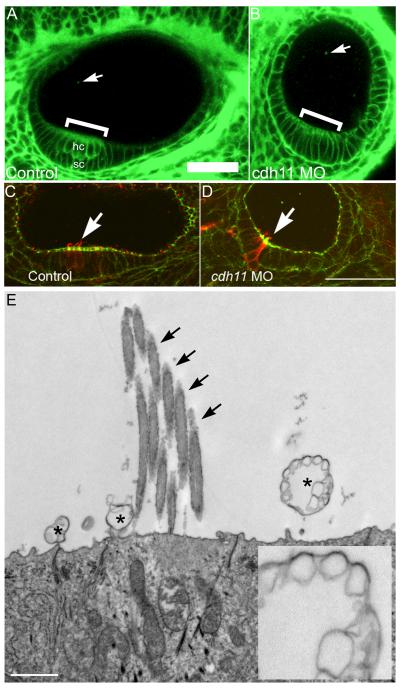
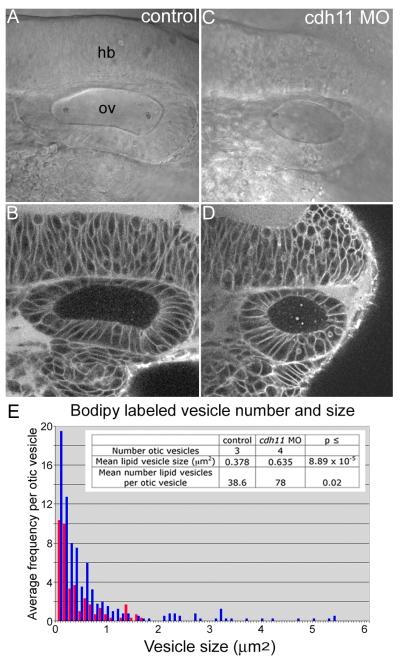
Using BODIPY-ceramide to label membranes throughout the embryo (that has few if any side effects in relatively long-term time-lapse experiments) (Cooper et al., 1999), a confocal optical section of the control embryo inner ear shows otic epithelial cells, including the stratified sensory patch (Fig. 4A). In Cdh11 knockdown embryos, hair cells and supporting cells in the sensory patch did not stratify and form two separate cell layers, but this area of the otic epithelium can be seen as a thickened region of the epithelium (Fig. 4B). Punctate BODIPY-ceramide fluorescent spots were also seen in the otolymph (Fig. 4A-B). Control and Cdh11 knockdown embryos (48 hpf) were labeled to detect actin and tubulin, and inner ear hair cells with stereocilia bundles and kinocilia in sensory patches were always seen in control and knockdown embryos (Fig. 4C-D). The altered shape of the sensory patch may suggest that numbers of hair cells or supporting cells were reduced, raising a possibility that reduced numbers of hair cells or supporting cells could affect otolith growth, but this was not measured. Because otolith assembly nucleation occurs at kinocilia (Riley et al., 1997), it is useful to note that Cdh11 morphant embryos had kinocilia (Fig. 4D).
Figure 4. Inner ear epithelial morphology and vesicular structures.
BODIPY-ceramide (fluorescent lipid) labeling of living 42 hpf zebrafish embryos and confocal microscopy (A, B) outlines cell surfaces, showing cell shapes of otic epithelial cells. Confocal microscope (Zeiss LSM 510, C-Apochromat 40×W 1.2NA objective, mounted in embryo medium) BODIPY-ceramide fluorescence image planes of a three-dimensional stack are shown (pixels 0.45 micron, z-step 2 microns). The double layered pseudostratified hair cell (hc) and supporting cell (sc) layers of the sensory patch in control morpholino oligonucleotide injected embryos (A, indicated by a bracket). The stratification was not detected in cdh11 antisense morpholino oligonucleotide injected embryos (B), but in these morphant embryos, sensory patches had a thickened appearance (bracket in B). Also, BODIPY-ceramide fluorescence was detected in vesicular structures within the otolymph (arrows in A and B, see also Fig. 5). For panels A and B, rostral is left, and dorsal is up. Control morpholino oligonucleotide injected embryo (42 hpf, panel C) and 42 hpf cdh11 antisense morpholino oligonucleotide injected embryo (panel D) were double-labeled using FITC-phalloidin (Molecular Probes) and anti-acetylated tubulin (Sigma) using TRITC-anti mouse secondary antibody (Jackson Labs). We examined several inner ears, and all control and morphant sensory patches had hair cells containing actin-labeled stereocilia and tubulin-labeled kinocilia extending above stereocilia. These are representative images (n=2). Arrows indicate hair cells within the sensory patch. These data also indicate that the kinocilia are present in the cdh11 antisense morpholino oligonucleotide injected embryo, which are required for attachment of membrane structures subsequent to otolith nucleation (initial attachment of glycogen particles to the kinocilia; Riley et al., 1997). Rostral is to the left, and dorsal is up in panels C and D. Scale bar in A represents 50 microns for panels A-D. Transmission electron microscopy of normal zebrafish otic epithelial cells (E) shows several vesicular structures at their apical surface (asterix). One vesicular structure that is seen (inset) is quite large and has a multivesicular appearance. Arrows indicate stereocilia of the hair cell bundle.
Extracellular membrane structures were frequently evident at the surfaces of otic epithelial cells near and within sensory patches, visualized by transmission electron microscopy (Fig. 4E). Many examples of these membrane-containing structures were observed in our experiments associated with the apical surfaces of otic epithelial cells in each embryo examined (all micrographs in this experiment had membrane structures in the otolymph like those shown). Other investigators first observed these structures (Takagi and Takahashi, 1999; Ernest et al., 2000; Nicolson, 2005). Several examples of membrane structures in the otolymph were large and multilaminar in appearance. Rather than perform electron microscopy to quantify these membrane structures, which would be subject to sampling error, we used BODIPY-ceramide labeling to quantify lipid-labeled puncta.
Cdh11 knockdown did not eliminate these BODIPY-ceramide labeled puncta in 20 hpf embryos, but instead, BODIPY-ceramide labeled puncta were more abundant and larger (compare Figs. 5B and 5D). Image analysis of otic vesicles from three control and four Cdh11 knockdown embryos was performed using ImageJ software to count puncta and measure their size (Abramoff et al., 2004). For this analysis, all planes through the entire otolymph volume were quantified for the first and fifth time point of each 100-second time-lapse sequence. Time lapse sequences were acquired by laser scanning confocal microscopy (Zeiss LSM 510) configured to obtain stacks of five optical slices, where each slice was 2 microns thick, spaced 2 microns apart. The time required for acquisition of each stack was 2 seconds. Because the particles did not move very rapidly, this acquisition setup made it possible to observe the motion of individual BODIPY-labeled puncta over time (Fig. 6). These data were plotted in a frequency distribution graph (Fig. 5), showing that there were more puncta and larger puncta in Cdh11 knockdown embryos. This indicates that normal Cdh11 expression reduces the number of vesicles. We hypothesize that there are fewer membrane structures because these vesicles are efficiently fusing with the growing otolith. When Cdh11 is knocked down, smaller vesicles may not fuse normally with the otolith, but may collide to form the larger structures; alternatively, the otic epithelium may produce abnormally large membrane structures as a consequence of cdh11 loss-of-function.
Figure 5. Total number and size of fluorescently labeled vesicular structures increased in Cdh11 knockdown embryos.
BODIPY-ceramide labeling of living 20 hpf zebrafish embryos injected with control morpholino oligonucleotide (A, B) and with cdh11 antisense morpholino oligonucleotide injected (C, D). Transmitted light, DIC microscope images (A, C) and the corresponding confocal optical sections (B, D) are shown, imaged using a Zeiss LSM 510, C-apochromat 63× W 1.2 NA objective, mounted in embryo medium. Single confocal image planes are shown. Fluorescent vesicular structures were quantified using ImageJ software to measure total numbers of vesicular structures and their sizes (for details see methods section). These numbers were plotted on a frequency distribution graph (E, average frequency of vesicles vs vesicle size). The table shown as an inset in panel E lists the number of otic vesicles that were analyzed, the mean lipid vesicle size and the mean number of lipid vesicles per otic vesicle in control morpholino oligonucleotide injected (control) and cdh11 antisense morpholino oligonucleotide injected (cdh11 MO) embryos. Results of student’s t-test comparisons of control and cdh11 antisense morpholino oligonucleotide injected embryo groups were statistically significant. Abbreviations: hb=hindbrain, ov=otic vesicle.
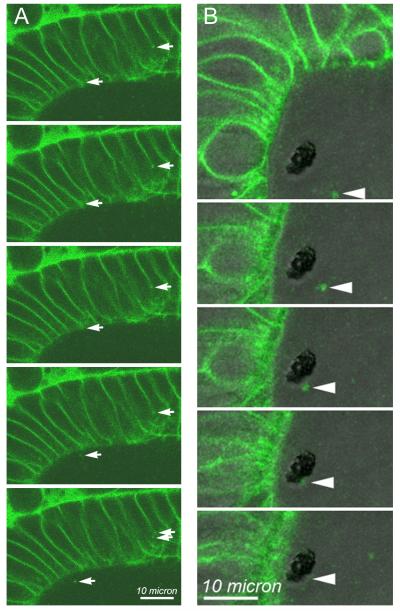
Figure 6. Time-lapse imaging of BODIPY-ceramide labeled vesicular structures suspended within the otolymph and fusing with the otolith.
Using the same time-lapse images used to determine size and numbers of vesicular structures within the otolymph of 22 hpf control embryos, movement of individual BODIPY-ceramide labeled structures were examined. BODIPY-labeled vesicles were seen moving within otic epithelial cells, being exported from otic epithelial apical cell surfaces into the otolymph, moving within the otolymph and fusing with the growing otolith. Examples of these events are shown in time-lapse sequence. DIC and BODIPY fluorescence images were collected every 2 seconds. The first image of each time sequence is shown at the top of the figure and proceeds every two seconds in sequential images down the figure. (A) Only the BODIPY fluorescence images are shown for this sequence. Arrows on the left side of each panel indicate a BODIPY-ceramide labeled structure at the lateral cell surface of an otic epithelial cell that moves in a generally apical direction. Arrows on the right side of each panel indicate the apico-lateral region of the cell that appears to generate a BODIPY-labeled punctate vesicle-like structure that enters the otolymph in the final two frames of this sequence. (B) DIC and BODIPY fluorescence images were combined into a single image shown in each frame. In panel B, the dark, unstained structure in the otolymph is the otolith. Single image planes are presented, and the specific image plane within the volume varied over the time sequence to optimize the visualization of the particular BODIPY-labeled puncta highlighted by the arrowheads in each panel. All other image planes were examined to insure that the indicated vesicular structure was not merely moving out of the plane of focus, but appeared to adhere and fuse at the surface of the otolith. A 10 micron scale bar is shown.
Increased number and size of BODIPY-labeled puncta detected in the otolymph appeared to persist in older (42 hpf) embryos. Representative single image planes through the inner ear showed BODIPY-labeled puncta in 42 hpf embryos, and these puncta appeared to be more numerous and larger in Cdh11 knockdown embryos (see Supplemental Data, Fig. S3). Because control otic vesicles in 42 hpf embryos were often too large to fit in a single image frame, measurement of puncta in time lapse would be complicated by the area of the otolymph selected and puncta moving in and out of the imaging field. Thus, we did not quantify experiments using 42 hpf embryos. Significantly, 7 dpf morphant larvae displayed reduced or absent otoliths (data not shown), arguing against the possibility that reduced or absent otolith phenotype is merely a developmental delay.
In our analysis of BODIPY-ceramide labeled puncta (Fig. 5), images were viewed as a time-lapse movie. We noted: (i) BODIPY-ceramide labeled puncta moved within otic vesicle epithelial cells in a general apical direction (Fig. 6A, arrows on right side of each image); (ii) BODIPY-ceramide labeled puncta appeared to be exported from otic vesicle epithelial cells (Fig. 6A, arrows on left side of each image); (iii) BODIPY-ceramide labeled puncta moved within otolymph (Fig. 6B, arrowheads on images); and, BODIPY-ceramide labeled puncta appeared to adhere to the growing otolith (Fig. 6B, arrowheads on images). Export of puncta from otic epithelial cells and puncta binding to the otolith were too infrequent within the large volume of time lapse data to quantify without new method development. Despite several examples being seen, these were isolated observations. The rate of particle movement was roughly compared to determine whether Cdh11 knockdown reduces mixing of otolymph contents. The time-lapse imaging in these experiments had excellent x-y resolution but very little z-resolution (2 micron z-steps), and thus, these measurements are only sufficient to compare with one another (relative movement, not absolute movement rate). Particle movement was nearly identical in control (1.01 microns/second, n=30) and Cdh11 (0.98 microns/second, n=47) knockdown embryos, indicating that particle movement did not limit otolith growth.
Hindbrain defects in Cdh11 knockdown embryos
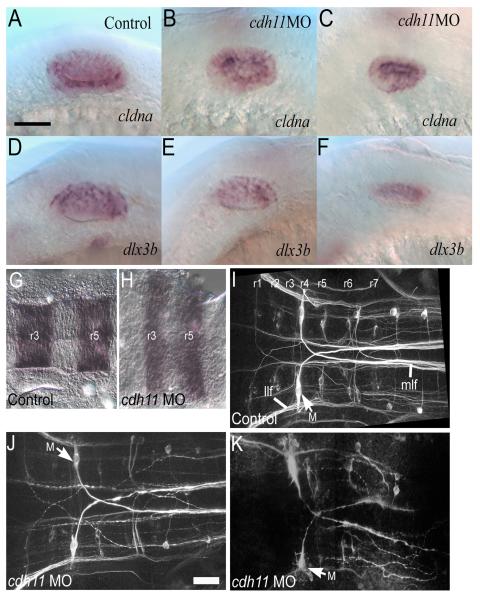
Hindbrain tissue induces otic placode formation and regulates otic vesicle development (Barald and Kelley, 2004; Nicolson, 2005). The strongest effect of the hindbrain is on otic induction, and otic induction proceeds relatively normally in cdh11 morphants. Hindbrain also influences otic vesicle patterning and morphogenesis. Specification and early patterning were examined by in situ hybridization using cldna (specification) and dlx3b (specification and dorsal otic vesicle pattern) (Fig. 7). Although more severely affected cdh11 knockdown embryos (Fig. 7C and F) had smaller otic vesicles, the expression pattern for these early specification and patterning markers was comparable with control embryos (Fig. 7A and D). Further evidence of otic vesicle patterning is illustrated by actin and acetylated tubulin labeling, showing that sensory patches assemble and differentiate sensory cells in Cdh11 knockdown embryos (Fig. 4C-D).
Figure 7. Otic specification, early otic vesicle patterning and hindbrain phenotype in Cdh11 knockdown embryos.
Control morpholino oligonucleotide injected (A, D) and cdh11 antisense morpholino oligonucleotide injected 24 hpf embryos (B-C and E-F) were labeled by in situ hybridization with cldna (A-C) and dlx3b (D-F) cRNA probes. Normal expression for cldna (which begins during otic specification and continues in all otic vesicle epithelial cells) was seen in both control and Cdh11 knockdown embryos, although the otic vesicles were smaller in Cdh11 knockdown embryos. Normal expression for dlx3b (which begins during otic specification, continues in the otic vesicle, then resolves into a dorsal pattern in the otic vesicle) was seen in both control and Cdh11 knockdown embryos. Again, otic vesicles were smaller in Cdh11 knockdown embryos. Scale bar in panel A represents 50 microns. In panels A-F, rostral is left, and dorsal is up. Hindbrain markers were examined in control morpholino oligonucleotide injected embryos (G, I) and cdh11 antisense morpholino oligonucleotide injected embryos (H, J, K) using for krox20 in situ hybridization (G, H) or neurofilament immunofluorescence (I-K). In situ hybridization labeled embryos at 18 hpf were imaged in flat mount using transmitted light DIC microscopy (G, H) using Nikon diaphot 200, 40× DIC 0.55 NA objective. Immunofluorescence of embryos at 48 hpf was imaged in flat mount (I-K) using multiphoton microscopy (BioRad MRC1024, 60× W 1.2 NA objective, 800 nm illumination). Projection images were made using Voxx Software in alpha blending mode. In severe Cdh11 knockdown embryos, lateral longitudinal fasciculus (llf) and medial longitudinal fasciculus (mlf) are severely reduced or absent. However, there is a small fiber tract labeling that might be part of the lateral longitudinal fasciculus, and Mauthner cells (M) axons still contribute to the medial longitudinal fasciculus in severely affected Cdh11 knockdown embryos. Scale bar in J represents 50 microns for panels H-K. In panels G-K, dorsal images are viewed with rostral at left. Representative images are shown. For all panels, rostral is left, and dorsal is facing the viewer.
In addition, Cdh11 knockdown effects on hindbrain development were examined. Hindbrain specification and patterning were examined using krox20 in situ hybridization (labeling rhombomeres 3 and 5), which was detected in a similar sized expression domain in the hindbrain cells of control and Cdh11 knockdown embryos, but this domain was thinner in the anterior-posterior axis and broader in the medial-lateral axis in Cdh11 knockdown embryos compared to control embryos (Fig. 7A-B). The altered shape of this hindbrain compartment probably reflects reduced convergence and extension cell movements during gastrulation (also see Fig. 1N-O) (Keller, 2005). This result shows rhombomeres are specified in Cdh11 knockdown embryos.
Neurofilament staining showed that moderately affected Cdh11 knockdown embryos had a similar organization and trajectory of hindbrain neurons to those of control embryos (Fig. 7C-D). In severely affected embryos, labeling of both cell bodies and fiber tracts was greatly reduced, the segmental pattern of the contralateral projecting neurons (which are organized as segments into rhombomeres) of the hindbrain were severely disrupted, although the Mauthner cells (a pair of the largest cell bodies in hindbrain) and their axons were present and projected contralaterally (Fig. 7E). We cannot completely discount possible hindbrain effects on otic vesicle patterning or otolith formation. However, otic induction, specification, initial otic vesicle patterning, and sensory patch differentiation were detected in Cdh11 knockdown embryos.
Discussion
Our evidence suggests that Cdh11 activity in otolith formation illustrates a novel extracellular cadherin function. Previously documented cadherin functions involve interactions between apposing cells, including cell-to-cell adhesion and cadherin-mediated intracellular signaling (Gumbiner, 2005; Halbleib and Nelson, 2006). In the developing inner ear, we have detected Cdh11 by immunofluorescence in puncta (presumably vesicular structures produced by otic epithelial cells) and cdh11 loss-of-function blocks otolith assembly.
In most epithelial cells, cadherins accumulate at sites of cell-cell contact (Gumbiner, 2005; Halbleib and Nelson, 2006). Cdh11 within otic epithelial cells was only faintly detected at sites of cell-cell contact; most Cdh11 protein within otic epithelial cells was localized within the cytoplasm. Stronger Cdh11 immunofluorescence was in puncta within the otolymph fluid within the otocyst, and some puncta were associated with the otolith surface. The strongest accumulation of Cdh11 detected by immunofluorescence was on the surface and within the forming otolith, arranged in an annular pattern. This annular pattern has been described for other proteins that are deposited in layers of growing otolith biomineral structures (Sollner et al., 2003). Although there may be other interpretations, our observations support the hypothesis that Cdh11 proteins synthesized in otic epithelial cells are packaged into vesicles that are exported from these cells, and these vesicles coalesce on the nucleating otolith at the kinocilium. Sustained deposition of vesicles mediates otolith growth. We hypothesize that adhesive properties of Cdh11 on these otic epithelium derived vesicles mediates otolith growth by more efficiently delivering packets containing appropriate building materials (minerals and proteins) to the growing otolith that permit normal biomineralization.
Morpholino oligonucleotide Cdh11 knockdown inhibited or prevented otolith growth; reduced Cdh11 expression in vesicle-like puncta in the otolymph; but did not block vesicle-like, BODIPY-ceramide labeled puncta production. The BODIPY-ceramide lipid labeling technique shows that living embryos produce BODIPY-ceramide labeled punctate structures that deposited in the otolymph. Large vesicle-like structures were also seen in transmission electron micrographs that were associated with otic epithelial cell apical surfaces and within the otolymph. Perhaps, otolymph vesicles in Cdh11 knockdown embryos less efficiently adhere and fuse with other vesicles or with the growing otolith. Larger vesicles found in the otolymph of Cdh11 knockdown embryos may be produced by collisions between the accumulating membrane structures in the otolymph, or perhaps another stochastic processes. In control embryos, normal adhesion and fusion of vesicles with the forming otolith could serve as a ‘sink’ for the vesicles, reducing the number of vesicles and reducing the possibility that small vesicles could collide to form larger structures. It is also interesting to note that the BODIPY-ceramide did not accumulate within the growing otolith, suggesting that incorporation into the otolith structure quenches BODIPY fluorescence or there may be a lipid retrieval mechanism.
Previous studies have observed membrane structures in the otolymph (Takagi and Takahashi, 1999; Ernest et al., 2000; Nicolson, 2005), suggesting that otic epithelial cells export vesicles during inner ear development, and these vesicles have been compared with matrix vesicles produced by osteoblast cells (Hughes et al., 2006). These matrix vesicles were hypothesized to contain packets of material used during biomineralization, including proteins that control materialization. Similar proteins control both otolith and bone biomineralization, and Cdh11 is expressed in both otic epithelial cells and osteoblasts (Sollner and Nicolson, 2005; Hughes et al., 2006). Starmaker protein regulates otolith biomineralization and is related to the human dentin sialophosphoprotein gene (DSPP), which encodes a protein required for biomineralization in teeth (Sollner et al., 2003). Starmaker protein was detected in otic epithelial cells, in punctate structures suspended in the otolymph and within the otolith in an annular pattern (Sollner et al., 2003). We speculate that Cdh11vesicles may also contain Starmaker and other materials that are delivered and concentrated together on the surface of the growing otolith to be used during biomineralization.
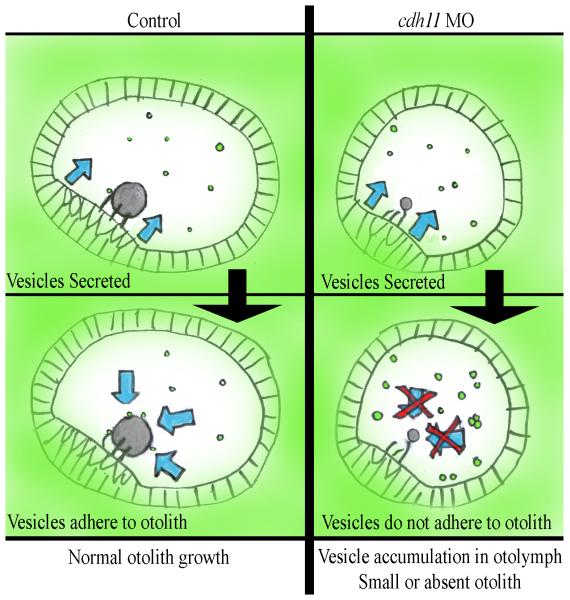
Hindbrain development was affected by cdh11 loss-of-function, which raised the possibility that normal hindbrain activities that induce and pattern the developing inner ear were defective. Moderate hypomorph cdh11 phenotype showed relatively normal hindbrain neuron patterning, but in severe (possibly null) cdh11 phenotype embryos, the hindbrain neurons were disorganized. Hindbrain influences on otic vesicle patterning are well documented (Barald and Kelley, 2004; Nicolson, 2005). We cannot completely eliminate the possibility that hindbrain defects lead to changes in otic vesicle patterning or otolith formation, but expression of cldna and dlx3b showed that cdh11 loss-of-function permits otic induction, otic specification and initial otic vesicle patterning. Also, sensory patches differentiate, even in severely affected Cdh11 knockdown embryos. In addition, BODIPY-ceramide labeling experiments and the Cdh11 immunofluorescence distribution pattern supports our hypothesis that Cdh11 accumulates on vesicles that were derived from otic epithelial cells, and these Cdh11 containing puncta adhere to the surface of the growing otolith. We were unable to detect any contribution of hindbrain disorganization on the cdh11 loss-of-function otolith phenotype. Evidence better supports our model that Cdh11 function in otolith assembly is intrinsic to the otic epithelium (see working model, Fig. 8).
Figure 8. A working model for Cdh11 activity that promotes efficient otolith assembly.
In normal embryos (Control, left panels) otic vesicle epithelial cells export vesicles that contain abundant Cdh11 protein and may contain other components that are needed for biocrystallization (Vesicles Secreted). Cdh11 mediated adhesion of these extracellular vesicles (Vesicles adhere to otolith) promotes efficient otolith assembly (blue arrows). In contrast cdh11 loss-of-function (cdh11 MO, right panels) does not interfere with vesicle export (Vesicles Secreted), but these vesicles have reduced Cdh11 adhesion molecule levels, which prevents adhesion (Vesicles do not adhere to otolith). Without Cdh11 adhesion (blue arrows with red Xs), vesicles that are exported do not adhere to one another or to a growing otolith biocrystal (Vesicle accumulation in otolymph, Small or absent otolith). Evidence supporting this model is outlined in the Discussion.
That Cdh11 may have an extracellular activity during inner ear development leads us to speculate that other Cdh11 mediated developmental events or physiological roles in the adult animal may also act through extracellular mechanisms. As mentioned above, osteoblast matrix vesicles may use a similar mechanism to adhere to growing bone surfaces (Hughes et al., 2006). Cdh11 also has effects on gastrulation cell movements and neural development, in addition to its inner ear development role. It is intriguing to consider that some of these Cdh11 effects during these processes could also be mediated by extracellular mechanisms.
In summary, our studies of Cdh11 function in otolith biomineralization point to a novel cadherin activity, namely, providing adhesion for vesicular packages that mediate rapid, efficient growth of the otolith, which is our working model (Fig. 8). Several experiments support this model. Punctate structures were frequently seen at the surface of otic epithelial cells and within the otolymph in electron micrographs and in confocal images of BODIPY-ceramide labeled living embryos. Cdh11 protein was detected in punctate structures within otic epithelial cells and in punctate structures suspended in the otolymph. These puncta also adhered to the otolith surface, and Cdh11 accumulated in annular patterns in the growing otoliths. Blocking Cdh11 expression did not interfere with BODIPY-labeled puncta accumulation in the otolymph, but otolith assembly was inhibited, perhaps by blocking otolymph vesicle adhesion to other vesicles and to the otolith surface. Our results point to a novel extracellular cadherin-mediated vesicle adhesion mechanism, which promotes efficient otolith assembly.
Experimental Procedures
Zebrafish husbandry and morpholino injection
Zebrafish (Danio rerio) were raised and kept under standard laboratory conditions (Westerfield, 2000) in accordance with Indiana University, University of Akron and University of Michigan policies on animal care and use. Morpholino oligonucleotides (cdh11MOA: 5′-CTA AAG AAG GTA AAG TGT GTG AAT G-3′; cdh11MOB: 5′-CCC CAT CAG GTA GAG TCT GCT TCC T-3′; and cdh11MOC: 5′-TCA AGG ACA ACA GAG GTG CGT GAC A-3′; standard control; Gene Tools, Corvallis OR) were microinjected into the yolk of one- to four-cell stage embryos (Ekker, 2000). These oligonucleotides designed to block splicing and thus mRNA maturation (Draper et al., 2001; Morcos, 2007; Eisen and Smith, 2008). Injected embryos were incubated at 28.5°C. For some experiments, phenylthiourea (PTU; 0.2 mM) was added to embryo medium to prevent melanization.
Synthetic cdh11 mRNA production and injection
Reverse transcriptase PCR using primers (sense: TAGGATCCATGTGGGAGGGACTGAGA; antisense: ACTCTAGATTAAGAGTTGTCATCGACGGAG) specific for cdh11 untranslated 5′ and 3′ sequences mRNA was performed according to manufacturers instructions (Roche, Indianapolis IN) using 24 hpf RNA purified using Trisol reagent (Invitrogen, Carlsbad CA). PCR product was cloned using Topo TA cloning kit (Invitrogen) according to manufactures instructions. Topo TA clones containing the appropriate insert were digested with BamHI and XbaI. The 3.7 kb insert was subcloned into BamHI and XbaI digested pCS2+ (originally constructed by David Turner and Ralph Rupp, Fred Hutchinson Cancer Center). The pCS2+/cdh11 construct was linearized by digesting with XbaI, and in vitro transcription of sense transcripts (capped and polyadenylated) was performed using Message Machine (Ambion, Austin TX). These synthetic transcripts were co-injected with morpholino oligonucleotides as described above.
Antibody production, immunohistochemistry and immunoblotting
Affinity purified, polyclonal, anti-peptide antibody was raised in rabbits using a peptide of zebrafish Cdh11 (CSATDVDEMAHRQHFSL) as an immunogen, and antibodies were affinity purified using the immunizing peptide. For Cdh11 immunostaining, embryos were fixed overnight in 4% paraformaldehyde in phosphate buffered saline at 4°C. The anti-Cdh11 antibody was used at 1:100 dilution and followed by TRITC-conjugated anti-rabbit (Jackson ImmunoResearch) at 1:50 dilution. For neurofilament staining, embryos were fixed using 2% trichloracetic acid in PBS for 3hr at room temperature. Mouse anti-neurofilament RMO-44 (Zymed) was used at a dilution of 1:25 and followed by TRITC-conjugated anti mouse (Jackson ImmunoResearch) at 1:50. Two-photon microscope image volumes were acquired using a Zeiss LSM-510 Meta Confocal microscope System (Göttingen Germany) equipped with a tunable Titanium-Sapphire laser at the Indiana Center for Biological Microscopy (www.nephrology.iupui.edu/imagingfacility). Projection images were produced from image volumes using Voxx, a voxel based three-dimensional near real time rendering program, developed at the Indiana Center for Biological Microscopy (Clendenon et al., 2002).
For immunoblotting, zebrafish embryos were solubilized in sodium dodecyl sulfate (SDS) buffer (10 mM Tris-HCl, pH 7.5, 2 mM EDTA, 1% SDS), separated using SDS-PAGE, transferred to nitrocellulose and blocked in 3% ovalbumin in TBST (10 mM Tris pH 7.5, 0.1 M NaCl, 0.1% Tween 20). Membranes incubated with antiserum (1:15,000 dilution) and anti-rabbit HRP-conjugated secondary antibody (1:10,000 dilution) (Amersham, Arlington Heights, IL) were processed for chemiluminescence (ECL kit, Amersham) and exposed to film (Kodak Bio-Max ML, Eastman Kodak, Rochester, NY). To test specificity, excess immunizing peptide (or irrelevant peptide) was preincubated with the antiserum prior to incubating with the membrane during immunoblotting.
BODIPY-ceramide labeling and image analysis
BODIPY-ceramide-Fl-C5 (Molecular Probes) stock solution (5 mM in dimethyl sulfoxide) was diluted to100 μM in embryos medium with 10 mM HEPES, and dechorionated living embryos were soaked in BODIPY-ceramide solution for 2 hr in the dark (Cooper et al., 1999). The embryos were washed, and confocal images were acquired using a Zeiss LSM-510 confocal microscope. Time-lapse sequences were acquired using a Zeiss LSM 510. Five 2-micron optical slices, spaced 2 microns apart were acquired, with the center slices positioned to include the greatest anterior-posterior distance within the otic vesicle. The time required for acquisition of each stack was 2 seconds. This acquisition speed made it possible to observe the motion of individual vesicles over time.
The number and size of BODIPY-ceramide labeled puncta within the otolymph were quantified using ImageJ software (Abramoff et al., 2004). Time lapse sequences from three 20 hpf control and four 20 hpf Cdh11 knockdown embryos (1 slightly affected, 2 moderately affected and 1 severely affected) were analyzed. All of the five image planes from the first and fifth time point of each 100 second time-lapse sequence were used. Thresholds for each image were adjusted in ImageJ to identify the BODIPY-ceramide positive puncta within the fluid filled space. All of these images were compared against the original images to insure accurate identification of puncta. Within each image, a region was drawn that included only the fluid filled space. ImageJ analysis tools were then used to count and measure the size of all BODIPY-ceramide positive puncta. Average lipid vesicle (puncta) size and average number of lipid vesicles (puncta) per otic vesicle were compared using students t-test.
Transmitted light microscopy
Images of whole, unstained embryos were collected using a Leica MZ12 dissecting microscope equipped with a SPOT RT camera (Diagnostic Instruments). Images of unstained otic vesicles and isolated otoliths were collected using a Nikon Diaphot microscope (Nikon, Inc., Melville, NY) equipped with diffraction interference contrast (DIC) optics and a SPOT RT camera (Diagnostic Instruments).
Transmission electron microscopy
Whole-mount 5 days postfertilization (dpf) embryos were anaesthetized with 0.02% 3-aminobenzoic acid ethyl ester then fixed in 2% paraformaldehyde/2% glutaraldehyde in 0.1 M Phosphate Buffer overnight at 4°C. Specimens were post-fixed in 1% OsO4 in phosphate buffer for 1 hour, dehydrated by incubating in a series of ethanol solutions and embedded in Embed 812 (Electron Microscopy Sciences). Ultrathin sections (70-90 nm) were cut then stained with uranyl acetate. Sections were viewed on a Tecnai BioTwin electron microscope (FEI, Hillsboro, OR), and digital images were acquired with an AMT CCD camera (Advanced Microscopy Techniques, Canvers, MA) in the Indiana University Electron Microscopy Center.
In situ hybridization
Whole-mount in situ hybridization of zebrafish embryos was performed as described (Liu et al., 1999b). Digoxigenin-labeled riboprobe for cldna, dlx3b and krox20 (Oxtoby and Jowett, 1993) was synthesized using the Genius System DIG RNA Labeling Kit (Roche, Indianapolis, IN) from clone cb427 (ZFIN, Eugene, OR).
Statistical Analysis
Comparisons were mane using Mantel-Haenszel chi-square tests for ordered categorical responses. Fisher’s protected least significant differences method was used to control the overall significance level of the tests.
Supplementary Material
Figure S1. Cdh11 antibody specificity and morpholino-induced expression reduction. (A) Immunoblot analysis using anti-Cdh11 rabbit polyclonal antibody (see Materials and Methods) to probe adult zebrafish brain homogenate (Cdh11 was abundantly detected in the adult brain) reacted with a single polypeptide (relative migration ~120 kD; right lane). Immunodetection was reduced by co-incubating the anti-Cdh11 antibody with the immunogen Cdh11 peptide (i. e., used to raise the antibody; middle lane), but immunodetection was not affected by co-incubating the anti-Cdh11 antibody with an identical concentration of an irrelevant peptide (right lane). Immunofluorescence using anti-Cdh11 rabbit polyclonal antibody in wholemount 30 hpf embryos. The typical inner ear Cdh11 expression pattern was detected in control embryos (B), but co-incubating the anti-Cdh11 antibody with the immunogen Cdh11 peptide eliminated all labeling (C). Laser power, gain settings, filters and image processing were identical for panels B and C. (D) Pools of protein homogenate from three embryos (24 hpf) per lane were immunoblotted to detect Cdh11. This was the smallest number of embryos that we could reliably detect Cdh11 expression in control embryos (left lane). A pool of three cdh11 morpholino oligonucleotides injected 24 hpf embryos showed reduced Cdh11 expression levels. Representative result from two experiments is shown.
Figure S2. Cdh11 morpholino-induced expression reduction through the entire otic vesicle. To determine whether optical section selection in Figure 1B-C minimized strongly labeled structures (like otoliths), we collected image volumes from the same zebrafish embryos labeled using Cdh11-specific antibodies shown in Figure 1B-C (i. e., injected with cdh11 splice-site blocking antisense morpholino oligonucleotide, as here, panels A and B). Instead of a projection of 5 optical sections as shown in Fig. 1B-C, an image volume was collected through entire otic vesicle of a slightly affected 30 hpf embryo (panel A; 45 optical sections, 0.3 micron z-step) and a moderately affected 30 hpf embryo (panel B; 25 optical sections, 0.3 micron z-step). Projection images were made in alpha blending mode using Voxx software. Adobe Photoshop 6 used to prepare figure. The otic vesicle was smaller in the moderately affected embryo, but both images (panels A and B) are projections of image volumes through entire inner ear region of the embryos. Panels A and B have identical orientation, with rostral and lateral as shown in panel A. These image volumes illustrate that only small amounts of Cdh11 labeling were detected in slightly affected phenotype embryos (panel A), and little or no Cdh11 labeling was detected above background in moderately affected (in panel B, background from 25 optical sections is combined) and severely affected (not shown) embryos. Laser power, gain settings, filters and image processing were identical for panels A and B. Two-photon imaging was performed using Zeiss LSM510 Meta NLO, C-Apochromatic 40×W 1.2NA objective, TRITC-labeled secondary antibody, 800 nm excitation, and mounted in PBS.
Figure S3. BODIPY-ceramide labeled vesicular structures in control and Cdh11 knockdown 42 hpf embryos. BODIPY-ceramide labeled living 42 hpf zebrafish embryos injected with control morpholino oligonucleotide (A) and with cdh11 antisense morpholino oligonucleotide injected (B). Confocal optical sections are shown, using a Zeiss LSM 510, C-apochromat 63× W 1.2 NA objective, mounted in embryo medium. Representative confocal image planes are shown. Due to technical difficulties described in the text, quantification would not be as reliable at this developmental stage. However, there was an apparent increase in the number and size of BODIPY-labeled puncta in Cdh11 knockdown embryos. Scale bar is 30 microns.
Acknowledgements
Dr. Victoria Prince (University of Chicago) generously provided the krox20 cDNA construct for in situ hybridization probe synthesis. Dr. Cunming Duan (University of Michigan) generously provided the cldna cDNA construct for in situ hybridization probe synthesis, and Dr. Monte Westerfield (University of Oregon) generously provided the dlx3b cDNA construct for in situ hybridization probe synthesis. Zebrafish handling and care was reviewed and approved by Institutional Review Boards at Indiana University, the University of Michigan and the University of Akron. Images were collected at the Indiana Center for Biological Microscopy. This work was supported by a grant from the NIH to JAM, KFB and QL (DC006436) and by grants from the DRF and the NIH to KFB (NIH DC05939 and DC04184).
This work was supported by a grant to JAM, KFB and QL (National Institutes of Health; DC006436) and by grants to KFB (Deafness Research Foundation and National Institutes of Health; DC05939; DC04184).
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Babb-Clendenon S, Shen YC, Liu Q, Turner KE, Mills MS, Cook GW, Miller CA, Gattone VH, 2nd, Barald KF, Marrs JA. Cadherin-2 participates in the morphogenesis of the zebrafish inner ear. J Cell Sci. 2006;119:5169–5177. doi: 10.1242/jcs.03299. [DOI] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Blasiole B, Canfield VA, Vollrath MA, Huss D, Mohideen MA, Dickman JD, Cheng KC, Fekete DM, Levenson R. Separate Na,K-ATPase genes are required for otolith formation and semicircular canal development in zebrafish. Dev Biol. 2006;294:148–160. doi: 10.1016/j.ydbio.2006.02.034. [DOI] [PubMed] [Google Scholar]
- Clendenon JL, Phillips CL, Sandoval RM, Fang S, Dunn KW. Voxx: a PC-based, near real-time volume rendering system for biological microscopy. Am J Physiol Cell Physiol. 2002;282:C213–218. doi: 10.1152/ajpcell.2002.282.1.C213. [DOI] [PubMed] [Google Scholar]
- Cooper MS, D’Amico LA, Henry CA. Confocal microscopic analysis of morphogenetic movements. Methods Cell Biol. 1999;59:179–204. doi: 10.1016/s0091-679x(08)61826-9. [DOI] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Ekker SC. Morphants: a new systematic vertebrate functional genomics approach. Yeast. 2000;17:302–306. doi: 10.1002/1097-0061(200012)17:4<302::AID-YEA53>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest S, Rauch GJ, Haffter P, Geisler R, Petit C, Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum Mol Genet. 2000;9:2189–2196. doi: 10.1093/hmg/9.14.2189. [DOI] [PubMed] [Google Scholar]
- Fekete DM. Developmental biology. Rocks that roll zebrafish. Science. 2003;302:241–242. doi: 10.1126/science.1091171. [DOI] [PubMed] [Google Scholar]
- Franklin JI, Sargent TD. Ventral neural cadherin, a novel cadherin expressed in a subset of neural tissues in the zebrafish embryo. Dev Dyn. 1996;206:121–130. doi: 10.1002/(SICI)1097-0177(199606)206:2<121::AID-AJA1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Hardison AL, Lichten L, Banerjee-Basu S, Becker TS, Burgess SM. The zebrafish gene claudinj is essential for normal ear function and important for the formation of the otoliths. Mech Dev. 2005;122:949–958. doi: 10.1016/j.mod.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Hughes I, Blasiole B, Huss D, Warchol ME, Rath NP, Hurle B, Ignatova E, Dickman JD, Thalmann R, Levenson R, Ornitz DM. Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev Biol. 2004;276:391–402. doi: 10.1016/j.ydbio.2004.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes I, Thalmann I, Thalmann R, Ornitz DM. Mixing model systems: using zebrafish and mouse inner ear mutants and other organ systems to unravel the mystery of otoconial development. Brain Res. 2006;1091:58–74. doi: 10.1016/j.brainres.2006.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsch M, Anken RH, Rahmann H. Calcium gradients in the fish inner ear sensory epithelium and otolithic membrane visualized by energy filtering transmission electron microscopy (EFTEM) Adv Space Res. 2004;33:1395–1400. doi: 10.1016/j.asr.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Kawaguchi J, Azuma Y, Hoshi K, Kii I, Takeshita S, Ohta T, Ozawa H, Takeichi M, Chisaka O, Kudo A. Targeted disruption of cadherin-11 leads to a reduction in bone density in calvaria and long bone metaphyses. J Bone Miner Res. 2001;16:1265–1271. doi: 10.1359/jbmr.2001.16.7.1265. [DOI] [PubMed] [Google Scholar]
- Keller R. Cell migration during gastrulation. Curr Opin Cell Biol. 2005;17:533–541. doi: 10.1016/j.ceb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Kerstetter AE, Azodi E, Marrs JA, Liu Q. Cadherin-2 function in the cranial ganglia and lateral line system of developing zebrafish. Dev Dyn. 2004;230:137–143. doi: 10.1002/dvdy.20021. [DOI] [PubMed] [Google Scholar]
- Lee DM, Kiener HP, Agarwal SK, Noss EH, Watts GF, Chisaka O, Takeichi M, Brenner MB. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007;315:1006–1010. doi: 10.1126/science.1137306. [DOI] [PubMed] [Google Scholar]
- Malicki J, Schier AF, Solnica-Krezel L, Stemple DL, Neuhauss SC, Stainier DY, Abdelilah S, Rangini Z, Zwartkruis F, Driever W. Mutations affecting development of the zebrafish ear. Development. 1996;123:275–283. doi: 10.1242/dev.123.1.275. [DOI] [PubMed] [Google Scholar]
- Mbalaviele G, Shin CS, Civitelli R. Cell-cell adhesion and signaling through cadherins: connecting bone cells in their microenvironment. J Bone Miner Res. 2006;21:1821–1827. doi: 10.1359/jbmr.060811. [DOI] [PubMed] [Google Scholar]
- Morcos PA. Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos. Biochem Biophys Res Commun. 2007;358:521–527. doi: 10.1016/j.bbrc.2007.04.172. [DOI] [PubMed] [Google Scholar]
- Murayama E, Herbomel P, Kawakami A, Takeda H, Nagasawa H. Otolith matrix proteins OMP-1 and Otolin-1 are necessary for normal otolith growth and their correct anchoring onto the sensory maculae. Mech Dev. 2005;122:791–803. doi: 10.1016/j.mod.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Nicolson T. The genetics of hearing and balance in zebrafish. Annu Rev Genet. 2005;39:9–22. doi: 10.1146/annurev.genet.39.073003.105049. [DOI] [PubMed] [Google Scholar]
- Novince ZM, Azodi E, Marrs JA, Raymond PA, Liu Q. Cadherin expression in the inner ear of developing zebrafish. Gene Expr Patterns. 2003;3:337–339. doi: 10.1016/s1567-133x(03)00030-9. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BB, Grunwald DJ. A mutation in zebrafish affecting a localized cellular function required for normal ear development. Dev Biol. 1996;179:427–435. doi: 10.1006/dbio.1996.0272. [DOI] [PubMed] [Google Scholar]
- Riley BB, Zhu C, Janetopoulos C, Aufderheide KJ. A critical period of ear development controlled by distinct populations of ciliated cells in the zebrafish. Dev Biol. 1997;191:191–201. doi: 10.1006/dbio.1997.8736. [DOI] [PubMed] [Google Scholar]
- Sollner C, Burghammer M, Busch-Nentwich E, Berger J, Schwarz H, Riekel C, Nicolson T. Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science. 2003;302:282–286. doi: 10.1126/science.1088443. [DOI] [PubMed] [Google Scholar]
- Sollner C, Nicolson T. The zebrafish as a genetic model to study otolith formation. In: Bauerlein E, editor. Biomineralization: Progress in Biology, Molecular Biology and Application. Wiley-VCH; Weinheim: 2005. pp. 229–241. [Google Scholar]
- Sollner C, Schwarz H, Geisler R, Nicolson T. Mutated otopetrin 1 affects the genesis of otoliths and the localization of Starmaker in zebrafish. Dev Genes Evol. 2004;214:582–590. doi: 10.1007/s00427-004-0440-2. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Takahashi A. Characterization of ootolith soluble-matrix producing cells in the saccular epithelium of rainbow trout (Oncorhynchus mykiss) inner ear. Anat Rec. 1999;254:322–329. doi: 10.1002/(SICI)1097-0185(19990301)254:3<322::AID-AR2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. The University of Oregon Press; Eugene, OR: 2000. [Google Scholar]
- Whitfield TT, Granato M, van Eeden FJ, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Mutations affecting development of the zebrafish inner ear and lateral line. Development. 1996;123:241–254. doi: 10.1242/dev.123.1.241. [DOI] [PubMed] [Google Scholar]
- Wilson AL, Shen YC, Babb-Clendenon SG, Rostedt J, Liu B, Barald KF, Marrs JA, Liu Q. Cadherin-4 plays a role in the development of zebrafish cranial ganglia and lateral line system. Dev Dyn. 2007;236:893–902. doi: 10.1002/dvdy.21085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cdh11 antibody specificity and morpholino-induced expression reduction. (A) Immunoblot analysis using anti-Cdh11 rabbit polyclonal antibody (see Materials and Methods) to probe adult zebrafish brain homogenate (Cdh11 was abundantly detected in the adult brain) reacted with a single polypeptide (relative migration ~120 kD; right lane). Immunodetection was reduced by co-incubating the anti-Cdh11 antibody with the immunogen Cdh11 peptide (i. e., used to raise the antibody; middle lane), but immunodetection was not affected by co-incubating the anti-Cdh11 antibody with an identical concentration of an irrelevant peptide (right lane). Immunofluorescence using anti-Cdh11 rabbit polyclonal antibody in wholemount 30 hpf embryos. The typical inner ear Cdh11 expression pattern was detected in control embryos (B), but co-incubating the anti-Cdh11 antibody with the immunogen Cdh11 peptide eliminated all labeling (C). Laser power, gain settings, filters and image processing were identical for panels B and C. (D) Pools of protein homogenate from three embryos (24 hpf) per lane were immunoblotted to detect Cdh11. This was the smallest number of embryos that we could reliably detect Cdh11 expression in control embryos (left lane). A pool of three cdh11 morpholino oligonucleotides injected 24 hpf embryos showed reduced Cdh11 expression levels. Representative result from two experiments is shown.
Figure S2. Cdh11 morpholino-induced expression reduction through the entire otic vesicle. To determine whether optical section selection in Figure 1B-C minimized strongly labeled structures (like otoliths), we collected image volumes from the same zebrafish embryos labeled using Cdh11-specific antibodies shown in Figure 1B-C (i. e., injected with cdh11 splice-site blocking antisense morpholino oligonucleotide, as here, panels A and B). Instead of a projection of 5 optical sections as shown in Fig. 1B-C, an image volume was collected through entire otic vesicle of a slightly affected 30 hpf embryo (panel A; 45 optical sections, 0.3 micron z-step) and a moderately affected 30 hpf embryo (panel B; 25 optical sections, 0.3 micron z-step). Projection images were made in alpha blending mode using Voxx software. Adobe Photoshop 6 used to prepare figure. The otic vesicle was smaller in the moderately affected embryo, but both images (panels A and B) are projections of image volumes through entire inner ear region of the embryos. Panels A and B have identical orientation, with rostral and lateral as shown in panel A. These image volumes illustrate that only small amounts of Cdh11 labeling were detected in slightly affected phenotype embryos (panel A), and little or no Cdh11 labeling was detected above background in moderately affected (in panel B, background from 25 optical sections is combined) and severely affected (not shown) embryos. Laser power, gain settings, filters and image processing were identical for panels A and B. Two-photon imaging was performed using Zeiss LSM510 Meta NLO, C-Apochromatic 40×W 1.2NA objective, TRITC-labeled secondary antibody, 800 nm excitation, and mounted in PBS.
Figure S3. BODIPY-ceramide labeled vesicular structures in control and Cdh11 knockdown 42 hpf embryos. BODIPY-ceramide labeled living 42 hpf zebrafish embryos injected with control morpholino oligonucleotide (A) and with cdh11 antisense morpholino oligonucleotide injected (B). Confocal optical sections are shown, using a Zeiss LSM 510, C-apochromat 63× W 1.2 NA objective, mounted in embryo medium. Representative confocal image planes are shown. Due to technical difficulties described in the text, quantification would not be as reliable at this developmental stage. However, there was an apparent increase in the number and size of BODIPY-labeled puncta in Cdh11 knockdown embryos. Scale bar is 30 microns.