Abstract
Regulatory T cells play important roles in the control of autoimmunity and maintenance of transplantation tolerance. Foxp3, a member of the forkhead/winged-helix family of transcription factors, acts as the master regulator for regulatory T-cell (Treg) development and function. Mutation of the Foxp3 gene causes the scurfy phenotype in mouse and IPEX syndrome (immune dysfunction, polyendocrinopathy, enteropathy, X-linked syndrome) in humans. Epigenetics is defined by regulation of gene expression without altering nucleotide sequence in the genome. Several epigenetic markers, such as histone acetylation and methylation, and cytosine residue methylation in CpG dinucleotides, have been reported at the Foxp3 locus. In particular, CpG dinucleotides at the Foxp3 locus are methylated in naive CD4+CD25− T cells, activated CD4+ T cells, and TGF-β–induced adaptive Tregs, whereas they are completely demethylated in natural Tregs. The DNA methyltransferases DNMT1 and DNMT3b are associated with the Foxp3 locus in CD4+ T cells. Methylation of CpG residues represses Foxp3 expression, whereas complete demethylation is required for stable Foxp3 expression. In this review, we discuss how different cis-regulatory elements at the Foxp3 locus are subjected to epigenetic modification in different subsets of CD4+ T cells and regulate Foxp3 expression, and how these mechanisms can be exploited to generate efficiently large numbers of suppressive Tregs for therapeutic purposes.
Introduction
Foxp3, a member of forkhead/winged-helix family of transcription factors acts as a “master” regulator for the development and suppressive function of regulatory T cells (Tregs). Its constitutive expression is necessary for the suppressive function of Tregs, and mutation or deficiency of Foxp3 leads to development of autoimmune diseases.1 In thymus, a subset of CD4+CD25+ T cells develop into Foxp3+CD4+ T cells known as natural regulatory Tregs (nTregs). However, in peripheral lymphoid organs, transforming growth factor-β (TGF-β) induces naive CD4+CD25− T cells to develop into Foxp3+CD4+ T cells known as adaptive or induced Tregs. In vitro culture of peripheral naive CD4+CD25− T cells in the presence of TGF-β induces expression of Foxp3 and provides a method to generate Foxp3+ Tregs ex vivo.2 However, nTregs and TGF-β–induced Tregs have different functional characteristics.3 Recent studies suggest that nTregs are more stable compared with TGF-β–induced Tregs, and this may be related to the epigenetic regulation of Foxp3.4–6
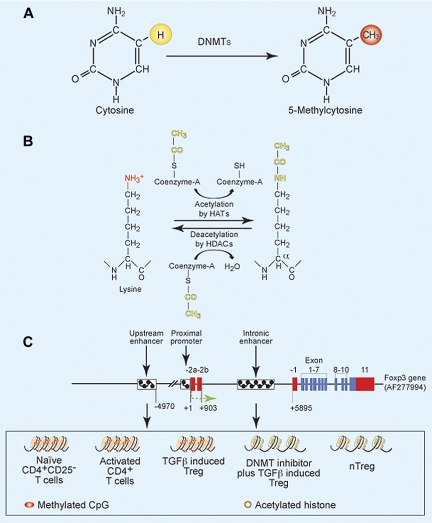
Epigenetics is defined by heritable changes in gene expression without changing the DNA sequence of the genome. These gene modifications can take place either in the chromosomal DNA or proteins that are directly linked with the chromosomal DNA. Epigenetic functional units involve either methylation of CpG residues (5′ position of cytosine of CpG) or covalent posttranslational modification of histones (Figure 1A-B). In mammals, approximately 60% of CpG dinucleotides are methylated,7 and DNA methyltransferases (DNMTs) control DNA methylation of CpG residues and maintenance of methylation during cell differentiation. There are 3 different DNMTs known: DNMT1, DNMT3a, and DNMT3b.7 DNA methylation can be divided in 2 types: de novo and maintenance methylation. DNMT3a and DNMT3b are involved in the de novo introduction of methyl group on cytosine residues, whereas DNMT1 is associated with replication machinery to maintain methylation in dividing cells.8 However, it has been reported that, in some CpG dense regions, DNMT1 is not sufficient to maintain the levels of methylated CpG during the replication; and in such somatic cells, the activity of DNMT3a and DNMT3b plays important roles in maintaining methylation.9 Methylated CpG recruits transcriptional repressors, such as the methyl-binding proteins MBD and MeCP, and histone deacetylases (HDACs). The HDACs introduce positive charges in the histone tail, which bind tightly to negatively charged nucleic acids and lead to the formation of closely compact nucleosomes that prevent transcription.
Figure 1.
Methylation and acetylation of DNA and histones, and gene structure of Foxp3. (A) Methylation of 5-position of cytosine by DNMTs. (B) Acetylation and deacetylation of lysine residues in histone molecules. (C) Gene structure of Foxp3 in different CD4+ T-cell subsets. Red bars represent noncoding exons; blue bars, coding exons; small boxes filled with black checks, cis-regulatory elements; +1, transcription start site. Upstream and intronic enhancers in naive T cells, activated T cells, and TGF-β–induced Tregs have methylated CpG (pink) and are transcriptionally inactive (marked as compact histone bundles), whereas these regions have unmethylated CpG (yellow) and are transcriptionally active (marked as separated histone bundles) in nTregs and DNMT inhibitor plus TGF-β–induced Tregs.
Core histone molecules are organized in octamers, consisting of 2 each of H2A, H2B, H3, and H4. The octamers cover approximately 140 DNA bp and form a complex structure called the nucleosome.10 Specific amino acids of histone tails are targets for several posttranslational modifications, including acetylation, phosphorylation, poly-ADP ribosylation, ubiquitination, and methylation.11 Acetylation of histones in the nucleosome increases their net negative charge, thereby interrupting their interaction with DNA and leading to open chromatin structure of negatively charged DNA (Figure 1B). Thus, open chromatin structure is permissive for recruitment of transcriptional machinery to initiate gene transcription and expression.
Several human diseases are associated with alterations in genome architecture, and genome-wide analysis suggests that epigenetic dysregulation contributes to the severity of these diseases.12 For example, the T-cell genome of systemic lupus erythematosus (SLE) patients is hypomethylated, resulting in overexpression of integrins, such as LFA-1 (CD11a/CD18).13,14 Demethylating drugs can induce overexpression of these same molecules and induce both autoreactive T cells and an SLE syndrome in mice.15 Demethylation of regulatory elements of the promoter also leads to overexpression of perforin in CD4+ T cells and cytotoxic activity in SLE.16 DNA methylation patterns of promoter regions are also linked with overexpression of CD70, CD3ζ chain, CD40L, CTLA4, ITGAL, interleukin-2 (IL-2), IL-4, IL-10, and interferon-γ (IFN-γ) in CD4+ T cells.17–23
Conditional DNMT1−/− mice, where DNA methylation is ablated from the CD4+CD8+ thymocyte stage onwards, show increased production of IFN-γ, IL-2, IL-3, and IL-4 in peripheral T cells after activation compared with littermate-matched controls.24 MBD2−/− mice, which lack another regulatory protein important for maintaining methylation and transcriptional repression, show ectopic expression of IL-4, which is sensitive to DNA methylation under Th1-polarized conditions.25,26 In contrast to naive T cells, effector T cells and Tregs express receptors for inflammatory chemokines and ligands for E- and P-selectin, which help in their recruitment into inflamed tissues.27–29 Treatment of naive CD4+ T cells with 5-aza-2′-deoxycytidine (Aza), which inhibits DNMTs, leads to expression of P-selectin ligand, suggesting that ligand expression is also controlled by DNA methylation.30
Structure of the Foxp3 gene
In Tregs, the Foxp3 gene acts as the master regulator for the development of Tregs, and its constitutive expression is required for Treg-suppressive function.1 Genetic defects in Foxp3 cause the scurfy phenotype in mice and IPEX syndrome (immune dysfunction, polyendocrinopathy, enteropathy, X-linked syndrome) in humans.1,31 The Foxp3 gene possesses 11 coding and 3 noncoding exons.32 The 2 extreme 5′-noncoding exons (−2a and −2b) are separated by 640 bp, and these 2 exons are spliced to a second common noncoding exon (−1). The −2b and −1 exons are separated by approximately 5000 bp and possess several regulatory cis-elements (Figure 1C). It has been noted that a 2-bp (amino acid [AA]) insertion in exon 8 leads to scurfy in mice.32 Sequencing of a large cohort of IPEX persons shows that 60% of patients have missense mutations mainly in exons 9, 10, and 11 (which together form a forkhead domain), and other mutations are distributed throughout the gene.1
Foxp3 protein is highly conserved in bovine, canine, feline, murine, macaque, and humans.33 Indeed, the Foxp3 protein sequences of humans (gene number NP_054728) and mouse (gene number NP_473380) have 86% identity and 91% similarity in their amino acids. Western blot analysis shows that human cells express 2 Foxp3 isoforms. The upper band is similar to the mouse Foxp3, whereas the lower band is unique to humans and lacks exon 2 (amino acids 71-105), which is part of the repressor domain in the Foxp3 protein. This region interacts and represses the function of retinoic acid–related orphan receptor-α (ROR-α)34 and ROR-γt.35 Expression of Foxp3 Δexon2 in human CD4+CD25−Foxp3− T cells leads to more IL-2 secretion and proliferation in response to T-cell receptor (TCR) stimulation compared with full-length Foxp3. It has been proposed that Foxp3 Δexon2 acts as a dominant- negative mutant Foxp3.1 Human Tregs also express a third isoform, which lacks both exon 2 and exon 7 (amino acids 239-260).36 Exon 7 encodes for a leucine zipper motif that acts as a dimerization structural element. However, it has been reported that the absence of exon 7 in natural Foxp3Δ2Δ7 does not affect dimerization but abrogates the suppressive function of Tregs.37
Regulation of Foxp3 expression by trans-acting transcription factors
Immediately upstream to the Foxp3 transcriptional start site (−511 to +176) is a region that possesses several important transcription factor-binding sites (AP-1, NFAT) with features indicative of eukaryotic promoters, including TATA, GC, and CAAT boxes.38 This region acts as a promoter for Foxp3 transcription and is referred to as the proximal promoter region.38 Other regulatory cis-elements are present between noncoding exons (−2b and −1) or far upstream of the transcriptional start site (∼ 5 kb), and these elements are referred to as the intronic enhancers or upstream enhancers, respectively (Figure 1C). Different extracellular stimuli and intracellular signaling molecules control the development and function of Tregs by regulating the transcription of Foxp3. For example, IL-2 signaling has an important role in the development and function of Tregs, and IL-2 receptor signaling is generally dependent on STAT5.39–41 Burchill et al identified 11 STAT consensus sites in Foxp3 (3 at the proximal promoter, 8 between intronic regions), yet only the proximal promoter conserved region shows binding with STAT5 and is dependent on IL-2Rβ signaling.42 Recently, it has been shown that phosphorylation of STAT1 in Th1 cells by IL-27 and IFN-γ leads to Foxp3 expression, and this is the result of the direct interaction of phosphorylated STAT1 with the Foxp3 proximal promoter in human CD4+ T cells.43 Conversely, IL-27 also inhibits Foxp3 expression in murine CD4+ T cells in a STAT3-dependent manner,44 and IL-27 induced murine CD4+ T cells to express IL-10 but not Foxp3.45 These studies suggest that IL-27 is dispensable for Treg generation and that STAT5 but not other STATs are required for Foxp3 expression.
Small guanosine triphosphate-binding protein superfamily molecules, such as N-Ras or K-Ras, are involved in the control of cell differentiation, proliferation, and apoptosis.46 Mor et al showed that inhibition of N- or K-Ras signaling in CD4+ T cells leads to induction of NFAT and Foxp3 expression and enhanced Treg-suppressive function.47 However, the mechanism of Ras signaling to regulate Foxp3 expression is not clearly defined.
In Th1, the IFN-γ–induced protein interferon regulatory factor 1 binds to the Foxp3 proximal promoter and inhibits Foxp3 expression.48 In Th2, IL-4 inhibits Foxp3 expression in peripheral naive CD4+CD25− T cells2 by stimulating phosphorylation of STAT6, which binds between exons −2b and −1 (+2459 to +2866 bp) and inhibits TGF-β–induced Foxp3 expression.2,49 TGF-β and IL-4 signaling together induce IL-9 secretion, leading to the newly identified IL-9+IL-10+Foxp3− (Th9) subset.50
The vitamin A metabolite retinoic acid (RA) inhibits Th17 cells and induces de novo generation of Foxp3+ Tregs.51–55 RA induction of Tregs from naive CD4 T cells may be the result of enhanced TGF-β–driven phosphorylation of SMAD3 along with the inhibition of IL-6 and IL-23 receptor expression.56 However, RA also enhances TGF-β–induced conversion of naive CD4+ T cells to Tregs, by inhibiting the CD4+CD44+ memory T cells that produce Foxp3 inhibitory cytokines IL-4, IL-21, and IFN-γ.57 Kang et al showed that all-trans-retinoic-acid induces histone H4 acetylation at the Foxp3 locus and Foxp3 expression in naive CD4+CD25− T cells.51 RA induces expression of CCR9 and CD103 (αE subunit of αEβ7 integrin) on Tregs, and these Tregs showed increased migration to gut-homing chemokine CCL25.51,53 Benson at al reported that, in the presence of TGF-β, CD80/86 knockout dendritic cells (DCs) induce increased Foxp3 expression compared with wild-type DCs.54 The reduced Foxp3 expression with wild-type DCs can be overcome with the addition of RA to culture.54 Furthermore, Coombes et al showed that mucosal CD103+ DCs are responsible for TGF-β and RA-induced Foxp3 expression.55 CD103+ DCs express high levels of retinal dehydrogenase (aldh 1a2) compared with CD103− DCs, which is required for the conversion of retinal to RA.55 CD103− DCs after stimulation with lipopolysaccharide or anti-CD40 produce large amounts of proinflammatory cytokines compared with CD103+ DCs.55 This suggests that RA together with cytokines and T-cell activation signals provided by different subsets of DCs regulate Foxp3 expression in CD4 T cells.
Phosphatidylinositol 3-kinase (PI3K) is induced by TCR and CD28 signaling and is required for cell-cycle progression, cell survival, and proliferation. PI3K signaling activates the Akt-mTOR pathway, Akt signaling interferes with Foxp3 expression, and Foxp3+ Tregs show reduced Akt phosphorylation.58 CD28 signals can interfere with adaptive Treg differentiation by TGF-β,2,54 whereas TCR plus CD28 signals increase Foxp3 enhancer activity and Foxp3 expression.59 Rapamycin, a chemotherapeutic drug, targets Akt-mTOR signaling and rapamycin-induced differentiation of Tregs and has been used to induce tolerance.58 Negative regulator molecules, such as SHIP, a lipid phosphatase, hydrolyze phosphatidylinositol 3,4,5-triphosphate, a second messenger of the PI3K pathway, which in turn inhibits Akt signaling, leading to enhanced differentiation of Foxp3+ Tregs.60 Sphingosine 1-phosphate (S1P) is a natural lysophospholipid, which signals through 5 known G protein–coupled receptors (S1P1-5) and is required for migration of immune cells.28 FTY720, an antagonist, acts as an immunosuppressant by sequestering T cells in lymphoid organs. We showed that receptor S1P1 causes tissue retention by inhibiting the entry of peripheral T cells to afferent lymphatics.28 Recently, Liu et al showed that the S1P1 receptor activates Akt-mTOR kinase signaling, thereby inhibiting the development and suppressive function of Foxp3+ Tregs.61 Sauer et al showed that PI3K/Akt/mTOR signaling antagonizes Foxp3 expression by differential modification of histone 3 (H3K4 dimethylation and trimethylation) at the Foxp3 locus, and continuous TCR stimulation leads to decreased H3K4 dimethylation and trimethylation, and reduced Foxp3 expression.62
Together, these finding demonstrate that there are several diverse intrinsic and extrinsic signals that regulate of Foxp3 expression and Treg function, yet detailed molecular mechanisms require further investigation.
Role of CpG DNA methylation in Treg development and function
Epigenetic regulation by CpG methylation at specific sites in T cells controls the differentiation of T helper cells.24,63,64 Different regulatory cis-elements are present in the Foxp3 locus. There are regulatory elements present upstream of the transcriptional start site (−600 to −1 bp) at the proximal promoter (Figure 1C). Apart from trans-acting factors binding to the proximal promoter, the methylation status of the CpG residues in the proximal promoter region has an essential role in Foxp3 expression. Zorn et al reported that demethylation induced by Aza in human NK cells leads to Foxp3 expression.39 Subsequently, Kim and Leonard reported that 10% to 45% of the CpG sites in the Foxp3 proximal promoter (−250 to +1) are methylated in naive CD4+CD25− T cells, whereas all were demethylated in nTregs,65 and TGF-β induces demethylation of CpG at this site in CD4+CD25− T cells. Janson et al showed that this region is approximately 70% methylated in CD4+CD25lo cells compared with approximately 5% in CD4+CD25hi T cells in humans.66 These studies demonstrate that methylation of the proximal promoter is an important regulator of Foxp3 expression.
There are also regulatory cis-elements present in between noncoding exons (−2b and −1) that act as enhancers and are defined as intronic enhancers (Figure 1C). This intronic region of Foxp3 is highly conserved and is responsible for the regulation of Foxp3 expression. It has been shown that CpG residues in this intronic region (+4201 to +4500) are completely methylated in naive CD4+CD25− T cells and fully demethylated in nTregs in mice5,65 and in human.67 Independent studies showed that this region has different levels of demethylation after TGF-β stimulation in mouse5,6,65 and human.67 The first intronic CpG containing region (+4393 to +4506 bp, conserved noncoding sequence 3) has decreased methylation of CpG residues after TGF-β signaling, and after TCR signaling has increased binding to the cyclic-AMP response element–binding protein/activating transcription factor, leading to increased Foxp3 expression.65 Another evolutionary conserved intronic region between exons −2b and −1 (+2177 to +2198, conserved noncoding sequence 2) binds SMAD3 and NFAT and possesses enhancer activity59 (Figure 1C). Because the SMAD3 and NFAT binding sites (CNS2) and the CpG methylation site (conserved noncoding sequence 3) are present in between the same noncoding exons and do not overlap with each other, this suggests that these cis-elements may be responsible for the regulation of Foxp3 as a result of different extracellular signaling environments. The interaction of these 2 elements, the signals that regulate them, and the interaction of these signals are areas that require further investigation.
We have shown that there is a CpG island approximately 5 kb upstream of the transcriptional start site, and we refer it as the upstream enhancer. The upstream enhancer (−5786 to −5558 bp) is methylated in naive CD4+CD25− T cells, activated CD4+ T cells, and TGF-β–induced Foxp3+ Tregs, but is demethylated in nTregs4 (Figure 1C). This region functions as an enhancer. In nTregs, but not TGF-β induced Tregs, this CpG island has acetylated histone 3 indicative of a transcriptionally active site and interacts with the transcription factors Sp1 and TGF-β–induced early 1 product (TIEG1). Conversely, this region is bound by the repressors DNMT1, DNMT3b, MeCP2, and MBD2 in naive CD4+CD25− T cells, activated CD4+ T cells, and TGF-β induced CD4+Foxp3+ T cells.4 Culture of CD4+CD25− T cells with Aza demethylates the upstream enhancer, leading to positive interactions with transcription factors and markedly increased Foxp3 mRNA and protein expression, and acquired suppressor activity.
TGF-β induces Foxp3 expression in peripheral naive CD4+CD25− T cells2; however, its activities are very complex in this regard. In addition to TGF-β receptor–induced SMAD3 signaling for Treg generation,59 TGF-β signaling may also act via TIEG1 and E3 ubiquitin ligase itch in a ubiquitin-dependent pathway.68 Venuprasad et al showed that itch−/− and TIEG1−/− naive CD4+ T cells have less Foxp3 induction after TGF-β stimulation, and these Tregs do not protect from antigen-induced airway inflammation.68 TGF-β also inhibits the phosphorylation of ERK leading to inhibition of DNMT expression69; and inhibition of DNMT with siRNA or DNMT inhibitors leads to Foxp3 expression in CD4+ T cells,4,6,65,70 suggesting that inhibition of DNMT activity plays an important role in Foxp3 expression. These CpG methylation epigenetic markers have been used to screen suppressive Tregs derived from the tissues of cancerous or noncancerous patients.71,72
The inflammatory cytokine IL-6 suppresses the development and function of Tregs.73–75 IL-6 induces DNMT1 expression and enhances its activity.76,77 IL-6 induces STAT3-dependent methylation of the upstream Foxp3 enhancer by DNMT1 in nTregs, leading to repression of Foxp3.4 Preactivated CD4+CD25− T cells or CD4+CD25−CD44hi memory T cells express very little Foxp3 after TGF-β stimulation.4,78 This is probably the result of high levels of DNMT1 activity in these cells77,79 because inhibition of DNMT with Aza or deficiency of DNMT1 in T cells leads to Foxp3 expression,80 suggesting that regulation of Foxp3 is tightly controlled by epigenetic modification in activated CD4+ T cells.
Together, these reports demonstrate that Foxp3 is regulated by complex mechanisms where many extracellular signals control transacting factors as well as chromatin remodeling through covalent modification of CpG DNA. Thus, TGF-β along with inflammatory cytokines has a different effect than TGF-β alone on the development of Tregs. Understanding these signals and their cumulative intracellular effect on Foxp3 cis-elements at different T-cell developmental stages will be key for manipulating T-cell responses therapeutically (Figure 2).
Figure 2.
Foxp3 regulation from the various extracellular signals. Red arrow represents negative signal; and green arrow, positive signals for Foxp3 expression.
Role of HDACs in Treg development and function
Acetylated histone is a marker for open chromatin structure. Acetylation of core histone molecules is catalyzed by histone acetyltransferase, and acetyl groups are removed by HDACs. Histone acetyltransferase acetylates the conserved ϵ-amino group of the lysine residue at the amino-terminus of the histone tail (Figure 1B). This decreases the overall positive charge, thereby decreasing histone affinity for negatively charged DNA, and providing a platform for the binding of transcription factors to the chromatin template.81 According to size, subcellular expression, number of enzymatic domains, and structure, HDACs are divided into 4 classes. Class I HDACs (HDACs 1, 2, 3, and 8) are detected in the nucleus and ubiquitously expressed in different tissues and cell lines. Class II HDACs (HDACs 4, 5, 6, 7, 9, and 10) shuttle between the nucleus and cytoplasm and are expressed in a tissue-specific manner. For example, human HDAC4 is more abundant in skeletal muscle, brain, heart, and ovary, but not detectable in liver, lung, spleen, and placenta. HDAC5 is expressed in mouse skeletal muscle, liver, and brain, but not in spleen.82 Class III HDACs are composed of NAD+-dependent deacetylases SIRT1 to SIRT7. Class III HDACs are structurally unrelated to class I and class II HDACs. They have a unique enzymatic mechanism of action that requires the cofactor NAD+ for their activity. HDAC11 is the only member of class IV,83 and its classification is still under debate. Phylogenetic analysis shows that HDAC11 is closely related to HDAC 3 and HDAC8, suggesting that it might be closer to class I than class II. After TCR stimulation, murine CD4+CD25− naive T cells and CD4+CD25+ nTregs do not show significant differences in the level of class I HDACs, whereas class II HDACs are mainly expressed in Tregs.84 Little else is known about the function and distribution of class II HDACs in nTregs, suggesting an area for productive research.
The N-terminal region of the Foxp3 protein is proline rich, which makes Foxp3 different from the other family members Foxp1, Foxp2, and Foxp4, and this region of Foxp3 recruits the corepressor lysine acetyltransferase TIP60. TIP60 acetylates the Foxp3 protein, and in turn acetylated Foxp3 protein binds to its target gene promoter, such as IL-2, to repress its transcription. The N-terminal region of Foxp3 can also recruit the class II deacetylase HDAC7.85 During T-cell activation, HDAC7 is recruited to the Foxp3 corepressor complex, deacetylates the Foxp3 protein, and thus inhibits Foxp3 function. It is important to note that HDAC7 can also deacetylate histones in the Foxp3 promoter and repress transcription. HDAC inhibitors enhance Foxp3 expression in CD4+CD25− and CD4+CD25+ T cells, suggesting that HDACs directly regulate both the Foxp3 gene and protein and enhance the suppressive function of Tregs.84,86,87 Another class II deacetylase, HDAC9, interacts with Foxp3 and down-regulates its acetylation. Treatment with the HDAC inhibitor trichostatin A enhances Foxp3 acetylation and Treg function.85
TGF-β induces chromatin binding and promoter occupancy of acetylated Foxp3 on the IL-2 promoter. Recently, it has been shown that TGF-β and IL-6 down-regulate the chromatin binding of acetylated Foxp3 to the human IL-2 promoter, and treatment with HDAC inhibitors under these conditions restores the binding of Foxp3 to the IL-2 promoter, suggesting that under inflammatory conditions HDACs play a role in the regulation of Foxp3 function.74
Role of histone methylation in Treg development and function
Apart from inheritance of methylated CpG residues in genomic DNA, parental histone molecules can also divide and be transmitted to the progeny cells. Among the 4 types of histones in the nucleosome, H3 and H4 histones are highly conserved and stably transmitted to progeny cells during replication. It has been proposed that methylated histones H3 and H4 act as epigenetic markers. Methylation of lysine residues in histones occurs at positions 4, 9, 27, and 36 in H3 and position 20 in H4. H3 is more heavily methylated and stable compared with H4. H3-K4 trimethylation is specific for the active state of transcription, and H3-K4 dimethylation exists in both active and repressed genes.88 These methylated histones recruit methyl-binding proteins (eg, MBD1) and other transcriptional repressors that maintain CpG DNA methylation and regulate the transcription.
TCR signaling controls Foxp3 expression in CD4+ T cells. Sauer et al showed that TCR stimulation induces H3-K4 dimethylation and trimethylation after 18 hours of stimulation of naive CD4+ T cells at the Foxp3 promoter (−295 to −100 bp) and intronic enhancer (+4505 to +4505, +6144 to +6280).62 These regions are also H3-K4 methylated in Tregs.62 Continued TCR stimulation leads to loss of these epigenetic marks and subsequent inhibition of Foxp3 expression, suggesting that these epigenetic marks provide a tool for determining the permissiveness for Foxp3 expression in specific cell types. Schmidl et al elegantly analyzed more than 100 differentially methylated regions, including the entire Foxp3 locus and associated upstream enhancers in human Tregs, and showed that lineage-specific CpG DNA methylation in T cells correlates with H3K4 methylation and its enhancer activity.23
Stability of Treg
The stability of CD4+Foxp3+ T cells is an interesting area in Treg biology that has been recently reviewed.89,90 nTregs possess demethylated CpG at the Foxp3 locus and show stable Foxp3 expression, whereas TGF-β induced Tregs show methylated CpG and do not maintain constitutive Foxp3 expression after restimulation in the absence of TGF-β.4,5 It has been reported that a fraction of Foxp3+CD4+ nTregs adoptively transferred into lymphophenic mice converted into Foxp3− T cells.91 Under inflammatory conditions, Foxp3+ Tregs lose Foxp3 expression and suppressive function in an IL-6–dependent manner.92 Yang et al reported that IL-6 and TCR signaling induced down-regulation of Foxp3 expression and led to development of Th17 cells.93 Using Foxp3-GFP-Cre X ROSA26-YFP dual-reporter mice, Zhou et al showed that approximately 10% of YFP+ T cells lose their Foxp3 expression over time.94 Recently, we showed that IL-6 induces remethylation of CpG DNA at the upstream enhancer and down-regulates Foxp3 expression in nTregs.4 It has been reported that Foxp3+CD4+ Tregs are a very heterogeneous population in mice91,95 and humans96 and different subsets of Tregs possess different levels of CpG DNA methylation at the Foxp3 locus.96 It has been shown that increased methylation of CpG nucleotides at the Foxp3 locus was linked with less Foxp3 expression, decreased Treg stability, and reduced suppressive Treg function.4,71,96 Epigenetic inheritance during the cell cycle is crucial in maintaining chromatin structure in cell lineages.97 The extrinsic and intrinsic signals that regulate CpG DNA methylation and perpetuate H3 methylation level at the Foxp3 locus from one cell cycle to another are not understood and require further investigation. These studies will help us to understand the maintenance of Foxp3 stability in Tregs.
Pharmacologic agents that alter epigenetically regulated genes
Epigenetic therapy is an emerging field in pharmacology that promises therapeutic agents for the control of various diseases.98 Epigenetic drugs can be divided into 2 groups: DNMT or HDAC inhibitors. Some of these inhibitors are shown in Table 1. The prototypic inhibitors 5-azacytidine and Aza were developed as cytotoxic agents and subsequently have been discovered to possess potent DNMT inhibitor activity. These drugs are converted into nucleotide triphosphates and are incorporated in place of cytosine into replicating DNA. Therefore, they are more active in the S-phase of cells. 5-Azacytosine is incorporated into both RNA and DNA, whereas Aza is incorporated only into DNA and is less toxic compared with 5-azacytosine. The disadvantages of azanucleosides are instability in aqueous solutions and strong toxicity. These disadvantages might be overcome by the use of other analogs, such as zebularine, procainamide, 5-fluoro-2′-deoxycytidine, and hydralazine. Procainamide, used to treat cardiac arrhythmias, is a specific inhibitor of DNMT1.99 Several natural products derived from tea and sponges, such as epigallocatechin-3-gallate, also show DNMT inhibitory activity.100 Antisense oligonucleotides complementary to the human DNMT1 mRNA are in clinical trials.101
Table 1.
DNA methylation and HDAC inhibitors
| Target/drugs |
|---|
| DNA methylation |
| Nucleoside analog inhibitors |
| 5-Azacytidine6,22,65 |
| 5-Aza-2′-deoxycytidine (Decitabine)4,6,39,69 |
| Zebularine102 |
| 5-fluoro-2′-deoxycytidine (FCDR)103 |
| Non-nucleoside analog inhibitors |
| Procainamide4,22,99 |
| Hydralazine4 |
| Procaine104 |
| Epigallocatechin-3-gallate (EGCG)100 |
| N-Phthalyl-L-tryptophan (RG108)4 |
| Antisense oligonucleotides |
| DNMT1 ASO101 |
| HDAC inhibitors |
| Hydroxamates |
| Trichostatin A87 |
| Suberoylanilide hydroxamic acid (Vorinostat; SAHA)105 |
| Oxamflatin106 |
| Scriptaid107 |
| Pyroxamide108 |
| M-carboxycinnamic acid bishydroxamide (CBHA)109 |
| Cyclic tetrapeptides |
| Desipeptide (FK228)110 |
| Apicidin111 |
| Trapoxin-A and trapoxin B112 |
| Aliphatic acids |
| Valproic acid113 |
| Phenyl butyrate114 |
| Benzamides |
| MS-275115 |
| N-acetyldinaline (CI-994)115 |
| Electrophilic ketones |
| Trifluoromethyl ketones116 |
| α-Ketoamides |
| Miscellaneous compounds |
| Depudecin117 |
There are several HDACs inhibitors available, and some are used clinically for the treatment of neurologic disorders and cancer (Table 1). Importantly, the hydroxamate compounds trichostatin A and suberoylanilide hydroxamic acid have been shown to induce Tregs and prolong allograft function in mice.84,87,118 Other HDAC inhibitors are not as potent as hydroxamates for enhancing the suppressive function of Tregs.119
Epigenetic regulators as immunotherapy
There are a variety of methods by which Tregs can be generated, although many are laborious, inefficient, and expensive.120,121 The stability of Foxp3 expression in Tregs is very important for their therapeutic use. The conversion of antigen-specific Tregs into effector T cells will have detrimental effects and limit clinical applicability.122 Epigenetic regulation may be an efficient therapeutic strategy for generation of stable Tregs and suppression. In vivo injection of HDAC inhibitors increases Treg numbers and function in mice87 and rhesus.118 DNMT inhibitors induce strong Foxp3 expression,4,6 but associated cell toxicity as well as induction of Th1 and Th2 cytokines limit its use. To overcome these problems, we cultured naive CD4+ T cells with low doses of DNMT inhibitors under TGF-β–induced Treg conditions. After 24 hours, DNMT inhibitor was removed and the T cells further cultured under TGF-β–induced Treg conditions. After this brief exposure, the Foxp3 enhancer was demethylated and stable Foxp3+ Tregs were generated, without indiscriminately activating other gene loci.4 These Tregs were more stable than conventional TGF-β–induced Tregs and were able to protect from autoimmune colitis and prolong allogeneic islet survival. It has been reported that prolonged in vitro culture of human Tregs leads to methylation of Foxp3 locus and decreased expression of Foxp3,123 and TGF-β–induced human Tregs are not stable and do not have efficient suppressive function.3,4 Using the epigenetic approach, we also generated enhanced Foxp3+-suppressive human Tregs that could be used as cell therapy.4 Human Foxp3+CD4+ T cells are very heterogeneous and, based on CD25, CD62L, CD45RA, HLA-DR, and ICOS expression, can be divided in different subsets. Miyara et al reported 3 deferent populations of Foxp3+CD4+ T cells in the peripheral blood mononuclear cells of a healthy man.96 CD45RA+Foxp3lo resting Tregs and CD45RA−Foxp3hi activated Tregs show completely demethylated CpG at the proximal promoter (−256 to −16 bp) and more than 85% demethylation of the intronic region promoter (+3824 to +3937 bp), and are suppressive.96 However, CD45RA−Foxp3lo Tregs show decreased demethylation at the proximal promoter and less than 50% demethylation at the intronic region, secrete cytokines such as IL-2 and IFN-γ, and do not show suppressive function.96 These finding suggest that epigenetic mechanisms provide better strategies to differentiate suppressive Tregs, understand the etiology of immunologic diseases resulting from reduced Treg-suppressive function, and help in designing better approaches to generate suppressive human Tregs.
Perspective on epigenetics in Treg biology
In humans, 1% to 3% of total T cells are CD4+CD25+Foxp3+ Tregs. Tregs have been used experimentally for the tolerance induction and the control of autoimmunity.2,121,124 These studies suggest that Treg adoptive therapy may provide an effective approach to control alloreactivity. Remaining to be elucidated is the regulation of Foxp3 under inflammatory conditions, such as bone marrow transplantation, where the conditioning regimen leads to enhanced inflammatory responses, or in solid organ transplantation, where inflammation is enhanced by ischemia and reperfusion injury.125,126 Recent advances in Treg biology, such as the role of S1P1 in T-cell migration28 as well as its interference in the suppressive function of Tregs,61 provide avenues for the development of therapeutic agents. Further studies about Akt-mTOR signaling on epigenetic modification at the Foxp3 locus may provide a better understanding for the use of chemotherapeutic agents, such as FTY720 and rapamycin in the generation of stable Foxp3+ Tregs. Similarly, the use of immunotherapeutic drugs that inhibit the down-regulation of Foxp3 under inflammatory condition may be potent strategies to enhance the function of Tregs. DNMT and HDAC inhibitors may be candidates as they enhance Foxp3 expression as well as inhibit the effect of the proinflammatory cytokine IL-6 on Foxp3 regulation and function.4,74 How epigenetic mechanisms work under different inflammatory and tolerance conditions and regulate Foxp3 expression are fertile areas for further investigation. Understanding what cytokines and surface receptors are involved in the epigenetic regulation of Treg development and function will provide a better approach to generate Tregs for therapeutic use. Global genome-wide analysis of epigenetic markers in Tregs will enrich our ability to design epigenetic therapy. A future challenge is the development of cell type–specific targeting of DNMTs and HDACs to provide better epigenetic therapeutics.
Acknowledgments
This work was supported by National Institutes of Health grants AI41428 and AI62765, and Juvenile Diabetes Research Foundation grant 1-2005-16 (all to J.S.B.).
Authorship
Contribution: G.L. and J.S.B wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan S. Bromberg, Mount Sinai School of Medicine, One Gustave L. Levy Pl, Box 1104, New York, NY 10029-6574; e-mail: Jon.Bromberg@mountsinai.org.
References
- 1.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 2.Fu S, Zhang N, Yopp AC, et al. TGF-beta induces Foxp3+ T-regulatory cells from CD4+ CD25- precursors. Am J Transplant. 2004;4(10):1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- 3.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110(8):2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lal G, Zhang N, van der Touw W, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floess S, Freyer J, Siewert C, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polansky JK, Kretschmer K, Freyer J, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 7.Bestor TH, Coxon A. Cytosine methylation: the pros and cons of DNA methylation. Curr Biol. 1993;3(6):384–386. doi: 10.1016/0960-9822(93)90209-7. [DOI] [PubMed] [Google Scholar]
- 8.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71(5):865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 9.Liang G, Chan MF, Tomigahara Y, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22(2):480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 11.Spencer VA, Davie JR. Role of covalent modifications of histones in regulating gene expression. Gene. 1999;240(1):1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 12.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Richardson BC, Strahler JR, Pivirotto TS, et al. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T-cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 1992;35(6):647–662. doi: 10.1002/art.1780350608. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi T, Amano K, Sekine H, Koide J, Abe T. Upregulated expression and function of integrin adhesive receptors in systemic lupus erythematosus patients with vasculitis. J Clin Invest. 1993;92(6):3008–3016. doi: 10.1172/JCI116924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yung R, Powers D, Johnson K, et al. Mechanisms of drug-induced lupus: II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupuslike disease in syngeneic mice. J Clin Invest. 1996;97(12):2866–2871. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172(6):3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174(10):6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 18.Mishra N, Brown DR, Olorenshaw IM, Kammer GM. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc Natl Acad Sci U S A. 2001;98(5):2628–2633. doi: 10.1073/pnas.051507098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuji-Takayama K, Suzuki M, Yamamoto M, et al. The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. J Immunol. 2008;181(6):3897–3905. doi: 10.4049/jimmunol.181.6.3897. [DOI] [PubMed] [Google Scholar]
- 20.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16(5):649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 21.Thomas RM, Gao L, Wells AD. Signals from CD28 induce stable epigenetic modification of the IL-2 promoter. J Immunol. 2005;174(8):4639–4646. doi: 10.4049/jimmunol.174.8.4639. [DOI] [PubMed] [Google Scholar]
- 22.Januchowski R, Jagodzinski PP. Effect of 5-azacytidine and procainamide on CD3-zeta chain expression in Jurkat T cells. Biomed Pharmacother. 2005;59(3):122–126. doi: 10.1016/j.biopha.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Schmidl C, Klug M, Boeld TJ, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19(7):1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PP, Fitzpatrick DR, Beard C, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 25.Hutchins AS, Mullen AC, Lee HW, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10(1):81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 26.Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol. 2003;109(1):37–45. doi: 10.1016/s1521-6616(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 27.Austrup F, Vestweber D, Borges E, et al. P- and E-selectin mediate recruitment of T-helper-1 but not T-helper-2 cells into inflamed tissues. Nature. 1997;385(6611):81–83. doi: 10.1038/385081a0. [DOI] [PubMed] [Google Scholar]
- 28.Ledgerwood LG, Lal G, Zhang N, et al. The sphingosine 1-phosphate receptor 1 causes tissue retention by inhibiting the entry of peripheral tissue T lymphocytes into afferent lymphatics. Nat Immunol. 2008;9(1):42–53. doi: 10.1038/ni1534. [DOI] [PubMed] [Google Scholar]
- 29.Zhang N, Schroppel B, Lal G, et al. Regulatory T cells sequentially migrate from inflamed tissues to draining lymph nodes to suppress the alloimmune response. Immunity. 2009;30(3):458–469. doi: 10.1016/j.immuni.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syrbe U, Jennrich S, Schottelius A, Richter A, Radbruch A, Hamann A. Differential regulation of P-selectin ligand expression in naive versus memory CD4+ T cells: evidence for epigenetic regulation of involved glycosyltransferase genes. Blood. 2004;104(10):3243–3248. doi: 10.1182/blood-2003-09-3047. [DOI] [PubMed] [Google Scholar]
- 31.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4(4):337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 32.Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 33.Lankford S, Petty C, LaVoy A, Reckling S, Tompkins W, Dean GA. Cloning of feline FOXP3 and detection of expression in CD4+CD25+ regulatory T cells. Vet Immunol Immunopathol. 2008;122(1):159–166. doi: 10.1016/j.vetimm.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Huang C, Zhou B, Ziegler SF. Isoform-specific inhibition of ROR alpha-mediated transcriptional activation by human FOXP3. J Immunol. 2008;180(7):4785–4792. doi: 10.4049/jimmunol.180.7.4785. [DOI] [PubMed] [Google Scholar]
- 35.Ichiyama K, Yoshida H, Wakabayashi Y, et al. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem. 2008;283(25):17003–17008. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- 36.Smith EL, Finney HM, Nesbitt AM, Ramsdell F, Robinson MK. Splice variants of human FOXP3 are functional inhibitors of human CD4+ T-cell activation. Immunology. 2006;119(2):203–211. doi: 10.1111/j.1365-2567.2006.02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mailer RK, Falk K, Rotzschke O. Absence of leucine zipper in the natural FOXP3Delta2Delta7 isoform does not affect dimerization but abrogates suppressive capacity. PLoS ONE. 2009;4(7):e6104. doi: 10.1371/journal.pone.0006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantel PY, Ouaked N, Ruckert B, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176(6):3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 39.Zorn E, Nelson EA, Mohseni M, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108(5):1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201(5):723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6(11):1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 42.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178(1):280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 43.Ouaked N, Mantel PY, Bassin C, et al. Regulation of the foxp3 gene by the Th1 cytokines: the role of IL-27-induced STAT1. J Immunol. 2009;182(2):1041–1049. doi: 10.4049/jimmunol.182.2.1041. [DOI] [PubMed] [Google Scholar]
- 44.Huber M, Steinwald V, Guralnik A, et al. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20(2):223–234. doi: 10.1093/intimm/dxm139. [DOI] [PubMed] [Google Scholar]
- 45.Batten M, Kljavin NM, Li J, Walter MJ, de Sauvage FJ, Ghilardi N. Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. J Immunol. 2008;180(5):2752–2756. doi: 10.4049/jimmunol.180.5.2752. [DOI] [PubMed] [Google Scholar]
- 46.Genot E, Cantrell DA. Ras regulation and function in lymphocytes. Curr Opin Immunol. 2000;12(3):289–294. doi: 10.1016/s0952-7915(00)00089-3. [DOI] [PubMed] [Google Scholar]
- 47.Mor A, Keren G, Kloog Y, George J. N-Ras or K-Ras inhibition increases the number and enhances the function of Foxp3 regulatory T cells. Eur J Immunol. 2008;38(6):1493–1502. doi: 10.1002/eji.200838292. [DOI] [PubMed] [Google Scholar]
- 48.Fragale A, Gabriele L, Stellacci E, et al. IFN regulatory factor-1 negatively regulates CD4+ CD25+ regulatory T cell differentiation by repressing Foxp3 expression. J Immunol. 2008;181(3):1673–1682. doi: 10.4049/jimmunol.181.3.1673. [DOI] [PubMed] [Google Scholar]
- 49.Takaki H, Ichiyama K, Koga K, et al. STAT6 Inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem. 2008;283(22):14955–14962. doi: 10.1074/jbc.M801123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dardalhon V, Awasthi A, Kwon H, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol. 2008;9(12):1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang SG, Lim HW, Andrisani OM, Broxmeyer HE, Kim CH. Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J Immunol. 2007;179(6):3724–3733. doi: 10.4049/jimmunol.179.6.3724. [DOI] [PubMed] [Google Scholar]
- 52.Sun CM, Hall JA, Blank RB, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204(8):1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mucida D, Park Y, Kim G, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 54.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204(8):1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204(8):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao S, Jin H, Korn T, et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181(4):2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill JA, Hall JA, Sun CM, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29(5):758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205(3):565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9(2):194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 60.Locke NR, Patterson SJ, Hamilton MJ, Sly LM, Krystal G, Levings MK. SHIP regulates the reciprocal development of T regulatory and Th17 cells. J Immunol. 2009;183(2):975–983. doi: 10.4049/jimmunol.0803749. [DOI] [PubMed] [Google Scholar]
- 61.Liu G, Burns S, Huang G, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10(7):769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sauer S, Bruno L, Hertweck A, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105(22):7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 64.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24(4):369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janson PC, Winerdal ME, Marits P, Thorn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Tregs population in humans. PLoS ONE. 2008;3(2):e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baron U, Floess S, Wieczorek G, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 68.Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9(3):245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo X, Zhang Q, Liu V, Xia Z, Pothoven KL, Lee C. Cutting edge: TGF-beta-induced expression of Foxp3 in T cells is mediated through inactivation of ERK. J Immunol. 2008;180(5):2757–2761. doi: 10.4049/jimmunol.180.5.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagar M, Vernitsky H, Cohen Y, et al. Epigenetic inheritance of DNA methylation limits activation-induced expression of FOXP3 in conventional human CD25-CD4+ T cells. Int Immunol. 2008;20(8):1041–1055. doi: 10.1093/intimm/dxn062. [DOI] [PubMed] [Google Scholar]
- 71.Stockis J, Fink W, Francois V, et al. Comparison of stable human Tregs and Th clones by transcriptional profiling. Eur J Immunol. 2009;39(3):869–882. doi: 10.1002/eji.200838807. [DOI] [PubMed] [Google Scholar]
- 72.Wieczorek G, Asemissen A, Model F, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69(2):599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]
- 73.Dominitzki S, Fantini MC, Neufert C, et al. Cutting edge: trans-signaling via the soluble IL-6R abrogates the induction of FoxP3 in naive CD4+CD25 T cells. J Immunol. 2007;179(4):2041–2045. doi: 10.4049/jimmunol.179.4.2041. [DOI] [PubMed] [Google Scholar]
- 74.Samanta A, Li B, Song X, et al. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci U S A. 2008;105(37):14023–14027. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doganci A, Eigenbrod T, Krug N, et al. The IL-6R alpha chain controls lung CD4+CD25+ Tregs development and function during allergic airway inflammation in vivo. J Clin Invest. 2005;115(2):313–325. doi: 10.1172/JCI22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001;276(43):39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102(19):6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178(7):4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108(3):1058–1064. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Josefowicz SZ, Wilson CB, Rudensky AY. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J Immunol. 2009;182(11):6648–6652. doi: 10.4049/jimmunol.0803320. [DOI] [PubMed] [Google Scholar]
- 81.Li B, Greene MI. FOXP3 actively represses transcription by recruiting the HAT/HDAC complex. Cell Cycle. 2007;6(12):1432–1436. [PubMed] [Google Scholar]
- 82.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol. 2001;79(3):243–252. [PubMed] [Google Scholar]
- 83.Brandl A, Heinzel T, Kramer OH. Histone deacetylases: salesmen and customers in the post-translational modification market. Biol Cell. 2009;101(4):193–205. doi: 10.1042/BC20080158. [DOI] [PubMed] [Google Scholar]
- 84.Tao R, de Zoeten EF, Ozkaynak E, et al. Histone deacetylase inhibitors and transplantation. Curr Opin Immunol. 2007;19(5):589–595. doi: 10.1016/j.coi.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li B, Samanta A, Song X, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104(11):4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lucas JL, Mirshahpanah P, Haas-Stapleton E, Asadullah K, Zollner TM, Numerof RP. Induction of Foxp3(+) regulatory T cells with histone deacetylase inhibitors. Cell Immunol. 2009;257(1):97–104. doi: 10.1016/j.cellimm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 87.Tao R, de Zoeten EF, Ozkaynak E, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13(11):1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 88.Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 89.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4(+) FoxP3(+) T cells. Curr Opin Immunol. 2009;21(3):281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Tregs cells. Curr Opin Immunol. 2009;21(3):274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 91.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A. 2009;106(6):1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299(5609):1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 93.Yang XO, Nurieva R, Martinez GJ, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29(1):44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205(9):1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fu S, Yopp AC, Mao X, et al. CD4+ CD25+ CD62+ T-regulatory cell subset has optimal suppressive and proliferative potential. Am J Transplant. 2004;4(1):65–78. doi: 10.1046/j.1600-6143.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 96.Miyara M, Yoshioka Y, Kitoh A, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30(6):899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 97.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10(3):192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 98.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 99.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280(49):40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–7570. [PubMed] [Google Scholar]
- 101.Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-alpha (ER) in ER-negative human breast cancer cell lines. Cancer Biol Ther. 2003;2(5):552–556. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- 102.Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321(4):591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 104.Villar-Garea A, Fraga MF, Espada J, Esteller M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63(16):4984–4989. [PubMed] [Google Scholar]
- 105.Kumagai T, Wakimoto N, Yin D, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer. 2007;121(3):656–665. doi: 10.1002/ijc.22558. [DOI] [PubMed] [Google Scholar]
- 106.Kim YB, Lee KH, Sugita K, Yoshida M, Horinouchi S. Oxamflatin is a novel antitumor compound that inhibits mammalian histone deacetylase. Oncogene. 1999;18(15):2461–2470. doi: 10.1038/sj.onc.1202564. [DOI] [PubMed] [Google Scholar]
- 107.Lee EJ, Lee BB, Kim SJ, Park YD, Park J, Kim DH. Histone deacetylase inhibitor scriptaid induces cell cycle arrest and epigenetic change in colon cancer cells. Int J Oncol. 2008;33(4):767–776. [PubMed] [Google Scholar]
- 108.Butler LM, Webb Y, Agus DB, et al. Inhibition of transformed cell growth and induction of cellular differentiation by pyroxamide, an inhibitor of histone deacetylase. Clin Cancer Res. 2001;7(4):962–970. [PubMed] [Google Scholar]
- 109.Coffey DC, Kutko MC, Glick RD, et al. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all-trans retinoic acid. Cancer Res. 2001;61(9):3591–3594. [PubMed] [Google Scholar]
- 110.Shaker S, Bernstein M, Momparler LF, Momparler RL. Preclinical evaluation of antineoplastic activity of inhibitors of DNA methylation (5-aza-2′-deoxycytidine) and histone deacetylation (trichostatin A, depsipeptide) in combination against myeloid leukemic cells. Leuk Res. 2003;27(5):437–444. doi: 10.1016/s0145-2126(02)00222-9. [DOI] [PubMed] [Google Scholar]
- 111.Darkin-Rattray SJ, Gurnett AM, Myers RW, et al. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc Natl Acad Sci U S A. 1996;93(23):13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J Biol Chem. 1993;268(30):22429–22435. [PubMed] [Google Scholar]
- 113.Saouaf SJ, Li B, Zhang G, et al. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87(2):99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Um SJ, Han HS, Kwon YJ, et al. In vitro antitumor potential of 4-BPRE, a butyryl aminophenyl ester of retinoic acid: role of the butyryl group. Oncol Rep. 2004;11(3):719–726. [PubMed] [Google Scholar]
- 115.Gediya LK, Belosay A, Khandelwal A, Purushottamachar P, Njar VC. Improved synthesis of histone deacetylase inhibitors (HDIs) (MS-275 and CI-994) and inhibitory effects of HDIs alone or in combination with RAMBAs or retinoids on growth of human LNCaP prostate cancer cells and tumor xenografts. Bioorg Med Chem. 2008;16(6):3352–3360. doi: 10.1016/j.bmc.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frey RR, Wada CK, Garland RB, et al. Trifluoromethyl ketones as inhibitors of histone deacetylase. Bioorg Med Chem Lett. 2002;12(23):3443–3447. doi: 10.1016/s0960-894x(02)00754-0. [DOI] [PubMed] [Google Scholar]
- 117.Kwon HJ, Owa T, Hassig CA, Shimada J, Schreiber SL. Depudecin induces morphologic reversion of transformed fibroblasts via the inhibition of histone deacetylase. Proc Natl Acad Sci U S A. 1998;95(7):3356–3361. doi: 10.1073/pnas.95.7.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson J, Pahuja A, Graham M, Hering B, Hancock WW, Bansal-Pakala P. Effects of histone deacetylase inhibitor SAHA on effector and FOXP3+ regulatory T cells in rhesus macaques. Transplant Proc. 2008;40(2):459–461. doi: 10.1016/j.transproceed.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3(+) regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol. 2009;87(3):195–202. doi: 10.1038/icb.2008.106. [DOI] [PubMed] [Google Scholar]
- 120.Sagoo P, Lombardi G, Lechler RI. Regulatory T cells as therapeutic cells. Curr Opin Organ Transplant. 2008;13(6):645–653. doi: 10.1097/MOT.0b013e328317a476. [DOI] [PubMed] [Google Scholar]
- 121.Roncarolo MG, Battaglia M. Regulatory T-cell immunotherapy for tolerance to self antigens and alloantigens in humans. Nat Rev Immunol. 2007;7(8):585–598. doi: 10.1038/nri2138. [DOI] [PubMed] [Google Scholar]
- 122.Riley JL, June CH, Blazar BR. Human T regulatory cell therapy: take a billion or so and call me in the morning. Immunity. 2009;30(5):656–665. doi: 10.1016/j.immuni.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hoffmann P, Boeld TJ, Eder R, et al. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur J Immunol. 2009;39(4):1088–1097. doi: 10.1002/eji.200838904. [DOI] [PubMed] [Google Scholar]
- 124.Chen D, Zhang N, Fu S, et al. CD4+ CD25+ regulatory T-cells inhibit the islet innate immune response and promote islet engraftment. Diabetes. 2006;55(4):1011–1021. doi: 10.2337/diabetes.55.04.06.db05-1048. [DOI] [PubMed] [Google Scholar]
- 125.Kruger B, Krick S, Dhillon N, et al. Donor Toll-like receptor 4 contributes to ischemia and reperfusion injury following human kidney transplantation. Proc Natl Acad Sci U S A. 2009;106(9):3390–3395. doi: 10.1073/pnas.0810169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang Y, Yin H, Han J, et al. Extracellular hmgb1 functions as an innate immune-mediator implicated in murine cardiac allograft acute rejection. Am J Transplant. 2007;7(4):799–808. doi: 10.1111/j.1600-6143.2007.01734.x. [DOI] [PubMed] [Google Scholar]