Abstract
Defining the rate at which T cells turn over has important implications for our understanding of T lymphocyte homeostasis and AIDS pathogenesis, yet little information on T cell turnover is available. We used the nucleoside analogue bromodeoxyuridine (BrdUrd) in combination with five-color flow cytometric analysis to evaluate T lymphocyte turnover rates in normal and simian immunodeficiency virus (SIV)-infected rhesus macaques. T cells in normal animals turned over at relatively rapid rates, with memory cells turning over more quickly than naive cells. In SIV-infected animals, the labeling and elimination rates of both CD4+ and CD8+ BrdUrd-labeled cells were increased by 2- to 3-fold as compared with normal controls. In normal and SIV-infected animals, the rates of CD4+ T cell BrdUrd-labeling and decay were closely correlated with those of CD8+ T cells. The elimination rate of BrdUrd-labeled cells was accelerated in both naive and memory T lymphocytes in SIV-infected animals. Our results provide direct evidence for increased rates of both CD4+ and CD8+ T cell turnover in AIDS virus infection and have important implications for our understanding of T cell homeostasis and the mechanisms responsible for CD4+ T cell depletion in AIDS.
Maintenance of T cell homeostasis reflects a complex balance between production of new T cells from thymic and extrathymic sources and death as the result of apoptosis and exogenous factors such as HIV infection (1, 2). Determination of the rate of T cell turnover provides an indirect measurement of the production and death rates of T cells and thus offers information that may advance our understanding of processes responsible for the depletion of CD4+ T cells during the course of HIV infection. However, little information on T cell turnover is available, especially for humans. In rodents, the nucleoside analogue BrdUrd has been widely used to evaluate T cell turnover (3–5). BrdUrd is incorporated into the DNA of dividing cells, is not reused after incorporation, and can be detected by flow cytometric analysis, allowing simultaneous determination of the presence of BrdUrd in different subsets of lymphocytes. Analysis of rates of BrdUrd-labeling in rodents have demonstrated relatively rapid rates of T cell turnover, with 30–50% of peripheral T cells labeling in <2 wk (2, 5) and memory cells dividing more frequently than naive phenotype cells (3). In humans, there are no studies using BrdUrd to study T cell turnover. However, analysis of radiation-induced chromosomal damage has suggested that human T cells turn over relatively slowly, on average once every 3.5 years for naive cells and every 22 wk for memory cells (6).
The limited information on T cell turnover represents a significant impediment to advancing our understanding of events that ultimately result in the depletion of CD4+ T lymphocytes in HIV infection. Over the past several years, widely differing estimates of CD4+ T lymphocyte turnover in HIV-infected people have been suggested, ranging from over a 78-fold increase (7) to no increase (8). Based on the kinetics of viral decay and rebound of CD4+ T cells after treatment of HIV-infected subjects with potent antiretroviral therapy, Ho, Shaw, and colleagues (7, 9) have proposed a relatively rapid turnover of CD4+ T cells equal to 1–2 × 109 cells/day or ≈5% of all CD4+ T cells. However, others have suggested the rebound of CD4+ T cells after antiretroviral therapy may be caused by redistribution of T cells between lymphoid tissues and peripheral blood (10–13). Similarly, based on the finding that telomere length, which progressively decreases during normal cell division (14), is shortened in CD8+ but not CD4+ T cells from HIV-infected subjects, Wolthers et al. (8) have proposed that there is no increased turnover of CD4+ T cells in HIV infection.
To directly evaluate T cell turnover in a nonhuman primate model of AIDS, we used the nucleoside analogue BrdUrd to label lymphocytes in normal and simian immunodeficiency virus (SIV)-infected rhesus macaques. Rhesus macaques infected with SIV develop disease that is quite similar to the course of HIV infection in people, both with respect to clinical manifestations and the kinetics of viral turnover (15, 16). By analyzing both the rate of BrdUrd-labeling and the elimination of BrdUrd-labeled cells, we provide direct evidence for a 2- to 3-fold increased rate of turnover of both CD4+ and CD8+ T cells in SIV-infected animals.
METHODS
Animals.
Rhesus monkeys used in this study were colony born animals maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the “NIH Guide for the Care and Use of Laboratory Animals.” Animals were anesthetized with ketamine HCl for blood drawing and BrdUrd injection. BrdUrd was administered for a total of 14–18 days (60 mg/kg/day i.p. for 5 days/wk and fed 120 mg/kg orally twice per day for 2 days/wk). Animals were infected with the pathogenic strains SIVmac239 (animal 377–90), SIVmac251 (23–96, 168–94, 196–94), or SIVmac239Δvpr (105–91) and had been infected for at least 9 mo at the time of study. The virologic and clinical course of infection with SIVmac239Δvpr is indistinguishable from animals infected with SIVmac239 (17). Quantitative assays for the measurement of SIV RNA were performed by using a branched chain DNA signal amplification assay specific for SIV (18). The lower quantitation limit of this assay is 10,000 copies of SIV RNA per milliliter of plasma.
Flow Cytometric Analysis.
Peripheral blood mononuclear cells were isolated by standard Ficoll–Hypaque density gradient centrifugation. After washing with PBS, 1 × 106 peripheral blood mononuclear cells were resuspended in 2% heat inactivated normal mouse serum in PBS and stained with the appropriate antibody. Antibodies used for these studies included fluorescein isothiocyanate-conjugated IgG1/PE-conjugated IgG2, PE-conjugated CD62L (Leu8), APC-conjugated CD45RA, PE- conjugated CD2, biotinylated CD4 (all Becton Dickinson Immunocytometry Systems), PE-Cy5-conjugated CD20 (Coulter), biotin-conjugated CD3 (6G12) (19) (kindly provided by J. Wong, Massachusetts General Hospital), and CD8 (51.1, American Type Culture Collection ref. #HB 230) custom conjugated to cascade blue (Chromaprobe, Mountain View, CA). After incubation on ice for 30 min in the dark, cells were washed with 2% normal mouse serum in PBS. One microliter of streptavidin-RED613 (GIBCO) was added to appropriate tubes and incubated on ice for 30 min in the dark. Cells were washed in 2% normal mouse serum in PBS and resuspended in 2% paraformaldehyde with 0.1% Tween 20 at 37° for 30 min and then at room temperature for another 30–60 min. After washing, cells were incubated for 30 min at 37° with 50 Kunitz units of DNase I (Sigma) in 0.15 M NaCl and 4.2 mM MgCl2, pH 5. Cells were washed with 2% normal mouse serum in PBS and resuspended in 150 μl of 0.5% Tween 20 in PBS. Twenty microliters of fluorescein isothiocyanate-conjugated anti-BrdUrd (Becton Dickinson Immunocytometry Systems) were added to appropriate tubes and incubated at room temperature for 30–45 min. Cells were washed twice with 0.1% Tween 20 in PBS and resuspended in PBS with 2% paraformaldehyde. Samples were analyzed on a Becton Dickinson FACS Vantage flow cytometer equipped with a Coherent Enterprise laser simultaneously emitting 360 nm and 488 nm and a Coherent Helium–Neon laser emitting 630 nm. Band pass and dichroic filters for each detector were set up as follows: 530/30 in FL1, 575/26 in FL2, a 560 SP dichroic separating FL1 and FL2, a 610 SP with a half-mirror separating FL3 and FL4, a 610/20 or a 675/20 in FL3, a 660/20 in FL4, a 424/44 in FL5, and a 640 LP dichroic separating FL4 and FL5. Twenty thousand events were collected and analyzed by using cellquest Software (Becton Dickinson). Fluorescent measurements were collected by using a logarithmic amplifier and forward scatter and side scatter by using a linear amplifier. Peripheral blood mononuclear cells from a normal, unlabeled animal were subjected to the same staining procedure and used to establish the negative gates for BrdUrd fluorescence.
Statistical Analysis.
BrdUrd uptake rates were determined using weighted linear regression. The decay of BrdUrd-labeled cells was modeled by using the equation y = a*e−mx, where y = % BrdUrd-labeled cells, a = initial % of BrdUrd-labeled cells, m = elimination rate constant, and x = time (days). The experimental data were fit to the model equation by weighted least squares analysis. Linear correlations and Mann–Whitney U tests were performed by using statview (Abacus Concepts, Berkeley, CA).
RESULTS
In Vivo Labeling of Rhesus Macaques with BrdUrd.
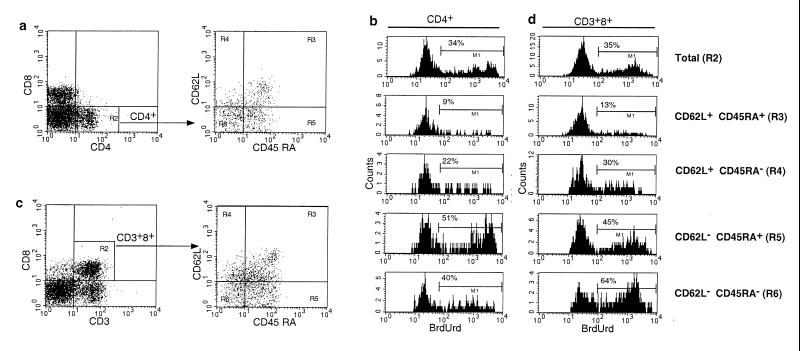
Rhesus macaques received daily doses of BrdUrd for a total of 14–18 days and then were observed during a washout phase of 28 days. A total of four normal and five SIV-infected macaques were studied; repeat cycles of BrdUrd labeling and washout were performed in two normal and three SIV-infected animals. SIV-infected animals used for these studies had moderately advanced disease with CD4+ T cell counts ranging from 295 to 861/mm3 and viral loads of 12,000–1,081,000 copies/ml of SIV RNA. Serial determinations of plasma SIV RNA revealed no effect of BrdUrd administration on SIV replication in vivo (data not shown). The percentage of BrdUrd-labeled cells during both labeling and washout phases was determined twice weekly by five-color flow cytometric analysis using antibodies to T cell markers (CD3, CD4, and CD8), markers of naive phenotype lymphocytes (CD45RA and CD62L) (20), and BrdUrd. Levels of BrdUrd labeling in CD4+ cells were analyzed in naive (CD45RA+CD62L+) and memory subsets (which lacked expression of either or both CD45RA or CD62L) (Fig. 1 a and b). For CD8+ T lymphocytes, cells were first defined based on coexpression of CD3 and CD8, thereby excluding CD3−CD8+ natural killer cells (21), and then the percentage of BrdUrd-labeled cells determined in naive and memory subpopulations (Fig. 1 c and d).
Figure 1.
Five-color flow cytometry analysis of in vivo BrdUrd labeling in an SIV-infected macaque. Peripheral blood lymphocytes from an SIV-infected rhesus macaque that had been labeled with BrdUrd were surface stained with antibodies to CD3, CD8, CD45RA, and CD62L or CD4, CD8, CD45RA, and CD62L, then permeabilized, and stained with antibodies to BrdUrd. (a) Gating of CD4+ T lymphocytes. Cells enclosed in the gate designated by R2 then were analyzed for expression of CD45RA and CD62L. (b) Analysis of BrdUrd uptake in memory and naive populations of CD4+ T cells. (c) Gating of CD8+ T lymphocytes. To exclude CD3–CD8+ natural killer cells, subsequent analysis of expression of CD45RA, CD62L, and BrdUrd was confined to CD3+CD8+ cells indicated in the R2 gate. (d) Analysis of BrdUrd uptake in memory and naive populations of CD8+ T cells. The gate corresponding to each population of naive and memory cells is indicated in parentheses. The percentage of BrdUrd-labeled cells for each gated cell population is shown in each histogram.
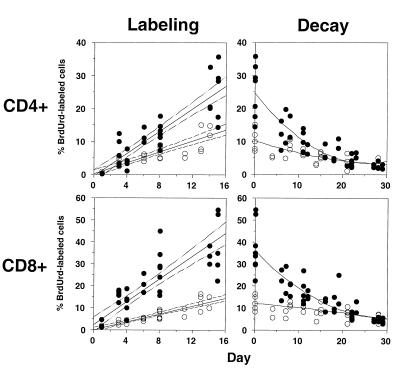
Composite data for the accumulation and decay of BrdUrd-labeled CD4+ and CD8+ T lymphocytes in all normal and SIV-infected animals studied are shown in Fig. 2. For both normal and SIV-infected animals, the accumulation of BrdUrd-labeled cells occurred at a linear rate, whereas the percentage of BrdUrd-labeled cells decayed exponentially. At the completion of BrdUrd administration, higher peak values of BrdUrd-labeled cells were observed in SIV-infected animals as compared with normal controls, for both CD4+ T cells (22% vs. 10%, P < 0.01) and CD8+ T cells, (30% vs. 11%, P < 0.005).
Figure 2.
Serial analysis of BrdUrd-labeled cells during BrdUrd labeling and decay in normal and SIV-infected macaques. Macaques were labeled with BrdUrd for ≈2 wk and then observed during a washout phase of 4 wk. The percentage of BrdUrd-labeled cells was determined in peripheral blood lymphocytes twice a week for the duration of labeling and washout phases. Each symbol represents an individual time point for a given animal; open circles are normal animals, and filled circles are SIV-infected animals. For the labeling phase, a linear regression of the percentage of BrdUrd-labeled cells with 95% confidence intervals is shown for normal and SIV-infected animals. For the decay phase, a nonlinear regression of the percentage of BrdUrd-labeled cells is shown for normal and SIV-infected animals.
Increased Turnover of CD4+ and CD8+ T lymphocytes in SIV-Infected Macaques.
We used two approaches to compare relative turnover rates of CD4+ and CD8+ T cells in normal and SIV-infected animals. We first analyzed the rate of accumulation of BrdUrd-labeled cells during BrdUrd administration using linear regression analysis. For both CD4+ and CD8+ T cells, the rates of BrdUrd-labeling were increased by ≈2-fold in SIV-infected animals as compared with normal controls (Table 1), a finding consistent with the higher peak levels of BrdUrd-labeled T cells observed in SIV-infected animals.
Table 1.
Turnover of CD4+ and CD8+ T lymphocytes in normal and SIV-infected macaques
| Animal | CD4+*, per mm3 | Viral load*, 103 per ml | Labeling rate, d−1†
|
Elimination rate, d−1‡
|
||
|---|---|---|---|---|---|---|
| CD4+ | CD8+ | CD4+ | CD8+ | |||
| Normal | ||||||
| 246-94 | 1150 | n/a | 0.008 | 0.010 | 0.034 | 0.018 |
| 247-94 | 1500 | n/a | 0.008 | 0.006 | 0.020 | 0.012 |
| 394-95 | 2120 | n/a | 0.007 ± 0.001 | 0.008 ± 0.002 | 0.038 ± 0.006 | 0.021 ± 0.007 |
| 361-95 | 1865 | n/a | 0.004 ± 0.002 | 0.007 ± 0.001 | 0.033 ± 0.006 | 0.022 ± 0.004 |
| Mean ± SD | 0.006 ± 0.002 | 0.008 ± 0.002 | 0.033 ± 0.008 | 0.019 ± 0.005 | ||
| SIV-infected | ||||||
| 23-96 | 295 | 1081 | 0.012 ± 0.001 | 0.020 ± 0.005 | 0.096 ± 0.023 | 0.076 ± 0.044 |
| 105-91 | 507 | 83 | 0.012 ± 0.003 | 0.012 ± 0.003 | 0.083 ± 0.010 | 0.071 ± 0.038 |
| 168-94 | 846 | 697 | 0.011 | 0.015 | 0.076 | 0.042 |
| 196-94 | 861 | 179 | 0.012 | 0.017 | 0.055 | 0.043 |
| 377-90 | 337 | 12 | 0.011 ± 0.004 | 0.010 ± 0.006 | 0.116 ± 0.049 | 0.067 ± 0 |
| Mean ± SD | 0.012 ± 0.002§ | 0.014 ± 0.006¶ | 0.090 ± 0.029§ | 0.064 ± 0.026§ | ||
CD4+ T cell counts and viral loads represent the average of all determinations during labeling and decay phases. Values for most animals represent the mean ± SD of two independent experiments.
The rate of accumulation of BrdUrd-labeled cells was determined by weighted linear regression. The mean correlation coefficient (R value) for curve fitting was 0.91.
The elimination rate of BrdUrd-labeled cells was calculated by nonlinear regression using weighted least squares analysis. The mean correlation coefficient (R value) for curve fitting was 0.86.
The difference between normal and SIV-infected animals is statistically significant (P < 0.002, Mann–Whitney U test).
The difference between normal and SIV-infected animals is statistically significant (P < 0.02, Mann–Whitney U test). n/a, not applicable.
We next examined the decay of BrdUrd-labeled cells by using nonlinear regression analysis with curve fitting done by weighted least squares analysis. The elimination rate of BrdUrd-labeled cells was then used to compare the turnover of each cell population. Using this approach, we observed an ≈3-fold increase in the elimination rate of BrdUrd-labeled cells for both CD4+ and CD8+ T cells in SIV-infected animals. Calculation of lymphocyte elimination rates was highly reproducible, with mean SD of 27% observed for repeat rounds of labeling in a given animal.
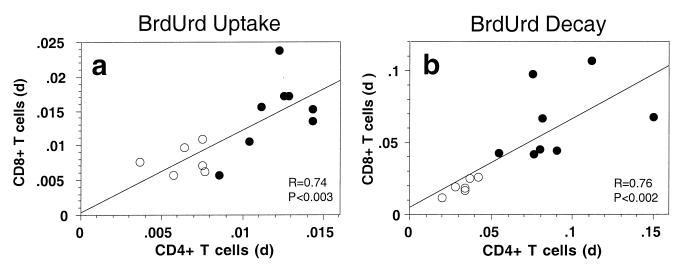
The observation that turnover rates of both CD4+ and CD8+ T cells were increased by a similar magnitude in SIV-infected animals was unexpected and prompted an analysis of whether the turnover of these lymphocyte populations might be linked. We therefore analyzed whether rates of BrdUrd-labeling of CD4+ and CD8+ lymphocytes were correlated for individual animals. Both in normal and SIV-infected animals, there was a statistically significant (P < 0.003) correlation between the rates of BrdUrd-labeling of CD4+ and CD8+ T cells (Fig. 3a). A similar relationship was observed for the elimination rate of BrdUrd-labeled cells (Fig. 3b), suggesting that processes affecting turnover of CD4+ and CD8+ T cells are linked, both in normal and SIV-infected animals.
Figure 3.
Correlation of BrdUrd labeling and decay rates of CD4+ and CD8+ T cells. (a) Correlation of the labeling coefficients for uptake of BrdUrd by CD4+ and CD8+ T lymphocytes during administration of BrdUrd to normal and SIV-infected animals. Labeling coefficients were calculated by linear regression analysis. (b) Correlation of the elimination rates for BrdUrd-labeled CD4+ and CD8+ T lymphocytes after stopping administration of BrdUrd to normal and SIV-infected animals. The elimination rate of BrdUrd-labeled CD4+ and CD8+ T lymphocytes was calculated by nonlinear regression analysis. Each symbol represents an individual animal; open circles are normal animals, and closed circles are SIV-infected animals. R and P values for linear regressions for both uptake and decay rates are shown.
Increased Elimination of BrdUrd-Labeled Naive and Memory T Cells in SIV Infection.
We also analyzed the decay of BrdUrd-labeled cells in naive and memory T lymphocyte subpopulations in normal and SIV-infected macaques. Because the incorporation of BrdUrd into cellular DNA occurs via the nucleotide salvage pathway, which is preferentially used by mature T lymphocytes, the efficiency of BrdUrd uptake varies significantly between thymus and lymphoid organs (and thus for naive and memory lymphocytes) (5). We therefore did not compare rates of BrdUrd labeling in naive and memory lymphocytes. Consistent with previous results in mice and humans (3, 6, 22), in normal animals, we observed lower elimination rates of BrdUrd-labeled naive CD4+ and CD8+ T cells as compared with memory T cells. For CD4+ T cells in normal animals, naive CD45RA+CD62L+ cells had the slowest elimination rate of any subpopulation analyzed, whereas CD4+ T cells lacking expression of one or both of these markers had elimination rates that were 2- to 3-fold higher (Table 2). For CD8+ T cells in normal animals, BrdUrd-labeled cells expressing CD45RA, regardless of expression of CD62L, had an elimination rate 4-fold lower than that of CD45RA− cells. In SIV-infected animals, increased elimination rates of BrdUrd-labeled T cells were observed in both naive and memory cells. The increased elimination rate was particularly remarkable for naive phenotype CD4+ and CD8+ cells, for which the elimination rate was increased by 7- to 9-fold in SIV-infected animals (P < 0.01) (Table 2). Increased elimination rates were observed for all subsets of memory and naive phenotype CD4+ and CD8+ T cells examined in infected animals as compared with uninfected controls.
Table 2.
Increased elimination rates of BrdUrd-labeled naive and memory T lymphocytes in SIV-infected macaques
| Population | CD45RA | CD62L | Elimination rate, d−1*
|
|
|---|---|---|---|---|
| Normal | SIV | |||
| CD4+ | ||||
| + | + | 0.007 ± 0.003 | 0.061 ± 0.047† | |
| + | − | 0.016 ± 0.003 | 0.055 ± 0.030† | |
| − | + | 0.016 ± 0.005 | 0.112 ± 0.090† | |
| − | − | 0.024 ± 0.004 | 0.107 ± 0.054† | |
| CD8+ | + | + | 0.007 ± 0.003 | 0.054 ± 0.037† |
| + | + | 0.007 ± 0.003 | 0.054 ± 0.037† | |
| + | − | 0.007 ± 0.002 | 0.042 ± 0.027† | |
| − | + | 0.032 ± 0.008 | 0.118 ± 0.076‡ | |
| − | − | 0.028 ± 0.013 | 0.088 ± 0.053‡ | |
Elimination rates for naive and memory T cells were calculated using nonlinear regression analysis of the washout phase in normal and SIV-infected macaques. Data are presented as the mean ± SD for normal and SIV-infected animals.
Significantly different than in normal animals, P < 0.01 (Mann–Whitney U test).
P < 0.05 (Mann–Whitney U test).
DISCUSSION
Our data provide direct evidence for increased rates of turnover of both CD4+ and CD8+ T lymphocytes in SIV-infected animals. This conclusion is supported by a higher peak percentage of BrdUrd-labeled cells, an increased rate of BrdUrd labeling, and a more rapid disappearance of BrdUrd-labeled cells in infected animals as compared with normal controls. Interestingly, rates of T cell turnover in SIV-infected animals were increased in both CD4+ and CD8+ T cells to a comparable degree, a finding reinforced by the close correlation of the BrdUrd-labeling and elimination rates for CD4+ and CD8+ T cells. Increased elimination rates of BrdUrd-labeled cells were observed in both naive and memory phenotype cells in SIV-infected animals, although the greatest relative increase in elimination rates was noted in the naive phenotype CD4+ and CD8+ T cells. This observation suggests the possibility that newly formed T cells, possibly of thymic origin, may contribute to the generation of T cells in response to SIV-induced lymphocyte depletion, a conclusion that also is supported by increased rates of BrdUrd labeling of the naive CD45RA+CD62L+ cells in SIV-infected animals compared with uninfected controls (data not shown).
Although depletion of CD4+ T lymphocytes is the hallmark immunologic abnormality of AIDS, our data also demonstrate an increased turnover (and by inference death) of CD8+ T lymphocytes in SIV-infected animals. These results are supported by reports that document a decrease in the telomere length of CD8+ T cells from HIV-infected subjects (8, 23, 24) and by the finding of an increased frequency of mutations in the hypoxanthine guanine phosphoribosyl transferase locus in both CD4+ and CD8+ T cells of individuals with advanced HIV disease (25). These observations are consistent with the large number of reports documenting increased rates of activation and apoptosis in both CD4+ and CD8+ T lymphocytes in HIV-infected people and SIV-infected macaques (26, 27), suggesting that increased T cell turnover may in part reflect the generalized activation of the immune system. However, our results contradict the conclusion that there is no increased turnover of CD4+ T cells in HIV-infected subjects, a finding based on the failure to observe a shortening of telomere lengths in CD4+ T cells in infected individuals (8, 24). This discrepancy may reflect limitations in the ability of telomere measurements to accurately assess rates of T cell turnover or the killing of dividing CD4+ T cells by HIV and SIV, leading to a selective loss of those cells with shortened telomeres.
Several limitations to our analysis of T cell turnover using BrdUrd should be noted. With respect to the use of BrdUrd to study cell turnover, although BrdUrd has been used widely for this purpose, some investigators have suggested that administration of BrdUrd may decrease cell proliferation rates (2). However, in macaques that received BrdUrd for periods of up to 3 wk (Fig. 2 and data not shown), no evidence for a decrease in the rate of labeling was observed, suggesting that BrdUrd did not have a cumulative antagonistic effect on T cell proliferation. With respect to our mathematical analysis, different factors affect the interpretation of the labeling and decay phases. During the labeling phase, the rate of accumulation of BrdUrd-labeled cells is dependent not only on the rate of cell division but also on the efficiency with which dividing cells incorporate BrdUrd and the death rate of labeled cells. Because of the relatively inefficient uptake of BrdUrd by cells in lymphoid organs other than the thymus (5) and the rapid clearance of BrdUrd (28), our calculations of BrdUrd uptake are likely to underestimate true proliferation rates. During the decay phase, loss of BrdUrd-labeled cells occurs not only by cell death but also by successive cell divisions leading to decay of the cell label. However, the relative contribution of cell division to the elimination rate will be diminished by the fact that on average four to seven cell divisions are necessary for a BrdUrd-labeled cell to lose the ability to be detected by flow cytometry (unpublished observation). In addition, our analysis of the elimination rates of naive and memory T cells may be skewed by interconversion among these subsets. Finally, because we have only labeled a minority of all CD4+ and CD8+ T cells, there may be a subpopulation of cells with a slower rate of T cell turnover that is not efficiently labeled by BrdUrd. Nonetheless, our mathematical analysis permits a valid comparison of the relative rates of T cell turnover between normal and SIV-infected animals. Further refinements of the mathematical analysis of these data may allow more precise calculations of T lymphocyte proliferation and death rates.
These observations also have implications for our understanding of T lymphocyte homeostasis. The significant correlation of the rates of BrdUrd-labeling and decay for CD4+ and CD8+ T cells in both normal and SIV-infected animals suggests that similar mechanisms may affect T cell turnover of both of these cell populations. This conclusion would not be anticipated based on the blind T cell homeostasis model, which proposes that homeostatic mechanisms control the total number of T cells in the body and do not independently regulate the percentage of CD4+ and CD8+ T lymphocytes (29). Our findings also suggest a more rapid turnover of T cells in normal primates than previously appreciated. Based on analysis of radiation-induced chromosomal damage, previous calculations of T cell turnover in humans have suggested that naive T cells divide on average once every 3.5 years, memory T cells divide every 22 wk, and T cells have an average life span of 20 years (6). Conversion of our calculated elimination rates to estimates of the average life spans of BrdUrd-labeled cells yields approximate values of 16 wk for naive (CD45RA+) T cells and 7 wk for memory (CD45RA–) T cells. Although these estimated life spans of BrdUrd-labeled cells reflect the combined contributions of cell death and cell division, regardless of the exact contribution of these two processes, these values represent a significant increases over previous estimates of T cell turnover rates in primates. Although these discrepant values for T cell turnover may reflect interspecies differences or different study populations, it is also possible that they may reflect the fact that there may be subpopulations of T cells that turn over at different rates (2) and these techniques may be sampling different populations of lymphocytes.
A variety of different models have been proposed to account for the progressive depletion of CD4+ T lymphocytes in AIDS. One model, the “tap and drain” hypothesis (7), proposes that CD4+ T cell depletion ultimately occurs as the result of the inability of the host capacity to generate new T cells (the “tap”) to keep pace with the rapid destruction of CD4+ T cells induced by HIV (the “drain”). An alternative hypothesis, termed “blind T cell homeostasis” (29), hypothesizes that because homeostatic mechanisms regulate only the total number of T cells, and not the individual levels of CD4+ and CD8+ T cells, destruction of CD4+ T cells by HIV progressively results in their replacement by CD8+ cells. Others (8) have proposed that depletion of CD4+ T cells results from insufficient production without increased rates of turnover. By documenting a clear increase in T cell turnover in SIV-infected animals, our results support the accelerated T cell turnover postulated by the tap and drain hypothesis. However, they advance our understanding of the model in two respects. First, they provide an estimate of the magnitude of the increased turnover. Previous critical assessments of the tap and drain hypotheses were hindered in part by the lack of knowledge of what constitutes normal rates of T cell turnover. Second, they document that increased turnover involves not only CD4+ T lymphocytes but also CD8+ T lymphocytes. Although our results in SIV-infected macaques suggest that turnover of CD4+ and CD8+ T cells is likely also to be increased in HIV-infected people because the time course of disease progression differs in these systems (average time to AIDS of 1–2 years in rhesus macaques infected with SIVmac vs. 10 years in humans infected with HIV-1), the magnitude of the increase in T cell turnover in humans may differ from that in macaques.
Many questions remain regarding the mechanisms responsible for increased rates of T cell turnover and progressive depletion of CD4+ T cells. Although we cannot currently assess to what extent SIV infection of CD4+ T cells directly contributes to the increased rate of T cell turnover, the relatively low percentage of infected cells in vivo (30, 31), and the parallel increase in the turnover rates of both CD4+ and CD8+ T cells, all support the hypothesis that indirect mechanisms of cell death may contribute to the destruction of CD4+ T cells. With respect to the increased turnover of CD8+ T cells, because CD8+ T cells are infected only to a limited extent in vivo by HIV or SIV (32), indirect mechanisms of CD8+ T cell death induced by these lentiviruses likely are to be operative, for example up-regulation of Fas ligand by infected cells (33). We cannot yet answer why selective depletion of CD4+ T lymphocytes occurs during the course of AIDS when the turnover rates of both CD4+ and CD8+ T lymphocytes are increased. The slow evolution of CD4+ T cell depletion may result from small imbalances in the rates of proliferation and death of CD4+ T cells or differences in the long-term regenerative capacities of these lymphocyte subsets, which may differ for CD4+ and CD8+ cells (34, 35). The end result of CD4+ T cell depletion in AIDS is therefore likely to reflect not only increased rates of destruction but also impaired regenerative capacity (13).
Acknowledgments
The authors thank David Ho for helpful discussions; Ron Desrosiers and Bruce Walker for review of the manuscript; Ron Desrosiers, Andrew Lackner, Mike Wyand, and Kelledy Manson for providing SIV-infected macaques; Peter Dailey for viral load determinations; Elaine Roberts, Mike Casto, and Mike O’Connell for technical assistance and animal care, and Carolyn A. O’Toole for manuscript preparation. Supported by National Institutes of Health Grants AI-36550 and RR-00168.
Note Added in Proof
After the submission of this manuscript, Mohri et al. (36) published their results using BrdUrd-labeling to demonstrate an increased turnover of CD4+ and CD8+ T lymphocytes in SIV-infected macaques.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: simian immunodeficiency virus (SIV).
References
- 1.Roederer M. Nat Med. 1995;1:621–622. doi: 10.1038/nm0795-621. [DOI] [PubMed] [Google Scholar]
- 2.Freitas A A, Rocha B B. Immunol Today. 1993;14:25–29. doi: 10.1016/0167-5699(93)90320-K. [DOI] [PubMed] [Google Scholar]
- 3.Tough D F, Sprent J. J Exp Med. 1994;179:1127–1135. doi: 10.1084/jem.179.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penit C. J Immunol. 1986;137:2115–2121. [PubMed] [Google Scholar]
- 5.Rocha B, Penit C, Baron C, Vasseur F, Dautigny N, Freitas A A. Eur J Immunol. 1990;20:1697–1708. doi: 10.1002/eji.1830200812. [DOI] [PubMed] [Google Scholar]
- 6.McLean A R, Michie C A. Proc Natl Acad Sci USA. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 8.Wolthers K J, Wisman G B A, Otto S A, de Roda Husman A-M, Schaft N, deWolf F, Goudsmit J, Coutinho R A, van der Zee A G J, Meyaard L, et al. Science. 1996;274:1543–1547. doi: 10.1126/science.274.5292.1543. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 10.Mosier D E. Nature (London) 1995;375:193–194. doi: 10.1038/375193b0. [DOI] [PubMed] [Google Scholar]
- 11.Sprent J, Tough D. Nature (London) 1995;375:194. doi: 10.1038/375194a0. [DOI] [PubMed] [Google Scholar]
- 12.Grossman Z, Herberman R B. Nat Med. 1997;3:486–490. doi: 10.1038/nm0597-486. [DOI] [PubMed] [Google Scholar]
- 13.Hellerstein M K, McCune J M. Immunity. 1997;7:583–589. doi: 10.1016/s1074-7613(00)80379-9. [DOI] [PubMed] [Google Scholar]
- 14.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 15.Kindt T J, Hirsch V M, Johnson P R, Sawasdikosol S. Adv Immunol. 1992;52:425–475. doi: 10.1016/s0065-2776(08)60880-9. [DOI] [PubMed] [Google Scholar]
- 16.Nowak M A, Lloyd A L, Vasquez G M, Wiltrout T A, Wahl L M, Bischofberger N, Williams J, Kinter A, Fauci A S, Hirsch V M, et al. J Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller C J, McChesney M B, Lu X, Dailey P J, Chutkowski C, Lu D, Brosio P, Roberts B, Lu Y. J Virol. 1997;71:1911–1921. doi: 10.1128/jvi.71.3.1911-1921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T, Wong J, MacLean J, Cosimi A B, Wee S. Transplant Proc. 1994;26:1845–1846. [PubMed] [Google Scholar]
- 20.Roederer M, Dubs J G, Anderson M T, Raju P A, Herzenberg L A, Herzenberg L A. J Clin Invest. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimann K A, Waite B C D, Lee-Parritz D E, Lin W, Uchanska-Ziegler B, O’Connell M J, Letvin N L. Cytometry. 1994;17:102–108. doi: 10.1002/cyto.990170113. [DOI] [PubMed] [Google Scholar]
- 22.Michie C A, McLean A, Alcock C, Beverley P C L. Nature (London) 1992;360:264–265. doi: 10.1038/360264a0. [DOI] [PubMed] [Google Scholar]
- 23.Effros R B, Allsopp R, Chiu C-P, Hausner M A, Hirji K, Wang L, Harley C B, Villeponteau B, West M D, Giorgi J V. AIDS. 1996;10:F17–F22. doi: 10.1097/00002030-199607000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Palmer L D, Weng N-P, Levine B L, June C H, Lane H C, Hodes R J. J Exp Med. 1997;185:1381–1386. doi: 10.1084/jem.185.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paganin C, Monos D S, Marshall J D, Frank I, Trinchieri G. J Clin Invest. 1997;99:663–668. doi: 10.1172/JCI119209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo G, Fauci A S. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 27.Gougeon M L, Garcia S, Heeney J, Tschopp R, Lecoeur H, Guetard D, Rame V, Dauguet C, Montagnier L. AIDS Res Hum Retroviruses. 1993;9:553–563. doi: 10.1089/aid.1993.9.553. [DOI] [PubMed] [Google Scholar]
- 28.Russo A, Gianni L, Kinsella T J, Klecker R W J, Jenkins J, Rowland J, Glatstein E, Mitchell J B, Collins J, Myers C. Cancer Res. 1984;44:1702–1705. [PubMed] [Google Scholar]
- 29.Adleman L M, Wofsy D. J Acquired Immune Defic Syndr. 1993;6:144–152. [PubMed] [Google Scholar]
- 30.Chun T W, Carruth L, Finzi D, Shen X, DiGuiseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, et al. Nature (London) 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 31.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z Q, et al. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 32.Tiran B, Heller I, Isakov A, Burke M, Topilsky M. Lancet. 1996;348:1527–1528. doi: 10.1016/S0140-6736(05)65959-4. [DOI] [PubMed] [Google Scholar]
- 33.Xu X-N, Screaton G R, Gotch F M, Dong T, Tan R, Almond N, Walker B, Stebbings R, Kent K, Nagata S, et al. J Exp Med. 1997;186:7–16. doi: 10.1084/jem.186.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackall C L, Hakim F T, Gress R E. Immunol Today. 1997;18:245–251. doi: 10.1016/s0167-5699(97)81664-7. [DOI] [PubMed] [Google Scholar]
- 35.Mackall C L, Fleisher T A, Brown M R, Andrich M P, Chen C C, Feuerstein I M, Magrath I T, Wexler L H, Dimitrov D S, Gress R E. Blood. 1997;89:3700–3712. [PubMed] [Google Scholar]
- 36.Mohri H, Bonhoeffer S, Monard S, Perelson A S, Ho D D. Science. 1998;279:1223–1227. doi: 10.1126/science.279.5354.1223. [DOI] [PubMed] [Google Scholar]