Abstract
Metagenomics has paved the way for cultivation-independent assessment and exploitation of microbial communities present in complex ecosystems. In recent years, significant progress has been made in this research area. A major breakthrough was the improvement and development of high-throughput next-generation sequencing technologies. The application of these technologies resulted in the generation of large datasets derived from various environments such as soil and ocean water. The analyses of these datasets opened a window into the enormous phylogenetic and metabolic diversity of microbial communities living in a variety of ecosystems. In this way, structure, functions, and interactions of microbial communities were elucidated. Metagenomics has proven to be a powerful tool for the recovery of novel biomolecules. In most cases, functional metagenomics comprising construction and screening of complex metagenomic DNA libraries has been applied to isolate new enzymes and drugs of industrial importance. For this purpose, several novel and improved screening strategies that allow efficient screening of large collections of clones harboring metagenomes have been introduced.
Keywords: Metagenomics, Metagenomic library, Biocatalysts, Function-based screens, Sequence-based screens
Introduction
Metagenomics has been defined as function-based or sequence-based cultivation-independent analysis of the collective microbial genomes present in a given habitat (Riesenfeld et al. 2004b). This rapidly growing research area provided new insights into microbial life and access to novel biomolecules (Banik and Brady 2008; Edwards et al. 2006; Frias-Lopez et al. 2008; Venter et al. 2004). The developed metagenomic technologies are used to complement or replace culture-based approaches and bypass some of their inherent limitations. Metagenomics allows the assessment and exploitation of the taxonomic and metabolic diversity of microbial communities on an ecosystem level.
Recently, advances in throughput and cost-reduction of sequencing technologies have increased the number and size of metagenomic sequencing projects, such as the Sorcerer II Global Ocean Sampling (GOS) (Biers et al. 2009; Rusch et al. 2007), or the metagenomic comparison of 45 distinct microbiomes and 42 viromes (Dinsdale et al. 2008a). The analysis of the resulting large datasets allows exploration of biodiversity and performance of system biology in diverse ecosystems.
So far, the main application area of metagenomics is mining of metagenomes for genes encoding novel biocatalysts and drugs (Lorenz and Eck 2005). Correspondingly, new sensitive and efficient high-throughput screening techniques that allow fast and reliable identification of genes encoding suitable biocatalysts from complex metagenomes have been invented.
In this review, an overview of the recent developments and achievements of bioprospecting and metagenomic analyses of microbial communities derived from different environments is given. In addition, novel metagenomic approaches are briefly discussed.
Exploring the phylogenetic diversity
Metagenomics is a powerful tool for assessing the phylogenetic diversity of complex microbial assemblages present in environmental samples such as soil, sediment, or water. The total number of prokaryotic cells on earth has been estimated to be 4–6 × 1030 comprising 106 to 108 separate genospecies (Sleator et al. 2008). The majority of these microbes is uncharacterized and represents an enormous unexplored reservoir of genetic and metabolic diversity. In recent years, high-throughput metagenomic approaches produced millions of environmental gene sequences, thereby, providing access to the so far hidden phylogenetic composition of complex environmental microbial communities (Sjöling and Cowan 2008).
To explore the microbial diversity of environmental samples, also termed “taxonomical binning,” different approaches can be applied (Richter et al. 2008). Usually, phylogenetic relationships are determined by analysis of conserved ribosomal RNA (rRNA) gene sequences (Woese 1987). Extensive sequencing of ribosomal RNA genes resulted in generation of several large reference databases, such as the ribosomal database project (RDP) II (Cole et al. 2003), Greengenes (DeSantis et al. 2006), or SILVA (Ludwig et al. 2004). These comprehensive databases allow classification and comparison of environmental 16S rRNA gene sequences. Traditional surveys of environmental prokaryotic communities are based on amplification and cloning of 16S rRNA genes prior to sequence analysis. However, some inherent disadvantages such as PCR bias, instability of the recombinant plasmids in the host strain, or the varying number of gene copies between taxa are limitations of this approach (Biddle et al. 2008; Venter et al. 2004). More comprehensive views of prokaryotic communities can be achieved by use of high-throughput shotgun sequencing of environmental samples. Direct sequencing of metagenomic DNA has been proposed to be the most accurate approach for assessment of the taxonomic composition (von Mering et al. 2007). The major advantage of this cloning-independent approach is the avoidance of bias introduced by amplification of phylogenetic marker genes and cloning. In addition, Manichanh et al. (2008) showed that evaluation of a shotgun sequencing-derived dataset provides a reliable estimate of the microbial diversity stored in metagenomic libraries. Venter et al. (2004) were the first to apply whole genome shotgun sequencing to samples of the Sargasso Sea in order to characterize the microbial community and identify new genes and species. The dataset included 1.66 million sequences comprising 1.045 billion base pairs. The taxonomic composition was evaluated by 16S rRNA gene analysis and employment of alternative phylogenetic markers such as RecA/RadA, heat shock protein Hsp70, elongation factor Tu, and elongation factor G. The assignment to phylogenetic groups was consistent among the different markers but the abundance of the encountered phylogenetic groups varied (Venter et al. 2004).
Determination of the taxonomic diversity by analysis of pyrosequencing- or shotgun-derived datasets has been applied to various environments, including an acid mine biofilm (Tyson et al. 2004), seawater samples (Angly et al. 2006; DeLong et al. 2006), the Soudan mine (Edwards et al. 2006), the Peru Margin subsea floor (Biddle et al. 2008), honey bee colonies (Cox-Foster et al. 2007), and deep-sea sediments (Hallam et al. 2004). To date, the largest metagenomic dataset was generated within the framework of the GOS expedition (Rusch et al. 2007; Yooseph et al. 2007). The GOS dataset extends the previously published Sargasso Sea dataset (Venter et al. 2004). Random insert libraries were constructed from DNA isolated from bacterioplankton derived from 41 surface marine environments and a few nonmarine aquatic samples. The phylogenetic diversity stored in this dataset, which comprises 7.7 million sequences (6.3 billion bp), was assessed by analysis of the 16S rRNA gene sequences present in the metagenomic libraries (Biers et al. 2009; Rusch et al. 2007). In general, the alphaproteobacteria were the dominant phylogenetic group in ocean surface waters, whereas, the abundance of other phyla differed depending on the type of environment (Biers et al. 2009).
Due to the enormous quantity of short DNA fragments in large shotgun sequencing-derived or pyrosequencing-derived metagenomic datasets, methods have been developed that are more suitable for taxonomic binning than the analysis of highly conserved phylogenetic marker genes. Phylogenetic classification of metagenomic fragments can be based on sequence composition, i.e., oligonucleotide frequencies, which vary significantly among genomes and exhibit weak phylogenetic signals (Abe et al. 2003; Karlin and Burge 1995; Pride et al. 2003; Teeling et al. 2004a). For a phylogenetic classification of complex microbial communities based on oligonucleotide frequencies, bioinformatic software tools such as TETRA or PhyloPythia have been developed (McHardy et al. 2007; Teeling et al. 2004b). These tools require training, employing known genomic sequences of different taxonomic origin. The accuracy of the phylogenetic classification depends on different factors such as fragment length of the environmental DNA and amount or origin of the genomic sequences used for training. The above-mentioned tools have been successfully employed for characterization of several habitats such as the Sargasso Sea and sludge used in industrial wastewater processing (Abe et al. 2005; McHardy et al. 2007). Recently, other software tools such as the metagenome analyzer MEGAN (Huson et al. 2007), CARMA (Krause et al. 2008), and the sequence ortholog-based approach for binning and improved taxonomic estimation of metagenomic sequences Sort-ITEMS (Monzoorul et al. 2009) have been invented for taxonomic binning of large metagenomic datasets that consist of short environmental DNA fragments. The algorithms differ in the method for phylogenetic classification. MEGAN (Huson et al. 2007; Huson et al. 2009) compares metagenomic datasets with one or more sequence databases, i.e., NCBI-NR, NCBI-NT, NCBI-ENV-NR, or NCBI-ENV-NT (Benson et al. 2006). Subsequently, the reads are assigned to the lowest common ancestor of the nearest relatives in the reference databases. In order to validate the algorithm, the authors applied MEGAN to the Sargasso Sea dataset and deduced species distribution, which is similar to that reported by Venter et al. (2004). Additionally, Poinar et al. (2006) analyzed a dataset derived from a mammoth bone using MEGAN. Approximately 50% of the analyzed sequences were identified as mammoth DNA, whereas, the remaining sequences were derived from endogenous bacteria and nonelephantid environmental contaminants (Poinar et al. 2006).
Krause et al. (2008) introduced the CARMA algorithm, which uses conserved domains and protein families of the protein families (Pfam) database (Finn et al. 2006) as phylogenetic markers for taxonomic classification of the environmental DNA sequences. These environmental gene tags (EGTs) are identified by employing the Pfam profile hidden Markov models. Subsequently, for each matching Pfam family a phylogenetic tree is reconstructed, and the metagenomic sequences are, thereby, assigned to phylogenetic groups. In this way, EGTs as short as 27 amino acids can be classified (Krause et al. 2008). CARMA has been shown to provide accurate results, but it is computationally expensive (Diaz et al. 2009; Krause et al. 2008).
The most recent binning algorithm Sort-ITEMS utilizes the bit score and alignment parameters of the basic local alignment search tool BLAST (Altschul et al. 1990) for an initial taxonomic classification. Subsequently, a higher resolution is achieved by an orthology-based approach (Monzoorul et al. 2009).
Phylogenetic classification of the metagenomic datasets relies on the use of the above-mentioned reference databases that contain sequences of known origin and gene function. To date, the common databases are biased towards model organisms or readily cultivable microorganisms. This is a major limitation for taxonomic classification of microbial communities in ecosystems. According to Huson et al. (2009), up to 90% of the sequences of a metagenomic dataset may remain unidentified due to the lack of a reference sequence.
Connecting function to phylogeny
Exploring the phylogenetic diversity and population structure of environmental samples is essential for the reconstruction of the metabolic potential of individual organisms or phylogenetic groups and the discovery of their interactions. The employment of metagenomics allows the discovery of interactions between microorganisms and the environment and the assignment of ecosystem functions to microbial communities (Lopez-Garcia and Moreira 2008; Sjöling and Cowan 2008).
Linking functional genes of uncultured organisms to phylogenetic groups can be accomplished by cloning and sequencing of large genomic DNA fragments containing phylogenetic markers or by reconstruction of genomes from metagenomic datasets (Sjöling and Cowan 2008). An illustrative example is the discovery of rhodopsin-like photoreceptors and proteorhodopsin-dependent phototrophy in marine bacteria by analyzing large-insert metagenomic libraries (Béjà et al. 2000). The open reading frame coding for proteorhodopsin was located in the vicinity of a 16S rRNA gene, which originated from a member of the gammaproteobacteria (Béjà et al. 2000). In additional datasets derived from aquatic samples, new and diverse rhodopsin-like genes were identified and indicated a widespread abundance and importance of this light-driven way of energy conservation (Rusch et al. 2007; Venter et al. 2004). Reconstruction of near complete and complete genomes of individual microorganisms derived from metagenomic datasets is restricted to low-diversity habitats, since the species-richness of high-diversity habitats such as soil and sediment would require enormous sequencing and assembly efforts. Recently, this approach has been successfully applied for low-diversity samples from acid mines (Tyson et al. 2004), an anaerobic ammonium-oxidizing community (Strous et al. 2006), and enrichments (Hallam et al. 2004).
Functional diversity of microbial communities
Large-scale sequencing of metagenomic DNA permits the identification of the most frequently represented functional genes and metabolic pathways that are relevant in a given ecosystem. In this way, the dominant biosynthetic pathways and primary energy sources can be assessed. Edwards et al. (2006) conducted the first study in which metabolic profiles of whole microbial communities based on a pyrosequencing-derived dataset were analyzed. The authors compared two different sampling sites in the Soudan mine (Minnesota, USA). Significant differences in the use of substrates and metabolic pathways were established. In addition, the geochemical conditions of the two analyzed sites and the microbial metabolism correlated (Edwards et al. 2006). The rapid identification of the metabolic capacity and genetic diversity of this habitat indicated the significance of metagenomics for functional analysis of ecosystems. Other examples for identification of the functional diversity and profiles by analysis of pyrosequencing-derived datasets include an obesity-associated gut microbiome (Turnbaugh et al. 2006), a coral-associated microbial community (Wegley et al. 2007), a comparison of nine biomes (Dinsdale et al. 2008a), ocean surface waters (Frias-Lopez et al. 2008), the Peru Margin subseafloor (Biddle et al. 2008), coral atolls (Dinsdale et al. 2008b), and stressed coral holobionts (Thurber et al. 2009).
For functional binning of metagenomic datasets, sequences are compared to reference databases, such as the clusters of orthologous groups of proteins (Tatusov et al. 2003), the Kyoto encyclopedia of genes and genomes (Kanehisa et al. 2004), Pfam, SEED (Overbeek et al. 2005), the search tool for the retrieval of interacting genes/proteins STRING (Jensen et al. 2009), or TIGRFAM (Haft et al. 2003), which contain known protein functions, families, and pathways (Richter et al. 2008). Bioinformatic analyses are crucial for linking function to phylogenetic diversity of ecosystems. Recently, Meyer et al. (2008) introduced the metagenome rapid annotation using subsystem technology (MG-RAST) server for analysis of metagenomic datasets. The server provides annotation of sequence fragments, phylogenetic classification, and metabolic reconstruction by implementing the SEED, the national microbial pathogen database resource (McNeil et al. 2007), Greengenes, RDP-II, SILVA, and the European ribosomal RNA database (Wuyts et al. 2004). In addition, this open-source online tool allows the comparison of metagenomic datasets derived from different environments (Meyer et al. 2008). Comparative metagenomics is useful for identification of differences in the ability of microbial communities to adapt to changing environmental conditions. Tringe et al. (2005) analyzed and compared metagenomic datasets from various environments and deduced habitat-specific functions and profiles of the sampled environments. Thus, profiling of the functions encoded by a microbial community rather than the types of organisms producing them provides a means to distinguish samples on the basis of the functions selected by the local environment and reveals insights into features of that environment. This gene-centric approach to environmental sequencing suggests that the functional profile predicted from environmental sequences of a community is similar to that of other communities whose environments of origin pose similar demands.
Nevertheless, the analysis of the taxonomic diversity, functional binning, and profiling of metagenomic datasets bears several limitations. The reference databases used for functional annotation of the sequences are inherently incomplete. Therefore, metagenomic analyses can only be as good as the quality of the reference databases (Meyer et al. 2008). To cope with the increasing number and size of metagenomic sequencing projects, improvement and development of bioinformatic tools for metagenomic data analysis is still required (Meyer et al. 2008).
Metatranscriptomics
Recently, sequencing and characterization of metatranscriptomes have been employed to identify expressed biological signatures in complex ecosystems. Metagenomic complementary DNA (cDNA) libraries have been constructed from messenger RNA (mRNA) that has been isolated from environmental samples (Bailly et al. 2007; Frias-Lopez et al. 2008; Gilbert et al. 2008, 2009; Grant et al. 2006). In contrast to libraries constructed from environmental DNA, cDNA libraries reflect the active metabolic functions of a microbial community. However, due to difficulties associated with RNA isolation, separation of mRNA from other RNA species, and instability of mRNA, constructing libraries derived from environmental mRNA is more challenging than generation of metagenomic DNA libraries (Sjöling and Cowan 2008). Frias-Lopez et al. (2008) constructed cDNA libraries from metagenomic microbial mRNA derived from ocean surface water. The cDNA libraries were subjected to pyrosequencing, and the resulting dataset was compared to diverse databases. Many of the identified genes were highly similar to genes previously identified in ocean samples. Approximately 50% of all detected transcripts were unique, indicating that a large unknown metabolic diversity is present in the ocean. The few published metatranscriptomic studies were mainly performed with samples from marine environments and soils. The microbial community transcriptome analyses revealed that the identification of indigenous gene- and taxon-specific patterns, and the identification of key metabolic functions are feasible. In addition, when paired with metagenomic data, detailed analyses of both structure and function of microbial communities are provided (Frias-Lopez et al. 2008; Gilbert et al. 2008; Urich et al. 2008).
Metagenomes as sources for novel biomolecules
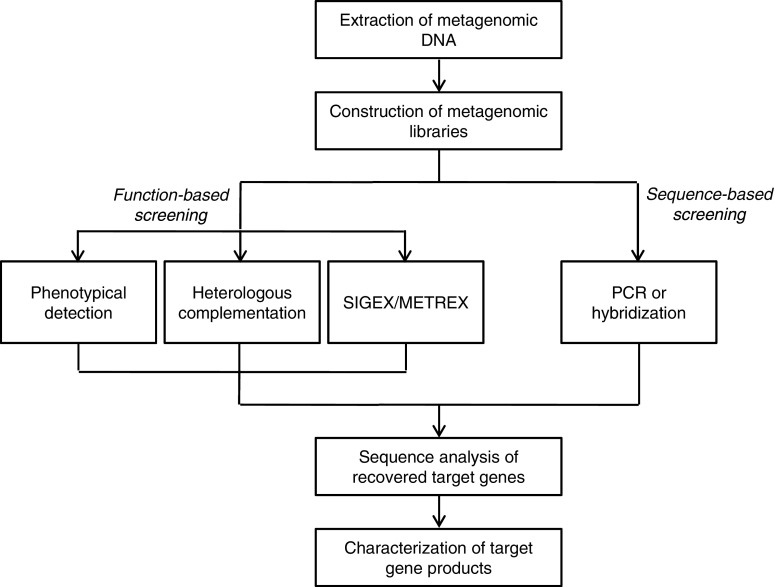
Most biocatalysts employed for biotechnological or industrial purposes are of microbial origin. This reflects the fact that the broadest genetic variety in the biosphere can be found in the different microbial communities present in the various ecosystems on earth (Ferrer et al. 2009). The application of culture-independent metagenomic approaches allows exploiting this almost unlimited resource of novel biomolecules (Sjöling and Cowan 2008). The work published in this field showed that the cloning of metagenomic DNA and the subsequent screening of the constructed complex environmental libraries bear the potential to encounter entirely new classes of genes for new or known functions, including genes encoding, i.e., lipases, antibiotics, antibiotic resistance genes, oxidoreductases, catabolic enzymes, and biotin synthesis (see Table 1). Several techniques have been used to identify and retrieve genes and gene clusters from metagenomic libraries. Due to the complexity of metagenomic libraries, high-throughput and sensitive screening approaches have been employed. Screens have been based either on nucleotide sequence (sequence-driven approach) or on metabolic activity (function-driven approach) (Fig. 1).
Table 1.
Recent examples for metagenome-derived biocatalysts and the employed screening strategy
| Target | Source | Number of screened clones | Sampling site | Screening technique | Reference |
|---|---|---|---|---|---|
| Lipase | Fosmids | >7,000 | Baltic sea sediment (Sweden) | Phenotypical detection | Hårdeman and Sjöling 2007 |
| Cosmids | 10,000 | Sequencing fed-batch reactor enriched with gelatin | Phenotypical detection | Meilleur et al. 2009 | |
| Plasmids | Not mentioned | Soil samples from different altitudes of Taishan (China) | Phenotypical detection | Wei et al. 2009 | |
| Cosmids | 1,532 | Soil from uncultivated field (Germany) | Phenotypical detection | Voget et al. 2003 | |
| Fosmids | 386,400 | Tidal flat sediments (Korea) | Phenotypical detection | Lee et al. 2006b | |
| Lipase/Esterase | Plasmids | 1,016,000 | Soil from a meadow, sugar beet field, and river valley (Germany) | Phenotypical detection | Henne et al. 2000 |
| Esterase | Fosmids | 5,000 | Hot springs and mud holes in solfataric fields (Indonesia) | Phenotypical detection | Rhee et al. 2005 |
| Phagemids | 385,000 | Wadi Natrun (Egypt), Lake Nakuru, and Crater Lake (Kenya) and enrichments | Phenotypical detection | Rees et al. 2003 | |
| Fosmids | 100,000 | Desert soil (Antarctica) | Phenotypical detection | Heath et al. 2009 | |
| Plasmids | 93,000 | Vegetable soil | Phenotypical detection | Li et al. 2008 | |
| BACs | 8,000 | Surface water microbes from Yangtze river (China) | Phenotypical detection | Wu and Sun 2009 | |
| Cellulase | Phagemids | 385,000 | Wadi Natrun (Egypt), Lake Nakuru, and Crater Lake (Kenya) and enrichments | Phenotypical detection | Rees et al. 2003 |
| Cosmids | 1,700 | Soil microbial consortia (Germany) | Phenotypical detection | Voget et al. 2006 | |
| Cosmids | 3,744 | Aquatic community and soil Germany) | Phenotypical detection | Pottkämper et al. 2009 | |
| Cosmids | 15,000 | Buffalo rumen | Phenotypical detection | Duan et al. 2009 | |
| Cosmids | 32,500 | Rabbit cecum | Phenotypical detection | Feng et al. 2007 | |
| Protease | Plasmids | 80,000 | Compost soil (Germany), soil from mining shaft (Germany), and mixed soil sample (Germany, Israel, and Egypt) | Phenotypical detection | Waschkowitz et al. 2009 |
| Fosmids | 30,000 | Deep-sea sediment from a clam bed community (Korea) | Phenotypical detection | Lee et al. 2007 | |
| Agarase | Cosmids | 1,532 | Soil from uncultivated field (Germany) | Phenotypical detection | Voget et al. 2003 |
| Oxidative coupling enzyme (OxyC) | Cosmids | 10,000,000 | Collection of soil samples (USA and Costa Rica) | Sequence-based | Banik and Brady 2008 |
| Alcohol oxidoreductase | Plasmids | 900,000 and 400,000 | Soil and enrichment cultures from a sugar beet field (Germany), river sediment (Germany), sediment from Solar Lake (Egypt), and sediment from the Gulf of Eilat (Israel) | Sequence-based and phenotypical detection | Knietsch et al. 2003b, c |
| Amidase | Plasmids | 193,000 | Soil and enrichment cultures from marine sediment, goose pond, lakeshore, and an agricultural field (Netherlands) | Heterologous complementation | Gabor et al. 2004 |
| Xylanase | Phagemids | 5,000,000 | Manure wastewater lagoon (USA) | Phenotypical detection | Lee et al. 2006a |
| Antibiotics | Cosmids | Not mentioned | Bromeliad tank water (Costa Rica) | Phenotypical detection | Brady and Clardy 2004 |
| Glycerol dehydratase and diol dehydratase | Plasmids | 158,000 and 560,000 | Soil from a sugar beet field (Germany), river sediment (Germany), and sediment from Solar Lake (Egypt) | Sequence-based and heterologous complementation | Knietsch et al. 2003a |
| Magnetosome island gene clusters | Fosmids | 5,823 | Different aquatic sediments (Germany) | Sequence-based | Jogler et al. 2009 |
| Benzoate 1,2-dioxygenase alpha subunit and chlorocatechol 1,2-dioxygenase | DNA | - | Soil from a conserved forest (Japan) | Sequence-based | Morimoto and Fuji 2009 |
| DNA polymerase I | Plasmids and fosmids | 230,000 and 4,000 | Glacier ice (Germany) | Heterologous complementation | Simon et al. 2009 |
| Multicopper oxidases | DNA | - | Not specified | Sequence-based | Meyer et al. 2007 |
| Blue light photoreceptor | Cosmids | 2,500 | Soil from a botanical garden (Germany), enrichment | Sequence-based | Pathak et al. 2009 |
| Na+/H+ antiporters | Plasmids | 1,480,000 | Soil from a meadow, sugar beet field, and river valley (Germany) | Heterologous complementation | Majernik et al. 2001 |
| Antibiotic resistance | BACs and plasmids | 28,200 and 1,158,000 | Plano silt loam (USA) | Heterologous complementation | Riesenfeld et al. 2004a |
| Poly-3-hydroxybutyrate metabolism | Cosmids | 45,630 | Activated sludge and soil microbial communities (Canada) | Heterologous complementation | Wang et al. 2006 |
| Lysine racemase | Plasmids | Not mentioned | Garden soil (Taiwan) | Heterologous complementation | Chen et al. 2009 |
| Aromatic-hydrocarbon catabolic operon fragments | Plasmids | 152,000 | Crude-oil contaminated groundwater microbial flora (Japan) | SIGEX | Uchiyama et al. 2005 |
| Quorum sensing inducer/inhibitor | BACs and fosmids | 52,500 and 300 | Soil on the floodplain of the Tanana River (Alaska) | METREX | Williamson et al. 2005 |
| Beta-lactamase | Fosmids | 8,823 | Cold-seep sediments of Edison seamount (Papua New Guinea) | Phenotypical detection | Song et al. 2005 |
| Chitinase | DNA | - | Water and sediment samples from aquatic environments (USA and Arctic ocean) | Sequence-based | LeCleir et al. 2004 |
| Cyclodextrinase | Phagemids | 200,000 | Cow rumen | Phenotypical detection | Ferrer et al. 2005 |
Fig. 1.
Strategies for recovery of novel biomolecules
Sequence-based screening
The sequence-based screening approach is limited to the identification of new members of known gene families. In general, target genes are identified either by PCR-based or hybridization-based approaches employing primers and probes derived from conserved regions of known genes and gene products (Daniel 2005; Handelsman 2004). Thus, only genes harboring regions with similarity to the sequences of the probes and primers can be recovered by this approach. In addition, sequence-driven screening is not selective for full-length genes and functional gene products. The advantage of this screening strategy is the independence on gene expression and production of foreign genes in the library host (Lorenz et al. 2002). Several novel functional enzymes such as chitinases, alcohol oxidoreductases, diol dehydratases, and enzymes conferring antibiotic resistance have been recovered by employing sequence-driven approaches (see Table 1). For example, Banik and Brady (2008) isolated two novel glycopeptide-encoding gene clusters from a large-insert megalibrary, which comprised 10,000,000 cosmid-containing clones derived from desert soil by a PCR-based screen. Degenerate primers were employed, which were deduced from OxyC, an oxidative coupling enzyme encoded by glycopeptide biosynthetic clusters. The isolation of these biosynthetic clusters is important for the development of novel glycopeptides analogs, which can serve as substitutes of currently used antibiotics such as vancomycin.
Another recent example for a screening based on sequence similarity was published by Jogler et al. (2009). After selective enrichment of magnetotactic bacteria (MTB), large DNA fragments from uncultured MTB derived from various aquatic habitats were cloned into fosmid vectors. Four fosmid libraries comprising 5,823 clones were screened by hybridization using mam genes of known magnetotactic alphaproteobacteria as probes. Two fosmids, which contain operons with similarity to magnetosome islands of cultured MTB, were detected, and the organization of the magnetosome island of uncultured MTB was elucidated.
A new approach to retrieve complete functional genes is PCR-denaturing gradient gel electrophoresis (DGGE) followed by metagenomic walking. Morimoto and Fujii (2009) conducted a PCR-DGGE targeting benA and tfdC, which encode the alpha subunits of benzoate 1,2-dioxygenase and chlorocatechol 1,2-dioxygenase, respectively. Two DGGE bands, which appeared after addition of 3-chlorobenzoate to the samples, were chosen for further analysis. The complete functional genes were recovered by metagenome walking (Morimoto and Fujii 2009).
Recently, Meyer et al. (2007) introduced subtractive hybridization magnetic bead capture as approach for recovery of multicopper oxidases from metagenomic DNA. Conserved regions of the target genes are amplified from a metagenomic DNA sample by PCR using biotinylated degenerated primers. Subsequently, the resulting amplified target gene fragments are immobilized on streptavidin-covered magnetic beads, which are then used as probes for capturing the full-length genes from metagenomic DNA by hybridization. In contrast to previously published PCR-based techniques, the subtractive hybridization approach allows the recovery of multiple gene targets in a single reaction. According to Meyer et al. (2007), the employment of immobilized large gene fragments as probes results in specificity, which is higher than that of other PCR-based approaches (Meyer et al. 2007).
In a few cases, microarray technology has been employed for sequence-driven screening of metagenomic DNA and libraries. A recent example is the recovery of genes encoding blue light-sensitive proteins (Pathak et al. 2009).
Function-based screening
Function-driven screening of metagenomic libraries is not dependent on sequence information or sequence similarity to known genes. Thus, this is the only approach that bears the potential to discover new classes of genes that encode either known or new functions (Heath et al. 2009; Rees et al. 2003). A significant limitation of this technique is the dependence on expression of the target genes and production of functional gene products in a foreign host, which is in most studies Escherichia coli. Thus, the incapability to discover functional gene products or a low detection frequency during function-based screens of metagenomic libraries might be a result of the inability of the host to express the foreign genes and to form active recombinant proteins. In addition, function-driven screening often requires the analysis of more clones than sequence-based screening for the recovery of a few positive clones (Daniel 2005). The major advantage of a function-based screening approach is that only full-length genes and functional gene products are detected. The following three different types of function-driven approaches have been employed for screening of metagenomic libraries: (1) direct detection of specific phenotypes of individual clones; (2) heterologous complementation of host strains or mutants; (3) induced gene expression (Fig. 1 and Table 1).
To identify enzymatic functions of individual clones, chemical dyes and insoluble or chromophore-containing derivatives of enzyme substrates can be incorporated into the growth medium (Daniel 2005; Ferrer et al. 2009; Handelsman 2004). Examples for this simple activity-based approach are the detection of recombinant E. coli clones exhibiting protease activity on indicator agar containing skimmed milk as protease substrate (Lee et al. 2007; Waschkowitz et al. 2009) or the detection of lipolytic activity by employing indicator agar containing tributyrin or tricaprylin as enzyme substrates (Hårdeman and Sjöling 2007; Heath et al. 2009; Lee et al. 2006b). Clones with proteolytic or lipolytic activity are identified by halo formation on solidified indicator medium.
A different approach is the use of host strains that require heterologous complementation by foreign genes for growth under selective conditions. Only recombinant clones harboring the targeted gene and producing the corresponding gene product in an active form are able to grow. In this way, a high selectivity of the screen is achieved. One recent example is the identification of DNA polymerase-encoding genes from metagenomic libraries derived from microbial communities present in glacier ice (Simon et al. 2009). An E. coli mutant, which carries a cold-sensitive lethal mutation in the 5′-3′ exonuclease domain of the DNA polymerase I, was employed as host for the metagenomic libraries. At a growth temperature of 20 °C only recombinant E. coli strains complemented by a gene conferring DNA polymerase-activity are able to grow. In this way, novel genes encoding DNA polymerases were recovered and almost no false positive clones were obtained (Simon et al. 2009). Further examples for this screening approach are the detection of genes encoding Na+/H+ antiporters (Majernik et al. 2001), antibiotic resistance (Riesenfeld et al. 2004a), enzymes involved in poly-3-hydroxybutyrate metabolism (Wang et al. 2006), and lysine racemases (Chen et al. 2009).
The third function-driven approach is based on induced gene expression. Uchiyama et al. (2005) introduced a substrate-induced gene expression screening system (SIGEX) for the identification of novel catabolic genes. An operon-trap expression vector, which contains the gene for a promoterless green fluorescent protein (gfp), was employed for cloning of environmental DNA. Catabolic operons are often adjacent to cognate transcriptional regulators and promoters that are induced by the substrate. If expression of a target gene is induced by the substrate, the gfp gene is coexpressed, and positive clones can rapidly be separated from other clones by fluorescent-activated cell sorting (Handelsman 2005; Uchiyama et al. 2005). This method was validated by the screening of a metagenomic library derived from groundwater microbial flora. Regulated by the induction substrates benzoate and naphthalene 58 and 4 positive clones, respectively, were identified. The major drawback of this high-throughput screening approach is the possible activation of transcriptional regulators by other effectors than the specific substrates. This may lead to the recovery of false-positives (Galvao et al. 2005). A similar screening strategy termed metabolite-regulated expression has been published by Williamson et al. (2005). In contrast to SIGEX, metagenomic clones producing small molecules are identified. A biosensor that detects small diffusible signal molecules, which induce quorum sensing, is inside the same cell as the vector harboring a metagenomic DNA fragment. When a threshold concentration of the signal molecule is exceeded, green fluorescent protein is produced. Subsequently, positive fluorescent clones are identified by fluorescence microscopy (Williamson et al. 2005).
Metagenomics of extreme environments with low microbial community size
Physicochemical extreme environments such as ice (Simon et al. 2009), highly polluted environments (Abulencia et al. 2006), or deep hypersaline anoxic basins (van der Wielen et al. 2005) contain a low microbial community size. These habitats represent a widely unexplored ecological niche with a vast potential of novel biocatalysts of industrial use (Abulencia et al. 2006; Sjöling and Cowan 2008). Microbes that are capable of living in these hostile environments have evolved special mechanisms for survival. Due to the low community size and biomass of these ecosystems, these habitats are not as easily accessible as other environments by metagenomic approaches (Ferrer et al. 2009). The major challenge is to extract a sufficient amount of high-quality DNA. To overcome this limitation, whole genomic amplification of environmental DNA using the φ29 polymerase can be applied. In this way, high-throughput metagenomic approaches from small quantities of DNA as starting material are feasible. Drawbacks of whole genome amplification are the formation of chimeric artifacts and amplification bias, which is a result of template inaccessibility or low priming efficiency (Abulencia et al. 2006). Nevertheless, this approach has been successfully employed in several metagenomic studies of different environments, including contaminated sediments (Abulencia et al. 2006), the Soudan mine (Edwards et al. 2006), scleratinian corals (Yokouchi et al. 2006), the marine viral metagenomes of four oceanic regions (Angly et al. 2006), and glacier ice (Simon et al. 2009).
Conclusions
Metagenomics is an important and indispensable tool for the identification of novel biomolecules and analysis of the genetic diversity and metabolic potential of microbial communities. New and efficient high-throughput screening techniques have been developed, which facilitated the recovery of a high amount of new biocatalysts and small molecules. One of the main hurdles with respect to bioprospecting is the limited production of active recombinant proteins in heterologous hosts. Progress in metagenomic sequence analysis has been driven by the development of next-generation sequencing technologies, which permit cloning-independent and low-cost sequencing analyses of metagenomes. The rapid development of high-throughput DNA sequencing technologies and the corresponding increase in large and complex environmental require permanent development of appropriate bioinformatic tools for their analysis. A combination of metagenomics, metatranscriptomics, and metaproteomics is necessary for a comprehensive understanding of complex microbial communities. In this way, the structure and function of microbial communities in complex environments can be unraveled, and the monitoring of in situ responses and activities of microbes on an ecosystem level is feasible.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe T, Kanaya S, Kinouchi M, Ichiba Y, Kozuki T, Ikemura T. Informatics for unveiling hidden genome signatures. Genome Res. 2003;13:693–702. doi: 10.1101/gr.634603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Sugawara H, Kinouchi M, Kanaya S, Ikemura T. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 2005;12:281–290. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- Abulencia CB, Wyborski DL, Garcia JA, Podar M, Chen W, Chang SH, Chang HW, Watson D, Brodie EL, Hazen TC, Keller M. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl Environ Microbiol. 2006;72:3291–3301. doi: 10.1128/AEM.72.5.3291-3301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Olson R, Parsons R, Rayhawk S, Suttle CA, Rohwer F. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly J, Fraissinet-Tachet L, Verner MC, Debaud JC, Lemaire M, Wesolowski-Louvel M, Marmeisse R. Soil eukaryotic functional diversity, a metatranscriptomic approach. Isme J. 2007;1:632–642. doi: 10.1038/ismej.2007.68. [DOI] [PubMed] [Google Scholar]
- Banik JJ, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Natl Acad Sci U S A. 2008;105:17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci U S A. 2008;105:10583–10588. doi: 10.1073/pnas.0709942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biers EJ, Sun S, Howard EC. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol. 2009;75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SF, Clardy J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J Nat Prod. 2004;67:1283–1286. doi: 10.1021/np0499766. [DOI] [PubMed] [Google Scholar]
- Chen I-C., Lin W.-D., Hsu S.-K., Thiruvengadam V., Hsu W.-H. Isolation and Characterization of a Novel Lysine Racemase from a Soil Metagenomic Library. Applied and Environmental Microbiology. 2009;75(15):5161–5166. doi: 10.1128/AEM.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, vanEngelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz NN, Krause L, Goesmann A, Niehaus K, Nattkemper TW. TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan Y, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, Feng JX. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol. 2009;107:245–256. doi: 10.1111/j.1365-2672.2009.04202.x. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander EC, Jr, Rohwer F. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Duan CJ, Pang H, Mo XC, Wu CF, Yu Y, Hu YL, Wei J, Tang JL, Feng JX. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl Microbiol Biotechnol. 2007;75:319–328. doi: 10.1007/s00253-006-0820-9. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Santos VA, Strompl C, Elborough K, Jarvis G, Neef A, Yakimov MM, Timmis KN, Golyshin PN. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol. 2005;7:1996–2010. doi: 10.1111/j.1462-2920.2005.00920.x. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Beloqui A, Timmis KN, Golyshin PN. Metagenomics for mining new genetic resources of microbial communities. J Mol Microbiol Biotechnol. 2009;16:109–123. doi: 10.1159/000142898. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, Delong EF. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci U S A. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor EM, de Vries EJ, Janssen DB. Construction, characterization, and use of small-insert gene banks of DNA isolated from soil and enrichment cultures for the recovery of novel amidases. Environ Microbiol. 2004;6:948–958. doi: 10.1111/j.1462-2920.2004.00643.x. [DOI] [PubMed] [Google Scholar]
- Galvao TC, Mohn WW, de Lorenzo V. Exploring the microbial biodegradation and biotransformation gene pool. Trends Biotechnol. 2005;23:497–506. doi: 10.1016/j.tibtech.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, Joint I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Thomas S, Cooley NA, Kulakova A, Field D, Booth T, McGrath JW, Quinn JP, Joint I. Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ Microbiol. 2009;11:111–125. doi: 10.1111/j.1462-2920.2008.01745.x. [DOI] [PubMed] [Google Scholar]
- Grant S, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S. Identification of eukaryotic open reading frames in metagenomic cDNA libraries made from environmental samples. Appl Environ Microbiol. 2006;72:135–143. doi: 10.1128/AEM.72.1.135-143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Sorting out metagenomes. Nat Biotechnol. 2005;23:38–39. doi: 10.1038/nbt0105-38. [DOI] [PubMed] [Google Scholar]
- Hårdeman F, Sjöling S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol. 2007;59:524–534. doi: 10.1111/j.1574-6941.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- Heath C, Hu XP, Cary C, Cowan D. Isolation and characterisation of a novel, low-temperature-active alkaliphilic esterase from an Antarctic desert soil metagenome. Appl Environ Microbiol. 2009;75:4657–4659. doi: 10.1128/AEM.02597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Schmitz RA, Bömeke M, Gottschalk G, Daniel R. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol. 2000;66:3113–3116. doi: 10.1128/aem.66.7.3113-3116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC. Methods for comparative metagenomics. BMC Bioinformatics. 2009;10(Suppl 1):S12. doi: 10.1186/1471-2105-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Lin W, Meyerdierks A, Kube M, Katzmann E, Flies C, Pan Y, Amann R, Reinhardt R, Schüler D. Towards cloning the magnetotactic metagenome: Identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl Environ Microbiol. 2009;75:3972–3979. doi: 10.1128/AEM.02701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- Knietsch A, Bowien S, Whited G, Gottschalk G, Daniel R. Identification and characterization of coenzyme B12-dependent glycerol dehydratase- and diol dehydratase-encoding genes from metagenomic DNA libraries derived from enrichment cultures. Appl Environ Microbiol. 2003;69:3048–3060. doi: 10.1128/AEM.69.6.3048-3060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knietsch A, Waschkowitz T, Bowien S, Henne A, Daniel R. Metagenomes of complex microbial consortia derived from different soils as sources for novel genes conferring formation of carbonyls from short-chain polyols on Escherichia coli. J Mol Microbiol Biotechnol. 2003;5:46–56. doi: 10.1159/000068724. [DOI] [PubMed] [Google Scholar]
- Knietsch A, Waschkowitz T, Bowien S, Henne A, Daniel R. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl Environ Microbiol. 2003;69:1408–1416. doi: 10.1128/AEM.69.3.1408-1416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F, Edwards RA, Stoye J. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCleir GR, Buchan A, Hollibaugh JT. Chitinase gene sequences retrieved from diverse aquatic habitats reveal environment-specific distributions. Appl Environ Microbiol. 2004;70:6977–6983. doi: 10.1128/AEM.70.12.6977-6983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Kibblewhite-Accinelli RE, Wagschal K, Robertson GH, Wong DW. Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles. 2006;10:295–300. doi: 10.1007/s00792-005-0499-3. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lee CH, Oh TK, Song JK, Yoon JH. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol. 2006;72:7406–7409. doi: 10.1128/AEM.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Jeon JH, Jang MK, Kim NY, Lee JH, Lee JH, Kim SJ, Kim GD, Lee SH. Screening and characterization of a novel fibrinolytic metalloprotease from a metagenomic library. Biotechnol Lett. 2007;29:465–472. doi: 10.1007/s10529-006-9263-8. [DOI] [PubMed] [Google Scholar]
- Li G, Wang K, Liu YH. Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the Metagenome. Microb Cell Fact. 2008;7:38. doi: 10.1186/1475-2859-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D. Tracking microbial biodiversity through molecular and genomic ecology. Res Microbiol. 2008;159:67–73. doi: 10.1016/j.resmic.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Lorenz P, Eck J. Metagenomics and industrial applications. Nat Rev Microbiol. 2005;3:510–516. doi: 10.1038/nrmicro1161. [DOI] [PubMed] [Google Scholar]
- Lorenz P, Liebeton K, Niehaus F, Eck J. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr Opin Biotechnol. 2002;13:572–577. doi: 10.1016/s0958-1669(02)00345-2. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar A, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majernik A, Gottschalk G, Daniel R. Screening of environmental DNA libraries for the presence of genes conferring Na+(Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J Bacteriol. 2001;183:6645–6653. doi: 10.1128/JB.183.22.6645-6653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Chapple CE, Frangeul L, Gloux K, Guigo R, Dore J. A comparison of random sequence reads versus 16S rDNA sequences for estimating the biodiversity of a metagenomic library. Nucleic Acids Res. 2008;36:5180–5188. doi: 10.1093/nar/gkn496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- McNeil LK, Reich C, Aziz RK, Bartels D, Cohoon M, Disz T, Edwards RA, Gerdes S, Hwang K, Kubal M, Margaryan GR, Meyer F, Mihalo W, Olsen GJ, Olson R, Osterman A, Paarmann D, Paczian T, Parrello B, Pusch GD, Rodionov DA, Shi X, Vassieva O, Vonstein V, Zagnitko O, Xia F, Zinner J, Overbeek R, Stevens R. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 2007;35:D347–353. doi: 10.1093/nar/gkl947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilleur C, Hupe JF, Juteau P, Shareck F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J Ind Microbiol Biotechnol. 2009;36:853–861. doi: 10.1007/s10295-009-0562-7. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer QC, Burton SG, Cowan DA. Subtractive hybridization magnetic bead capture: a new technique for the recovery of full-length ORFs from the metagenome. Biotechnol J. 2007;2:36–40. doi: 10.1002/biot.200600156. [DOI] [PubMed] [Google Scholar]
- Monzoorul HM, Tarini S, Dinakar K, Sharmila SM. SOrt-ITEMS : sequence orthology based approach for improved taxonomic estimation of metagenomic sequences. Bioinformatics. 2009;25:1722–1730. doi: 10.1093/bioinformatics/btp317. [DOI] [PubMed] [Google Scholar]
- Morimoto S, Fujii T. A new approach to retrieve full lengths of functional genes from soil by PCR-DGGE and metagenome walking. Appl Microbiol Biotechnol. 2009;83:389–396. doi: 10.1007/s00253-009-1992-x. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak G. P., Ehrenreich A., Losi A., Streit W. R., Gärtner W. Novel blue light-sensitive proteins from a metagenomic approach. Environmental Microbiology. 2009;11(9):2388–2399. doi: 10.1111/j.1462-2920.2009.01967.x. [DOI] [PubMed] [Google Scholar]
- Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, Rampp M, Miller W, Schuster SC. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- Pottkämper J, Barthen P, Ilmberger N, Schwaneberg U, Schenk A, Schulte M, Ignatiev N, Streit WR. Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green chemistry. 2009;11:957–965. [Google Scholar]
- Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13:145–158. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HC, Grant S, Jones B, Grant WD, Heaphy S. Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries. Extremophiles. 2003;7:415–421. doi: 10.1007/s00792-003-0339-2. [DOI] [PubMed] [Google Scholar]
- Rhee JK, Ahn DG, Kim YG, Oh JW. New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl Environ Microbiol. 2005;71:817–825. doi: 10.1128/AEM.71.2.817-825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS ONE. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–989. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Herath J, Rockstroh S, Daniel R. Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl Environ Microbiol. 2009;75:2964–2968. doi: 10.1128/AEM.02644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöling S, Cowan DA. Metagenomics: microbial community genomes revealed. In: Margesin R, Schinner F, Marx J-C, Gerday C, editors. Psychrophiles: from biodiversity to biotechnology. Berlin Heidelberg: Springer; 2008. pp. 313–332. [Google Scholar]
- Sleator RD, Shortall C, Hill C. Metagenomics. Lett Appl Microbiol. 2008;47:361–366. doi: 10.1111/j.1472-765X.2008.02444.x. [DOI] [PubMed] [Google Scholar]
- Song JS, Jeon JH, Lee JH, Jeong SH, Jeong BC, Kim SJ, Lee JH, Lee SH. Molecular characterization of TEM-type beta-lactamases identified in cold-seep sediments of Edison Seamount (south of Lihir Island, Papua New Guinea) J Microbiol. 2005;43:172–178. [PubMed] [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- Teeling H, Waldmann J, Lombardot T, Bauer M, Glöckner FO. TETRA: a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber Rebecca Vega, Willner-Hall Dana, Rodriguez-Mueller Beltran, Desnues Christelle, Edwards Robert A., Angly Florent, Dinsdale Elizabeth, Kelly Linda, Rohwer Forest. Metagenomic analysis of stressed coral holobionts. Environmental Microbiology. 2009;11(8):2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzen A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Wielen PW, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L, D’Auria G, de Lange GJ, Huebner A, Varnavas SP, Thomson J, Tamburini C, Marty D, McGenity TJ, Timmis KN. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science. 2005;307:121–123. doi: 10.1126/science.1103569. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR. Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol. 2003;69:6235–6242. doi: 10.1128/AEM.69.10.6235-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voget S, Steele HL, Streit WR. Characterization of a metagenome-derived halotolerant cellulase. J Biotechnol. 2006;126:26–36. doi: 10.1016/j.jbiotec.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- Wang C, Meek DJ, Panchal P, Boruvka N, Archibald FS, Driscoll BT, Charles TC. Isolation of poly-3-hydroxybutyrate metabolism genes from complex microbial communities by phenotypic complementation of bacterial mutants. Appl Environ Microbiol. 2006;72:384–391. doi: 10.1128/AEM.72.1.384-391.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschkowitz T, Rockstroh S, Daniel R. Isolation and characterization of metalloproteases with a novel domain structure by construction and screening of metagenomic libraries. Appl Environ Microbiol. 2009;75:2506–2516. doi: 10.1128/AEM.02136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;11:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wei P, Bai L, Song W, Hao G. Characterization of two soil metagenome-derived lipases with high specificity for p-nitrophenyl palmitate. Arch Microbiol. 2009;191:233–240. doi: 10.1007/s00203-008-0448-5. [DOI] [PubMed] [Google Scholar]
- Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol. 2005;71:6335–6344. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Sun B. Identification of novel esterase from metagenomic library of Yangtze river. J Microbiol Biotechnol. 2009;19:187–193. doi: 10.4014/jmb.0804.292. [DOI] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi H, Fukuoka Y, Mukoyama D, Calugay R, Takeyama H, Matsunaga T. Whole-metagenome amplification of a microbial community associated with scleractinian coral by multiple displacement amplification using phi29 polymerase. Environ Microbiol. 2006;8:1155–1163. doi: 10.1111/j.1462-2920.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, Strausberg RL, Frazier M, Venter JC. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]