Abstract
This study demonstrates that methyl-CpG-binding PCR (MB-PCR) is a rapid and simple method for detecting fragile X syndrome (FXS) in males, which is performed by verifying the methylation status of the FMR1 promoter in bloodspots. Proteins containing methyl-CpG-binding (MB) domains can be freeze-stored and used as stocks, and the entire test requires only a few hours. The minimum amount of DNA required for the test is 0.5 ng. At this amount, detection sensitivity is not hampered, even mixing with excess unmethylated alleles up to 320 folds. We examined bloodspots from 100 males, including 24 with FXS, in a blinded manner. The results revealed that the ability of MB-PCR to detect FMR1 promoter methylation was the same as that of Southern blot hybridization. Since individuals with 2 or more X chromosomes generally have methylated FMR1 alleles, MB-PCR cannot be used to detect FXS in females.
1. Introduction
Fragile X syndrome (FXS) is the most common form of inherited mental retardation. This syndrome is caused by mutations in the fragile X mental retardation-1 (FMR1) gene (OMIM *309550); in most patients, this gene contains an expanded CGG repeat in the 5′-untranslated region [1]. Alleles with a repeat number less than 59 do not expand to full mutation upon a single transmission [2]. It was initially thought that there are no phenotypic effects in individuals with repeats in the premutation range, that is, between 59 and 200. However, the recent reports have identified a degenerative disease caused by such repeats, namely, fragile X-associated tremor/ataxia syndrome in older individuals and premature ovarian failure in women [3]. The premutation allele is unstable, and the repeat region tends to expand while being transmitted to the next generation via a carrier mother. Once there is a transition to full mutation, that is, the FMR1 has a repeat region of more than 200 CGG repeats, the repeat region and the surrounding CpG islands become hypermethylated, rendering the gene transcriptionally inactive [1, 2].
To date, there is no curative treatment for this disease. Nonetheless, establishment of a correct diagnosis of FXS can not only reduce the stress and frustration of a parent at not knowing the cause of a child's developmental delay, but also facilitate early intervention, thereby benefiting the patients [4–6]. Due to its X-linked nature, FXS is predominantly observed in males. It is rather difficult to clinically diagnose FXS in young boys because its phenotypic manifestations are nonspecific and variable [7]. Therefore, some clinicians prefer to use a simple molecular test to screen boys suspected of having this disease. One of the frequently used tests is based on polymerase chain reaction (PCR) amplification of the FMR1 CGG-repeat region. Moreover, because it is relatively easy to collect and transport bloodspot samples on filter paper, they are often used for molecular screening purposes. If the PCR results are inconclusive, confirmation tests need to be performed [7–9]. However, in such cases, it is difficult to verify whether the bloodspot samples contain the methylated FMR1 promoter; this is mainly because of the limited amount of DNA that can be isolated from bloodspots.
Southern blot hybridization (SBH) is currently the golden standard for confirmation diagnosis of FXS. It can not only detect/exclude the fully expanded repeats, but also check the methylation status of the CGG repeats and the surrounding CpG dinucleotides in the FMR1 gene. Methylation-sensitive PCR is another commonly used method for verifying the aforementioned methylation status. However, in addition to being extremely labor and time consuming, these methods often require an amount of DNA more than what can be obtained from bloodspot samples. Generally, more than 1 μg of high-molecular-weight DNA is required for SBH. Methylation-sensitive PCR is a very specific method; although the sensitivity of this method has been reported to be as high as 1 ng [10], many studies have reported the use of more than 500 ng of DNA for a reliable assessment [11–14]. This is because the DNA sample is subject to significant degradation during the bisulfite conversion process [15], and additional sample loss occurs during the subsequent removal of the bisulfite and desulfonation steps [16].
Recently, 3 articles reported a novel assay for detecting methylated sequences of interest in samples with 1 ng or less DNA [17–19]. Polypeptides containing methyl-CpG-binding domains (MBDs) have been used in the abovementioned studies to specifically retain methylated DNA sequences in a solid matrix. The methylated sequence(s) of interest could then be detected using a variety of techniques. We thus developed a similar assay, namely, methyl-CpG-binding PCR (MB-PCR) [18]. In this study, we first determined the optimal conditions for this assay for reliably distinguishing between the control DNA samples with and without methylated FMR1 promoters. Next, we tested the reliability of this assay in a blinded manner by examining bloodspot samples obtained from 100 males, including 24 with FXS.
2. Materials and Methods
2.1. Strategy and Samples
Universally methylated or unmethylated genomic DNAs were used as positive and negative methylation controls, respectively, to determine the optimal conditions critical for the success of MB-PCR. Universally methylated DNA was prepared by treating lymphocyte DNA with M.SssI [20], while universally unmethylated DNA was prepared using nested whole genome amplification of the same DNA sample with phi29 DNA polymerase [21]. Then, we applied the optimized MB-PCR to examine the bloodspot samples from 100 males in a blinded manner. These subjects had been well characterized by SBH. Finally, comparing the SBH results, we estimated the sensitivity, specificity, and accuracy of MB-PCR by using the formula described at the following html address: http://www.rapid-diagnostics.org/accuracy.htm. The Institutional Ethics Committee at Chi Mei Medical Center approved this study (IRB09602-002).
Between 2002 and 2007, 1399 males submitted blood samples to our laboratory for molecular analysis of the FMR1 mutation. Of the 229 males who submitted 3 mL of venous blood for SBH, 10 were successfully diagnosed with FXS (Table 1). The procedure of SBH used in this study has been published elsewhere [22]. Another 1170 subjects mailed bloodspot samples on a filter paper to our laboratory for a free-of-charge screening PCR, in which the FMR1 CGG-repeat region was amplified. The protocol of screening PCR used in this study has been described elsewhere [9]. Most of these samples showed a strong positive PCR result, indicating that they were unlikely affected with FXS. The amplification results of 30 samples were either weak (n = 16) or negative (n = 14). These subjects were then encouraged to undergo another free-of-charge SBH for confirmation.
Table 1.
Molecular testing of 1399 males suspected of having fragile X syndrome.
| Methylation (+) | Methylation (–) | |||||
|---|---|---|---|---|---|---|
| Screening PCR | SBH MB-PCR | SBH MB-PCR | ||||
| Venous blood | Strong | 219§ | 0 | 0 | 219 | 60 |
| Weak | 0 | |||||
| Failed | 10* | 10 | 10 | 0 | 0 | |
| Bloodspot | Strong | 1140 | ||||
| Weak # | 16* | 2 | 2 | 14 | 14 | |
| Failed # | 14* | 12 | 12 | 2 | 2 | |
| Total | 1399 | 24 | 24 | 235 | 76 | |
Methylation (+) and Methylation (–): presence and absence of methylated FMR1 promoter, respectively; SBH: Southern blot hybridization; MB-PCR: methyl-CpG-binding PCR; §: of which 60 cases were randomly selected for the blinded test; *: all cases included for the blinded test; #: SBH was performed using resubmitted venous blood samples.
As indicated by markers in Table 1, from the 259 cases that had been confirmed by SBH, 100 cases were selected to evaluate the reliability of the MB-PCR procedure described below. The 100 samples screened included all the 24 FXS cases, all the 16 non-FXS cases that had shown a weak or negative PCR result, and another 60 non-FXS cases that were randomly selected from the 219 cases that presented a strong amplification yield in the screening PCR. After eliminating personal identifiers, the residual bloodspots from these 100 cases were randomly labeled and analyzed using MB-PCR in a blinded manner.
2.2. Preparation of Recombinant MBD2b and MBD3L1 Polypeptides
The expression plasmids of glutathione-S-transferase- (GST-) tagged MBD2b protein and histidine (HIS-) tagged MBD3L1 protein were provided by Professor GP Pfeifer [17]. After transforming Escherichia coli BL21 (DE3) with the plasmids, expression was induced with 1 mM isopropylthio-β-galacoside (GIBCO BRL, Carlsbad, CA, USA) for 3 hours at 37°C. Bacterial pellets were resuspended in ice-cold STE buffer (10 mM Tris-HCl, pH 7.8, 150 mM NaCl, and 1 mM or 0.1 mM EDTA for MBD2b and MBD3L1, resp.) containing 1 mM phenulmethylsulfonyl fluoride (GIBCO BRL, Carlsbad, CA, USA) and 200 μL Protease Inhibitor Cocktail Set II or VII (Calbiochem, Merck KgaA, Darmstadt, Germany) for MBD2b and MBD3L1, respectively. After incubating the cells for 10 minutes on ice, 0.3% of N-lauroylsarcosine (Sigma, St. Louis, MO, USA) was added, and a pressure of 1000 atm was applied through a French pressure cell press (Thermo Spectronic, Madison, WI, USA) for bacterial lysis. The lysate was cleared by centrifugation and loaded onto a GSTrap 4B column (GE Healthcare, Uppsala, Sweden) or HiTrap chelating HP column (GE Healthcare, Uppsala, Sweden), as appropriate. The columns and the tagged proteins were washed extensively and eluted according to the manufacturer's recommendations. The eluted proteins were dialyzed against 2 L of PBS at 4°C for 5 hours, followed by further overnight dialysis against storage buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM 2-mercaptoethanol, and 50% glycerol) at 4°C. After dialysis, the proteins were stored in aliquots at –20°C; their activity was unaffected for at least 6 months. The purity and concentrations of the recombinant proteins were assessed by 12.5% SDS-polyacrylamide gel electrophoresis and staining with Coomassie blue, as shown in supplementary material available online at doi:10.1155/2009/643692.
2.3. Cleavage of Genomic DNA
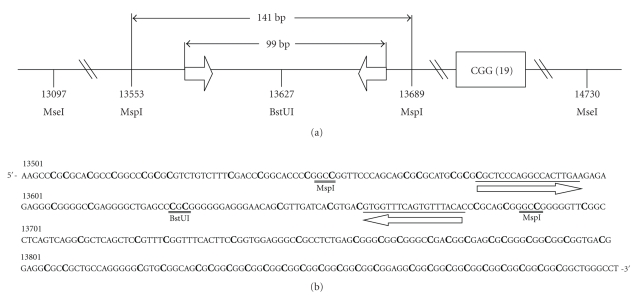
The procedures used for DNA isolation from venous blood and dried bloodspots have been described elsewhere [9]. To determine the best method for DNA cleavage, we compared the results of restriction enzyme digestion of the same DNA samples with MseI (T/TAA), MspI (C/CGG), or methylation-sensitive BstUI (C/GCG) (New England BioLabs, Beverly, MA, USA), individually or in combination. The relative locations of the recognition sites of these enzymes and MB-PCR primers are shown in Figure 1. It has been reported that the cytosines in the 14 CpG dinucleotides between the MspI recognition sites are methylated in FXS males and unmethylated in non-FXS males [23, 24].
Figure 1.
(a) Relative locations of recognition sites of restriction enzymes MseI, MspI, and BstUI and MB-PCR primers. (b) Sequence of FMR1 promoter region. The methylated cytosine of CpG dinucleotides is indicated by an enlarged letter “C”; the recognition sites of MspI and BstUI are double underlined; the primers are indicated by arrows.
2.4. Methyl-CpG-binding PCR
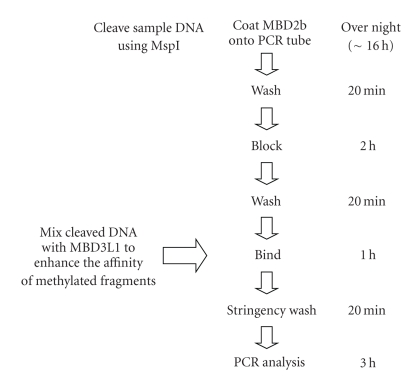
Figure 2 shows a simple flow diagram of MB-PCR. A day before the MB-PCR procedure was performed, 50 μL of recombinant MBD2b protein (15 μg/mL in 10 mM Tris-HCl (pH 7.5)) was applied to the wells of the heat-stable TopYield Strip (Nunc, Roskilde, Denmark) and incubated at 4°C overnight [18]. The polycarbonate TopYield Strip has a high-binding affinity for proteins. After washing 3 times with 200 μL of TBS (20 mM Tris (pH 7.5) and 170 mM NaCl), each well with the recombinant protein was incubated with 100 μL of blocking solution (10 mM Tris (pH 7.5), 170 mM NaCl, 5% skim milk powder, 5 mM EDTA, and 1 μg/mL of each poly (dI-dC), poly (dA-dT), and poly (dC-dG) (Amersham, Piscataway, NJ, USA)) at 4°C for 2 h. Each well was then washed twice with 200 μL of TBST (20 mM Tris (pH 7.5), 170 mM NaCl, and 0.05% Tween-20) and then with the binding buffer [20 mM Tris (pH 7.5), 2 mM MgCl2, 0.5 mM EDTA, 0.05% Tween-20, and NaCl at different concentrations ranging from 100 mM to 700 mM]. A total of 20 μL of predigested DNA was mixed with 50 μL of binding solution and 13 μL of recombinant MBD3L1 protein solution (60 μg/mL in 10 mM Tris-HCl (pH 7.5)) and loaded onto the wells. After a 1-hours incubation on a shaker (250 rpm) at room temperature, the wells were washed twice with 200 μL of binding buffer and once with 200 μL of 10 mM Tris-HCl (pH 8.0) to remove the unmethylated fragments.
Figure 2.
Flow diagram of methyl-CpG-binding PCR.
In each batch of MB-PCR assay, either universally methylated or unmethylated DNA samples were included as positive or negative control, respectively. Each sample was routinely examined twice in the same batch of MB-PCR. One received the whole stringency washes as above. The other did not receive the washes because the cleaved DNA samples were added last and therefore escaped the stringent washing steps [18]. Subsequently, these wells were subjected to PCR amplification by adding 30 μL of PCR mixture, which contained 72.5 mM Tris HCl (pH 9.0), 20 mM (NH4) 2SO4, 1.5 mM MgCl2, 0.01% (w/v) Tween-20, 200 nM dNTP, 2.5% glycerol, 200 nM of both primers (Figure 1), and 0.75 U Super-Therm DNA polymerase (Laboratory Product International, Kent, UK). The reaction conditions were as follows: initial denaturation at 95°C for 5 minutes; 36, 40, or 45 cycles at 95°C for 1 minutes; 59°C for 1 minutes; 72°C for 45 seconds a final extension at 72°C for 10 minutes. Subsequently, amplified products were resolved on a 5% polyacrylamide gel and visualized with ethidium bromide stain and UV illumination.
3. Results
3.1. Specificity of MB-PCR
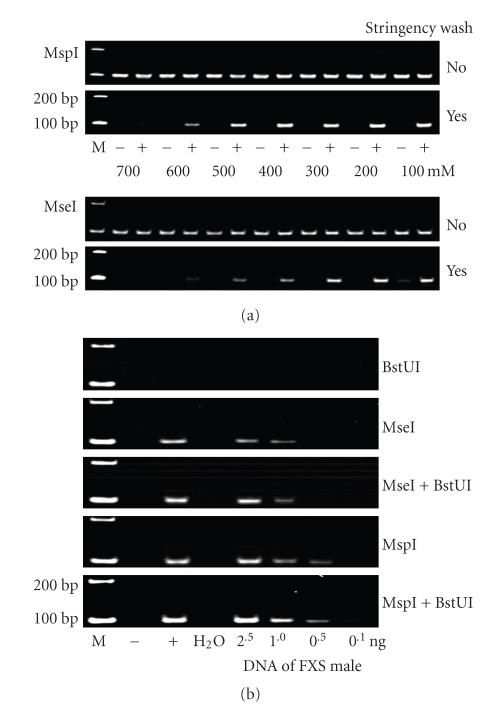
The most important step in this experiment was to determine the optimal NaCl concentration of the binding buffer; this is critical for enhancing the specificity of the MBD complex to distinctly bind with FMR1 alleles with different methylation statuses. We used 2.5 ng of the universally methylated or unmethylated DNA samples for the optimization experiments. After MspI or MseI digestion, each cleaved DNA sample was added to a binding buffer that contains different concentrations of NaCl ranging from 100 mM to 700 mM; it was then loaded in an MBD-coated PCR well.
The results of PCR amplification are shown in Figure 3(a). Where no stringency washes were performed, the PCR products of all samples digested by the same enzyme showed similar intensities. The MspI-digested samples exhibit slightly stronger intensity of the amplification products than MseI-digested ones. Where stringency washes were performed, the PCR products of all the universally unmethylated samples were not discernible, except in the case of a week yield in an MseI-digested sample for which a low-stringency wash with 100 mM NaCl was performed. On the contrary, the PCR products of many of the universally methylated samples showed variable intensities. In general, the intensity of PCR products decreases gradually with an increase in NaCl concentration in the stringency wash. The optimal NaCl concentrations in the stringency washes that helped in reliably distinguishing between methylated and unmethylated samples ranged from 100 nM to 500 nM in the case of the MspI-digested samples. However, the range for the MseI-digested samples narrowed down to 200–300 nM.
Figure 3.
Factors critical for the detection specificity of MB-PCR. (a) NaCl concentration in the stringency wash and enzymatic cleavage of the test DNA are the major factors that can critically determine the capability of MB-PCR in distinguishing between samples with methylated and unmethylated FMR1 alleles. (b) Under the same MB-PCR condition, including a stringency wash with 400 mM NaCl and 45 PCR cycles of amplification, the samples cleaved by different restriction enzyme(s) varied considerably in the detection sensitivity. (–) Control DNA with universally unmethylated sequences; (+): control DNA with universally methylated sequences; (M): molecular-size markers
Next, we examined the results of using a methylation-sensitive enzyme (BstUI) alone and in combination with MspI or MseI. Addition of BstUI can remove most of the unmethylated alleles in test samples, thus reducing the chances of false-positive results in MB-PCR [19]. However, when BstUI alone is used, no discernible PCR yield was observed (Figure 3(b), top panel). These results are consistent with the MB-PCR results shown in Figure 3(a), suggesting that the BstUI-digested fragments were most likely too long to be easily trapped by the MBD complex under such washing conditions. On the other hand, PCR using DNA cleaved with MseI or MspI yielded rather specific results, regardless of whether BstUI digestion was performed. Importantly, the sensitivity of PCR with MspI digestion was higher than that with MseI digestion.
3.2. Sensitivity of MB-PCR
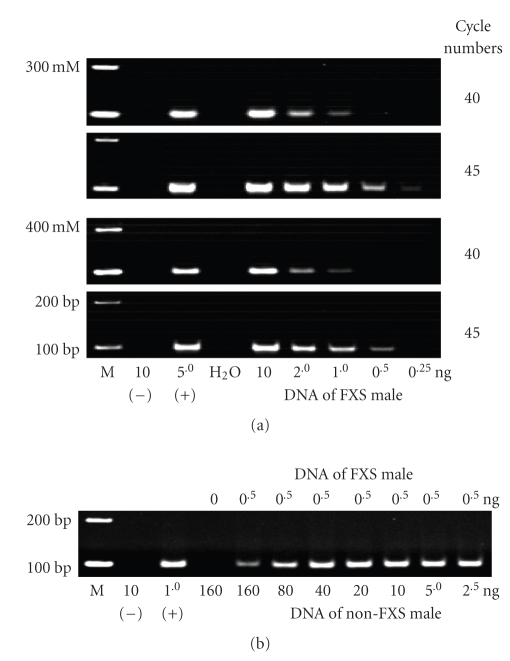
As expected, when DNA was cleaved using MspI alone, the detection sensitivity of methylated FMR1 promoter in the FXS samples improved gradually with increasing number of MB-PCR cycles (Figure 4(a)). Moreover, the NaCl concentrations in the stringency washes also affect the detection sensitivity. The results revealed that the minimum amount of DNA required for unequivocal detection of specific products in 45 cycles of MB-PCR amplification is 0.5 ng and 1.0 ng when stringency washes are performed with 300 mM and 400 mM NaCl, respectively.
Figure 4.
Detection sensitivity of MB-PCR. (a) MB-PCR analysis using different amounts of input DNA, cycle numbers of the PCR amplification, and NaCl concentration in the stringency washes. (b) MB-PCR analysis of the DNA samples simulated as mosaic FXS by mixing 0.5 ng of DNA sample from an FXS male with varying amounts of DNA sample from a non-FXS male. (–): Control DNA with universally unmethylated sequences; (+): control DNA with universally methylated sequences; (M): molecular-size markers.
Next, we evaluated if the optimized MB-PCR can reliably detect males with the mosaic form of FXS who often have expanded FMR1 alleles in either the methylated or unmethylated states. We thus simulated such mosaic samples by mixing 0.5 ng of the DNA from an FXS male having a nonmosaic methylation pattern with different amounts of DNA from a non-FXS male, ranging from 2.5 ng to 160 ng. Using the optimized MB-PCR conditions, including an NaCl concentration of 300 mM in the binding buffer, digestion of DNA with MspI, and 45 cycles of PCR amplification, we found that the methylated FMR1 promoter could be detected in mosaic samples, even when the amount of non-FXS sample was 160 ng (Figure 4(b)).
3.3. Reliability of MB-PCR
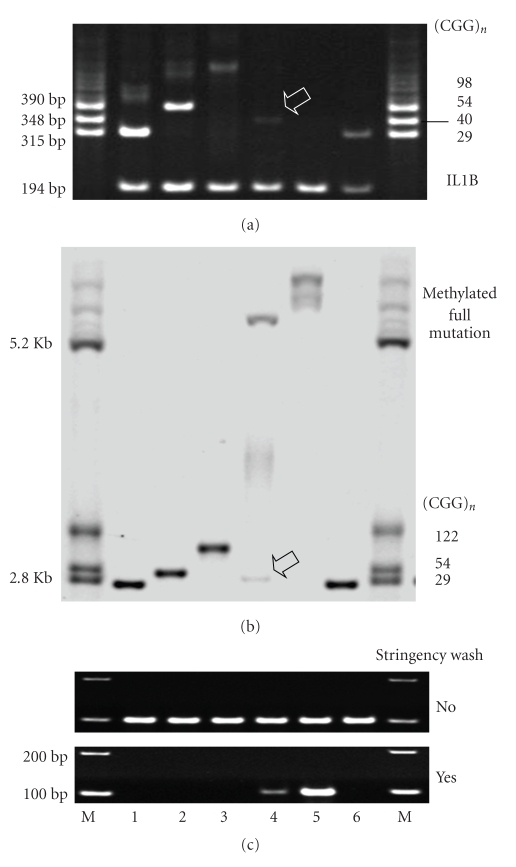
Before the blinded screening of the 100 bloodspot samples, we first performed MB-PCR on 6 representative samples, which were from 4 non-FXS males and 2 FXS males. The corresponding results of the screening PCR and SBH for these 6 samples are shown in Figures 5(a) and 5(b), respectively. The MB-PCR analysis of the 6 representative cases yielded a positive result only for 2 FXS samples (Figure 5(c)).
Figure 5.
Representative males examined with (a) screening PCR, (b) Southern blot hybridization, and (c) methyl-CpG-binding PCR. For the non-FXS samples, the CGG repeat numbers are 29 (lanes 1 and 6), 54 (lane 2), and 98 (lane 3). For the FXS samples, lane 4 is a mosaic pattern comprising expanded FMR1 alleles with and without methylation, and unmethylated alleles with CGG-repeats in the normal range (indicated by arrows), and lane 5 has a nonmosaic pattern. (M): molecular size markers.
Next, we applied the optimized MB-PCR to examine the residual bloodspot samples from the aforementioned 100 males, including 24 with FXS, in a blinded manner. A total of 5 μL of DNA sample from each case—approximately 0.5–12 ng DNA—was used for the test. The results are summarized in Table 1. Compared with the SBH results, the MB-PCR could detect all 24 FXS males, with 100% sensitivity, specificity, and accuracy.
4. Discussion
MBD polypeptides are the core elements of this MB-PCR assay and other similar methods. The family of vertebrate MBD proteins comprises MeCP2, MBD1, MBD2, MBD3, and MBD4 [25, 26]. MeCP2 was the first protein applied to an affinity matrix to specifically retain the sequences that are highly methylated [27]. However, since the use of MeCP2 requires A/T-rich sequences adjacent to the methylated CpG dinucleotides for efficient DNA binding, all methods based on MeCP2 affinity might be biased towards certain CpG motifs [28]. The main MBD protein used in this study was MBD2b, because MBD2 exhibits a higher affinity for CpG-methylated DNA than other MBD proteins [29]. MBD2b is the isoform of MBD2a that lacks the N-terminal region of the 152 amino acids proximal to the MBD [28]. MBD3L1 itself lacks the ability to bind methylated DNA but can interact with MBD2b and enhance its binding affinity for methylated sequences [29]. Because the MBD3L1-interaction domain has been mapped to the C-terminal end of MBD2b, the entire MBD2b protein (29.1 kD) should be used in MB-PCR [17], instead of the truncated MBD domain (7 kD) used in the other related studies [18, 19].
Other factors critically determining the sensitivity and specificity of this assay are the NaCl concentration of the binding buffer and the method used for DNA cleavage. The former must be determined by performing a series of titration tests. In this study, we found that the best optimal NaCl concentration of the binding buffer was between 300 and 400 mM. The optimal NaCl concentration values reported in 3 other similar studies analyzing different targets ranged from 700 mM [17] and 400 mM [18] to 50 mM [19]. DNA cleavage has often been performed by sonication or restriction enzyme digestion. The former appears to be ineffective in samples with limited amount of DNA; hence, we compared the results of the MB-PCR assay by using 3 restriction enzymes alone or in combination with others. The results of this comparison experiment reveal that, for groups of samples with diminished amounts of DNA, the MspI-based group provides consistently stronger product signals than the group based on MseI digestion. When DNA is cleaved using MseI or MspI digestion, the FMR1 fragments that cover the primers used in this study are 1633 bp and 141 bp long, respectively (Figure 1). It seems likely that the longer DNA fragments are more easily removed from the MBD complex during subsequent washing steps.
Under the conditions optimized previously, the MB-PCR could reliably distinguish DNA samples of the FXS group from those of the non-FXS group. However, this assay cannot be used to detect FXS in females, because the FMR1 allele in the inactivated X chromosome(s) of females is mostly methylated. In-house preparation of MBD proteins makes the test more cost effective, provided that the requisite facilities are available. The proteins can be freeze-stored and used as stocks, and the entire test can be completed within a few hours. Notably, the minimum amount of DNA required for the test is only 0.5 ng, which can be easily obtained from a bloodspot sample. Even when mixing 0.5 ng of DNA from an FXS male with a 320-fold-concentrated DNA sample (160 ng) from a non-FXS male, the detection sensitivity and specificity of MB-PCR remains unchanged. On the basis of the results of an FXS case (Figure 5, lane 4), it is plausible to believe that this assay is reliable in detecting FXS males with mosaic methylation patterns, who account for approximately 12–41% of male FXS patients [30, 31]. Interestingly, these results also suggest that the reliability of MB-PCR is retained when it is used to screen DNA samples pooled from many male subjects.
As demonstrated in this study, MB-PCR is a simple and affordable test to rapidly elucidate the methylation status of the FMR1 promoter in the bloodspots. This test would be particularly useful for the samples that show a negative or decreased PCR signal during initial screening. The bloodspot samples that showed negative results in the PCR screening test (e.g., lane 5, Figure 5) were often confirmed to be FXS-positive by using either SBH or MB-PCR (Table 1). Negative PCR results are also possibly observed in samples carrying a large-sized premutation in or deletion of a related FMR1 region, although such cases were not observed in our study. Decreased PCR signals may be attributed to the presence of a mosaic form of FXS or to a medium-sized premutation or simply to an artifact of the sample DNA. Taking into consideration the cases we have confirmed with SBH, we find that the interpretational uncertainty of these cases could be greatly reduced, when the IL1B gene is included as a comparative PCR control for the evaluation of the quantity and quality of bloodspot DNA. Those samples that exhibit decreased PCR yield intensity for FMR1 but not for IL1B, like that in lane 4, are often confirmed to be FXS-positive. If the subjects carry a medium-sized premutation allele (e.g., lane 3, Figure 5), the PCR products of FMR1 often have a decreased intensity but increased size; the corresponding amplification intensity of IL1B is often strong. On the other hand, the cases exhibiting similar decrease in yields for both FMR1 and IL1B, (e.g., lane 6, Figure 5) are often FXS-negative.
According to the Best Practice Guidelines for Molecular Analysis in Fragile X Syndrome provided by The European Molecular Genetics Quality Network (http://www.emqn.org/emqn/), the golden standard of the diagnosis of the FXS is SBH, which detects not only methylation but also methylation mosaics, expansions of the FMR1 CGG repeat, and deletions of the FMR1 promoter. Compared with the SBH results, we demonstrated that the MB-PCR could detect all 24 FXS males in the blind examination of 100 bloodspot samples, with 100% sensitivity, specificity, and accuracy. Similar detection power has been reported in methylation-sensitive PCR using methylated FMR1 alleles [11–14]. However, this method could not be easily applied to the bloodspot samples. Further, SBH cannot be used to analyze samples with DNA at a nanogram level thus far. Hence, in combination with screening PCR, MB-PCR can accelerate and simplify the detection of FXS in males. In the case of negative results in both tests, the subjects may have a large-sized premutation or deletion of a related FMR1 region. For the case of the former condition, many studies have used different PCR methods to verify the expanded CGG repeats [32–36].
In conclusion, we demonstrated for the first time that the MB-PCR is a simple and inexpensive test and can accelerate and simplify the detection of FXS in males by rapidly verifying the FMR1 promoter methylation in bloodspots. In-house preparation of MBD proteins makes the test more cost-effective. The freeze-stored proteins can be used as stocks, and the entire test can be completed within a few hours. Under the optimized conditions, the minimum amount of DNA required for the test is only 0.5 ng, and the detection sensitivity is not hampered at this concentration even after mixing it with a 320-fold-concentrated sample containing unmethylated alleles (160 ng). Since individuals with 2 or more X chromosomes generally have methylated FMR1 alleles, MB-PCR cannot be used to detect FXS in females.
Supplementary Material
Expression and purification of the GST-tagged MBD2b and HIS-tagged MBD3L1 proteins were run on a 12.5% SDS-PAGE gel and stained with Coomassie blue. M, size marker; N, before IPTG induction; Lysate, cell lysates after induction.
Acknowledgments
The authors thank Professor Gerd P. Pfeifer and Dr. Tibor A. Rauch, Beckman Research Institute of the City of Hope, Duarte, CA, USA, for providing expression plasmids of MBD2b and MBD3L1 proteins. This work was supported by a Grant (CMFHR9606) from Chi Mei Medical Center, Taiwan.
References
- 1.Hagerman RJ. The molecular biology of the fragile X mutation. In: Hagerman RJ, Cronister A, editors. Fragile X Syndrome: Diagnosis, Treatment and Research. Baltimore, Md, USA: Johns Hopkins University Press; 1996. pp. 88–113. [Google Scholar]
- 2.Nolin SL, Brown WT, Glicksman A, et al. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. The American Journal of Human Genetics. 2003;72(2):454–464. doi: 10.1086/367713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. American Journal of Human Genetics. 2004;74(5):805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranek GT, Roberts JE, David FJ, et al. Developmental trajectories and correlates of sensory processing in young boys with fragile X syndrome. Physical and Occupational Therapy in Pediatrics. 2008;28(1):79–98. doi: 10.1300/j006v28n01_06. [DOI] [PubMed] [Google Scholar]
- 5.Torrioli MG, Vernacotola S, Peruzzi L, et al. A double-blind, parallel, multicenter comparison of L-acetylcarnitine with placebo on the attention deficit hyperactivity disorder in fragile X syndrome boys. American Journal of Medical Genetics A. 2008;146(7):803–812. doi: 10.1002/ajmg.a.32268. [DOI] [PubMed] [Google Scholar]
- 6.Hagerman RJ, Berry-Kravis E, Kaufmann WE, et al. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123(1):378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saul RA, Friez M, Eaves K, et al. Fragile X syndrome detection in newborns: pilot study. Genetics in Medicine. 2008;10(10):714–719. doi: 10.1097/GIM.0b013e3181862a76. [DOI] [PubMed] [Google Scholar]
- 8.Larsen LA, Grønskov K, Nørgaard-Pedersen B, Brøndum-Nielsen K, Hasholt L, Vuust J. High-throughput analysis of fragile X (CGG)n alleles in the normal and premutation range by PCR amplification and automated capillary electrophoresis. Human Genetics. 1997;100(5–6):564–568. doi: 10.1007/s004390050552. [DOI] [PubMed] [Google Scholar]
- 9.Tzeng C-C, Lin S-J, Chen Y-J, et al. An effective strategy of using molecular testing to screen mentally retarded individuals for fragile X syndrome. Diagnostic Molecular Pathology. 2001;10(1):34–40. doi: 10.1097/00019606-200103000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Graff JR, Myöhänen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Kubota T, Song M, et al. Methylation analysis of the fragile X syndrome by PCR. Genetic Testing. 1998;1(3):151–155. doi: 10.1089/gte.1997.1.151. [DOI] [PubMed] [Google Scholar]
- 12.Panagopoulos I, Lassen C, Kristoffersson U, Aman P. A methylation PCR approach for detection of fragile X syndrome. Human Mutation. 1999;14(1):71–79. doi: 10.1002/(SICI)1098-1004(1999)14:1<71::AID-HUMU9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Weinhäusel A, Haas OA. Evaluation of the fragile X (FRAXA) syndrome with methylation-sensitive PCR. Human Genetics. 2001;108(6):450–458. doi: 10.1007/s004390100519. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Lum JMS, Yeo G-H, Kiing J, Tay SKH, Chong SS. Simplified molecular diagnosis of fragile X syndrome by fluorescent methylation-specific PCR and GeneScan analysis. Clinical Chemistry. 2006;52(8):1492–1500. doi: 10.1373/clinchem.2006.068593. [DOI] [PubMed] [Google Scholar]
- 15.Raizis AM, Schmitt F, Jost J-P. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Analytical Biochemistry. 1995;226(1):161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- 16.Munson K, Clark J, Lamparska-Kupsik K, Smith SS. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive QPCR. Nucleic Acids Research. 2007;35(9):2893–2903. doi: 10.1093/nar/gkm055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Laboratory Investigation. 2005;85(9):1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 18.Gebhard C, Schwarzfischer L, Pham TH, Andreesen R, Mackensen A, Rehli M. Rapid and sensitive detection of CpG-methylation using methyl-binding (MB)-PCR. Nucleic Acids Research. 2006;34(11, article e82) doi: 10.1093/nar/gkl437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Research. 2006;34(3, article e19) doi: 10.1093/nar/gnj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renbaum P, Abrahamove D, Fainsod A, Wilson GG, Rottem S, Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1 (M·SssI) Nucleic Acids Research. 1990;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochemical and Biophysical Research Communications. 2005;329(1):219–223. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- 22.Tzeng C-C, Tzeng P-Y, Sun HS, Chen RM, Lin S-J. Implication of screening for FMR1 and FMR2 gene mutation in individuals with nonspecific mental retardation in Taiwan. Diagnostic Molecular Pathology. 2000;9(2):75–80. doi: 10.1097/00019606-200006000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Hornstra IK, Nelson DL, Warren ST, Yang TP. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Human Molecular Genetics. 1993;2(10):1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- 24.Genç B, Müller-Hartmann H, Zeschnigk M. Methylation mosaicism of 5′-(CGG)(n)-3′ repeats in fragile X, premutation and normal individuals. Nucleic Acids Research. 2000;28(10):2141–2152. doi: 10.1093/nar/28.10.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Molecular and Cellular Biology. 1998;18(11):6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Research. 2003;31(6):1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nature Genetics. 1994;6(3):236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 28.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Molecular Cell. 2005;19(5):667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 29.Jiang C-L, Jin S-G, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. The Journal of Biological Chemistry. 2004;279(50):52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 30.Nolin SL, Glicksman A, Houck GE, Jr., Brown WT, Dobkin CS. Mosaicism in fragile X affected males. American Journal of Medical Genetics. 1994;51(4):509–512. doi: 10.1002/ajmg.1320510444. [DOI] [PubMed] [Google Scholar]
- 31.Rousseau F, Heitz D, Tarleton J, et al. A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. American Journal of Human Genetics. 1994;55(2):225–237. [PMC free article] [PubMed] [Google Scholar]
- 32.Hećimović S, Barišić I, Müller A, et al. Expand long PCR for fragile X mutation detection. Clinical Genetics. 1997;52(3):147–154. doi: 10.1111/j.1399-0004.1997.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 33.Saluto A, Brussino A, Tassone F, et al. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. The Journal of Molecular Diagnostics. 2005;7(5):605–612. doi: 10.1016/S1525-1578(10)60594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strom CM, Huang D, Li Y, et al. Development of a novel, accurate, automated, rapid, high-throughput technique suitable for population-based carrier screening for fragile X syndrome. Genetics in Medicine. 2007;9(4):199–207. doi: 10.1097/gim.0b013e31803d3ac9. [DOI] [PubMed] [Google Scholar]
- 35.Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. The Journal of Molecular Diagnostics. 2008;10(1):43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodds ED, Tassone F, Hagerman PJ, Lebrilla CB. Polymerase chain reaction, nuclease digestion, and mass spectrometry based assay for the trinucleotide repeat status of the fragile X mental retardation 1 gene. Analytical Chemistry. 2009;81(13):5533–5540. doi: 10.1021/ac9008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression and purification of the GST-tagged MBD2b and HIS-tagged MBD3L1 proteins were run on a 12.5% SDS-PAGE gel and stained with Coomassie blue. M, size marker; N, before IPTG induction; Lysate, cell lysates after induction.