Abstract
The goal of this study was to develop an automated method to segment breast masses on dynamic contrast-enhanced (DCE) magnetic resonance (MR) scans and to evaluate its potential for estimating tumor volume on pre- and postchemotherapy images and tumor change in response to treatment. A radiologist experienced in interpreting breast MR scans defined a cuboid volume of interest (VOI) enclosing the mass in the MR volume at one time point within the sequence of DCE-MR scans. The corresponding VOIs over the entire time sequence were then automatically extracted. A new 3D VOI representing the local pharmacokinetic activities in the VOI was generated from the 4D VOI sequence by summarizing the temporal intensity enhancement curve of each voxel with its standard deviation. The method then used the fuzzy c-means (FCM) clustering algorithm followed by morphological filtering for initial mass segmentation. The initial segmentation was refined by the 3D level set (LS) method. The velocity field of the LS method was formulated in terms of the mean curvature which guaranteed the smoothness of the surface, the Sobel edge information which attracted the zero LS to the desired mass margin, and the FCM membership function which improved segmentation accuracy. The method was evaluated on 50 DCE-MR scans of 25 patients who underwent neoadjuvant chemotherapy. Each patient had pre- and postchemotherapy DCE-MR scans on a 1.5 T magnet. The in-plane pixel size ranged from 0.546 to 0.703 mm and the slice thickness ranged from 2.5 to 4.5 mm. The flip angle was 15°, repetition time ranged from 5.98 to 6.7 ms, and echo time ranged from 1.2 to 1.3 ms. Computer segmentation was applied to the coronal T1-weighted images. For comparison, the same radiologist who marked the VOI also manually segmented the mass on each slice. The performance of the automated method was quantified using an overlap measure, defined as the ratio of the intersection of the computer and the manual segmentation volumes to the manual segmentation volume. Pre- and postchemotherapy masses had overlap measures of 0.81±0.13 (mean±s.d.) and 0.71±0.22, respectively. The percentage volume reduction (PVR) estimated by computer and the radiologist were 55.5±43.0% (mean±s.d.) and 57.8±51.3%, respectively. Paired Student’s t test indicated that the difference between the mean PVRs estimated by computer and the radiologist did not reach statistical significance (p=0.641). The automated mass segmentation method may have the potential to assist physicians in monitoring volume change in breast masses in response to treatment.
Keywords: magnetic resonance imaging, segmentation, level set, fuzzy c means, computer-aided diagnosis
INTRODUCTION
Neoadjuvant or presurgical chemotherapy can reduce the size of locally advanced masses and increase the chance of electing breast conserving surgery.1, 2, 3 Results of several randomized trials comparing adjuvant (postoperative) to neoadjuvant chemotherapy for operable breast cancer indicated no statistically significant difference in the long-term overall survival between the adjuvant and neoadjuvant arms.4, 5, 6 Neoadjuvant chemotherapy may therefore be a safe approach in reducing the mastectomy rate for women with locally advanced breast cancer.7
During neoadjuvant chemotherapy it is useful to monitor the changes in the tumor to assess treatment efficacy and to intervene based on response so that the patient can be spared the associated morbidity by early termination of an ineffective regimen or benefit from effective regimens sooner.8 In addition, there is evidence that compared to poor responders, good responders to neoadjuvant chemotherapy have superior outcome both in terms of disease-free survival and overall survival.7 This indicates that the response to neoadjuvant chemotherapy may also be used as an early surrogate of long-term outcome. Evaluation of tumor response to neoadjuvant chemotherapy is therefore an important clinical task.
Clinical trials indicate that the effect of neoadjuvant chemotherapy on breast masses may fall into pathologic responses ranging from complete response to progressive disease.9 Mass diameter has been used widely to describe mass extent and to monitor the response to therapy3, 10, 11 because it can be measured conveniently by physicians. For a solitary tumor, the World Health Organization criterion for partial response is a 50% or more decrease in the product of two measurements (the maximum diameter and the largest diameter perpendicular to this maximum diameter).12 The Response Evaluation Criteria in Solid Tumors (RECIST), introduced in 2000, defines partial response as a 30% or more decrease in the longest diameter of the lesion.13 However, both of these criteria are based on the assumption that the tumor is spherical and has a circular cross section. For masses with an irregular shape and those that may shrink nonuniformly in different directions, dual or single diameter measurements may be inadequate for response monitoring.14 Partridge et al. found that mass volume in dynamic contrast-enhanced (DCE) breast magnetic resonance (MR) imaging may provide a more sensitive assessment of neoadjuvant chemotherapy efficacy than mass diameter.15 One obstacle in adopting three-dimensional (3D) volume is that manual segmentation of 3D MR scans is time consuming and may have large inter- and intraobserver variabilities. Automated or semiautomated 3D segmentation methods potentially will be more efficient and reproducible.
Investigators have presented a number of segmentation methods14, 15, 16, 17, 18, 19, 20, 21 for lesions in breast MR scans. Lucas-Quesada et al.16 designed two semiautomatic techniques for segmentation of breast masses on DCE-MR images. Method 1 used the correlation of the enhancement curve of a pixel in the image to that of a user-defined ROI to generate a similarity map. A user-defined threshold was then used on the similarity map for segmentation. Method 2 used a feature map generated from a scatter plot of pixel intensities in the pre- and postcontrast images. They found that although both methods have similar accuracy indices, Method 1 required less user interaction and was less affected by image noise. Chen et al. proposed a fuzzy c-means (FCM) based method,17 which focuses on temporal intensity information of DCE-MR scans. Experimental results based on 121 breast masses indicated that the FCM clustering was more effective and efficient than their previously developed volume growing method. One drawback of their FCM implementation is that it utilizes mainly intensity enhancement in the temporal domain without including spatial constraints among neighboring voxels. Szabo et al.18 also performed pixel-by-pixel segmentation of breast tumors based on the signal enhancement curves in time. However, no comparison was made to manual segmentation. Wu et al. segmented DCE breast MR images based on the Markov random field (MRF) model.19 The challenge of this method is how to appropriately design and optimize the parameters of the MRF model to fit various mass structures. Alderliesten et al.14 used a volume growing method with a voxel-based stopping criterion automatically derived from the enhanced mass volume. The mass diameter was also manually measured by two radiologists on DCE-MR images. The ground truth was established by measuring the tumor volume at histopathology. The investigators found that semiautomatic volumetric measurement at DCE-MR provided a more accurate assessment of tumor extent with respect to histopathology. Although their results supported the usefulness of semiautomatic measurement in improving tumor size estimation, they did not analyze postchemotherapy masses, which is often more challenging, so that its role in assessing volume change in response to treatment is not known. Partridge et al.15 used a threshold-based semiautomatic method for tumor volume measurement on DCE-MR images and showed that both initial MRI volume and final change in volume were strong predictors of recurrence-free survival for breast cancer patients who underwent neoadjuvant chemotherapy. Bulow et al.20 designed a segmentation method that included lesion selection, automated threshold estimation, hole closing, and leakage removal steps. They compared computer segmentation with manual segmentation by two radiologists on a data set of five breast masses and investigated the effect of lesion selection on computer segmentation. The commercial software CADstream (Confirma, Seattle, WA) also provides a segmentation function, but it is based on global thresholding without considering the spatial structures of mass.21
Although previous studies evaluated the performance of automated or semiautomated segmentation methods for breast masses on DCE-MR images, very few studies included postchemotherapy images. Partridge et al.15 investigated semiautomated measurement of tumor volume in both pre- and postchemotherapy DCE-MR images, but they did not compare the segmented lesion boundaries and volume to manual segmentation. With the increased use of neoadjuvant chemotherapy and the increase in radiologists’ workload, automated and semiautomated segmentation methods for the evaluation of tumor response to treatment will become more appealing. It is therefore important to evaluate the performance of computer segmentation methods on both pre- and postchemotherapy images and compare the performance with manual segmentation by experienced radiologists. The goal of our study was to develop a segmentation method for breast masses on DCE-MR images and to evaluate its potential for estimating tumor volume on pre- and postchemotherapy images and tumor change in response to treatment.
We have previously designed a level set (LS) method for mass segmentation in mammograms and obtained encouraging results.22 In a preliminary study,23 we extended our implementation from 2D to 3D for breast MR exams and evaluated the method on the pre- and postchemotheraphy scan of ten lesions. The initial segmented volume in our preliminary study was provided by k-means clustering. In this study, we improved the performance by using FCM-based clustering for initial mass segmentation and adding a new propagation term that used FCM clustering as an additional external force to regulate the LS evolution. The newly proposed mass segmentation method takes advantage of both mass morphology (3D spatial structure) and mass pharmacokinetics (voxelwise 1D temporal intensity enhancement curve) information on DCE-MR scans. The LS method can flexibly handle the variation of 3D mass structures. Our method for breast mass segmentation was validated by comparing its results to manual segmentation by an experienced radiologist on pre- and postchemotherapy DCE-MR scans of 25 patients who underwent neoadjuvant chemotherapy. The capability of the computerized method in estimation of tumor volume change was also evaluated. The performance of the newly proposed segmentation method was compared to that in our preliminary study by using the radiologist’s manual segmentation as the reference standard. The same radiologist repeated the segmentation of all breast mass images after 1 year to estimate the intraobserver variability in manual segmentation.
MATERIALS AND METHODS
Figure 1 shows the flowchart for our breast mass segmentation method. A new 3D volume of interest (VOI) representing the local pharmacokinetic activities in the mass VOI, referred to as the “spatial-pharmacokinetic VOI” in the following discussion, was generated from the 4D VOI sequence by summarizing the temporal intensity enhancement curve of each voxel with its standard deviation. The FCM clustering was applied to the spatial-pharmacokinetic VOI, together with one 3D postcontrast VOI in which the average voxel intensity enhancement was the highest. The segmented volume resulting from the FCM clustering was adjusted using morphological filtering. The LS method was initialized using the filtered FCM clustering result. It evolved in the same 3D postcontrast VOI as that used by the FCM clustering and was regulated by the FCM membership function. With this approach, both spatial and time (4D) information in the input VOI sequence from the DCE-MR scan was utilized in the computer processing to produce the 3D output VOI.
Figure 1.
Flowchart for mass segmentation on breast DCE-MR scans.
Data set
DCE-MR scans were performed for 37 patients who underwent neoadjuvant chemotherapy as part of a previous study at our institution.24 We collected data from this previous study with Institutional Review Board approval. DCE-MR scans were available and evaluable for 25 patients. Each patient had one pre- and one postchemotherapy DCE-MR scans performed on a 1.5 T General Electric (GE) magnet with the Medrad dual-phased array dedicated breast coil for optimal resolution and signal-to-noise ratio. Three-dimensional volume acquisition gradient echo sequences were performed with fat saturation before and after the intravenous administration of 0.15 mmol∕kg gadolinium diethylenetriamine penta-acetic acid (Gd-DTPA). A hybrid sequence was used to combine rapid dynamic imaging with high resolution imaging. Dynamic imaging was preformed in 26.7 s intervals for about 4 min. Gd-DTPA was administered in the first 30–45 s of the scan; therefore, the first 3D volume in the scan sequence was contrast-free. Each DCE-MR scan sequence in the data set included ten consecutively acquired volumes. Computer segmentation was applied to the coronal T1-weighted images. The in-plane pixel size ranged from 0.546 to 0.703 mm and the slice thickness ranged from 2.5 to 4.5 mm. The flip angle was 15°, repetition time ranged from 5.98 to 6.7 ms, and echo time ranged from 1.2 to 1.3 ms.
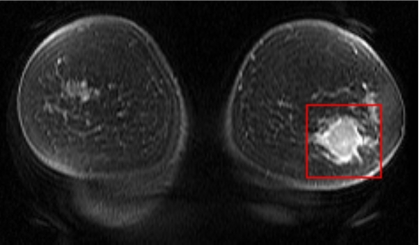
For each MR sequence, an expert radiologist, with more than 25 years of experience in breast imaging and 7 years of experience in interpreting breast MR scans, located and defined a 3D cuboid VOI that enclosed the mass on one of the postcontrast scans. An in-house developed graphical user interface was used to define the mass VOI in both pre- and postchemotherapy scans for all 25 patients in our data set. As an example, Fig. 2 shows one postcontrast coronal slice that included both left and right breasts. The radiologist marked the mass with a rectangle on the slice where it was best visualized and indicated the top and bottom slices where the mass was visible. By stacking the rectangles with the same size and the same in-plane location on the other slices containing the mass, a 3D cuboid VOI was obtained. A total of 50 VOIs (25 prechemotherapy masses and 25 postchemotherapy masses) were thus collected. The same radiologist also manually segmented each mass on each slice of the VOI twice in two reading sessions, so that the intraobserver variability could be estimated. To minimize the potential effect associated with memorization, the two readings were separated by more than 1 year. The manually segmented contours were used as the reference standard to compare with our automated segmentation method.
Figure 2.
A 2D postcontrast breast MRI slice. The prechemotherapy mass was enclosed by the rectangular ROI drawn by the radiologist.
Analysis of pharmacokinetic curves
Pharmacokinetic assessment from breast DCE-MR scans has been used for mass detection, diagnosis, and staging.25, 26, 27, 28, 29 Figure 3 illustrates four typical pharmacokinetic curves that characterize the uptake and washout properties of contrast agent in fat, parenchyma, malignant tissue, and benign tissue, respectively. Normal tissue typically has a flat curve, while malignant tissue typically shows a rapid uptake and washout.30 Benign masses may have various patterns of the uptake and washout curve. Because all masses undergoing neoadjuvant chemotherapy are malignant, we need only to differentiate normal from malignant tissue. A number of researchers have been investigating pharmacokinetic modeling methods to quantify physiological parameters that describe the dynamics of tissue enhancement. However, the estimated parameters from experimental data may show a large variation depending on the mathematical model and the experimental design.30, 31, 32
Figure 3.
Illustration of typical pharmacokinetic curves for various types of breast tissue in DCE-MR scans.
Ten volumes corresponding to the ten time points in each DCE-MR scan sequence in our data set provided samples for initial voxelwise statistical analysis, which was performed by computing the standard deviation of temporal intensity enhancement of each voxel. Voxels of a malignant mass typically had large standard deviations, while most voxels of the normal breast tissue had small standard deviations. Some normal voxels, such as those containing vessels, may also demonstrate large standard deviations. These voxels are excluded from the segmented mass in subsequent processing steps.
In this study, the LS segmentation method was applied to the most enhanced postcontrast VOI, which was selected from the DCE-MR VOI sequence by the following steps: (1) Define the precontrast (the first time point) VOI as the reference and one of the remaining nine volumes in the sequence as the search VOI, (2) subtract the reference VOI from the search VOI, (3) average all non-negative voxels, i.e., voxels with enhancement, in the difference VOI, and (4) perform steps (2) and (3) for each of the nine postcontrast time points and choose the VOI that has the maximum average.
Figure 4a shows ten 3D VOIs at ten sequential time points acquired before, during, and after contrast injection. The top row represents the 3D VOI acquired at the precontrast time point and the bottom row represents the last postcontrast time point. Figure 4b shows the spatial-pharmacokinetic VOI, in which the gray-level value of each voxel is proportional to the standard deviation of the temporal intensity variation of that voxel among the ten time points shown in Fig. 4a. The analysis of standard deviation at each voxel summarizes the spatial-variant pharmacokinetic 4D information in a 3D ROI, which enhances the malignant mass while suppressing most background tissue. The spatial-pharmacokinetic VOI shown in Fig. 4b and the 3D VOI with the highest average enhancement [the sixth row in Fig. 4a for the mass in this example] were then used as inputs to the FCM clustering. For each voxel vi in the 3D VOI to be segmented, a two-element vector xi was defined such that the first element corresponded to the standard deviation and the second element to the gray-level in the VOI with the highest enhancement.
Figure 4.
Breast mass VOIs defined from a DCE-MR scan sequence. (a) Slices of a sequence of 3D VOIs at ten time points from the top row to the bottom row. The first time point is a precontrast scan. For each row (time point), the leftmost slice is the closest to the nipple. (b) The 3D spatial-pharmacokinetic VOI derived by using the voxelwise standard deviation of temporal intensity enhancement over the ten scans.
Initial segmentation
In a preliminary study,23 we had implemented a simpler LS model that used k-means clustering for initialization. In this study, FCM clustering was used both for initial segmentation and for LS evolution. The previous implementation with the k-means initialization was compared to the current implementation in this study. We first describe the FCM initialization below, followed by a short description of the k-means initialization used in our previous preliminary study.
FCM is an unsupervised machine learner.33 It has been used widely in data analysis and medical image segmentation.34 Similar to k-means clustering, the FCM clustering is an iterative process that optimizes a specific objective function. However, unlike k-means clustering where each data sample is exclusively assigned to one cluster, the FCM clustering assigns each data sample to all predefined clusters with respective likelihoods.
For breast mass segmentation on DCE-MR scans, the objective function for the FCM clustering was the generalized least-squares error defined as
| (1) |
subject to
where N is the number of voxels in the 3D VOI, xi is a two-element vector at voxel i described above, cj (j=1 and 2) are two-element vectors obtained from weighted averaging of xi for the background tissue and the mass, respectively, uij is the likelihood that the voxel xi belongs to cluster j, and ‖⋅‖ stands for the Euclidean distance. Starting with random assignments for c1 and c2, we alternately updated the likelihoods:
| () |
and
| (2) |
and the cluster centers cj:
| (3) |
This iterative optimization stopped when the objective function J changed very little (in this study, ∣Jn+1−Jn∣≤0.01) between two consecutive iterations or when a predetermined number of iterations was reached (N=100).
For each voxel vi in the 3D VOI, FCM clustering resulted in a likelihood ui2 that vi belonged to the malignant mass. To binarize the FCM clustered VOI, we experimentally determined a threshold T=0.3 on ui2 which was then applied consistently to all masses in the data set. A voxel vi was assigned to the mass if and only if ui2 was larger than T. Within the binary 3D VOI, the largest 3D connected component based on 26 connectivity was selected, and the other voxels were grouped into the background. Since breast masses on DCE-MR scans may demonstrate rim enhancement (i.e., the enhancement is more pronounced at the periphery than the internal region of the mass), the object segmented by the FCM clustering algorithm may contain holes, formed by groups of voxels assigned to the background tissue that were completely surrounded by mass voxels. These holes were filled using morphological filling operation. The final step for the initial mass segmentation was the morphological opening operation. It slightly reduced the size of the initial segmented volume so that the initial segmentation can be inside the desired mass region. Morphological opening may also split one connected component into multiple isolated components, which can be handled effectively using the LS segmentation algorithm described below.
The clustering method described above utilized the pharmacokinetic information, because the standard deviation of the voxel value over the time sequence was one of the elements used in FCM clustering vector xi. In contrast, the k-means clustering utilized in our preliminary study made use of only the difference in voxel values between the post- and precontrast volumes as the class feature.23 After k-means clustering, any holes that might exist in the volume were filled using morphological filtering to obtain an initial segmented volume.
Level set segmentation
The initial segmentation was refined by the 3D LS method on the most enhanced postcontrast VOI. The LS method is a deformable model, which can capture object shape or surface by numerically solving a well-designed partial differential equation (PDE). It has several advantages over its predecessor, the explicit active contour model. For example, the LS method provides a flexible mechanism allowing topological change in target objects including object splitting and merging. If a connected object is split into multiple components by morphological opening in the previous step, the components that are determined to be parts of the same object can be reconnected during LS iteration. It can be extended easily from 2D to 3D or even higher dimensions. The LS numerical technique for evolving curves in 2D images or surfaces in 3D volumes has been used in various applications of image processing, computer graphics, and computational geometry.35, 36, 37 We have previously utilized the LS method for the segmentation of breast masses on mammograms.22 In this paper, we extended our implementation of the 2D LS segmentation on mammograms to 3D on breast MRI scans. We also employed the FCM clustering result as an additional external force to regulate the LS evolution. The use of the FCM clustering result to guide the LS was originally proposed by Suri et al.,36 although our implementation is different.
The LS formulation in this study is common to that in the literature.22, 35, 36, 37 Let ∇φ=(∂φ∕∂x,∂φ∕∂y,∂φ∕∂z) be the gradient of the distance function and the velocity field that guides the surface evolution. The success of the LS segmentation is subject to appropriate design of . In analogy to the explicit active contour model, we may incorporate internal forces to guarantee the smoothness of the surface and external forces to attract the zero LS to a desired mass surface. The mean curvature κ=−∇(∇φ∕∣∇φ∣) is a popular term35 to constrain the surface evolution and the edge information of the original volume I(x,y,z) has been used to construct external force in our 2D mass segmentation on mammograms. For 3D MR scans, we included a new external force u2(x,y,z), the FCM membership function which quantitatively defines the likelihood that a voxel at (x,y,z) belongs to a mass. Note that u2 is derived based on the voxel intensity of the most enhanced VOI and the voxelwise standard deviation in the spatial-pharmacokinetic VOI, and hence includes the dynamic enhancement information. The LS PDE for breast mass segmentation is formulated as
| (4) |
where and . The free parameter α regulates the external force u2. As α increases, dynamic enhancement will have a stronger impact on the LS evolution. We chose α=0.5 experimentally for the segmentation task in this study. The intuitive idea behind Eq. 4 is that the LS should move fast on a flat surface, while it should evolve slowly as it approaches object edges. Because boundaries of breast masses may be vague, segmentation leak is a challenge for breast mass segmentation. We used the homogeneity constraint to reduce segmentation leak.22 During evolution of the 3D zero LS surface, outer border voxels were examined and added to the existing mass region only if high homogeneity of mass gray level can be maintained.
Our preliminary LS implementation did not use the external force u2 and was initialized by k-means clustering. The LS method described above included the external force u2 and was initialized by FCM clustering. In the following discussion, when the two methods are compared, the former method is referred to as LS(k means) and the latter as LS(FCM). For simplicity, the latter method is also referred to only as LS, since it is the main focus of this study.
Performance measure
The mass volumes extracted by FCM-based initial segmentation, LS(FCM), and LS(k means) were compared to the reference standard, i.e., the radiologist’s manual segmentation. Pearson’s correlation coefficient was used to measure the correlation between the computer segmentation and the reference standard. Paired Student’s t test was used to evaluate the significance of the differences between the extracted volumes. An alpha level of 0.05 was defined as the significance criterion. To study the intraobserver variability, volumes obtained from the radiologist’s manual segmentation in two different reading sessions were compared using Pearson’s correlation coefficient and paired Student’s t test.
Physicians commonly compare mass sizes in pre- and postchemotherapy images to monitor cancer response to neoadjuvant chemotherapy. For example, in a previous study at our institution, a 70% decrease in volume was defined as pathological partial response and 70% increase in volume as pathological progressive response.24 Since the change in mass volume is an important indication of chemotherapy response, we evaluated the difference between the change of mass volume estimated from computer segmentation and radiologist’s manual segmentation. We defined the percentage volume reduction (PVR) as follows:
| (5) |
where Vpostchemo and Vprechemo denote the volumes of the same mass before and after chemotherapy, respectively. Pearson’s correlation coefficient was used to measure the correlation between the computer estimation and the reference standard. Paired Student’s t test was used to evaluate the statistical significance of the difference in PVRs estimated by the radiologist and the computer.
We also compared the computer segmentation results with the radiologist’s manual segmentation in terms of volume overlap ratios. Let VC and VR denote the mass volumes estimated by computer and by the radiologist, respectively. Several definitions have been used and discussed in the literature for measuring how well two objects overlap. In this study, we used two volume overlap ratios defined as
| (6) |
and
| (7) |
which respectively represent the fraction of the radiologist’s volume that overlaps with the computer’s volume and the overlap relative to the union of the two volumes. The minimum value of VOR1 and VOR2 is zero when VC does not overlap with VR. The maximum value of VOR1 and VOR2 is 1 when VC completely coincides with VR. We computed VOR1 and VOR2 for the FCM-based initial segmentation, LS(FCM), and LS(k means). Paired Student’s t test was used to evaluate whether the LS refinement improved the FCM-based initial segmentation when the radiologist’s manual segmentation was used as the reference standard. Likewise, paired Student’s t test was used to compare the overlap between the radiologist’s manual segmentation and LS(FCM) to that between the radiologist’s manual segmentation and LS(k means).
RESULTS
Segmentation
Figure 5a shows the slices of a postcontrast VOI that contained a prechemotherapy breast mass at the time point of maximum enhancement, which was automatically segmented using the FCM clustering and morphological filtering as shown in Fig. 5b. The LS refinement of FCM clustering and manual segmentation by the radiologist are shown in Figs. 5c, 5d, respectively. The refinement by the LS method resulted in computer-estimated mass boundaries that were closer to radiologist-estimated boundaries. The mass in this example became slightly oversegmented after the LS refinement if the manual segmentation is used as reference standard. The mass boundaries generated by computer segmentation were less polygonal than those drawn by the radiologist, indicating that computer segmentation had the potential to include more boundary details of breast masses.
Figure 5.
Mass segmentation on a breast DCE-MR scan. (a) Original postcontrast VOI containing a prechemotherapy breast mass at the time of maximum enhancement [the sixth time point in Fig. 4a]. (b) Initial segmentation by the FCM clustering using the information from the VOI at maximum enhancement and the spatial-pharmacokinetic VOI [Fig. 4b] and the morphological filters. (c) Automated refinement by the LS method. (d) Manual segmentation by the radiologist.
Intraobserver variability
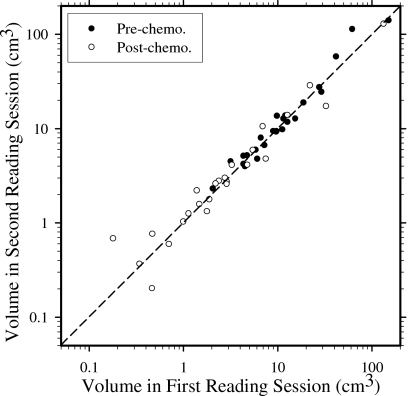
Figure 6 shows the log-log scatter plot of pre- and postchemotherapy breast mass volumes estimated by the same radiologist in two reading sessions separated by over 1 year. The difference between the volumes segmented in the two reading sessions was analyzed using the paired t test and the correlation coefficient (Table 1). For the pre- and postchemotherapy mass sets, the p values were 0.310 and 0.824, respectively, and the correlation coefficients were 0.946 and 0.991, respectively, indicating that the tumor volumes segmented in the two reading sessions were highly correlated and their difference did not demonstrate statistical significance. Thus, in the following analysis of segmentation performance of the computer algorithm, we only discussed the results using the radiologist’s first reading as the reference. The results using the second reading as reference were similar and not shown to avoid redundancy.
Figure 6.
Log-log scatter plot of pre- and postchemotherapy breast mass volumes estimated by the same radiologist in two reading sessions separated by over 1 year.
Table 1.
Intraobserver variability of manual volume segmentation by the same radiologist in two reading sessions separated by over 1 year for the prechemotherapy and postchemotherapy masses.
| Prechemo. (mean±s.d.) | Postchemo. (mean±s.d.) | PVR (mean±s.d.) | |
|---|---|---|---|
| Session 1 volume | 19.1±30.5 | 10.1±26.7 | 57.8±51.3 |
| Session 2 volume | 21.5±34.0 | 9.8±25.7 | 60.2±44.0 |
| p value | 0.310 | 0.824 | 0.912 |
| Correlation coefficient | 0.946 | 0.991 | 0.836 |
Volume of breast masses
Table 2 summarizes volumes of breast masses before and after chemotherapy and the PVRs estimated by the FCM-based initial segmentation, LS(FCM), and LS(k means). The radiologist’s manual segmentation is shown in Table 1.
Table 2.
Volumes (cm3) of breast masses before and after neoadjuvant chemotherapy and the percentages of volume reduction estimated using the initial FCM segmentation, LS(FCM), and LS(k means).
| Segmentation method | Prechemo. (mean±s.d.) | Postchemo. (mean±s.d.) | PVR (mean±s.d.) |
|---|---|---|---|
| Initial segmentation (FCM) | 16.6±26.3 | 4.5±9.0 | 70.4±35.1 |
| LS(FCM) | 19.3±28.5 | 8.8±22.7 | 55.5±43.0 |
| LS(k means) | 18.9±28.8 | 7.7±20.2 | 62.7±41.6 |
Figure 7a shows the log-log scatter plot of the volumes of prechemotherapy masses by the FCM-based initial segmentation and by the radiologist’s segmentation (solid circles) and the log-log scatter plot of the volumes of the same masses by LS(FCM) and by the radiologist’s segmentation (empty circles). The log scale is used to show the data points in the small volume range more clearly. The 25 empty circles are distributed close to the dotted line which represents the ideal trend line. Table 3 summarizes the statistical tests for mass volumes estimated by computer segmentation relative to the radiologist’s manual segmentation as the reference standard. The Pearson product-moment correlation coefficient between the mass volumes estimated by the FCM-based initial segmentation and radiologist segmentation was 0.993, and the paired t test achieved a p value of 0.028. This indicates that for prechemotherapy breast masses, the volume extracted by the FCM method was highly correlated with that by the radiologist. However, the two extracted volumes were significantly different on the average, with the FCM method underestimating the mass volume relative to the radiologist-extracted reference standard. The corresponding correlation coefficient and p value for the mass volumes estimated by LS(FCM) were 0.981 and 0.918, respectively. This indicated that the volume extracted by the LS method was also highly correlated with that by the radiologist. Although the initial segmentation volume based on the FCM clustering was significantly different from that extracted by the radiologist, the volume based on the LS(FCM) method did not show a significant difference compared to the radiologist-extracted volume. The corresponding correlation coefficient and p value for the mass volumes estimated by LS(k means) were 0.978 and 0.873, respectively.
Figure 7.
Breast mass volumes. (a) Log-log scatter plot of volume of prechemotherapy masses segmented by computer and the radiologist. (b) Log-log scatter plot of volume of postchemotherapy masses segmented by computer and the radiologist. (c) Percentages of volume reduction of 25 masses after neoadjuvant chemotherapy estimated by the FCM-based initial computer segmentation, the LS refinement, and the radiologist.
Table 3.
Statistical comparison of the pre- and postchemotherapy breast mass volumes estimated by the FCM-based initial segmentation, LS(FCM), and LS(k means) relative to the radiologist’s first manual segmentation as reference standard.
| Segmentation method | Prechemo. | Postchemo. | % Vol. Reduction (PVR) | |||
|---|---|---|---|---|---|---|
| t-test p value | Correl. coeff. | t-test p value | Correl. coeff. | t-test p value | Correl coeff. | |
| Initial segmentation (FCM) | 0.028 | 0.993 | 0.153 | 0.912 | 0.066 | 0.774 |
| LS(FCM) | 0.918 | 0.981 | 0.185 | 0.994 | 0.641 | 0.876 |
| LS(k means) | 0.873 | 0.978 | 0.095 | 0.994 | 0.418 | 0.816 |
Figure 7b shows the log-log scatter plot of the volumes of the same set of breast masses as in Fig. 7a but after chemotherapy. As in Fig. 7a, the solid and empty circles represent FCM-based initial segmentation and LS(FCM), respectively. The correlation coefficient and p value for the mass volumes estimated by the FCM-based initial segmentation were 0.912 and 0.153, respectively, while those for the mass volumes estimated by the LS(FCM) were 0.994 and 0.185, respectively (Table 3). The results indicate that, for this data set, the difference in the average mass volumes estimated by the radiologist and the computer did not reach statistical significance for postchemotherapy masses, and the LS segmentation slightly improved the FCM-based initial segmentation. The corresponding correlation coefficient and p value for the mass volumes estimated by LS(k means) were 0.994 and 0.095, respectively. Thus, for post chemotherapy masses, the mass volume extracted by LS(k means) was also highly correlated with that extracted by the radiologist. The average volumes were different, however, although the difference did not reach statistical significance. As can be seen from Tables 1, 2, all three computer methods underestimated the average volumes, but LS(FCM) had the smallest underestimation.
One purpose of measuring the volume of breast masses in both pre- and postchemotherapy DCE-MR scans was to evaluate the change in the mass volume. Figure 7c illustrates the PVRs of 25 breast masses in our data set based on the FCM-based initial segmentation (black bar), the LS refinement of FCM clustering (light-gray bar), and the radiologist segmentation (dark-gray bar). Table 3 shows that the correlation coefficient of the PVRs estimated by the radiologist and the FCM-based initial segmentation was 0.774, which was improved to 0.876 after the FCM segmentation was refined by the LS. Paired t test for the difference between the FCM-based initial segmentation and the radiologist segmentation achieved a p value of 0.066, while that between LS(FCM) and the radiologist segmentation achieved a p value of 0.641, indicating that the difference in the mean PVRs estimated by LS(FCM) and by the radiologist’s manual segmentation did not reach statistical significance for this data set. The difference in the PVRs estimated by LS(k means) and by the radiologist’s manual segmentation did not reach statistical significance either (p=0.418), but LS(k means) had a lower correlation with the radiologist’s PVR (r=0.816) compared to LS(FCM).
Volume overlap ratios
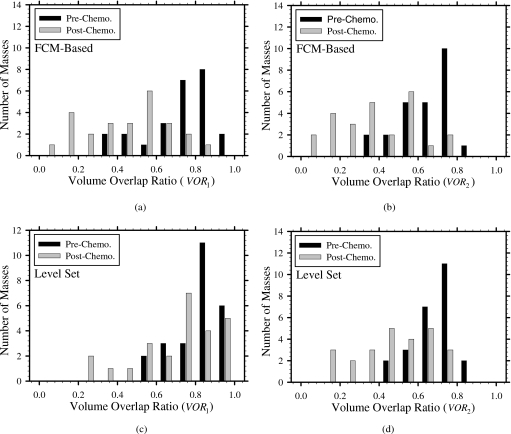
Figures 8a, 8b, 8c, 8d show the histograms of the volume overlap ratios obtained from the FCM-based initial segmentation and LS refinement of the FCM using the radiologist’s manual segmentation as reference. Table 4 summarizes the mean and standard deviation of the volume overlap ratios for the pre- and postchemotheraphy mass sets. The overlap ratios of postchemotheraphy lesions are consistently smaller than those of prechemotherapy lesions. After the LS refinement of the FCM, all prechemotherapy masses had VOR1 over 0.5, while four postchemotherapy masses had VOR1 under 0.5. Only 2 prechemotherapy masses had VOR2 under 0.5, while 13 postchemotherapy masses had VOR2 under 0.5.
Figure 8.
Volume overlap ratios of the prechemotherapy and the postchemotherapy masses.
Table 4.
Average overlap ratios of computer automated segmentation relative to the radiologist’s first manual segmentation. The statistical significance of the difference between FCM-based initial segmentation and the LS(FCM) was evaluated by the paired t test. Likewise, the statistical significance of the difference between LS(FCM) and LS(k means) was evaluated by the paired t test.
| Segmentation method | Overlap ratio VOR1 | Overlap ratio VOR2 | ||
|---|---|---|---|---|
| Prechemo. | Postchemo. | Prechemo. | Postchemo. | |
| Initial segmentation (FCM) | 0.72±0.16 | 0.44±0.21 | 0.64±0.14 | 0.38±0.20 |
| LS(FCM) | 0.81±0.13 | 0.71±0.22 | 0.69±0.10 | 0.47±0.20 |
| LS(k means) | 0.79±0.13 | 0.58±0.26 | 0.68±0.10 | 0.41±0.20 |
| p value between LS(FCM) and FCM | 4.3×10−7 | 1.0×10−6 | 6.1×10−3 | 1.8×10−3 |
| p value between LS(FCM) and LS(k means) | 3.3×10−3 | 1.3×10−3 | 0.51 | 0.014 |
The overlap ratio from the initial segmentation was compared to that from the final segmentation using the paired t test. The results (Table 4) indicate that the LS method improved the FCM-based initial segmentation significantly for both pre- and postchemotherapy masses in terms of either VOR1 or VOR2. LS(FCM) performed significantly better than LS(k means) in terms of VOR1 for both pre- and postchemotherapy masses. When VOR2 was used as the overlap criterion, LS(FCM) was significantly better than LS(k means) for postchemotherapy masses, but the difference for prechemotherapy masses did not reach statistical significance.
DISCUSSION
Accurate prediction of response to chemotherapy by using the noninvasive DCE-MR technique would have high clinical relevance. If DCE-MR is used to assess mass response to treatment, cancer size before and after chemotherapy in the MR volume will likely be one of the important measures of change. Manual slice-by-slice segmentation by a radiologist to estimate the tumor volume is both time consuming and subject to inter- and intraobserver variabilities. In this study, we designed a new automated segmentation method to delineate breast masses on DCE-MR scans.
Our results indicate that the proposed method may have the potential to assist physicians in the assessment of volume changes. Both the FCM-based initial segmentation volume and the LS refinement of the FCM segmentation volume had a high correlation with the manually extracted volume. The FCM-based initial segmentation consistently underestimated the mass volume compared to manual segmentation. The refinement by LS segmentation reduced this underestimation. In addition, LS refinement significantly improved the volume overlap between the computer segmentation and manual segmentation.
A comparison of the segmentation results of pre- and postchemotherapy masses indicated that our automated segmentation method performed worse for the latter. The average mass volume difference between the LS and manual methods was larger for postchemotherapy masses (Table 2). In addition, the volume overlaps were smaller for postchemotherapy masses (Table 4). One possible reason is that breast masses after chemotherapy are topologically more complicated and often have a fragmented appearance.
In a preliminary study, we analyzed segmentation results in ten pre- and postchemotherapy DCE-MR images containing breast masses.23 Our previous results demonstrated the feasibility of using the LS method for the segmentation of breast masses on DCE-MR images. In the current study, we further developed the segmentation technique by introducing a new energy term that incorporated the spatial-pharmacokinetic information of DCE-MR scans and applied the new technique to an enlarged data set. Important differences between the current and the preliminary studies, in addition to the difference of the segmentation technique and data set size, included the volume change assessment (PVR) and the intraobserver variability estimation. In a comparison of our previous technique to the current technique using the entire data set of 25 pre- and postchemotherapy scans, the results indicated that the current technique achieved higher overlap ratios on average than the previous technique for both the pre- and postchemotherapy scans.
One advantage of using breast DCE-MR scans for mass detection, diagnosis, and staging is that they provide both mass morphology (3D spatial structure) and mass pharmacokinetics (1D temporal intensity enhancement curve) information. The method developed in this study takes advantage of both morphology and pharmacokinetics of a breast mass for segmentation using FCM clustering and the LS method. The FCM clustering was designed to group voxels in a VOI in terms of their pharmacokinetic properties.17 It results in a 3D membership function that can effectively identify voxels that are likely to belong to the malignant mass. The FCM clustering output may contain multiple objects with jagged boundaries, so morphological filtering is applied to the clustering results. The LS method then refines the FCM-based initial segmentation by incorporating 3D mass intensity, mass spatial constraints, and its FCM membership function. The mass intensity constraint helps the zero LS evolve to the desired mass surface, the spatial constraints helps the zero LS maintain its smoothness, and the FCM membership function helps improve segmentation accuracy. Because the boundaries may be indistinct for many breast masses in DCE-MR scans, the zero LS may evolve into background tissue structure. We designed a method to reduce segmentation leak by adapting a region growing technique that uses a homogeneity constraint to decide whether to add a voxel to the existing mass region.
DCE-MR scans include 4D information, i.e., three spatial dimensions and one temporal dimension. It is thus plausible to model a 4D LS method for mass segmentation. Because the mass shape in spatial domain remains mostly the same for all MR time points within the 4D scan, this study applied the 3D LS method to the postcontrast VOI that was estimated to contain the most enhanced mass. Implementing the LS method in 3D was computationally more efficient than in 4D. The temporal information of DCE-MR scans was exploited partially by FCM-based clustering, where the pharmacokinetic curves were summarized by the voxelwise standard deviation of enhancement over time. Further investigation is warranted to explore other methods that could effectively and efficiently exploit the 4D information.
Manual segmentation may introduce inter- and intraobserver variabilities.38 To evaluate intraobserver variability, the same radiologist was asked to segment each mass on each slice of the VOI twice, in two reading sessions separated by more than 1 year. Statistical comparison (Table 1) shows that the intraobserver variability in this study did not reach statistical significance. Further investigation might compare computer segmentation to manual segmentation by multiple radiologists (interobserver variability) for breast mass volume estimation on DCE-MR scans.
Performance analysis of computer segmentation algorithms in medical imaging is difficult because an objective ground truth may not exist in most applications. In this investigation, we used an expert radiologist’s manual segmentation as the reference standard. As discussed above, to investigate whether the computer segmentation performance in this study may be generalized to a population of readers, it would be necessary to compare the computer segmentation to those from a group of experts and test if the computer segmentation may be acceptable as a substitute for the experts. Some previous studies estimated the probability that each voxel belongs to the lesion of interest based on the expert readings using methods such as p-map (Ref. 38) or STAPLE.39 When multireader data are available, it would be of interest to use these probabilistic methods to evaluate the computer segmentation. Other areas that warrant further investigations include comparison with other segmentation methods, experiments with larger data sets, assessment of segmentation stability with respect to VOI selection, and design of a user-friendly interface to reduce operator time.
CONCLUSION
The segmentation method developed in this study may have potential to assist physicians in the assessment of volume changes and may serve as an initial step in computerized image analysis methods for predicting breast cancer response to neoadjuvant chemotherapy. Our automated mass segmentation method achieved a promising performance for prechemotherapy volumes as measured by the overlap between the automated and manual segmentations. The segmentation performance on postchemotherapy volumes was lower likely due to the smaller size or the fragmented appearance of the masses after chemotherapy. The percentage volume reduction estimated from automated segmentation was in good agreement with that from radiologist’s manual segmentation. Further work is underway to improve the segmentation performance, especially for the postchemotherapy DCE-MR scans, and to validate the system performance on a larger data set.
ACKNOWLEDGMENT
This work was supported in part by USPHS Grant Nos. CA120234, CA118305, and CA95153.
References
- Fisher B. et al. , “Effect of preoperative chemotherapy on the outcome of women with operable breast cancer,” J. Clin. Oncol. 16, 2672–2685 (1998). [DOI] [PubMed] [Google Scholar]
- Newman L. A., Buzdar A. U., Singletary S. E., Kuerer H. M., Buchholz T., Ames F. C., Ross M. I., and Hunt K. K., “A prospective trial of preoperative chemotherapy in resectable breast cancer: Predictors of breast-conservation therapy feasibility,” Ann. Surg. Oncol. 9, 228–234 (2002). [DOI] [PubMed] [Google Scholar]
- Esserman L., Kaplan E., Partridge S., Tripathy D., Rugo H., Park J., Hwang S., Kuerer H., Sudilovsky D., Lu Y., and Hylton N., “MRI phenotype is associated with response to doxorubicin and cyclophosphamide neoadjuvant chemotherapy in stage III breast cancer,” Ann. Surg. Oncol. 8, 549–559 (2001). 10.1007/s10434-001-0549-8 [DOI] [PubMed] [Google Scholar]
- Broet P., Scholl S. M., de la Rochefordiere A., Fourquet A., Moreau T., De Rycke Y., Asselain B., and Pouillart P., “Short and long-term effects on survival in breast cancer patients treated by primary chemotherapy: An updated analysis of a randomized trial,” Breast Cancer Res. Treat. 58, 151–156 (1999). 10.1023/A:1006339918798 [DOI] [PubMed] [Google Scholar]
- van Nes J. G. H., Putter H., Julien J. P., Tubiana-Hulin M., van de Vijver M., Bogaerts J., de Vos M., and van de Velde C. J. H., “Preoperative chemotherapy is safe in early breast cancer, even after 10 years of follow-up; clinical and translational results from the EORTC trial 10902,” Breast Cancer Res. Treat. 115, 101–113 (2009). 10.1007/s10549-008-0050-1 [DOI] [PubMed] [Google Scholar]
- van der Hage J. H., van de Velde C. C., and Mieog S. J. S. D., “Preoperative chemotherapy for women with operable breast cancer,” Cochrane Database of Systematic Reviews, Issue 2 (2007). [DOI] [PMC free article] [PubMed]
- Cleator S. J., Makris A., Ashley S. E., Lal R., and Powles T. J., “Good clinical response of breast cancers to neoa uvant chemoendocrine therapy is associated with improved overall survival,” Ann. Oncol. 16, 267–272 (2005). 10.1093/annonc/mdi049 [DOI] [PubMed] [Google Scholar]
- Yu H. J., Chen J. H., Mehta R. S., Nalcioglu O., and Su M. Y., “MRI measurements of tumor size and pharmacokinetic parameters as early predictors of response in breast cancer patients undergoing neoadjuvant anthracycline chemotherapy,” J. Magn. Reson. Imaging 26, 615–623 (2007). 10.1002/jmri.21060 [DOI] [PubMed] [Google Scholar]
- Makris A., Powles T. J., Ashley S. E., Chang J., Hickish T., Tidy V. A., Nash A. G., and Ford H. T., “A reduction in the requirements for mastectomy in a randomized trial of neoadjuvant chemoendocrine therapy in primary breast cancer,” Ann. Oncol. 9, 1179–1184 (1998). 10.1023/A:1008400706949 [DOI] [PubMed] [Google Scholar]
- Rosen E. L., Blackwell K. L., Baker J. A., Soo M. S., Bentley R. C., Yu D. H., Samulski T. V., and Dewhirst M. W., “Accuracy of MRI in the detection of residual breast cancer after neoadjuvant chemotherapy,” AJR, Am. J. Roentgenol. 181, 1275–1282 (2003). [DOI] [PubMed] [Google Scholar]
- Yeh E., Slanetz P., Kopans D. B., Rafferty E., Georgian-Smith D., Moy L., Halpern E., Moore R., Kuter I., and Taghian A., “Prospective comparison of mammography, sonography, and MRI in patients undergoing neoadjuvant chemotherapy for palpable breast cancer,” AJR, Am. J. Roentgenol. 184, 868–877 (2005). [DOI] [PubMed] [Google Scholar]
- “WHO handbook for reporting results of cancer treatment,” World Health Organization Offset Publication No. 48, World Health Organization, Geneva, Switzerland, 1979.
- Therasse P., Arbuck S. G., Eisenhauer E. A., Wanders J., Kaplan R. S., Rubinstein L., Verweij J., Van Glabbeke M., van Oosterom A. T., Christian M. C., and Gwyther S. G., “New guidelines to evaluate the response to treatment in solid tumors,” J. Natl. Cancer Inst. 92, 205–216 (2000). 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- Alderliesten T., Schlief A., Peterse J., Loo C., Teertstra H., Muller S., and Gilhuijs K., “Validation of semiautomatic measurement of the extent of breast tumors using contrast-enhanced magnetic resonance imaging,” Invest. Radiol. 42, 42–49 (2007). 10.1097/01.rli.0000248849.99014.7e [DOI] [PubMed] [Google Scholar]
- Partridge S. C., Gibbs J. E., Lu Y., Esserman L. J., Tripathy D., Wolverton D. S., Rugo H. S., Hwang G. E., Ewing C. A., and Hylton N. M., “MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival,” AJR, Am. J. Roentgenol. 184, 1774–1781 (2005). [DOI] [PubMed] [Google Scholar]
- Lucas-Quesada F. A., Sinha U., and Sinha S., “Segmentation strategies for breast tumors from dynamic MR images,” J. Magn. Reson. Imaging 6, 753–763 (1996). 10.1002/jmri.1880060508 [DOI] [PubMed] [Google Scholar]
- Chen W. J., Giger M. L., and Bick U., “A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images,” Acad. Radiol. 13, 63–72 (2006). 10.1016/j.acra.2005.08.035 [DOI] [PubMed] [Google Scholar]
- Szabo B. K., Aspelin P., and Wiberg M. K., “Neural network approach to the segmentation and classification of dynamic magnetic resonance images of the breast: Comparison with empiric and quantitative kinetic parameters,” Acad. Radiol. 11, 1344–1354 (2004). 10.1016/j.acra.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Wu Q., Salganicoff M., Krishnan A., Fussell D. S., and Markey M. K., “Interactive lesion segmentation on dynamic contrast enhanced breast MRI using a Markov model,” Proc. SPIE 6144, 4M1–4M8 (2006). [Google Scholar]
- Bulow T., Meinel L. A., Wiemker R., Kose U., Shimauchi A., and Newstead G. M., “Segmentation of suspicious lesions in dynamic, contrast-enhanced breast MR images,” Proc. SPIE 6514, 0T1–0T12 (2007). [Google Scholar]
- Joe B. N., Vahidi K., Gibbs J., Lu Y., Proctor E., and Hylton N., “Automated breast MR tumor volume measurements for prediction of outcome in breast cancer patients on neoadjuvant chemotherapy,” RSNA 2006 (Radiological Society of North America, Chicago, IL, 2006), pp. 651–652.
- Shi J., Sahiner B., Chan H. P., Ge J., Hadjiiski L., Helvie M. A., Nees A., Wu Y. T., Wei J., Zhou C., Zhang Y., and Cui J., “Characterization of mammographic masses based on level set segmentation with new image features and patient information,” Med. Phys. 35, 280–290 (2008). 10.1118/1.2820630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Sahiner B., Chan H. -P., Paramagul C., Hadjiiski L. M., Helvie M. A., Wu Y. -T., Ge J., Zhang Y., Zhou C., and Wei J., “Breast segmentation on dynamic contrast-enhanced magnetic resonance scans using the level set method,” Proc. SPIE 6915, 2A1–2A7 (2008). [Google Scholar]
- Schott A. F., Roubidoux M. A., Helvie M. A., Hayes D. F., Kleer C. G., Newman L. A., Pierce L. J., Griffith K. A., Murray S., Hunt K. A., Paramagul C., and Baker L. H., “Clinical and radiologic assessments to predict breast cancer pathologic complete response to neoadjuvant chemotherapy,” Breast Cancer Res. Treat. 92, 231–238 (2005). 10.1007/s10549-005-2510-1 [DOI] [PubMed] [Google Scholar]
- Kuhl C., “The current status of breast MR imaging—Part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice,” Radiology 244, 356–378 (2007). 10.1148/radiol.2442051620 [DOI] [PubMed] [Google Scholar]
- Kuhl C. K., “Current status of breast MR imaging—Part 2. Clinical applications,” Radiology 244, 672–691 (2007). 10.1148/radiol.2443051661 [DOI] [PubMed] [Google Scholar]
- Orel S. G. and Schnall M. D., “MR imaging of the breast for the detection, diagnosis, and staging of breast cancer,” Radiology 220, 13–30 (2001). [DOI] [PubMed] [Google Scholar]
- Padhani A. R., Hayes C., Assersohn L., Powles T., Makris A., Suckling J., Leach M. O., and Husband J. E., “Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: Initial clinical results,” Radiology 239, 361–374 (2006). 10.1148/radiol.2392021099 [DOI] [PubMed] [Google Scholar]
- Williams T. C., DeMartini W. B., Partridge S. C., Peacock S., and Lehman C. D., “Breast MR imaging: Computer-aided evaluation program for discriminating benign from malignant lesions,” Radiology 244, 94–103 (2007). 10.1148/radiol.2441060634 [DOI] [PubMed] [Google Scholar]
- Armitage P., Behrenbruch C., Brady M., and Moore N., “Extracting and visualizing physiological parameters using dynamic contrast-enhanced magnetic resonance imaging of the breast,” Med. Image Anal. 9, 315–329 (2005). 10.1016/j.media.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Heilmann M., Kiessling F., Enderlin M., and Schad L. R., “Determination of pharmacokinetic parameters in DCE MRI: Consequence of nonlinearity between contrast agent concentration and signal Intensity,” Invest. Radiol. 41, 536–543 (2006). 10.1097/01.rli.0000209607.99200.53 [DOI] [PubMed] [Google Scholar]
- Lopata R. G. P., Backes W. H., van den Bosch P. P. J., and van Riel N. A. W., “On the identifiability of pharmacokinetic parameters in dynamic contrast-enhanced imaging,” Magn. Reson. Med. 58, 425–429 (2007). 10.1002/mrm.21336 [DOI] [PubMed] [Google Scholar]
- Bezdek J. C., Pattern Recognition with Fuzzy Objective Function Algorithms (Kluwer Academic, Dordrecht, 1981). [Google Scholar]
- Masulli F. and Schenone A., “A fuzzy clustering based segmentation system as support to diagnosis in medical imaging,” Artif. Intell. Med. 16, 129–147 (1999). 10.1016/S0933-3657(98)00069-4 [DOI] [PubMed] [Google Scholar]
- Osher S. J. and Fedkiw R. P., Level Set Methods and Dynamic Implicit Surfaces (Springer, New York, 2003). [Google Scholar]
- Suri J. S., Liu K. C., Singh S., Laxminarayan S. N., Zeng X. L., and Reden L., “Shape recovery algorithms using level sets in 2-D/3-D medical imagery: A state-of-the-art review,” IEEE Trans. Inf. Technol. Biomed. 6, 8–28 (2002). 10.1109/4233.992158 [DOI] [PubMed] [Google Scholar]
- Sethian J. A., Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision and Materials Sciences (Cambridge University Press, Cambridge, 1996). [Google Scholar]
- Meyer C. R., Johnson T. D., McLennan G., Aberle D. R., Kazerooni E. A., MacMahon H., Mullan B. F., Yankelevitz D. F., van Beek E. J. R., and S. G.ArmatoIII, “Evaluation of lung MDCT nodule annotation across radiologists and methods,” Acad. Radiol. 13, 1254–1265 (2006). 10.1016/j.acra.2006.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield S. K., Zou K. H., and Wells W. M., “Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation,” IEEE Trans. Med. Imaging 23, 903–921 (2004). 10.1109/TMI.2004.828354 [DOI] [PMC free article] [PubMed] [Google Scholar]